Significance
Herpes simplex virus-1 (HSV-1) is transmitted by contact between the infected tissues of a transmitter and those of an uninfected recipient. Effective spread requires that the lesions be small and not repulsive in appearance. For successful transmission HSV must control both its replication and its spread from infected to uninfected cells. Here we report that two HSV microRNAs (miRNAs) made late in productive infection are exported in exosomes. Ectopic expression in transfected cells before infection reduces the synthesis of viral gene products and the spread from infected to uninfected cells. The miRNAs accumulate in neurons after latent virus is induced to reactivate. The miRNAs conform to the expectations of gene products that control viral replication and spread.
Keywords: exosome, latency, reactivation, trigeminal ganglia
Abstract
We report on the properties and function of two herpes simplex virus-1 (HSV-1) microRNAs (miRNAs) designated “miR-H28” and “miR-H29.” Both miRNAs accumulate late in productive infection at a time when, for the most part, viral DNA and proteins have been made. Ectopic expression of miRNA mimics in human cells before infection reduced the accumulation of viral mRNAs and proteins, reduced plaque sizes, and at vey low multiplicities of infection reduced viral yields. The specificity of the miRNA mimics was tested in two ways. First, ectopic expression of mimics carrying mutations in the seed sequence was ineffective. Second, in similar tests two viral miRNAs made early in productive infection also had no effect. Both miR-H28 and miR-H29 are exported from infected cells in exosomes. A noteworthy finding is that both miR-H28 and miR-H29 were absent from murine ganglia harboring latent virus but accumulated in ganglia in which the virus was induced to reactivate. The significance of these findings rests on the principle that the transmission of HSV from person to person is by physical contact between the infected tissues of the donor and those of uninfected recipient. Diminished size of primary or recurrent lesions could be predicted to enhance person-to-person transmission. Reduction in the amount of reactivating latent virus would reduce the risk of retrograde transport to the CNS but would not interfere with anterograde transport to a site at or near the site of initial infection.
Herpes simplex viruses (HSVs) infect and multiply in cells at the portal of entry, i.e., mouth or genitals (1). They then are exported by retrograde transport to sensory or autonomic neurons where they establish latent (silent) infection. In response to neuronal stress, the virus reactivates and is transported anterograde to a site at or near the site of initial infection. At that site the virus is available for transmission by physical contact between the infected tissues of the donor and those of the uninfected recipient (1, 2). In principle it would be expected that extensive lesions on initial infection and on reactivation would seed more neurons with latent virus and increase the transmission of virus from person to person. This report challenges the hypothesis that viral gene products uniformly enhance viral replication and spread.
This report centers on the function of two newly detected viral microRNAs (miRNAs). Specifically HSV-1 and HSV-2 have been reported to express at least 27 miRNAs (3–13). Analyses of 17 of these miRNAs have shown that they differ in their requirements for synthesis in productively infected cells. Thus, some are made in higher amounts in the absence of viral protein synthesis, but others require viral protein synthesis for their synthesis (14). A confounding problem associated with the function of viral miRNAs is that miRNAs made in the course of latent infections in neurons are also made in productively infected cells, whereas some, but not all, miRNAs accumulating in neurons in which latent virus is in the process of reactivation are not made in appreciable amounts in productively infected cells (14). Here we report the properties of two newly identified miRNAs designated “miR-H28” and “miR-H29.” The key properties of these miRNAs are (i) they are made late in infection, i.e., after viral DNA and structural proteins have been made; (ii) they are exported in exosomes; and (iii) they accumulate in neurons in which the virus is in the process of reactivation from the latent state but are absent from neurons harboring latent virus. Additional studies showed that ectopic expression of mimics of these miRNAs in cells before infection reduces the rate of accumulation of viral proteins, decreases plaque size, and at the same time reduces viral yields in cells infected at a very low multiplicity of infection.
One hypothesis that could explain the evolution of these miRNAs is the need to block excessive replication of reactivating virus for two reasons. First, repulsive lesions on the mouth or genitals would reduce physical contact between the infected tissues of the transmitter and the uninfected tissue of the recipient. A second reason may be to reduce the amount of virus made on reactivation to ensure that the reactivated virus is transmitted anterograde to the mouth or genitals rather than retrograde to the CNS. Retrograde transport would likely kill the host and block further viral spread. In essence these studies support the conclusion that HSV-1 down-regulates its replication to enhance its spread and that this process is executed by viral gene products.
Results
The Isolation, Sequence, and Properties of HSV-1 miR-H28 and miR-H29.
Deep-sequencing analyses of cells infected with HSV-1(F) [the prototype HSV-1 strain used in our laboratories (15)] led to the identification of two heretofore unreported viral miRNAs. The critical parameters of miR-H28 and miR-H29 are listed in Table 1. In brief miR-H28 and miR-H29 were isolated from both infected HEK293T and HEp-2 cells. The miR-H28 sequence is antisense to and maps close to the N-terminal domain of unique long 4 (UL4) (Table 1). miR-H29 maps in the same sense as the coding sequence of unique long 30 (UL30) (Table 1). The function of UL4 is unclear. UL30 encodes the viral DNA polymerase (1).
Table 1.
