Significance
Emerging evidence suggest that the cellular composition of tumors is highly heterogeneous. Subclonal species of malignant cells may account for variability in therapeutic responses and for relapse following treatments. However, little is known about the molecular drivers of specific subsets of cancer cells. Herein, we identify expression of platelet-derived growth factor receptor beta (PDGFRβ) as a previously unrecognized feature of a minor malignant cell population in pancreatic neuroendocrine tumors. By the use of mice genetically deficient for Pdgfd, we reveal a crucial and nonredundant function for signaling by platelet-derived growth factor (PDGF)-DD in promoting functional tumor heterogeneity by providing growth-stimulatory cues. Taken together, the use of drugs targeting PDGFRβ signaling, such as the approved targeted therapy sunitinib, may affect the functional intratumoral cross talk in pancreatic neuroendocrine tumors.
Keywords: tumor heterogeneity, platelet-derived growth factor-DD, neuroendocrine tumor
Abstract
Intratumoral heterogeneity is an inherent feature of most human cancers and has profound implications for cancer therapy. As a result, there is an emergent need to explore previously unmapped mechanisms regulating distinct subpopulations of tumor cells and to understand their contribution to tumor progression and treatment response. Aberrant platelet-derived growth factor receptor beta (PDGFRβ) signaling in cancer has motivated the development of several antagonists currently in clinical use, including imatinib, sunitinib, and sorafenib. The discovery of a novel ligand for PDGFRβ, platelet-derived growth factor (PDGF)-DD, opened the possibility of a previously unidentified signaling pathway involved in tumor development. However, the precise function of PDGF-DD in tumor growth and invasion remains elusive. Here, making use of a newly generated Pdgfd knockout mouse, we reveal a functionally important malignant cell heterogeneity modulated by PDGF-DD signaling in pancreatic neuroendocrine tumors (PanNET). Our analyses demonstrate that tumor growth was delayed in the absence of signaling by PDGF-DD. Surprisingly, ablation of PDGF-DD did not affect the vasculature or stroma of PanNET; instead, we found that PDGF-DD stimulated bulk tumor cell proliferation by induction of paracrine mitogenic signaling between heterogeneous malignant cell clones, some of which expressed PDGFRβ. The presence of a subclonal population of tumor cells characterized by PDGFRβ expression was further validated in a cohort of human PanNET. In conclusion, we demonstrate a previously unrecognized heterogeneity in PanNET characterized by signaling through the PDGF-DD/PDGFRβ axis.
Undeniably, cancer progression is the consequence of dynamic, and yet poorly understood, cell–cell interactions driven by frequently deregulated signaling pathways (1). Further complexity arises from the notion that tumors are composed of phenotypically and functionally distinct subsets of both malignant and stromal cells (2, 3). Therefore, accounting for intratumoral heterogeneity poses an additional challenge when designing therapies that can efficiently control or eliminate tumors. An improved understanding of the functional contribution of different signaling pathways to genetic and phenotypic variation within tumors is therefore highly warranted.
Members of the platelet-derived growth factor (PDGF) family and their receptors (PDGFRs) have been extensively investigated and shown to be critical for cellular processes such as proliferation, survival, and motility during tumor growth and invasion (4). The roles of PDGF isoforms and their target cells in tumor development have been charted in different tumor types (5), and as a result, pharmacological blockade of PDGF signaling is now routinely used for the treatment of diverse malignancies, such as gastrointestinal stromal tumors and chronic myelomonocytic leukemia, among others (6, 7). The PDGF family is composed of four polypeptide chains that assemble into five dimeric isoforms (PDGF-AA, PDGF-BB, PDGF-AB, PDGF-CC, and PDGF-DD) that bind and activate two receptor tyrosine kinases (PDGFRα and PDGFRβ) expressed mainly by cells of mesenchymal origin (8). PDGF-DD is the most recently identified member of the family (9, 10), and unlike the other ligands, the role of PDGF-DD in normal development and pathology is largely a conundrum.
Herein, we report the use of a Pdgfd knockout mouse to explore the specific role of PDGF-DD in malignant growth. By monitoring tumorigenesis in the RIP1-TAg2 mouse model of pancreatic neuroendocrine tumors (PanNET), we found that disruption of PDGF-DD signaling significantly delayed tumor growth. In the absence of PDGF-DD, functional compensation by PDGF-BB was apparent in the stromal compartment. Unexpectedly, however, we identified a subpopulation of malignant cells expressing PDGFRβ with accompanying responsiveness to PDGF-DD. By modulating PDGFRβ+ malignant cells, PDGF-DD contributes to the maintenance of functional malignant cell heterogeneity in experimental PanNET.
Results
Pdgfd Is Predominantly Expressed in the Endothelial Cell Compartment of Tumors from RIP1-TAg2 Mice.
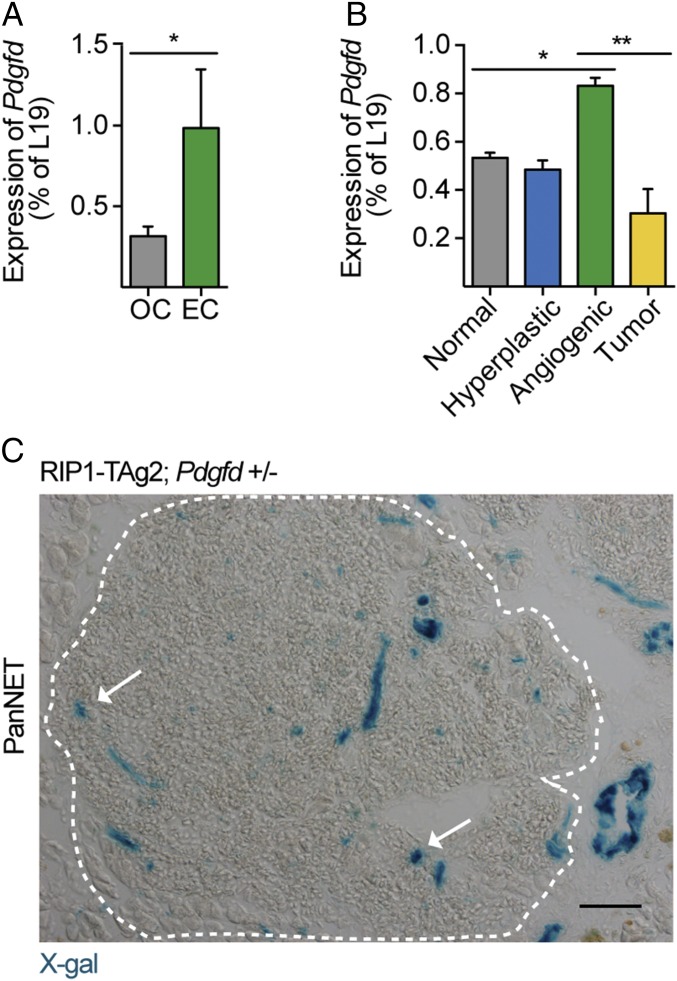
To study the effect of Pdgfd depletion in tumor development, we made use of the RIP1-TAg2 transgenic mouse model of multistage PanNET (11). Briefly, pancreatic β-cells in the islets of Langerhans of RIP1-TAg2 mice are engineered to express the oncogenic SV40 T antigens, under the control of the rat insulin promoter, leading to the formation of hyperproliferative islets that progress by activating angiogenesis and ultimately resulting in locally invasive and metastatic tumors. Previous expression profiling of PDGF ligands and receptors in tumors from RIP1-TAg2 mice found Pdgfd to be expressed exclusively by endothelial cells (ECs) (12). Consistent with these results, we observed a significant enrichment of Pdgfd mRNA in isolated ECs of tumors from RIP1-TAg2 mice, compared with non-ECs (Fig. 1A). In addition, when we analyzed the expression pattern of Pdgfd during tumorigenesis in RIP1-TAg2 mice, we found Pdgfd to be significantly up-regulated in angiogenic islets, compared with other stages of normal or malignant islets (Fig. 1B), an expression pattern previously found to be characteristic for genes expressed by ECs (13). Next, we generated a mouse line in which the Pdgfd exon 1 was substituted for a LacZ reporter cassette, allowing for monitoring of gene expression by X-gal staining. Using tumor tissue sections from compound RIP1-TAg2;Pdgfd+/− mice, we detected robust LacZ activity in large vessels, as well as weaker signal in microvascular structures within the tumor parenchyma (Fig. 1C, arrows). Taken together, these observations suggest that in the RIP1-TAg2 tumor model, ECs are the predominant source for PDGF-DD.
Fig. 1.
