Abstract
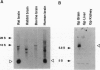
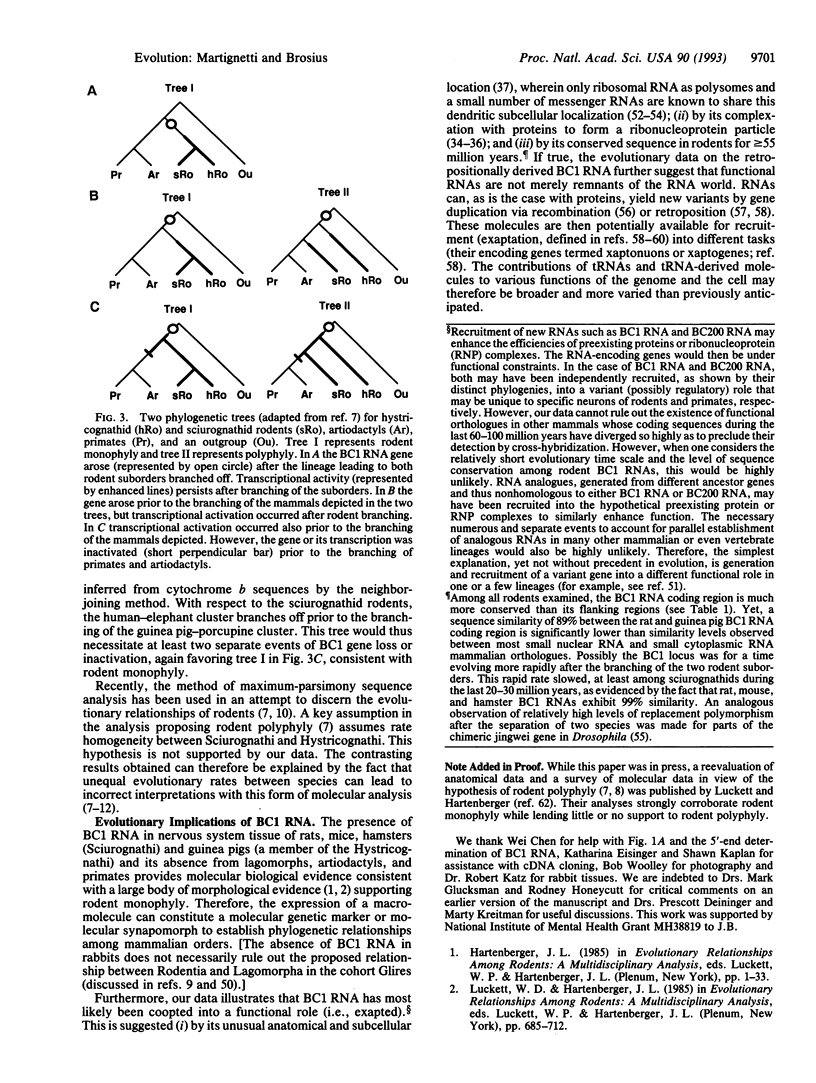
The traditional morphologically grounded placement of South American guinea pig-like rodents (Caviomorpha) within one of the two rodent suborders, Hystricognathi, has been disputed by recent analysis of protein and nucleic acid sequence data. The Caviomorpha and possibly all Hystricognathi would be considered a separate order, distinct from the other rodent suborder, Sciurognathi, and thus of the order Rodentia, and would be placed closer phylogenetically to other mammals [Graur, D., Hide, W. A. & Li, W.-H. (1991) Nature (London) 351, 649-652]. To address the discrepancy between morphological comparisons and sequence analyses, we have applied an alternative form of molecular analysis. We demonstrate that BC1 RNA, a neural-specific small cytoplasmic RNA that is the product of a retropositionally generated gene (a gene derived by reverse transcription of RNA followed by insertion of the DNA copy into the genome), is present in Sciurognathi and guinea pig but not in other mammalian orders including Lagomorpha, Artiodactyla, and Primates. The species-confined, tissue-specific expression of a retroposed sequence therefore supports the morphological evidence for monophyly of Rodentia inclusive of guinea pig and demonstrates the usefulness of such molecular genetic markers. Furthermore, the conservation and tissue-specific expression of the BC1 RNA gene in the two divergent rodent suborders suggests that this macromolecule has been exapted into a functional role (i.e., coopted into a variant or novel function) in the rodent nervous system.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allard M. W., Miyamoto M. M., Honeycutt R. L. Tests for rodent polyphyly. Nature. 1991 Oct 17;353(6345):610–611. doi: 10.1038/353610b0. [DOI] [PubMed] [Google Scholar]
- Beintema J. J., Neuteboom B. Origin of the duplicated ribonuclease gene in guinea-pig: comparison of the amino acid sequences with those of two close relatives: capybara and cuis ribonuclease. J Mol Evol. 1983;19(2):145–152. doi: 10.1007/BF02300752. [DOI] [PubMed] [Google Scholar]
- Boyer S., Montagutelli X., Gomès D., Simon-Chazottes D., Guénet J. L., Dupouey P. Recent evolutionary origin of the expression of the glial fibrillary acidic protein (GFAP) in lens epithelial cells. A molecular and genetic analysis of various mouse species. Brain Res Mol Brain Res. 1991 May;10(2):159–166. doi: 10.1016/0169-328x(91)90106-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Gould S. J. Molecular constructivity. Nature. 1993 Sep 9;365(6442):102–102. doi: 10.1038/365102a0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Gould S. J. On "genomenclature": a comprehensive (and respectful) taxonomy for pseudogenes and other "junk DNA". Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin K. E., Waxham M. N., Rickling S., Westgate S. A., Mobley W. C., Kelly P. T. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990 Jun;10(6):1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. J., Episkopou V., Zeitlin S., Karathanasis S. K., MacKrell A., Steiner D. F., Efstratiadis A. Guinea pig preproinsulin gene: an evolutionary compromise? Proc Natl Acad Sci U S A. 1984 Aug;81(16):5046–5050. doi: 10.1073/pnas.81.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson R. A., Tomarev S. I., Piatigorsky J. Taxon-specific recruitment of enzymes as major soluble proteins in the corneal epithelium of three mammals, chicken, and squid. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4004–4008. doi: 10.1073/pnas.89.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T. M., Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci U S A. 1987 May;84(9):2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozzo G., Guardiola J. A SINE insertion provides information on the divergence of the HLA-DQA1 and HLA-DQA2 genes. Immunogenetics. 1990;31(4):229–232. doi: 10.1007/BF00204892. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djian P., Green H. Involucrin gene of tarsioids and other primates: alternatives in evolution of the segment of repeats. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5321–5325. doi: 10.1073/pnas.88.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djian P., Green H. Vectorial expansion of the involucrin gene and the relatedness of the hominoids. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8447–8451. doi: 10.1073/pnas.86.21.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili U., Swanson K. Gene sequences suggest inactivation of alpha-1,3-galactosyltransferase in catarrhines after the divergence of apes from monkeys. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7401–7404. doi: 10.1073/pnas.88.16.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C. C., Tucker R. P., Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988 Dec 15;336(6200):674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- Graur D., Hide W. A., Li W. H. Is the guinea-pig a rodent? Nature. 1991 Jun 20;351(6328):649–652. doi: 10.1038/351649a0. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Cao Y., Adachi J., Yano T. Rodent polyphyly? Nature. 1992 Feb 13;355(6361):595–595. doi: 10.1038/355595a0. [DOI] [PubMed] [Google Scholar]
- Kelley M. J., Pech M., Seuanez H. N., Rubin J. S., O'Brien S. J., Aaronson S. A. Emergence of the keratinocyte growth factor multigene family during the great ape radiation. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9287–9291. doi: 10.1073/pnas.89.19.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido Y., Aono M., Yamaki T., Matsumoto K., Murata S., Saneyoshi M., Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Goto S., Anzai K. Brain-specific small RNA transcript of the identifier sequences is present as a 10 S ribonucleoprotein particle. J Biol Chem. 1991 Mar 15;266(8):4726–4730. [PubMed] [Google Scholar]
- Kobayashi S., Higashi N., Suzuki K., Goto S., Yumoto K., Anzai K. The 10 S BC-1 ribonucleoprotein particle contains identifier sequence-binding proteins that interact with an array of GCAAG/CTTGC motifs between split promoter sequences for RNA polymerase III. J Biol Chem. 1992 Sep 15;267(26):18291–18297. [PubMed] [Google Scholar]
- Koop B. F., Goodman M., Xu P., Chan K., Slightom J. L. Primate eta-globin DNA sequences and man's place among the great apes. Nature. 1986 Jan 16;319(6050):234–238. doi: 10.1038/319234a0. [DOI] [PubMed] [Google Scholar]
- Li W. H., Hide W. A., Zharkikh A., Ma D. P., Graur D. The molecular taxonomy and evolution of the guinea pig. J Hered. 1992 May-Jun;83(3):174–181. doi: 10.1093/oxfordjournals.jhered.a111188. [DOI] [PubMed] [Google Scholar]
- Long M., Langley C. H. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993 Apr 2;260(5104):91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- Ma D. P., Zharkikh A., Graur D., VandeBerg J. L., Li W. H. Structure and evolution of opossum, guinea pig, and porcupine cytochrome b genes. J Mol Evol. 1993 Apr;36(4):327–334. doi: 10.1007/BF00182180. [DOI] [PubMed] [Google Scholar]