Genomic location and sequence of HSV-1(F) miR-H28, miR-H29, and derivatives
| miRNAs and derivatives | miR-H28 | miR-H29 |
| Position in HSV-1 DNA | 12,316–12,462 | 64,820–64,994 |
| UL4 | UL30 | |
| (antisense) | (sense) | |
| miRNA precursor | ||
| auaggcuauacgcgauggu | ccaggguccuugaccccacu | |
| cgucuguggauuggacauc | uccggguuucaugugaaccc | |
| ucgcgguggguagugagucc | cguggugguguucgacuuu | |
| cccgggccggguucggugga | gccagccuguaccccagcau | |
| acuguaaggggacggcgggu | cauccaggcccacaaccugu | |
| uaauauacaaugaccacgu | gcuucagcacgcucucccug | |
| ucggaucgcgcagagccgau | agggccgacgcaguggcgca | |
| aguaugugcu | ccuggaggcgggcaaggacu | |
| accuggagaucgaggu | ||
| Mature miRNA | CGAUGGUCGUCUG | CUGGAGGCGGGCAAG |
| UGGAU | GACUACC | |
| Total reads [HSV-1(F), 0.1 pfu per cell, 16 h] | ||
| HEK293T | 487 | 415 |
| HEp-2 | 432 | 505 |
| NT | UUCUCCGAACGUGUCACGUTT | |
| M1-H29 | CAGCUGCGGGGCAAGGACUACC | |
| M2-H29 | CACCUCCGGGGCAAGGACUACC | |
Boldface indicates the mature miRNA sequence and its location.
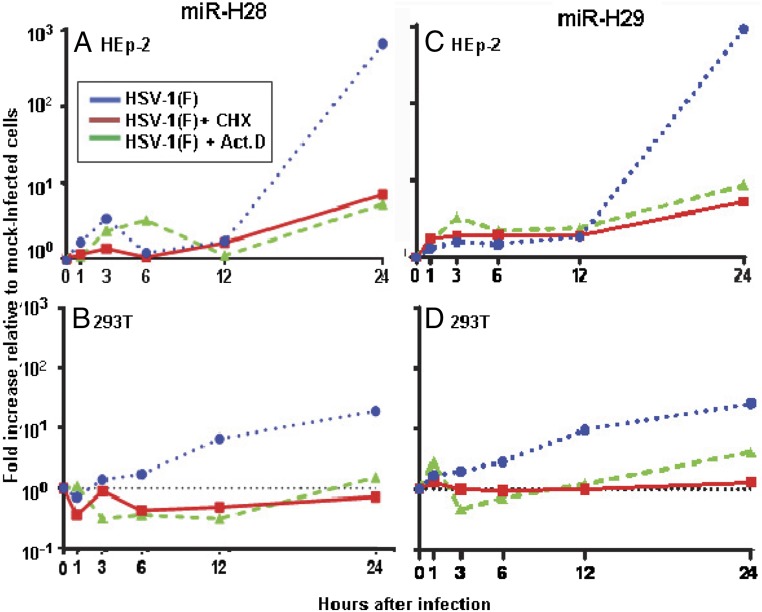
The patterns of accumulation of miR-H28 and miR-H29 in the course of productive infection in HEK293T and HEp-2 cell lines are shown in Fig. 1. In these studies, the cells were exposed to 1 pfu of wild-type virus per cell and were incubated in standard medium devoid of drugs or medium containing cycloheximide or actinomycin D. The measurements obtained were normalized with respect to the U6 small cellular RNA present in mock-infected untreated cells (0 h). The results show that expression of both miR-H28 and miR-H29 requires prior viral protein synthesis in both cell lines. The accumulation of miR-H28 and miR-H29 increased above base levels at or after 12 h postinfection.
Fig. 1.
Accumulation of miR-H28 and miR-29 in infected cells. Replicate cultures of HEp-2 cells (A and C) and HEK293T cells (B and D) were exposed to 1 pfu of HSV-1(F) per cell and were mock-treated or incubated in medium containing 100 µg/mL cycloheximide (CHX) or 10 µg/mL actinomycin D (Act.D). The cells were harvested at 1, 3, 6, 12, or 24 h postinfection. miR-H28 (A and B) and miR-H29 (C and D) were normalized with respect to U6 small cellular RNA detected in mock-infected cells (0 h).
Ectopic Expression of miR-H28 or miR-H29 Mimics in HEp-2, HEK293T, or Vero Cells Before Infection Results in Decreased Accumulation of Viral RNA and Proteins, Reduced Yields of Virus, and Smaller Plaques.
In this series of experiments, HEp-2 or HEK293T cells were transfected with miRNA corresponding to either a nontarget sequence (NT) or mimics of miR-H28 or miR-H29. We examined the accumulation of mRNAs encoding key viral proteins and infectious virus and the size of plaques formed by HSV-1(F) in Vero cells transfected with a miR-H29 mimic. The sequences of the NT miRNA and of miR-H28 and miR-H29 mimics are shown in Table 1. The results may be summarized as follows:
-
i)
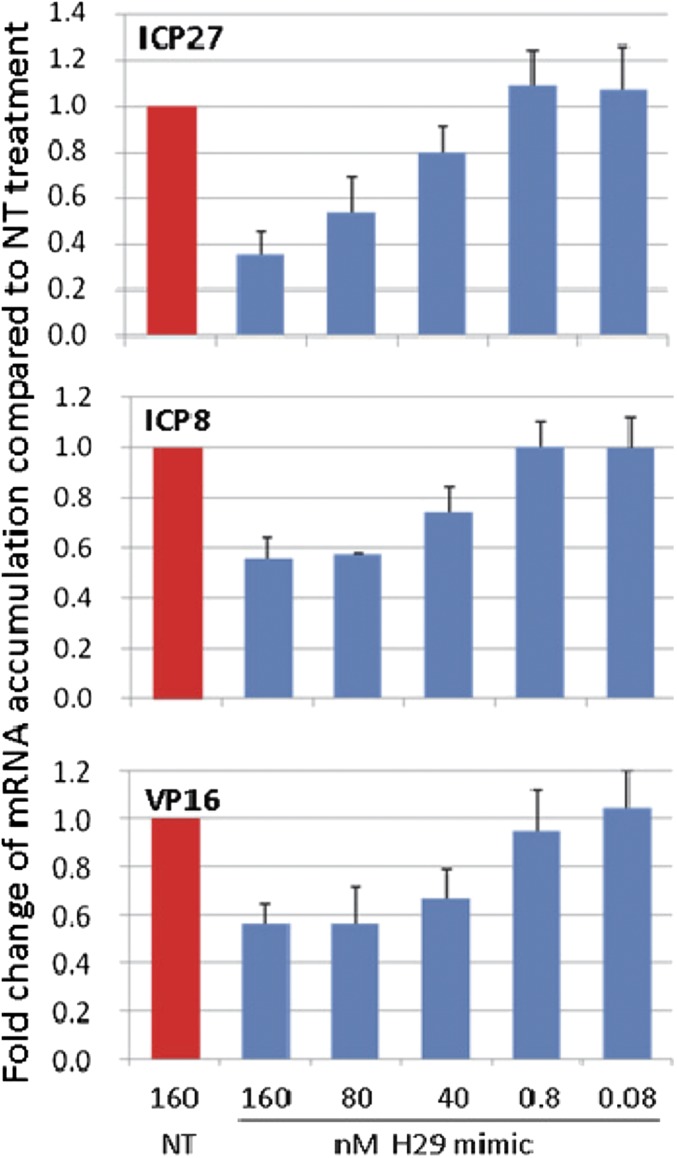