Pdgfd is expressed primarily by endothelial cells in tumors from RIP1-TAg2 mice. (A) Quantitative RT-PCR analysis of the expression of Pdgfd in endothelial cell (EC) fraction and other cell (OC) fraction isolated from tumors of RIP1-TAg2 mice. Error bars show the mean ± SD. (B) qRT-PCR analysis of the expression of Pdgfd in pancreatic islets from progressive tumor stages in RIP1-TAg2 mice (material pooled from >20 mice per tumor stage). (C) X-gal staining of islet tumor section from RIP1-TAg2;Pdgfd+/− mouse. A dashed line delineates the angiogenic islet lesion area. *P < 0.05, **P < 0.01. (Scale bar, 50 μm.)
Pdgfd Deficiency Delays Tumor Growth, Leading to Prolonged Survival.
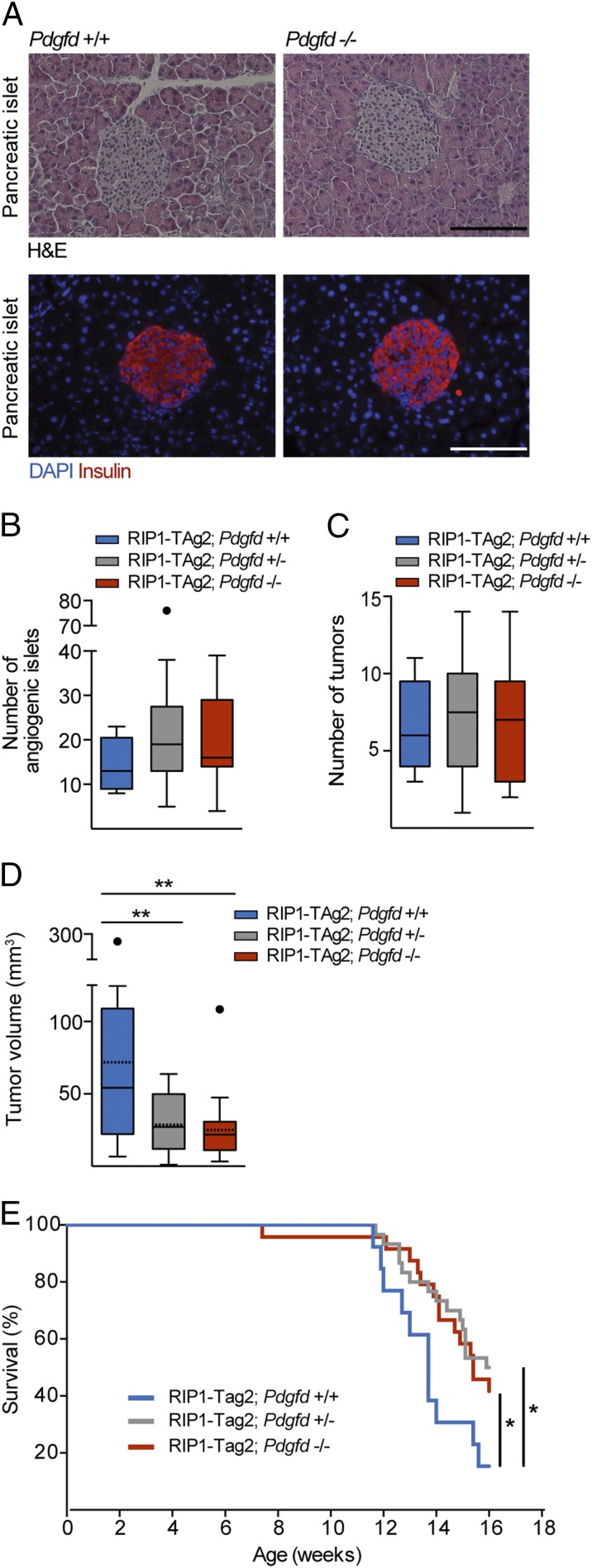
Mice homozygous for the inactivated Pdgfd allele (Pdgfd−/− mice) are viable and fertile and do not display any obvious discrepancies in the histology or insulin secretion of the islets of Langerhans, compared with Pdgfd+/+ littermates (Fig. 2A). We next evaluated the tumorigenic progression of RIP1-TAg2;Pdgfd+/− and RIP1-TAg2;Pdgfd−/− mice to that of RIP1-TAg2;Pdgfd+/+ littermates. First, we examined the effect of impaired Pdgfd expression on the activation of the angiogenic switch by quantifying the number of angiogenic islets and tumors present in the pancreas of 12-wk-old RIP1-TAg2 mice. Our analysis revealed a similar number of both angiogenic islets and tumors regardless of genotype (Fig. 2 B and C), suggesting that PDGF-DD does not affect the progression of tumors from premalignant angiogenic lesions into overt tumors. In sharp contrast, both RIP1-TAg2;Pdgfd+/− and RIP1-TAg2;Pdgfd−/− mice presented with a significantly reduced total tumor burden (29.5 ± 18 mm3 and 25.7 ± 21.1 mm3, respectively) compared with RIP1-TAg2;Pdgfd+/+ mice (71.9 ± 68 mm3) (Fig. 2D). Consistent with the decrease in tumor burden, RIP1-TAg2;Pdgfd+/− and RIP1-TAg2;Pdgfd−/− mice also showed significantly prolonged median survival (15.9 wk and 15.4 wk, respectively) compared with RIP1-TAg2;Pdgfd+/+ littermates (13.7 wk) (Fig. 2E).
Fig. 2.
Pdgfd deficiency delays tumor growth, leading to prolonged survival. (A) Representative images of islets of Langerhans from Pdgfd+/+ and Pdgfd−/− mice used for assessment of histology, demonstrated by H&E staining, and functionality, shown by immunostaining for insulin (red) to visualize secretion and distribution. Nuclei were stained with DAPI in Lower. (Scale bar, 100 μm.) (B) Quantification of the number of angiogenic islets, (C) the number of tumors, and (D) total tumor burden in 12-wk-old RIP1-TAg2;Pdgfd+/+ (n = 17), RIP1-TAg2;Pdgfd+/− (n = 26), and RIP1-TAg2;Pdgfd−/− (n = 25) mice. Boxes represent the interquartile range, and the bars represent the full range. Solid lines represent median values and dashed lines represent mean values. Full circles denote statistical outliers. (E) Survival of RIP1-TAg2;Pdgfd+/+ (blue line; median survival = 13.7 wk, n = 13), RIP1-TAg2;Pdgfd+/− (gray line; median survival = 15.9 wk, n = 30, P < 0.05), and RIP1-TAg2;Pdgfd−/− (red line; median survival = 15.4 wk, n = 24, P < 0.05) mice. Error bars show the mean ± SD. *P < 0.05, **P < 0.01.
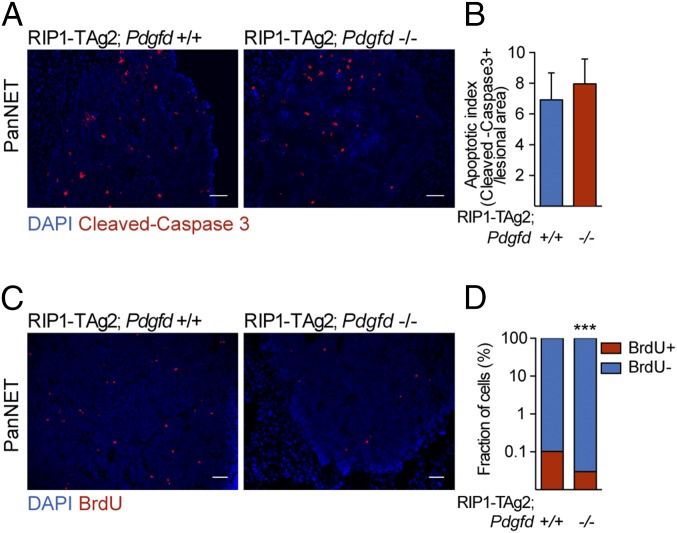
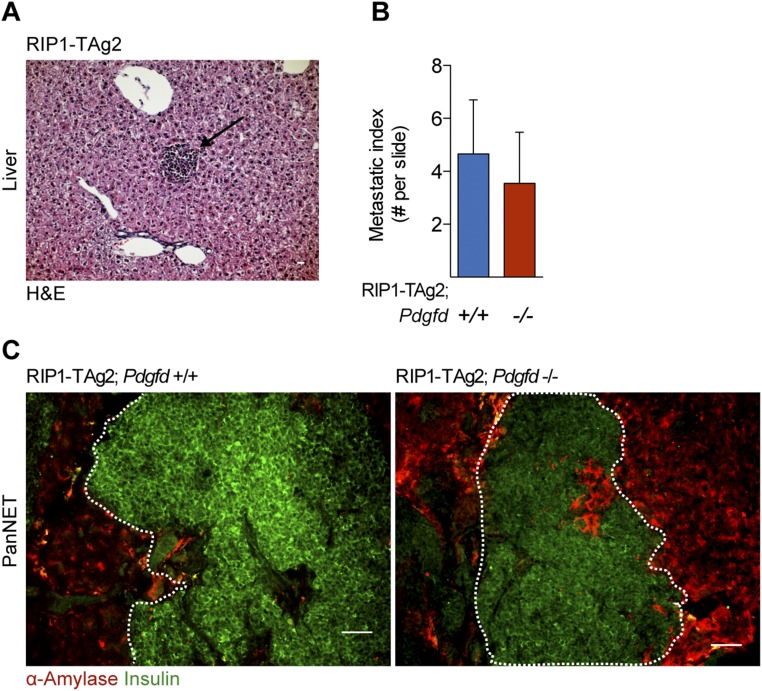
Pdgfd Ablation Reduces Tumor Cell Proliferation but Does Not Affect the Invasive or Metastatic Properties of Tumors from RIP1-TAg2 Mice.