- McConnell T. J., Talbot W. S., McIndoe R. A., Wakeland E. K. The origin of MHC class II gene polymorphism within the genus Mus. Nature. 1988 Apr 14;332(6165):651–654. doi: 10.1038/332651a0. [DOI] [PubMed] [Google Scholar]
- Minghetti P. P., Dugaiczyk A. The emergence of new DNA repeats and the divergence of primates. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1872–1876. doi: 10.1073/pnas.90.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi M., Kawai T., Yagi K. Guinea pigs possess a highly mutated gene for L-gulono-gamma-lactone oxidase, the key enzyme for L-ascorbic acid biosynthesis missing in this species. J Biol Chem. 1992 Oct 25;267(30):21967–21972. [PubMed] [Google Scholar]
- Novacek M. J. Mammalian phylogeny: shaking the tree. Nature. 1992 Mar 12;356(6365):121–125. doi: 10.1038/356121a0. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J., Wistow G. The recruitment of crystallins: new functions precede gene duplication. Science. 1991 May 24;252(5009):1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- Russo T., Costanzo F., Oliva A., Ammendola R., Duilio A., Esposito F., Cimino F. Structure and in vitro transcription of tRNA gene clusters containing the primers of MuLV reverse transcriptase. Eur J Biochem. 1986 Aug 1;158(3):437–442. doi: 10.1111/j.1432-1033.1986.tb09772.x. [DOI] [PubMed] [Google Scholar]
- Ryan S. C., Dugaiczyk A. Newly arisen DNA repeats in primate phylogeny. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9360–9364. doi: 10.1073/pnas.86.23.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., St-Jacques B. 'Brain-specific' transcription and evolution of the identifier sequence. 1986 Jan 30-Feb 5Nature. 319(6052):418–420. doi: 10.1038/319418a0. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Koeberl D. D., Sommer S. S. Direct sequencing of the activation peptide and the catalytic domain of the factor IX gene in six species. Genomics. 1990 Jan;6(1):133–143. doi: 10.1016/0888-7543(90)90458-7. [DOI] [PubMed] [Google Scholar]
- Steward O., Banker G. A. Getting the message from the gene to the synapse: sorting and intracellular transport of RNA in neurons. Trends Neurosci. 1992 May;15(5):180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Gottesfeld J. M., Reynolds W. Control of neuronal gene expression. Science. 1984 Sep 21;225(4668):1308–1315. doi: 10.1126/science.6474179. [DOI] [PubMed] [Google Scholar]
- Tiedge H., Chen W., Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993 Jun;13(6):2382–2390. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toneguzzo F., Glynn S., Levi E., Mjolsness S., Hayday A. Use of a chemically modified T7 DNA polymerase for manual and automated sequencing of supercoiled DNA. Biotechniques. 1988 May;6(5):460–469. [PubMed] [Google Scholar]
- Tronik D., Ekker M., Rougeon F. Structural analysis of 5'-flanking regions of rat, mouse and human renin genes reveals the presence of a transposable-like element in the two mouse genes. Gene. 1988 Sep 15;69(1):71–80. doi: 10.1016/0378-1119(88)90379-4. [DOI] [PubMed] [Google Scholar]
- Watson J. B., Sutcliffe J. G. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol Cell Biol. 1987 Sep;7(9):3324–3327. doi: 10.1128/mcb.7.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]