The reduction in the accumulation of viral mRNAs as a function of the amounts of miR-H29 transfected into HEp-2 cells. The objectives of these experiments were twofold: to determine whether the miR-H29 mimic is effective in reducing the accumulation of mRNAs of representative α (ICP27), β (ICP8), or γ (VP16) genes and whether the effects are dependent on the concentration of mimics transfected into cells. Replicate cultures of HEp-2 cells were transfected with 160 nM of NT miRNA or varying concentrations of miR-H29 ranging from 0.08–160 nM. After 24 h the cultures were infected with 0.1 pfu of wild-type virus per cell. After an additional 24 h the cells were harvested, RNAs were extracted, and the selected mRNAs were measured by RT-PCR as previously described (16). The results (Fig. 2) show the amounts of viral mRNA normalized with respect to 18s rRNA present in cells transfected with NT miRNAs at 24 h postinfection. The results show that the minimally effective concentration of the mimics was 80 nM and resulted in a two- to threefold decrease in the concentration of viral mRNAs.
-
ii)
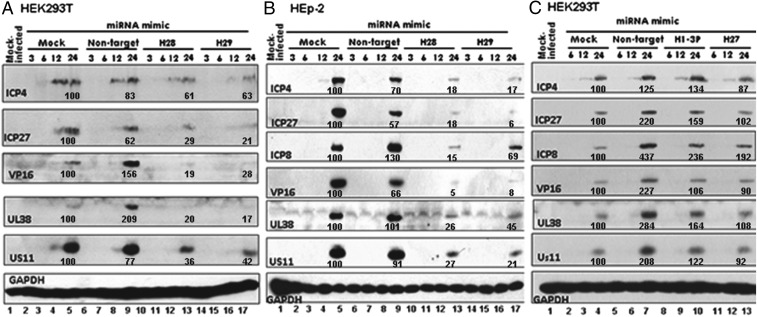
Viral protein accumulation is reduced in HEK293T or HEp-2 cells transfected with miR-H28 or miR-H29 mimics. Replicate cultures of HEK293T or HEp-2 cells were mock transfected or transfected with 150 nM of NT miRNA, miR-H28, or miR-H29 mimics. After 24 h, the cells were exposed to 0.1 pfu of wild-type virus per cell. The cultures were harvested at the indicated times postinfection and then were solubilized, subjected to electrophoresis in denaturing gels, and reacted with antibodies to ICP4, ICP27, ICP8, VP16, UL38, or US11. The proteins selected for study are the products of viral α-, β-, and γ genes. GAPDH served as a loading control. The protein bands were scanned with the aid of an ImageJ scanner and were normalized with respect to the optical density of bands produced in mock-treated infected cells. The results show that, as is consistent with the low multiplicity of infection in mock-transfected or NT miRNA-transfected cells, viral proteins were first detected in appreciable quantities 12 h postinfection and in higher amounts 24 h postinfection. In cells transfected with miR-H28 or miR-H29 mimics the amounts of the viral proteins were consistently lower than in mock-transfected HEK293T cells, infected HEK293T cells transfected with NT miRNA (Fig. 3A), or HEp-2 cells (Fig. 3B).
Fig. 2.
The miR-H29 mimic suppresses expression of viral mRNA. Replicate cultures of HEp-2 cells in 24-well plates were transfected with an miR-H29 mimic at the indicated concentrations or were treated with NT miRNA for 24 h. The cells then were exposed to 0.1 pfu of HSV-1(F) per cell for 1 h. Total RNA was extracted at 24 h postinfection. Five hundred nanograms of total RNA was reverse-transcribed to cDNA by random primers. ICP27, ICP8, and VP16 were quantified and normalized with respect to 18s rRNA and are shown as fold change compared with RNA from NT-treated cells at 24 h.
Fig. 3.
Accumulation of viral proteins in cells transfected with miR-H28, miR-H29, miR-H1-3P, and miR-H27 mimics. HEK293T cells (2 × 106) (A and C) or 2 × 106 HEp-2 cells (B) in a flask at 50–60% confluency were mock treated or were transfected with 150 nM of mimics or nontarget miRNA as indicated. After 24 h the cells were either mock infected or exposed to 0.1 pfu of HSV-1(F) per cell for 1 h; then medium was replaced with fresh medium. The cells were collected 3, 6, 12, and 24 h postinfection. The proteins were electrophoretically separated in a 10% denaturing gel and reacted with antibodies against ICP4, ICP27, ICP8, VP16, UL38, US11, or GAPDH. The protein bands were scanned with the aid of an ImageJ scanner. The optical density of the bands was normalized with respect to the optical density of corresponding bands generated from mock-treated infected cells.