To investigate whether the diminished tumor size in tumors from RIP1-Tag2;Pdgfd−/− mice was due to an increase in apoptosis or a decrease in proliferation, we stained tumor tissue sections for cleaved caspase-3, an apoptotic cell marker (Fig. 3 A and B), and assessed the proliferative rate by injecting mice with BrdU (Fig. 3 C and D). No change was observed when we quantified apoptotic cells in tumors from RIP1-TAg2;Pdgfd−/− mice compared with RIP1-TAg2;Pdgfd+/+ controls (Fig. 3B). In contrast, we detected a considerable decrease of 69% in proliferating BrdU+ cells in tumors from RIP1-TAg2;Pdgfd−/− compared with RIP1-TAg2;Pdgfd+/+ littermates (Fig. 3D). An increasing number of studies propose that PDGF-DD regulates the process of epithelial-to-mesenchymal transition (EMT), an event preceding metastatic spread (14). Hematoxylin/eosin (H&E) staining of liver tissue sections (Fig. S1A) revealed that the number of hepatic micrometastatic lesions was not different in RIP1-TAg2;Pdgfd−/− mice compared with RIP1-TAg2;Pdgfd+/+ mice (Fig. S1B). Furthermore, visualization of local tumor invasion, as determined by the border of the pancreatic endocrine lesion (assessed by immunostaining for insulin) with the surrounding exocrine tissue (assessed by immunostaining for α-amylase) demonstrated that tumors invaded the adjacent exocrine tissue to the same extent, regardless of Pdgfd expression (Fig. S1C).
Fig. 3.
Pdgfd deficiency does not affect cell apoptosis, but reduces tumor cell proliferation in tumors from RIP1-TAg2 mice. (A and B) Apoptotic index assessed by cleaved-caspase 3 immunostaining and (C and D) proliferation assessed by BrdU immunostaining of tumor sections from RIP1-TAg2;Pdgfd+/+ (n = 4) and RIP1-TAg2;Pdgfd−/− (n = 6). Seven to twelve islet lesions were assessed for each mouse. The number of apoptotic and proliferating cells was determined by quantification of the positively stained cells in relation to the total tumor lesion area (mm2) or total number of cells/lesion, respectively. Error bars show the mean ± SD. ***P < 0.001. (Scale bars, 50 μm.)
Fig. S1.
Local tumor invasion and metastatic dissemination are not affected in the absence of PDGF-DD in tumors of RIP1-TAg2 mice. (A) Representative images of hematoxylin/eosin staining of a hepatic micrometastatic lesion and (B) quantification of micrometastatic foci in the liver of RIP1-TAg2;Pdgfd+/+ and RIP1-TAg2;Pdgfd−/− mice at 12 wk of age (n = 6 mice per group). Error bars show the mean ± SD. (C) Representative images of tumor islets from RIP1-TAg2;Pdgfd+/+ and RIP1-TAg2;Pdgfd−/− depicting tumor cells (insulin; green) invading through acinar exocrine tissue (amylase; red). (Scale bars, 50 μm.)
Angiogenesis, Pericyte Recruitment, and Immune Cell Infiltration Are Not Affected by PDGF-DD Inhibition.
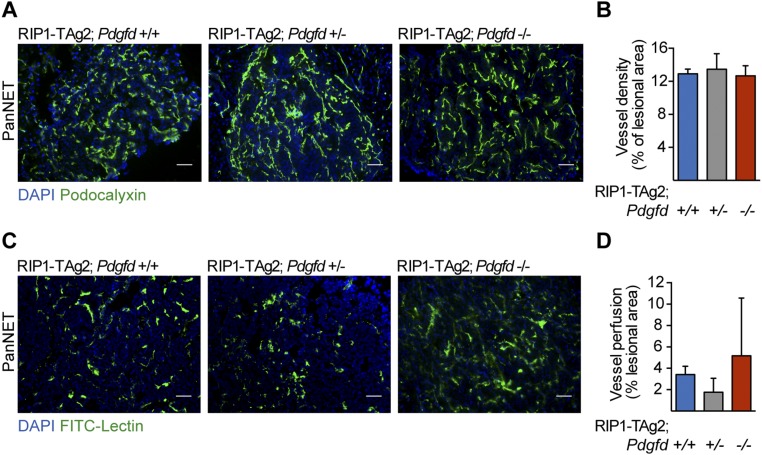
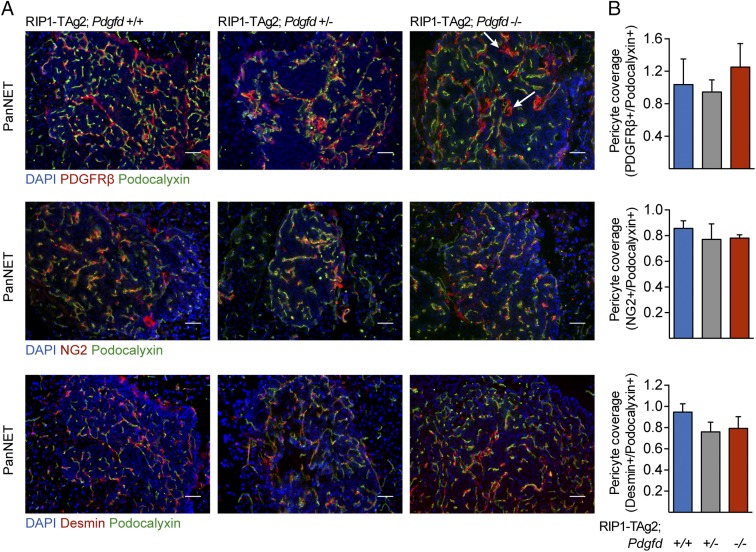
Given the reported effect of PDGF ligands on tumor angiogenesis in general and pericyte recruitment in particular (15), we characterized the vascular phenotype of tumors in RIP1-TAg2 mice following Pdgfd disruption. The vascular density, as shown by immunostaining for the luminal vessel marker podocalyxin (Fig. S2A), was unchanged upon blunted Pdgfd expression (Fig. S2B). Similarly, tumor vessel perfusion, measured in mice that were systemically administered with fluorescein-coupled tomato lectin before sacrifice, was not significantly affected by the absence of PDGF-DD (Fig. S2 C and D). The role of PDGF-BB, the prototypical ligand for PDGFRβ, in recruitment of pericytes to the tumor vasculature in RIP1-TAg2 mice has been previously described (16). Therefore, we asked whether PDGF-DD would have a similar effect on pericyte recruitment to tumor blood vessels. By immunostaining, we analyzed tumor sections from RIP1-TAg2 mice for the expression of well-characterized markers denoting different subsets of pericytes, i.e., PDGFRβ, NG2, and desmin (2, 17, 18) (Fig. 4A). Unexpectedly, pericyte coverage was unchanged in the tumor vasculature of RIP1-TAg2;Pdgfd+/− and RIP1-TAg2;Pdgfd−/− mice compared with RIP1-TAg2;Pdgfd+/+ mice (Fig. 4B). However, we observed in rare malignant lesions that PDGFRβ+ peri-vascular cells in tumors from RIP1-TAg2;Pdgfd−/− mice appeared more detached from the abluminal endothelial surface in blood vessels, compared with tumors from RIP1-TAg2;Pdgfd+/+ mice (Fig. 4A, arrows). Given that the increased detachment did not correlate with changes in vessel density or functionality (Fig. S2 A–D), we concluded that pericyte detachment was most likely not sufficient to account for the differences in tumor size observed upon impairment of Pdgfd expression.
Fig. S2.
