The greatest obstacle to curing HIV-1 infection is a latent reservoir comprising resting CD4+ T cells that contain an unexpressed, replication-competent copy of the HIV-1 genome integrated into the host DNA. These latently infected cells are essentially indistinguishable from uninfected ones and therefore cannot be selectively targeted for elimination. The latent reservoir is characterized by a notable stability that explains why it is a lifelong barrier to a cure. Longitudinal studies in HIV-1–infected individuals measuring changes in the frequency of latently infected cells over time have estimated the half-life of reservoir decay to be ∼3.6 y (1, 2). At this rate, a reservoir containing 106 cells would take more than 70 y to decay naturally. Because even a single replication-competent latent virus can lead to rebound viremia, HIV-1–infected individuals must endure lifelong treatment with antiretroviral therapy (ART). The disturbing possibility that the clonal expansion of latently infected cells contributes to the stability of the latent reservoir is explored in an important paper by Simonetti et al. in PNAS (3).
Current ART regimens are extremely effective and have far fewer side effects than their predecessors. The HIV treatment guidelines now recommend continuous ART for all infected individuals (4). However, variability in treatment access, the risks of inconsistent drug adherence, the unknown consequences of lifelong ART, and the unsustainable cost of lifelong treatment of all infected individuals have provided impetus to the search for a cure. The primary approach has been a “shock-and-kill” strategy in which expression of latent proviruses is induced (“shock”) by a latency reversing agent and then the infected cells are eliminated by viral cytopathic effects or by immune mechanisms (“kill”). In considering whether shock-and-kill is likely to be successful, the stability of the reservoir is germane. It is generally assumed that as long as an HIV-1–infected individual continues ART, the size of the reservoir should not increase, because new infection events are precluded. Hence, sequential rounds of this cure approach should gradually decrease the size of the reservoir and ultimately eliminate it. Unfortunately, it is coming to light that the apparent stability of the reservoir may be masking an underlying proliferative process. Whereas the overall size of the reservoir is stable in the setting of suppressive ART (1, 2), recent reports have raised the specter that previously underappreciated clonal expansion of HIV-1–infected cells may, in fact, allow some subpopulations of infected cells to increase in frequency (5–7). It was not known, though, whether these clonally expanded HIV-1–infected cells contained replication-competent virus. Simonetti et al. now provide evidence for clonal expansion of HIV-1–infected cells harboring replication-competent virus (3).
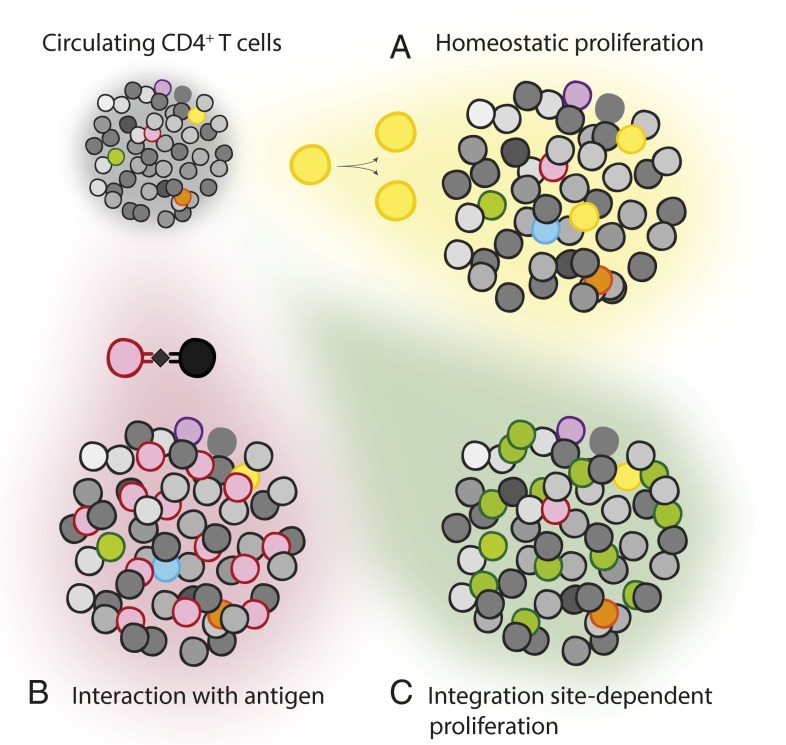
Clonal expansion (Fig. 1) is a feature of healthy immune systems and occurs during the response of activated naïve or memory T cells to antigen. Clonal expansion can also result from homeostatic proliferation, a process driven by cytokines that is important for the normal maintenance of size and diversity in the total pool of T cells (8). Whether HIV-1–infected cells can proliferate has been less clear. The half-life of productively infected cells in vivo is very short (t1/2 < 2 d) (9, 10) and the HIV-1 protein Vpr causes cell cycle arrest (11). In an in vitro model of HIV-1 latency, homeostatic proliferation can occur without up-regulation of HIV-1 gene expression (12). Early evidence for clonal expansion of HIV-1–infected cells in vivo came from studies of residual viremia in patients on ART (13, 14). Patients on ART have trace levels of viremia (∼one copy per milliliter) that appears to represent release of virus from