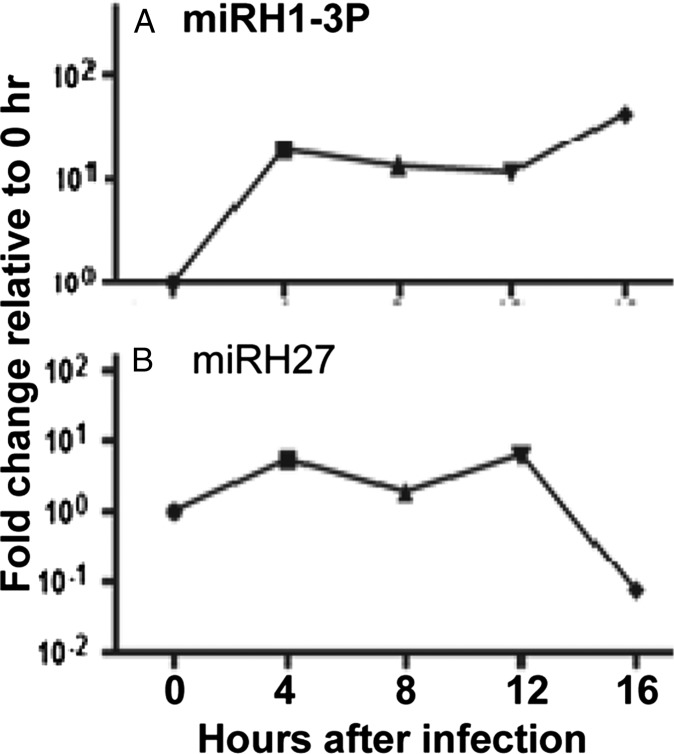
A key hypothesis underlying this report is that the miRNAs involved in restricting the expression of viral genes are those made late in infection. To test this hypothesis, we tested in the same fashion the mimics of two miRNAs made early in infection. The temporal patterns of accumulation of miR-H1-3P and miR-H27 are shown in Fig. 4. The temporal patterns of accumulation of the two miRNAs were determined as described in Materials and Methods. The results (Fig. 3C) of studies performed as described above show that the mimics of the two miRNAs made early in productively infected cells had no effect on the accumulation of viral proteins.
Fig. 4.
Accumulation of miR-H1-3P (A) and miR-H27 (B) in productively infected cells. Replicate cultures of HEK293T cells were exposed either to mock infection or to 0.1 pfu of HSV-1(F) per cell and were collected 4, 8, 12, and 16 h postinfection. miR-H1-3P and miR-H27 were quantified and normalized with respect to cellular small RNA U6 in mock-infected cells.
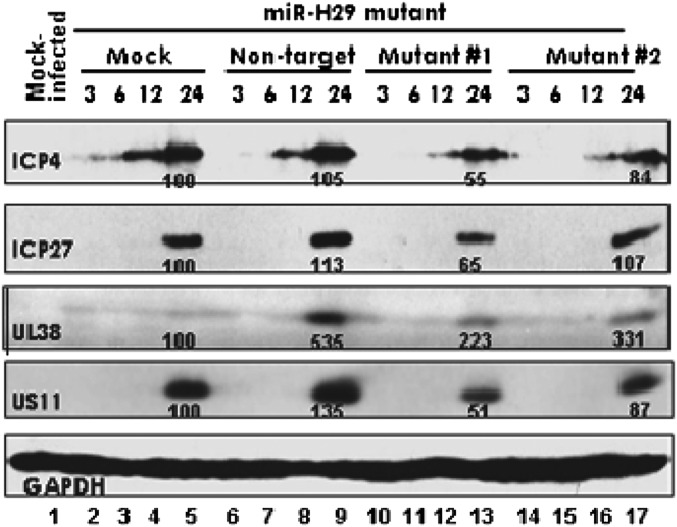
To test whether the reduction in the accumulation of viral proteins was the consequence of transfection of the mimic, we constructed two mutants, one, designated “M1,” with five replaced nucleotides in its seed sequence (Table 1) and one, designated “M2,” in which the nucleotides of the entire seed sequence were replaced (Table 1). Replicate cultures of HEK293T cells were mock treated or transfected with 150 nM of NT miRNA or M1 or M2 miRNAs. After 24 h, the cells were exposed to 0.1 pfu of HSV-1(F) per cell, were harvested at the indicated hours postinfection, and were reacted with antibodies to ICP4, ICP27, UL38, or US11. Again GAPDH served as a loading control. The results (Fig. 5) show that the pattern of accumulation of viral proteins in cells transfected with the M1 or M2 miRNAs could not be differentiated from those of mock-treated or NT miRNA-transfected cells.
-
iii)
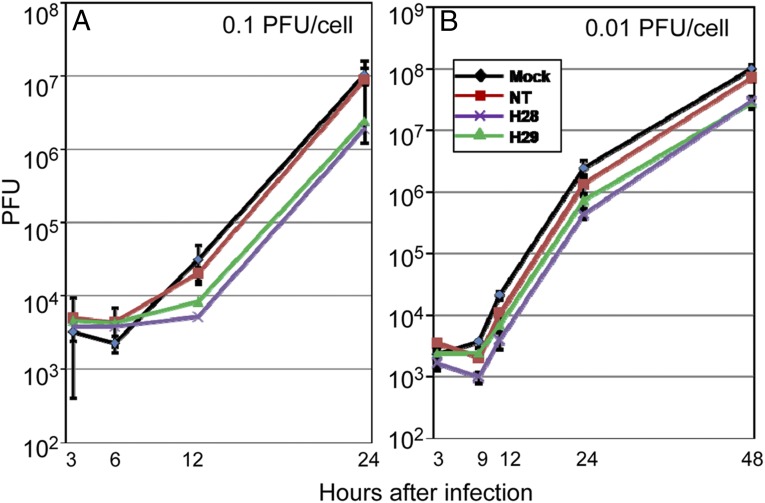
Effect of prior exposure to miR-H28 or miR-H29 on the accumulation of infectious virus in HEK293T or HEp-2 cells. In these experiments, replicate cultures of HEp-2 cells were mock treated or transfected with 150 nM of NT miRNA or with miR-H28 or miR-H29 mimics. At 24 h posttransfection the cells were exposed for 1 h to 0.1 or 0.01 pfu of the HSV-1(F) per cell. The cells then were rinsed and harvested at the times shown postinfection. Viral progeny were titrated on Vero cells. The results shown in Fig. 6 indicate that the accumulation of virus in cells transfected with miR-H28 or miR-H29 and then infected with 0.1 pfu of virus per cell was not significantly different from that in cells transfected with control miRNAs. The virus yields from cells transfected with miR-H28 and miR-H29 at 48 h postinfection were significantly lower (P = 0.02) than those obtained from cells transfected with control miRNAs.