Angiogenesis is not affected by PDGF-DD ablation in tumors from RIP1-Tag2 mice. (A and B) Quantification of the vascular density (podocalyxin immunostaining) and (C and D) vessel perfusion (FITC-conjugated lectin) as a percentage of the lesion area in RIP1-TAg2;Pdgfd+/+, RIP1-TAg2;Pdgfd+/−, and RIP1-TAg2;Pdgfd−/− mice. Cell nuclei were visualized using DAPI (blue) (n = 3 mice per group). Error bars show the mean ± SD. (Scale bars, 50 μm.)
Fig. 4.
Pericyte recruitment is not affected by PDGF-D ablation in tumors of RIP1-TAg2 mice. (A and B) Pericyte coverage quantification in tumor sections from RIP1-TAg2;Pdgfd+/+, RIP1-TAg2;Pdgfd+/−, and RIP1-TAg2;Pdgfd−/− mice based on immunostaining for pericyte markers NG2, PDGFRβ, and desmin (red) in relation to the endothelial cell marker podocalyxin (green). Cell nuclei were visualized using DAPI (blue) (n = 3 mice per group). Error bars show the mean ± SD. (Scale bars, 50 μm.)
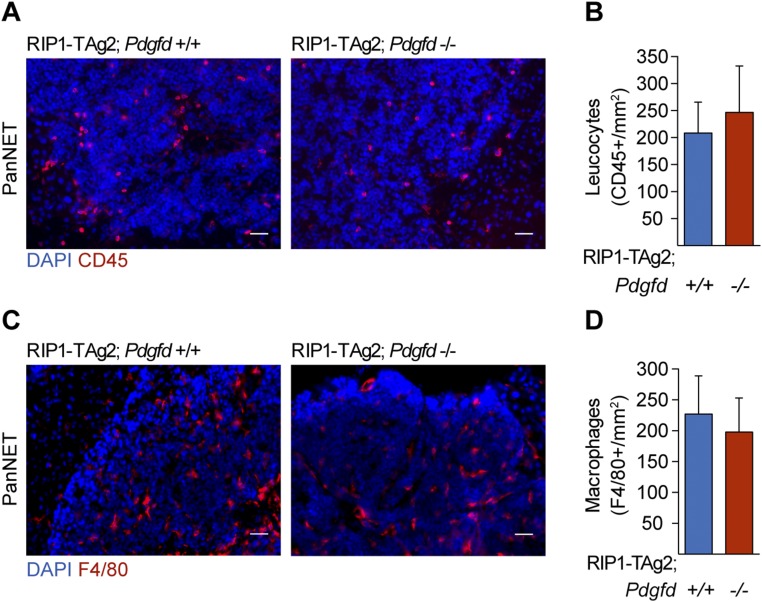
Additionally, in a mouse model of wound healing, Pdgfd overexpression was accompanied by an increased recruitment of macrophages (19), a cell type associated with the angiogenic phenotype in tumors from RIP1-TAg2 mice (20). However, the immune profile of RIP1-TAg2 mice characterized by immunostaining of tumor tissue sections for a general leukocyte marker (CD45) and a macrophage marker (F4/80) did not give any evidence for gross differences in the infiltration of inflammatory cells upon Pdgfd deletion (Fig. S3 A–D).
Fig. S3.
Immune cell infiltration is not affected upon inhibition of PDGF-DD. Immune cell infiltration in tumor sections from RIP1-TAg2;Pdgfd+/+ and RIP1-TAg2;Pdgfd−/− mice assessed by staining for (A and B) leukocytes (CD45; red) and (C and D) macrophages (F4/80; red). The number of tumor-infiltrating immune cells was determined by quantifying positively stained cells in relation to the total tumor lesion area (n = 3 mice per group, 7–12 lesions per mice). Error bars show the mean ± SD. (Scale bars, 50 μm.)
Identification of a Subset of Malignant Pancreatic β-Cells Expressing PDGFRβ in RIP1-TAg2 Mice.
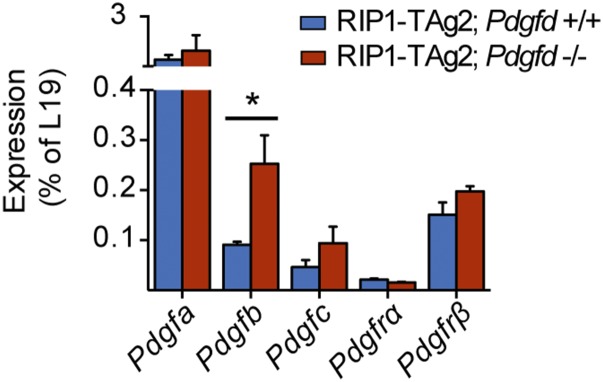
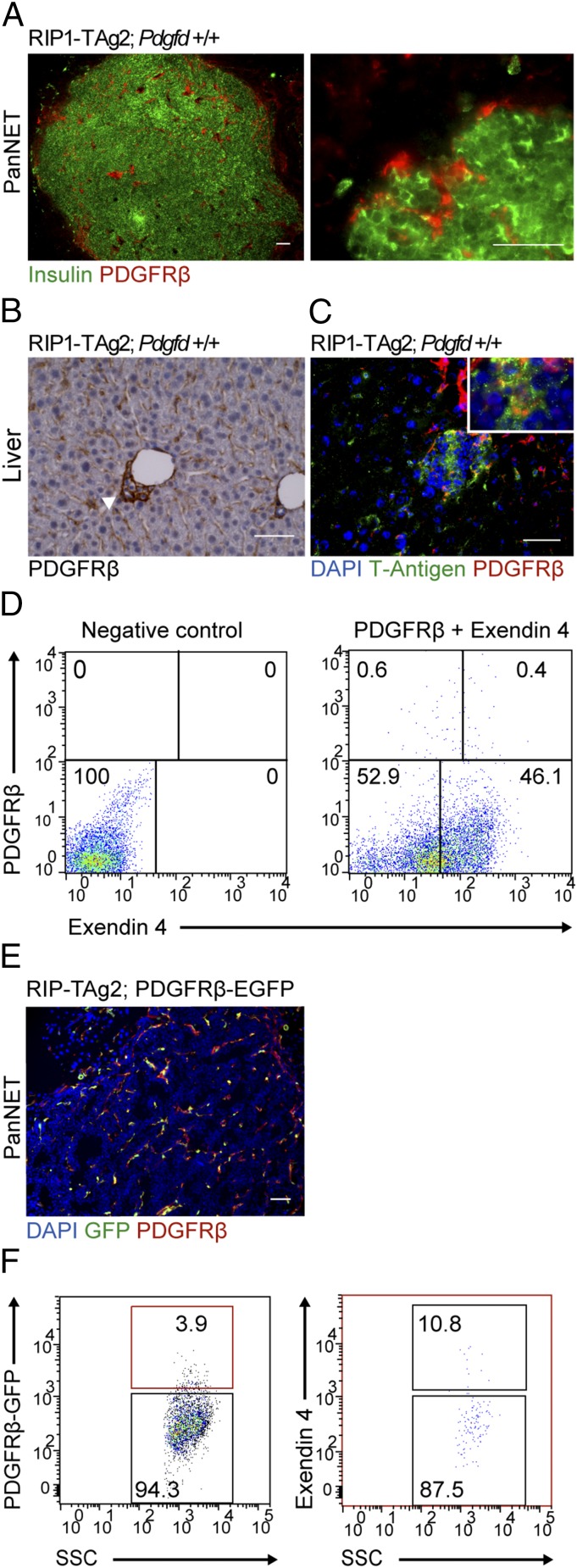
Because we did not detect major alterations in the tumor stroma of PanNET lesions from RIP1-TAg2 mice following Pdgfd depletion, we profiled the expression of PDGF family members by quantitative RT-PCR (qRT-PCR). Notably, we found that the level of Pdgfb transcript was significantly increased in tumors from RIP1-TAg2;Pdgfd−/− mice compared with RIP1-TAg2;Pdgfd+/+ mice (Fig. 5), suggesting a compensatory effect due to the lack of PDGF-DD. Nevertheless, the up-regulation of Pdgfb was still unable to rescue the observed reduction in overall tumor size, indicative of a specific role for PDGF-DD signaling during PanNET development. Therefore, we explored alternative mechanisms that could account for the significant reduction in tumor size. We speculated that there might exist cell types outside of the perivascular compartment responsive to PDGF-DD by expression of PDGFRβ. Consequently, we performed immunostaining of tumor and liver tissue sections from RIP1-TAg2 mice for PDGFRβ and insulin or T-Antigen to label malignant cells. We identified rare cells coexpressing PDGFRβ and insulin in primary tumors (Fig. 6A, Right), consistent with previous reports of activated PDGFRβ in whole tumor lysates of human PanNET lesions (21). Malignant cells expressing PDGFRβ were more prevalent in micrometastatic lesions in the liver (Fig. 6 B and C). To further confirm the observation of PDGFRβ+ malignant β-cells, we prepared a single-cell suspension from tumors of RIP1-TAg2 mice and ex vivo labeled the cells with fluorescently coupled Exendin 4, a peptide ligand for the glucagon-like peptide 1 receptor (GLP1R), which is selectively expressed by β-cells in the endocrine pancreas (22). By analyzing the cells using fluorescence activated cell sorting (FACS), we detected a subpopulation of cells (∼0.3–0.8% depending on the tumor) coexpressing GLP1R and PDGFRβ (Fig. 6D). We also made use of double transgenic RIP1-Tag2;PDGFRβ-EGFP mice (23), which faithfully produce the fluorescent marker in cells expressing PDGFRβ (Fig. 6E). Analysis of single-cell suspensions of PanNET from compound RIP1-TAg2;PDGFRβ-EGFP mice by FACS corroborated the occurrence of a minor population of malignant cells expressing PDGFRβ (Fig. 6F).