stable reservoirs (15). Analysis of residual viremia showed that in some patients, a particular HIV-1 clone repeatedly appeared in the plasma, with no sequence evolution over periods of months to years. This result is consistent with virus production by an expanded cellular clone rather than ongoing cycles of replication. Because these analyses were based on sequences from plasma virions, it was not possible to determine whether these identical viral genomes were produced by a single cell, by the clonal progeny of a single infected cell, or by multiple unrelated cells that were all infected by an identical viral variant.
Fig. 1.
Possible causes of clonal expansion of infected CD4+ T cells. Uninfected CD4+ T cells (in grayscale) circulate with rare CD4+ T cells that carry an integrated HIV-1 provirus (various colors). CD4+ T lymphocytes can undergo clonal expansion as a result of (A) homeostatic proliferation, (B) interaction with antigen from antigen presenting cell (in black), or (C) altered expression of host genes affecting cell growth because of effects of proviral integration into these genes.
Recent studies, including that of Simonetti et al., have implemented a novel protocol that confirms the existence of clonally expanded, HIV-1–infected T cells (3, 5, 7). Specific amplification and deep sequencing of proviral integration sites in the human genome provide a way to make this determination. HIV-1 typically integrates randomly into actively expressed genes throughout the human genome (16, 17). Thus, the integration site can distinguish proviruses that arose from independent infection events, even if the proviral sequences themselves are identical. In addition, the method of analysis used by Maldarelli et al. (5) can distinguish proviruses from different cells, even if the integration sites are identical. This is accomplished through the identification of different DNA breakpoints at the ends of provirus-containing fragments, resulting from unbiased shearing of genomic DNA. Thus, two proviruses that have the same sequence and the same integration site must have originated from two different cells if the DNA breakpoints are different. (If the DNA breakpoints are identical, the proviral sequence arose from the same DNA template in the PCR step of the protocol.) In this manner, it is possible to distinguish identical proviruses that came from different daughter cells generated by clonal expansion of a single infected cell.
Using this approach, Maldarelli et al. (5) carried out integration site analysis on peripheral blood mononuclear cells (PBMCs) and purified CD4+ T cells from five HIV-1–infected patients receiving ART. The authors found that in all five patients there was evidence of clonal expansion of HIV-1–infected cells. One patient in particular (patient 1) presented another very interesting result: two genes, for the transcription factors MKL2 and BACH2, which are associated with cell growth and have been linked to human cancers (18, 19), showed a disproportionately large number of integration sites. This was surprising because, given the general randomness of integration site selection, one would not expect to find any marked imbalance in the genomic distribution of proviruses in populations of infected cells. Many of the integration sites in these two genes were distinct (i.e., represented independent integrations) and many also were clonally expanded. Most notable was that all of the proviruses found in these two loci were in the same transcriptional orientation as the host gene. The involvement of these two genes in cell growth raises the possibility that the integration of the provirus in these locations altered expression of the respective host gene in a way that favorably affected cell survival and proliferation. Very similar results were also reported by Wagner et al. (6) using an elegant, alternative approach.