-
iv)
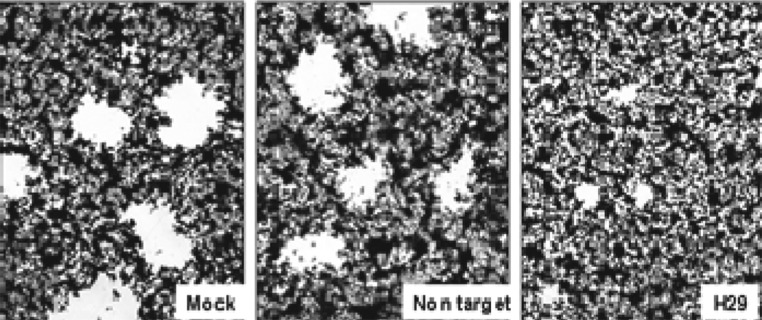
In Vero cells transfected with the miR-H29 mimic, the spread of wild-type virus from cell to cell is reduced, as indicated by plaque sizes. Vero cell monolayer cultures were mock transfected or transfected with 150 nM of NT miRNA or the miR-H29 mimic. After 24 h the cells were exposed to 0.001 pfu of HSV-1(F) per cell. The cultures were fixed and stained with Giemsa at 48 h postinfection as described in Materials and Methods. Representative plaques photographed at the same magnification are shown in Fig. 7. Examination of replicate cultures treated in the same fashion showed that the plaques formed in cells transfected with miR-H29 were significantly smaller than those formed in mock-treated cells or in monolayer cultures transfected with NT miRNA.
Fig. 5.
Mutated miR-H29 mimics have no effect on the expression of viral proteins. HEK293T cells (2 × 106) were mock treated with transfection reagent or were transfected with 150 nM of two different mutants of miR-H29 mimics or with nontarget miRNAs (NT). The sequences of mutants #1 and #2 (M1 and M2, respectively) are shown in Table 1. After 24 h the cells were either mock infected or exposed to 0.1 pfu of HSV-1(F) per cell for 1 h; then medium was replaced with fresh medium. The cells were collected 3, 6, 12, and 24 h postinfection. The proteins were electrophoretically separated in a 10% denaturing gel and reacted with indicated antibodies. The protein bands were scanned as described in the legend of Fig. 3.
Fig. 6.
Virus yields from miR-H28– and miR-H29–transfected cells. Replicate cultures of HEp-2 cells were mock treated (Mock) or treated with 150 nM of miR-H28, miR-H29, or the NT miRNA mimic. After 24 h of incubation, the cells were exposed to 0.1 pfu (A) or 0.01 pfu (B) of HSV-1(F) per cell for 1 h. The inoculum then was replaced with fresh medium. The virus progeny were harvested at the times shown and were titered in Vero cells.
Fig. 7.
The miR-H29 mimic blocks virus spreading. Vero cells grown in six-well plates were mock treated with transfection reagent or were transfected with 150 nM of NT miRNA or the miR-H29 mimic. After 24 h the cells were exposed to 0.001 pfu of HSV-1(F) per cell for 1 h and then were rinsed and overlaid with medium 199V (22). At 48 h postinfection the cells were fixed with 4% (wt/vol) paraformaldehyde for 30 min, stained with Giemsa for 30 min, and photographed at 5× magnification with the aid of an inverted microscope.
miR-H28 and miR-H29 Are Exported from Infected Cells via Exosomes.
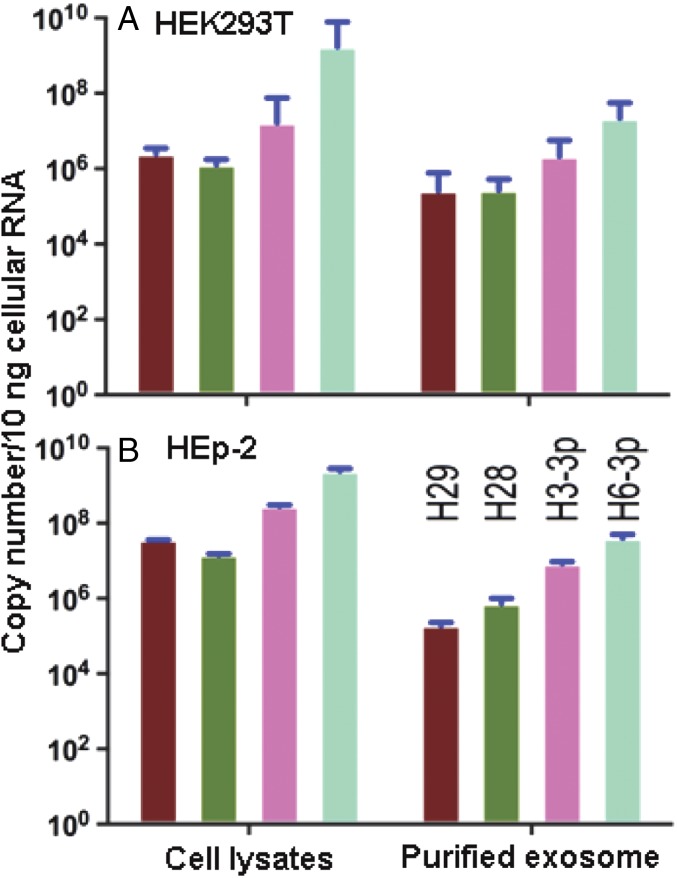
To reduce the synthesis of viral gene products and viral spread from cell to cell, miR-H28 and miR-H29 would have to be exported from infected cells to uninfected cells before or concurrently with the entry of the virus into newly infected cells. Elsewhere we have described the export of miRNA in exosomes (17). The objective of this series of experiments was to determine whether miR-H28 and miR-H29 are exported from infected cells in exosomes. In this series of experiments, HEK293T or HEp-2 cell cultures were either mock-infected or exposed to 10 pfu of HSV-1(F) per cell. After 24 h the exosomes were purified from extracellular medium, and total RNA was extracted both from exosomes and from the infected cells as described in Materials and Methods. In earlier studies (17) we have shown that miR-H3 and miR-H6 are present both in infected cells and in exosomes derived from the cultures (18). In the present study we quantified miR-H28 and miR-H29 as well as miR-H3 and miR-H6 contained in cell lysates and extracellular fluid. Analyses of miRNAs performed in duplicate cultures (Fig. 8) show that the miRNAs are present both in infected cells and in the exosomes exported from infected cells.