Fig. 5.
Pdgfb is up-regulated in tumors of RIP1-TAg2;Pdgfd−/− mice. Analysis of Pdgfa, Pdgfb, Pdgfc, Pdgfrα, and Pdgfrß mRNA expression by qRT-PCR in tumors of RIP1-TAg2;Pdgfd+/+ and RIP1-TAg2;Pdgfd−/− mice (n = 3 mice per group). Error bars show the mean ± SD. *P < 0.05.
Fig. 6.
Identification of a subset of malignant cells expressing PDGFRβ in RIP1-TAg2 tumors. (A) Immunostaining of tumor from RIP1-TAg2 mouse for malignant tumor cells (insulin; green) and PDGFRβ (red). (B and C) Expression of PDGFRβ by malignant cells in liver metastatic lesions of RIP1-TAg2 mice by (B) immunohistochemistry (arrow) and (C) immunofluorescence by costaining with T-Antigen (T-Ag). (D) Quantification of PDGFRβ+/GLP1R+ cell populations in RIP1-TAg2 pancreatic tumors. Tumors were dispersed into single cells, incubated with fluorescently labeled antibody for PDGFRβ (APC) and peptide-ligand, Exendin 4 (FAM), and analyzed by FACS. (E) Immunostaining of tumors from RIP1-TAg2;PDGFRβ-EGFP compound mice with PDGFRβ antibody (red) to determine colocalization with EGFP (green) expressed by PDGFRβ cells. (F) Flow cytometry gating strategy to analyze double positive Exendin 4+/PDGFRβ-EGFP+ subpopulation of tumor cells in single-cell suspension prepared from tumors of RIP1-TAg2;PDGFRβ-EGF mice. (Scale bars, 50 μm.)
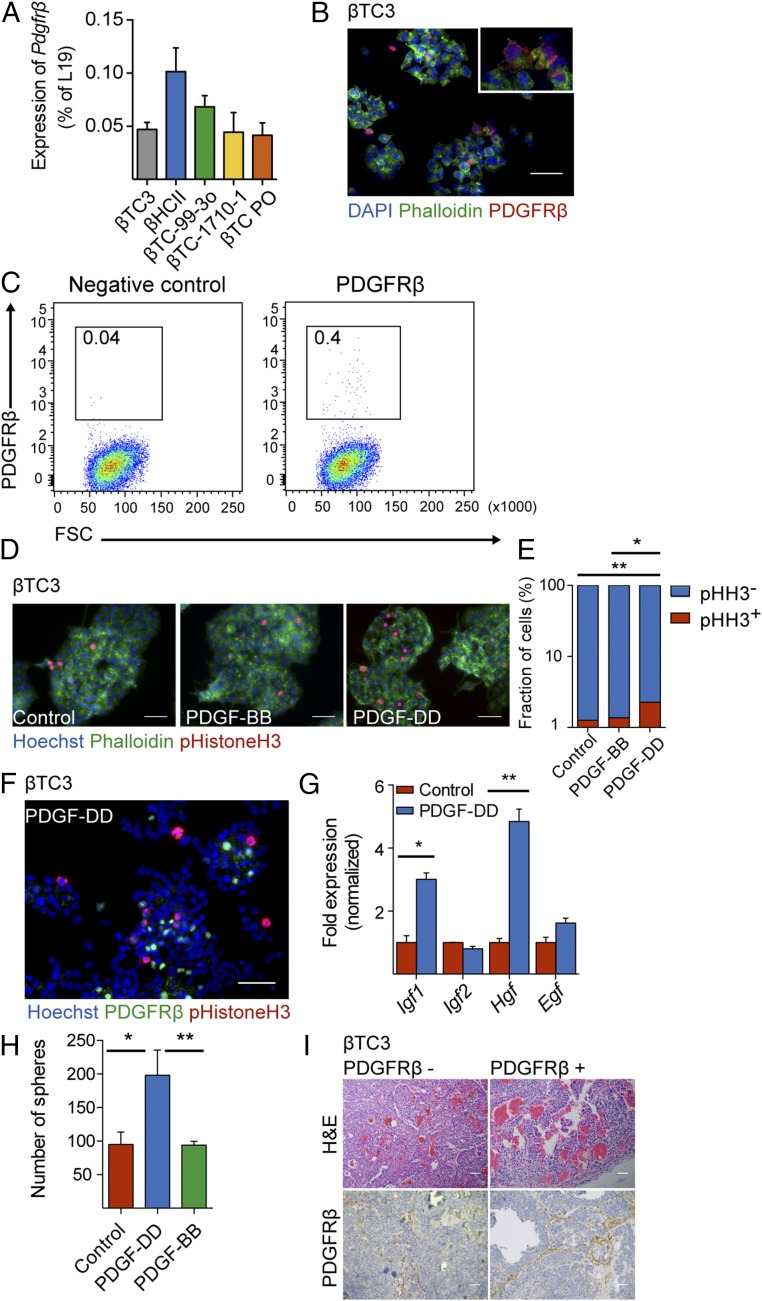
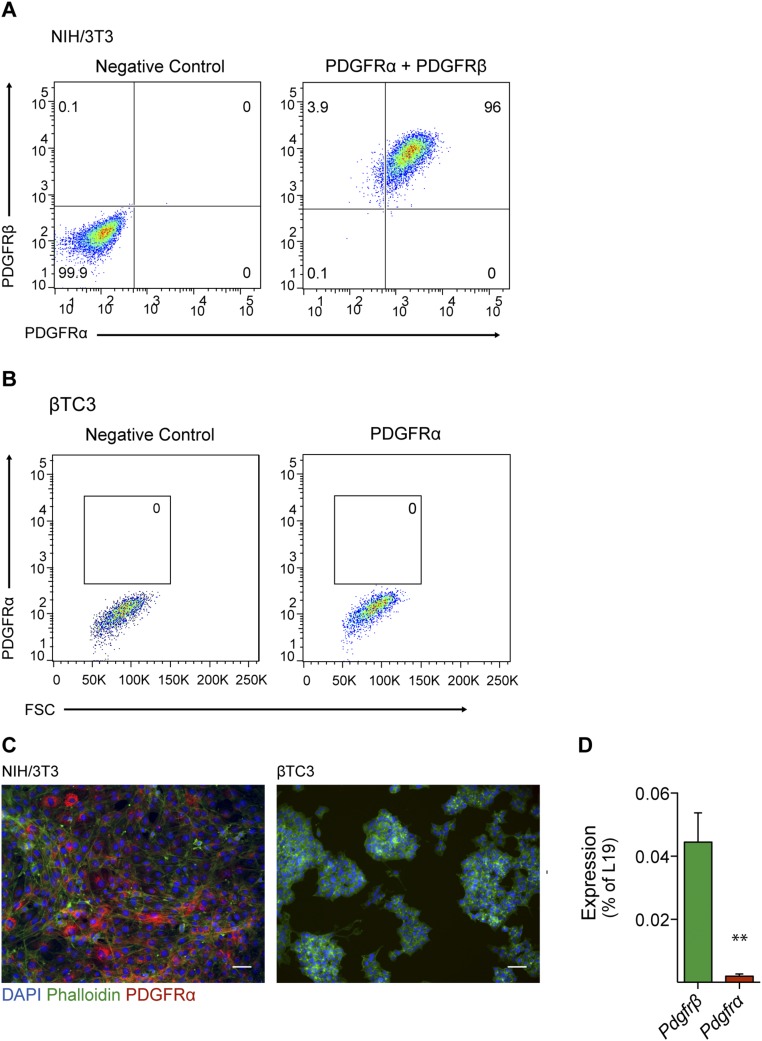
In parallel with the in vivo characterization, we validated our findings using various pancreatic β-tumor cell lines. First, we performed qRT-PCR analysis and detected Pdgfrß transcripts in malignant βTC3 cells (24), premalignant βHCII cells, and additional cell lines established from tumors of RIP1-TAg2 mice (βTC PO, βTC-99-3o, and βTC-1710-1) (Fig. 7A). Furthermore, to confirm the expression of PDGFRβ, we immunostained βTC3 cells and detected strongly positive staining on rare cells, indicating high expression of PDGFRβ by a subpopulation of cells, rather than a widespread low expression (Fig. 7B). Finally, we established the coexistence of PDGFRβ+ and PDGFRβ− cells in βTC3 cultures by FACS (Fig. 7C). In parallel analyses, no cells expressing PDGFRα were detected by flow cytometry or immunostaining, illustrating the specific expression of PDGFRβ (Fig. S4 A–C). Similarly, at the mRNA level, βTC3 express very low levels of Pdgfrα, compared with Pdgfrβ (Fig. S4D). Taken together, both our in vivo and in vitro analyses indicate that although most malignant β-cells are PDGFRβ−, a subset of pancreatic β-tumor cells readily expresses PDGFRβ.