What was not clear in the work of Maldarelli et al. (5), however, was whether the clonally expanded proviruses were replication-competent. We now know that the vast majority of HIV-1 proviruses are defective and exhibit substantial deletions and hypermutation introduced by APOBEC3G during reverse
Simonetti et al. now provide evidence for clonal expansion of HIV-1–infected cells harboring replication-competent virus.
transcription (20). Unfortunately, in integration site analyses, only a portion of the 5′ or 3′ end of the proviral DNA can be captured when flanking host DNA is also sought. Thus, one cannot conclude definitively that a provirus whose integration site is identified is also intact without determining its full sequence by some other means or demonstrating that it can replicate. Simonetti et al. (3) address this issue in their examination of one of the expanded clones identified by Maldarelli et al. (5) (also in patient 1). The integration site of this clone (eponymously named AMBI-1) was ambiguous and could not be conclusively identified because of integration in a region of repetitive DNA. To determine whether AMBI-1 was intact, Simonetti et al. (3) amplified the full-length AMBI-1 provirus and transfected the DNA into 293T cells to generate virus. The authors used the 293T supernatant (which contains virus) to infect healthy donor PBMCs and a luciferase indicator cell line and demonstrated the recovery of infectious virus. They also used viral outgrowth assays to recover viruses matching AMBI-1 in sequence, which could be sequentially passaged in vitro. Thus, the authors show that the AMBI-1 provirus, which is clonally expanded, can produce replication-competent virus.
Whereas the cells containing proviruses integrated in the MLK2 and BACH2 genes may have undergone clonal expansion as a result of altered expression of these genes, the mechanism governing expansion of cells carrying AMBI-1 is a matter of conjecture. One hypothesis is that this T-cell clone proliferated as a tumor-infiltrating lymphocyte. The patient from whom AMBI-1 was isolated was diagnosed with metastatic squamous cell carcinoma. In measurements of HIV-1 DNA in autopsy tissue, Simonetti et al. (3) found a more than fourfold enrichment of AMBI-1 DNA in metastatic lesions relative to lymphoid tissue. This observation could be explained by antigen-driven clonal expansion (Fig. 1).
Although the Simonetti et al. (3) study focuses on a single patient, the implications of this work are multiple and foreboding. First, it is now clear that at least some cells carrying replication-competent HIV-1 can undergo dramatic clonal expansion. Second, it cannot be assumed that reductions in the latent reservoir will be stable. At least some of the infected cells may be able to proliferate. In this situation, not only must we suppress viral replication and eliminate the current cells in the reservoir, we must also be able to block clonal expansion of any remaining latently infected cells. Whether these important findings are generalizable beyond patient 1 remains to be seen.
Footnotes
The authors declare no conflict of interest.
See companion article on page 1883.
References
- 1.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 2.Crooks AM, et al. Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Günthard HF, et al. International Antiviral Society-USA Panel Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312(4):410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 5.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seddon B, Tomlinson P, Zamoyska R. Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat Immunol. 2003;4(7):680–686. doi: 10.1038/ni946. [DOI] [PubMed] [Google Scholar]
- 9.Perelson AS, et al. Decay characteristics of HIV-1–infected compartments during combination therapy. Nature. 1997;387(6629):188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 10.Wei X, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373(6510):117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 11.Planelles V, et al. Fate of the human immunodeficiency virus type 1 provirus in infected cells: a role for vpr. J Virol. 1995;69(9):5883–5889. doi: 10.1128/jvi.69.9.5883-5889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosque A, Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113(1):58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobin NH, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J Virol. 2005;79(15):9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinoso JB, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2009;106(23):9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schröder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 17.Han Y, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78(12):6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi S, et al. Identification of IGHCδ-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes Chromosomes Cancer. 2011;50(4):207–216. doi: 10.1002/gcc.20845. [DOI] [PubMed] [Google Scholar]
- 19.Muehlich S, et al. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene. 2012;31(35):3913–3923. doi: 10.1038/onc.2011.560. [DOI] [PubMed] [Google Scholar]
- 20.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]