Fig. 8.
Analyses of exosomes for the presence of miR-H28 and miR-H29. HEK293T (A) or HEp-2 (B) cells grown in T150 flasks were exposed to 10 pfu of HSV-1(F) for 24 h. Exosomes were isolated from supernatant fluid, and total RNAs from purified exosomes were extracted as described in Materials and Methods. Total RNAs from cell lysates also were extracted as described in Materials and Methods. miR-H29, -H28, -H3-3P, and -H6 present in exosomes and cell lysates were quantified and normalized to the U6 cellular small RNA present in mock-infected cells.
miR-H28 and miR-H29 Are Expressed in Ganglionic Organ Cultures During Reactivation but Not During the Latent State.
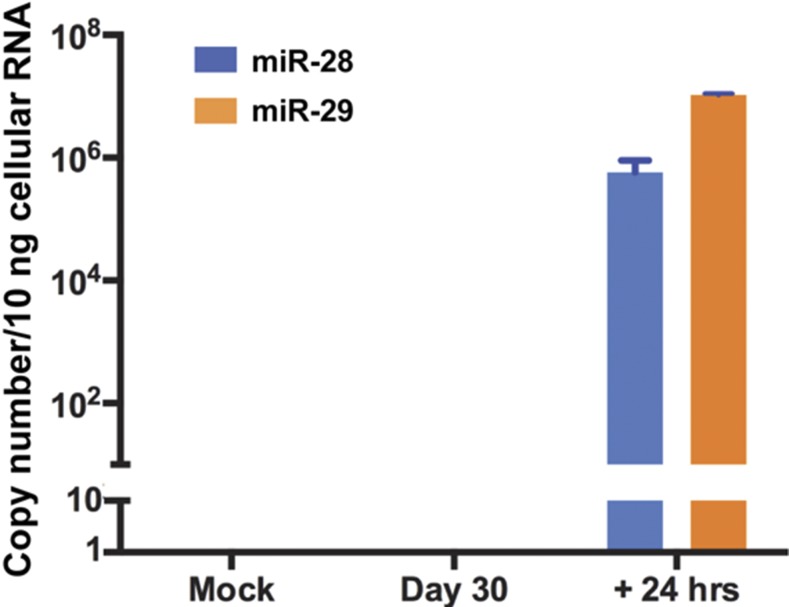
Elsewhere we reported that some miRNAs accumulate in murine trigeminal ganglia harboring latent virus but that these miRNAs decrease in amount during reactivation (14). In contrast a set of viral miRNAs is detectable only after the latent viruses undergo reactivation. The question we posed is whether miR-H28 and miR-H29 accumulate during latency or on reactivation. The protocol for these studies has been described in detail elsewhere (19). In brief, trigeminal ganglia were excised 30 d after inoculation by corneal scarification. On day 30 postinfection the trigeminal ganglia were excised and were processed immediately or were incubated for 24 h in medium containing nerve growth factor (NGF) or for the same length of time in medium containing anti-NGF antibody. The virus reactivates in medium containing anti-NGF antibody but is maintained in a latent state for at least 18–24 h in medium containing NGF. Previous studies have shown that most viral miRNAs expressed in ganglia during reactivation from the latent state are absent in ganglia harboring latent virus.
To determine whether miR-H28 or miR-H29 is expressed during latency and reactivation, we repeated the protocols described in detail elsewhere (14). Briefly, 5-wk-old CBA/J mice were inoculated by the corneal route with 105 pfu of wild-type virus per eye. The ganglia were excised 30 d postinfection and were analyzed for the presence of miR-H28 and miR-H29 either immediately or after incubation for 24 h in medium containing anti-NGF antibody. The results (Fig. 9) show that miR-H28 and miR-H29 were absent from freshly excised ganglia but were present in ganglia induced to reactivate by incubation in medium containing anti-NGF antibody.
Fig. 9.
Accumulation of miR-H28 and miR-H29 in murine trigeminal ganglia during latency and on reactivation from the latent state. At 30 d after inoculation by corneal scarification the trigeminal ganglia were excised and were analyzed immediately or were incubated in medium containing anti-NGF antibody for 24 h as described in Materials and Methods. miR-H28 and miR-H29 were quantified and normalized with respect to the U6 cellular small RNA present in ganglia excised from mock-infected mice.
Discussion
This report consists of two parts. In the first we report on the properties of two newly discovered miRNAs, miR-H28 and miR-H29. The second part stems from the observation that these miRNAs are synthesized near the end of the viral replicative cycle, that is after the synthesis of viral DNA and proteins and at a time when it could be expected that HSV had assumed full control of the infected cell. This finding raises questions regarding the function of these miRNAs.