Fig. 7.
βTC3 cells expressing PDGFRβ are responsive to PDGF-DD stimulation in vitro. (A) Quantitative RT-PCR analysis of the expression of Pdgfrß in different βTC lines derived from tumors of RIP1-TAg2 mice. (B) Immunostaining of βTC3 with a PDGFRβ antibody (red) and phalloidin (green). Cell nuclei were stained with DAPI. (C) Flow cytometry analysis of expression of PDGFRβ in βTC3 cell line using an APC fluorescently labeled antibody. (D) Quantification of proliferating βTC3 upon stimulation with PDGF-DD and PDGF-BB assessed by immunostaining with phospho-Histone H3 antibody (red) and phalloidin (green). Cell nuclei were stained with Hoechst. (E) The number of proliferating cells was determined by counting the number of phosho-Histone H3+ cells in relation to the total number of cells (Hoechst+). (F) Costaining of βTC3 cells with antibodies against phospho-Histone H3 (red) and PDGFRβ (green). Cell nuclei were stained with Hoechst. (G) Quantitative RT-PCR analysis of growth factors expressed by βTC3 upon 6 h of stimulation with PDGF-DD. (H) Quantification of tumor spheroids formed by βTC3, seeded in anchorage-independent conditions, upon treatment with PDGF-DD and PDGF-BB. (I) Analysis of tumors formed from βTC3 cells transplanted into NOD-SCID mice. Hematoxylin/eosin stainings (Upper) and immunohistochemistry staining of PDGFRβ (Lower) of tumors from PDGFRβ− βTC3 and PDGFRβ+ βTC3 transplanted cells. Error bars show the mean ± SD. Scale bars, 50 μm. *P < 0.05, **P < 0.01.
Fig. S4.
βTC3 cells do not express PDGFRα. (A and B) Flow cytometry analysis of expression of PDGFRβ and PDGFRα in control NIH/3T3 cells (A) and βTC3 cells (B). (C) Representative images of PDGFRα (red) immunostaining in control NIH/3T3 cells (Left) and βTC3 cells (Right). (D) Quantitative RT-PCR analysis of Pdgfrα and Pdgfrß expression in βTC3 cells. Error bars show the mean ± SD. **P < 0.01. (Scale bars, 50 μm.)
βTC3 Cells Respond to PDGF-DD, but Not PDGF-BB, Stimulation with Increased Proliferation.
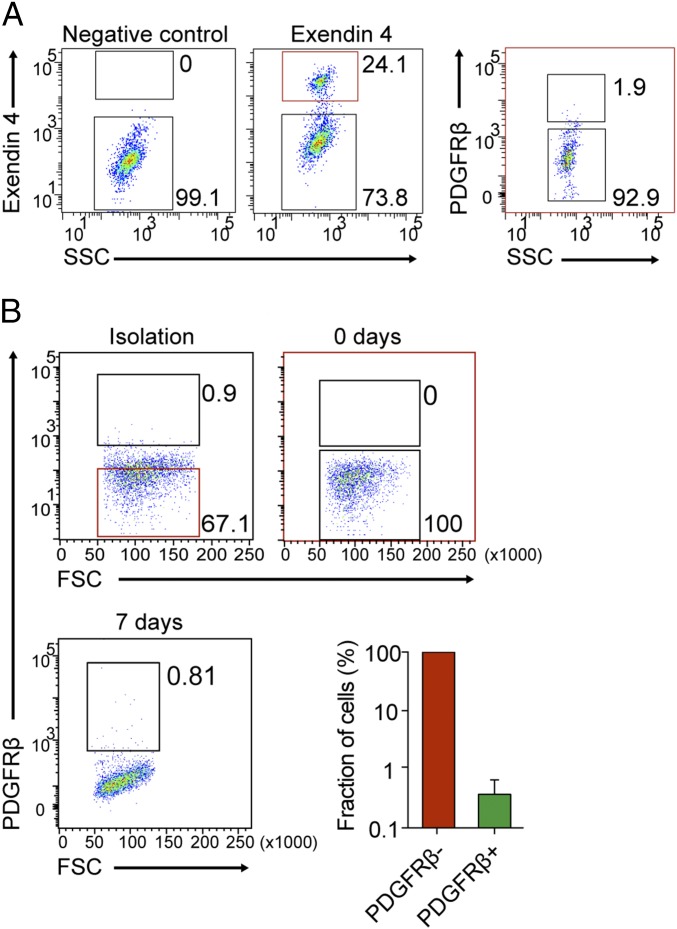
Next, we tested whether PDGFRβ+ βTC3 cells responded to PDGF-DD. We treated βTC3 cells with recombinant PDGF-DD protein and assessed cell proliferation by phospho-Histone H3 immunostaining (Fig. 7 D and E). We found that PDGF-DD induced a significant increase in the total number of cells by 37%, and the number of cells positively stained for phospho-Histone H3 was 2.4-fold higher compared with control cultures (Fig. 7E). Surprisingly, the prototypical ligand for PDGFRβ, i.e., PDGF-BB, did not augment the proliferation of βTC3 cells (Fig. 7E). Because costaining for PDGFRβ and phospho-Histone H3 demonstrated that it was predominantly PDGFRβ− cells in the vicinity of quiescent PDGFRβ+ cells that were engaged in mitosis (Fig. 7F), we reasoned that the effect may be indirect through paracrine secretion of mitogenic factors by PDGFRβ+ cells in response to PDGF-DD stimulation. To test this proposition, we assessed the expression of candidate known mitogens for malignant PanNET cells from RIP1-TAg2 mice following stimulation with PDGF-DD. Indeed, the expression of Igf1 and Hgf, but not of Igf2 or Egf, was significantly induced in βTC3 cells stimulated with PDGF-DD (Fig. 7G). Subsequently, to define the characteristics of PDGFRβ-expressing malignant β-cells, we tested whether PDGF-DD induced tumor-initiating capacity. We found that βTC3 cells stimulated by PDGF-DD, but not by PDGF-BB, in anchorage-independent conditions formed a significantly higher number of tumor spheroids, compared with untreated βTC3 control cells (Fig. 7H). Additionally, to investigate the tumorigenic properties of PDGFRβ+ βTC3 cells, we sorted cells by FACS based on their expression of PDGFRβ and transplanted subcutaneously (sc) into NOD-SCID mice. Injection of as few as 200 PDGFRβ− or PDGFRβ+ βTC3 cells resulted in tumor establishment and progressive growth in three out of four mice and two out of four mice, respectively. The resulting histology of tumors from PDGFRβ+ cells was indistinguishable from that of tumors established from PDGFRβ− βTC3 cells (Fig. 7I, Upper). In addition, the expression of PDGFRβ in the parenchyma of tumors was similar regardless of cell of origin, indicating interconversion between the different subsets of βTC3 cells (Fig. 7I, Lower). Furthermore, FACS analysis revealed that tumors established from PDGFRβ− cells reestablished the original relationship between PDGFRβ− and PDGFRβ+ malignant cells (Fig. 8A). To corroborate this finding, we investigated the prevalence of PDGFRβ− and PDGFRβ+ malignant cells in βTC3 cultures. We isolated PDGFRβ− PanNET cells from βTC3 cultures, and after immediately verifying the purity of the cell suspension, cells were propagated for 7 d. Flow cytometric analysis of the resulting culture for PDGFRβ demonstrated the occurrence of a mixed population of cells with the original frequencies, indicative of rapid conversion of PDGFRβ− malignant cells into PDGFRβ+ (Fig. 8B).
Fig. 8.
Rapid conversion of PDGFRβ− cells to PDGFRβ+ cells occurs in vitro and in vivo. (A) Flow cytometry analysis of tumor cells coexpressing GLP1R and PDGFRβ from tumors originated from transplanted PDGFRβ− βTC3 cells. (B) Flow cytometry-gating strategy for sorting PDGFRβ− tumor cells from parental βTC3 cells. The proportion of PDGFRβ+ βTC3 was analyzed immediately after sorting (0 d, dot plot) and after 7 d of culture (dot plot and column chart). Error bars show the mean ± SD.
Altogether, our comprehensive analyses of the first Pdgfd-deficient mouse model of cancer to our knowledge demonstrate that functional malignant cell heterogeneity in experimental PanNET is reinforced by PDGF-DD by stimulation of a subset of tumor cells expressing PDGFRβ that engages in paracrine cross talk with neighboring malignant cells. However, although PDGF-DD stimulates some features of cancer stem cells in PDGFRβ+ PanNET cells, the tumor-initiating capacity is not exclusive to this subset of malignant cells.
Human PanNET Harbor a Subset of PDGFRβ+ Malignant Cells.
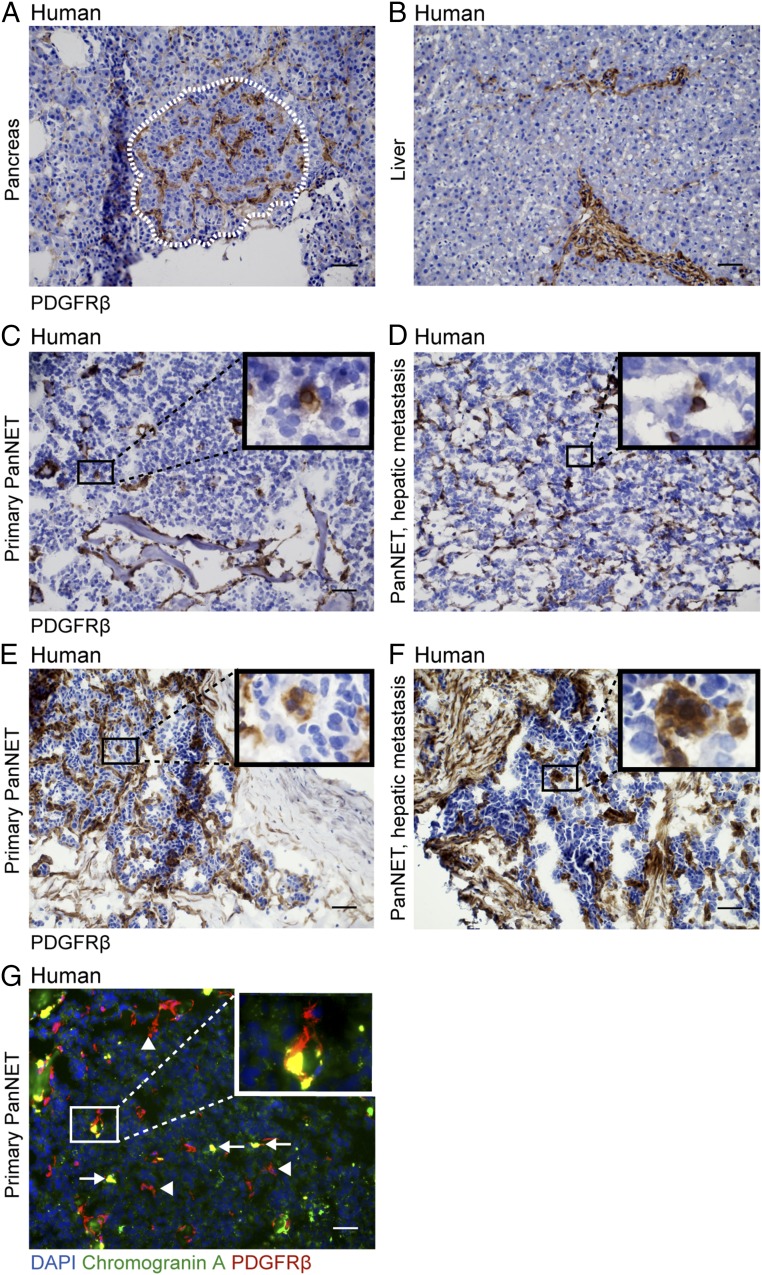
Finally, we analyzed the expression of PDGFRβ in human PanNET and their hepatic metastases by immunostaining. The normal pancreatic islet and liver parenchyma displayed expression of PDGFRβ exclusively in perivascular locations (Fig. 9 A and B). In contrast, all primary PanNET and hepatic metastases analyzed (n = 5 of each) exhibited a subset of malignant cells readily expressing PDGFRβ evenly distributed in the tissue (Fig. 9 C–F). Although all malignant lesions harbored PDGFRβ+ tumor cells, the relative abundance was variable with some lesions containing more (Fig. 9 E and F) and others fewer (Fig. 9 C and D). The PDGFRβ+ tumor cells were distinguishable from pericytes (Fig. 9G, arrowheads) by the coexpression of chromogranin A, a widely used marker for malignant cells of neuroendocrine origin (Fig. 9G, arrows) (25).
Fig. 9.
Human PanNETs harbor a subset of PDGFRβ+ malignant cells. PDGFRβ expression assessed by IHC in (A) normal human pancreatic islet and (B) liver and in primary and hepatic PanNET expressing (C and D) low to (E and F) moderate levels of PDGFRβ in tumor cells. (G) Costaining of a primary PanNET with antibodies against PDGFRβ (red) and a neuroendocrine tumor cell marker, Chromogranin A (green). Cell nuclei were stained with DAPI. Representative images are shown. Arrows point out PDGFRβ/chromogranin A double-positive malignant cells; arrowheads point out PDGFRβ single-positive pericytes. (Scale bars, 50 μm.)
Discussion
An emergent number of preclinical reports suggest that PDGF-DD is a key player in tumor formation by regulating various cellular processes, such as macrophage and stromal cell recruitment (19, 26), EMT (14, 27, 28), tumor cell proliferation, and invasion (26, 29, 30). In human cancers, PDGFD up-regulation has been documented for prostate, lung, renal, ovarian, brain, breast, and pancreatic cancers (31, 32). However, the mechanisms underlying the effect of PDGF-DD on tumor growth are still largely unknown.
Our study revealed that disruption of PDGF-DD signaling greatly delayed PanNET development in RIP1-TAg2 mice. Genetic loss of a single copy of the gene encoding Pdgfd was sufficient to significantly retard tumor growth. The reduction in tumor burden was associated with decreased tumor cell proliferation and prolonged survival of Pdgfd-deficient RIP1-TAg2 mice.
As previously reported for PDGF-BB (33, 34), the other known ligand for PDGFRβ, ECs in tumors from RIP1-TAg2 mice appear to be the major source for PDGF-DD. The effect of EC-secreted factors on the regulation of tissue and tumor growth has recently been highlighted (35–37), emphasizing the potential benefit of targeting the cross talk between ECs and tumor cells to supplement conventional antiangiogenic therapies. However, we do not exclude that there may be additional cells in the tumor microenvironment that express low levels of Pdgfd. Indeed, PDGFD expression has also been documented by malignant cells in ovarian, lung, breast, prostate, renal, and brain tumor-derived cell lines (10, 29, 38, 39).
In tumors, PDGFRβ is expressed mainly by mesenchymal cells, i.e., fibroblasts and pericytes (2, 40). The role of PDGF-BB in the recruitment of pericytes to blood vessels has been well established in tumors from RIP1-TAg2 mice (16). Also, Pdgfd overexpression in an orthotopic model of renal cell carcinoma resulted in increased perivascular cell coverage (26). In our model, however, there was no measurable effect on stromal-cell (pericyte) recruitment upon disruption of PDGF-DD signaling. Up-regulation of Pdgfb in tumors from RIP1-TAg2;Pdgfd−/− mice compared with the wild-type littermates suggests that PDGF-BB exerts a compensatory effect for the loss of PDGF-DD in the present context. The apparent need for compensation by PDGF-BB implies a yet uncharted functional role for PDGF-DD in the tumor stroma. In addition, the sharp decrease in tumor burden caused by Pdgfd deficiency strongly supports that compensation by PDGF-BB is only partially attained and that the functions of PDGF-BB and PDGF-DD within the context of tumorigenesis are only partly overlapping. Indeed, stimulation of a PanNET cell line with PDGF-DD, but not with PDGF-BB, increased proliferation and improved sphere-forming capacity. Given the fact that genetic studies have demonstrated closely related phenotypes of mice deficient for Pdgfb and Pdgfrß (41, 42), it is tempting to speculate that the unique functions of PDGF-DD are the result of binding to a distinct (co)-receptor that modulates the response of PDGFRβ.