One hypothesis that could account for the late synthesis of miR-H28 and miR-H29 stems from considerations of biologic properties that are quasi unique to HSV. Specifically, HSV spreads from person to person by direct (generally consensual) physical contact between the tissues of an infected individual and those of an uninfected individual. HSV is an exceptionally successful pathogen; in many countries the penetration rate approaches 100% of the population. Given the mode of transmission, it could be argued that people with extensive lesions on the face or genitals would be less likely to transmit the virus than individuals with barely visible lesions. In short, one hypothesis that could explain the late synthesis of miR-H28 and miR-H29 is that their function is to restrict virus spread.
To test this hypothesis, we used miR-H28 and miR-H29 mimics. In this report we show that the yields of viral mRNAs and viral proteins were reduced in cells transfected with the miR-H28 and miR-H29 mimics. In contrast, mimics of miRNAs made early postinfection had no effect on the accumulation of representative viral proteins. Finally, the results also show that in cell cultures transfected with miR-H28 and miR-H29 mimics before infection the size of plaques—a direct measure of yields and cell-to-cell spread of virus—is also diminished.
The mimics are surrogates of miRNAs, and hence their relevance needs to be qualified. Foremost, we show that mutants in the seed sequence of the mimics have no effect. The targets of the mimics are unknown. They could be viral or cellular mRNAs encoding a protein required for efficient transcription, protein synthesis, or extracellular transport.
Finally, the concept that HSV encodes functions that limit its own replication and spread is supported by numerous other observations. Specifically,
-
i)
Two components of innate immunity, protein kinase R (PKR) and NF-κB, are important cellular defense mechanisms against viral infections. Indeed, activation of PKR in the absence of ICP34.5 can result in shutoff of protein synthesis and cell death (20, 21). ICP34.5 precludes the shutoff of protein synthesis but does not block the antiviral effects of PKR because the depletion of PKR before infection results in significantly higher virus yields (22). Similarly, activated NF-κB cannot be depleted without a loss in virus yields (22). One hypothesis to explain the data is that HSV has evolved functions that depend on host defense mechanisms to control viral replication and spread.
-
ii)
STING (stimulator of interferon genes) is an important component of the signaling cascade that leads to the activation of IFN (23, 24). Recent studies show that STING is actively maintained and exported via exosomes from infected cells to uninfected cells (17). The response to STING depletion varies. In immortalized euploid cells, STING depletion results in higher virus yields (25). Although the role of exported STING remains to be verified, the data support the hypothesis that it is a means by which the virus controls its spread.
-
iii)
Viral DNA, on entering the nucleus, is coated by repressive marks. Included among the repressors is a complex consisting of HDAC-1 or 2, CoREST, and REST (16, 18, 19, 26, 27). Insertion into the viral genome of a dominant-negative REST that binds to REST response elements in viral DNA but cannot bind associated repressors (e.g., CoREST) results in a virus that is more lethal to mice than the wild-type parent (27). If increased viral yields were beneficial to virus spread, HSV would have evolved a genome free of REST response elements.
In the light of these reports, the discovery that miR-H28 and miR-H29 can be found in ganglia harboring viruses in the process of reactivation from latent state but not in ganglia harboring latent virus may be interpreted as an indication that HSV controls the amount of virus produced during reactivation. The objective may well be to block the transmission of virus to satellite cells but not the anterograde transport of reactivated virus to mucous membranes for transmission to uninfected individuals.
The strategy evolved by HSV, namely control of replication and spread in the infected individual in return for enhanced spread in the human population, does not eliminate the occurrence of severe and even lethal infections. The severity of these infections could reflect mutatios in the viral genome or a host’s impaired innate or adaptive immunity. The hypothesis and supporting data presented here argue that in these cases the probability of transmission to an uninfected individual is reduced.
Materials and Methods
Cells and Virus Strains.
HEK293T, HEp-2, or Vero cells were obtained from the American Type Culture Collection and were maintained in 10% (vol/vol) FBS, 5% (vol/vol) FBS, or 5% (vol/vol) newborn bovine serum, respectively. HSV-1(F) is the prototype HSV-1 strain used in our laboratories (15).
High-Throughput Sequencing.
HEK293T and HEp-2 cells were either mock infected or exposed to 0.1 pfu of HSV-1(F) per cell. The cells were harvested 16 h postinfection. Small RNAs were isolated and subjected to high-throughput sequencing by Capital Bio Corporation to identify HSV-1–derived miRNAs.
miRNA Mimics.
Mimics of miR-H28, miR-H29, and mutated miR-H29 mimics were designed and purchased from GenPharma. The sequences are shown in Table 1.
Antibodies.
Antibodies against to ICP4 (28), ICP27 (28), ICP8 (Rumbaugh-Goodwin Institute for Cancer Research, Inc.), VP16 (29), UL38 (30), and US11 (31) have been reported elsewhere. The antibody against GAPDH was purchased from Cell Signaling Technology.
RNA Extraction.
Total RNAs were extracted from murine trigeminal ganglia (TG) harboring latent viruses or from infected cells by the mirVana miRNA Isolation Kit (Ambion) or TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and earlier reports (32, 33). Murine model studies of HSV-1 latent infection and reactivation were performed at the University of Chicago as described elsewhere (32), according to protocols approved by the Institutional Animal Care and Use Committee of the University of Chicago.
Purification of Exosomes and Total Exosomal RNA.
Exosomes were purified from mock- or HSV-1(F)–infected cells with the aid of the Exosome Isolation Kit (Life Technology). Then total RNA was extracted from the purified exosome with the aid of the Total Exosomal RNA & Protein Isolation Kit (Life Technology) according to the manufacturer’s instructions.
Quantitative RT-PCR for miRNAs and Viral Genes.
The miRNAs tested in this study were reverse-transcribed in duplicate for real-time PCR after stem–loop reverse transcription as described in detail in the TaqMan miRNA reverse transcription kit (Applied Biosystems, Inc.). TaqMan MGB probes of miR-H27, -H3-3p, -H6-3p, -H28, and -H29 for real-time PCR assays were purchased from Applied Biosystems, Inc. A pair of locked nucleic acid-modified primers for real-time quantification of miR-H1-3P were purchased from Exiqon, Inc. The mRNAs of viral genes ICP27, ICP8, VP16, and 18s rRNA were quantified by quantitative RT-PCR using the SYBR Green Real-time PCR Master Mix Kit purchased from ToYoBo Life Science.
Virus Titration.
HEp-2 cells were transfected with miR-H28 or miR-H29 mimics. After 24 h the cells were exposed to 0.1 or 0.01 pfu of HSV-1(F) per cell. The cells were harvested at indicated time of postinfection. Viral progeny were titrated on Vero cells.
Plaque Assay.
Vero cells in six-well plates were transfected with 150 nM of the miR-H29 mimic or nontargeting miRNA for 24 h and then were exposed to 0.001 pfu of HSV-1(F) per cell for 1 h and were maintained in complete DMEM for 48 h. The cells were fixed with 4% (wt/vol) of paraformaldehyde for 30 min, rinsed three times with PBS, and stained with Giemsa.
Acknowledgments
We thank Lindsay Smith for technical assistance. These studies were supported by a grant from the Joseph Regenstein Foundation to the University of Chicago and by National Nature Science Foundation of China Grant NSFC 81472826 to Guangzhou Medical University. X.L. is the recipient of Grant 2015M572292 from the Postdoctoral Science Foundation of China.
Footnotes
The authors declare no conflict of interest.
References
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses. Fields' Virology, eds Knipe DM, et al. (Wolters Kluwer/Lippincott-Williams and Wilkins, New York), 6th Ed., pp 1823–1897.
- 2.Roizman B, Zhou G. The 3 facets of regulation of herpes simplex virus gene expression: A critical inquiry. Virology. 2015;479-480:562–567. doi: 10.1016/j.virol.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui C, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80(11):5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurak I, et al. Expression of herpes simplex virus 1 microRNAs in cell culture models of quiescent and latent infection. J Virol. 2014;88(4):2337–2339. doi: 10.1128/JVI.03486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurak I, et al. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;84(9):4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer MF, et al. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology. 2011;417(2):239–247. doi: 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munson DJ, Burch AD. A novel miRNA produced during lytic HSV-1 infection is important for efficient replication in tissue culture. Arch Virol. 2012;157(9):1677–1688. doi: 10.1007/s00705-012-1345-4. [DOI] [PubMed] [Google Scholar]
- 8.Flores O, et al. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol. 2013;87(12):6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Li Q. The miRNAs of herpes simplex virus (HSV) Virol Sin. 2012;27(6):333–338. doi: 10.1007/s12250-012-3266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H, Zhang C, Hou G, Song J. MicroRNA-H4-5p encoded by HSV-1 latency-associated transcript promotes cell proliferation, invasion and cell cycle progression via p16-mediated PI3K-Akt signaling pathway in SHSY5Y cells. Int J Clin Exp Med. 2015;8(5):7526–7534. [PMC free article] [PubMed] [Google Scholar]
- 12.Yun SJ, et al. Urinary microRNAs of prostate cancer: virus-encoded hsv1-miRH18 and hsv2-miR-H9-5p could be valuable diagnostic markers. Int Neurourol J. 2015;19(2):74–84. doi: 10.5213/inj.2015.19.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X, et al. A herpes simplex virus type 1 mutant disrupted for microRNA H2 with increased neurovirulence and rate of reactivation. J Neurovirol. 2015;21(2):199–209. doi: 10.1007/s13365-015-0319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du T, Han Z, Zhou G, Roizman B. Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation. Proc Natl Acad Sci USA. 2015;112(1):E49–E55. doi: 10.1073/pnas.1422657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 16.Zhou G, Te D, Roizman B. The CoREST/REST repressor is both necessary and inimical for expression of herpes simplex virus genes. MBio. 2011;2(1):e00313–10. doi: 10.1128/mBio.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA. 2014;111(46):E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roizman B. The checkpoints of viral gene expression in productive and latent infection: The role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol. 2011;85(15):7474–7482. doi: 10.1128/JVI.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou G, Du T, Roizman B. HSV carrying WT REST establishes latency but reactivates only if the synthesis of REST is suppressed. Proc Natl Acad Sci USA. 2013;110(6):E498–E506. doi: 10.1073/pnas.1222497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94(3):843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou J, Chen JJ, Gross M, Roizman B. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5- mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92(23):10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taddeo B, Zhang W, Lakeman F, Roizman B. Cells lacking NF-kappaB or in which NF-kappaB is not activated vary with respect to ability to sustain herpes simplex virus 1 replication and are not susceptible to apoptosis induced by a replication-incompetent mutant virus. J Virol. 2004;78(21):11615–11621. doi: 10.1128/JVI.78.21.11615-11621.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227):aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalamvoki M, Roizman B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci USA. 2014;111(5):E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu H, Roizman B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Virol. 2009;83(9):4376–4385. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du T, Zhou G, Khan S, Gu H, Roizman B. Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc Natl Acad Sci USA. 2010;107(36):15904–15909. doi: 10.1073/pnas.1010741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackermann M, Braun DK, Pereira L, Roizman B. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52(1):108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKnight JL, Kristie TM, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc Natl Acad Sci USA. 1987;84(20):7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward PL, Ogle WO, Roizman B. Assemblons: Nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70(7):4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roller RJ, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66(6):3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du T, Zhou G, Roizman B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci USA. 2011;108(46):18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du T, Zhou G, Roizman B. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci USA. 2013;110(28):E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]