PDGF-DD and PDGF-BB share a conserved growth factor core domain motif, but contrary to PDGF-DD, PDGF-BB carries a basic retention motif domain, allowing binding to heparan sulfate proteoglycans present in the extracellular matrix (15). In contrast, the latent full-length form of PDGF-DD has been suggested to be freely diffusible (43). It is thus reasonable to assume that PDGF-DD has a distinct distribution in the tumor stroma, being able to reach non-vessel-associated PDGFRβ-expressing cells. Therefore, we explored the possibility that nonvascular cell types in the tumor parenchyma of RIP1-TAg2 mice expressed PDGFRβ and constituted potential targets for PDGF-DD. A study of pancreatic islet regeneration demonstrated that immature mouse pancreatic β-cells express PDGFRα and PDGFRβ, signaling by which is required for sustained cell proliferation and islet expansion (44). In agreement, we identified a rare population of malignant cells expressing PDGFRβ in primary tumors and metastatic lesions of RIP1-TAg2 mice. Notably, PDGFRβ+ tumor cells were more abundant in early metastatic lesions in the liver, suggesting that this particular subset of malignant cells may be involved in establishing distant metastases. However, PDGF-DD evidently did not contribute to dissemination as such, because we did not detect any difference in the incidence of hepatic metastatic lesions.
When sorted and transplanted into NOD-SCID mice at quantities down to 200 cells, βTC3 cells were able to recapitulate the morphology and heterogeneity found in tumors from RIP1-TAg2 mice, regardless of the expression of PDGFRβ, evidencing the conserved malignant phenotype of all subsets of cancer cells within this cell line. Although PDGFRβ+ malignant cells held some features of tumor-initiating cells, we could not demonstrate that this was an exclusive or general trait of the subpopulation expressing PDGFRβ. The fact that we were able to identify minor subpopulations of β-cells even in late passages of βTC cell lines led us to consider that the presence of PDGFRβ–expressing βTC3 cells is necessary for the maintenance of the bulk tumor cell population. The coexistence of subpopulations of tumor cells with distinct phenotypic and functional properties within the same tumor points to the existence of uncharted cellular interactions or pathways that contribute to tumor growth and dissemination. Studies in renal cell carcinoma (45) and lung adenocarcinoma (46) have revealed substantial intratumoral heterogeneity as a result of subclonal driver events. Additional studies establish that interactions between genetically distinct subclones of tumor cells are necessary for maintaining tumor heterogeneity (47) and that even minor subpopulations of tumor cells may promote bulk tumor growth via mechanisms of non-cell–autonomous stimulation (48). Thus, one might hypothesize that the cross talk between PDGFRβ+ and PDGFRβ− malignant clones is necessary for more efficient tumor propagation. Indeed, we found that stimulation of PanNET cells with PDGF-DD in vitro engaged PDGFRβ+ cells in paracrine cross talk with neighboring PDGFRβ− cells through the induction of mitogenic factors, the identity of which should be corroborated in vivo. A recent study using βTC cells from RIP1-TAg2 mice as a model demonstrates that artificial tumor heterogeneity is the result of stable coexistence of different clones, even without a strict interdependence between subclones (49). We should also consider the prospect that expression of PDGFRβ in malignant cells is due to interconversion between malignant cell populations, e.g., as a result of PDGF-DD-stimulated EMT. In support of this train of thought, maintenance of a pure PDGFRβ− cell culture led to rapid reestablishment of the original frequencies of PDGFRβ− and PDGFRβ+ subclones, in close agreement with studies of subsets of breast cancer cells that hold the ability to spontaneously interconvert (50).
Consistently, we were able to confirm the presence of a rare population of tumor cells expressing PDGFRβ also in human primary and metastatic PanNET. Our findings are supported by previous studies showing that besides stromal cells, human primary PanNET and metastatic cells express high levels of PDGFRβ compared with normal tissue (51, 52). Moreover, clinical reports show evidence of PDGFR activation and copy number alteration in small intestine, gastroenteropancreatic, and pancreatic NET, although the use of whole tumor cell lysates precludes identification of the cell type expressing PDGFRβ (21, 53). The identification of PDGFRβ+ tumor cells in human PanNET has important clinical implications, considering that sunitinib, a PDGFR/VEGFR small molecule inhibitor, was recently approved to treat patients with progressive well-differentiated PanNET (54). Preclinical studies suggest that the therapeutic efficacy of sunitinib is derived from dual targeting of endothelial cells and pericytes (12, 55), but given our finding of the functional importance of PDGF-DD/PDGFRβ for maintaining a functional malignant cell heterogeneity, direct inhibitory effects on tumor cells cannot be excluded.
Altogether, we provide strong evidence for the importance of PDGF-DD for PanNET growth. Moreover, we have identified a subpopulation of PDGFRβ+ malignant β-cells responsive to PDGF-DD in PanNET from an instructive mouse model, and in human primary and hepatic metastatic lesions. Our study thus provides evidence for a functional heterogeneity that needs to be explored to fully understand the tumorigenic process in PanNET.
Materials and Methods
A detailed description of additional procedures can be found in Supporting Information.
Animal Care.
All experimental procedures involving mice were approved by the Stockholm North and Malmö/Lund committees for animal care (permits N96/11 and M142/13). RIP1-TAg2 transgenic mice were crossed with Pdgfd−/− and Pdgfd+/− mice or with PDGFRβ-EGFP mice (a kind gift from Christer Betsholtz, Uppsala University, Uppsala, Sweden) on the C57BL/6 background. RIP1-TAg2;Pdgfd+/+ littermates were used as controls. From 10 wk of age, all RIP1-TAg2 mice received 5% (wt/vol) sugar water to counteract symptoms of hypoglycemia.
Assessment of Angiogenic Islets, Tumor Burden, and Lesion Frequency.
Pancreata of 12-wk-old mice were analyzed for the number of angiogenic islets, counted under a stereological microscope, and defined as islets with red patches and <1 × 1 mm. Tumors were defined as >1 × 1 mm and were excised, counted, and measured with a caliper to obtain the total number of tumors and total tumor volume in each RIP1-TAg2 mouse. Tumor volume was calculated as length × width2 × π/6.
Mouse Tissue Preparation.
Mice were anesthetized, the heart perfused with PBS followed by 4% (wt/vol) paraformaldehyde (PFA). For paraffin embedding, pancreata and livers were kept in PFA at 4 °C overnight followed by paraffin embedding. For cryopreservation, organs were kept in 30% sucrose at 4 °C overnight followed by embedding in optimal cutting temperature (OCT) cryomount (Histolab).
Quantification of Tumor Cell Proliferation.
Mice were injected intraperitoneally with 100 μg/g body weight of 5-Bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich) and kept for 2 h before being euthanized. Pancreata were sectioned and stained with primary antibody against BrdU (1:100; Accurate Chemical). Cells with incorporated BrdU were quantified as the fraction of positively stained cells per total number of cells.
Fluorescence-Activated Cell Sorting and Analysis.
For procedures of preparation of cells for flow cytometry, please refer to Supporting Information. Cells were sorted using a BD FACSARIAIII sorter with BD FACSDiva software or analyzed on BD FACSCantoII or BD FACSVerse flow cytometers (all from Beckton Dickinson Immunocytometry Systems). Further analysis of acquired cells was performed using FlowJo software (FlowJo LLC).
RNA Isolation and Gene Expression Profiling.
Total RNA from cultured cells and tumor lysates was isolated using the RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions followed by cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad Laboratories). Quantitative RT-PCR was performed as described before (56). Expression levels were calculated relative to the ribosomal housekeeping gene RPL19 as 100 × 2-ΔCt. For primers, refer to Supporting Information.
Statistical Analysis.
Data are shown as mean ± SD. Statistical analysis comparing means was performed using the unpaired, two-tailed Student’s t test; analysis of proportions was performed using the χ2 test; analysis of survival was performed using the log rank test; and in all cases, statistical significance was defined as P < 0.05.
SI Materials and Methods
Processing of Cryosections.
Immunofluorescence.
Frozen sections (10–12 μm) were air-dried and postfixed in cold acetone followed by blocking with serum-free protein block (Dako) for 60 min at room temperature (RT). Primary antibodies (for specific clones, dilutions, and manufacturer, refer to Supporting Information, Antibodies for Immunostaining) were incubated overnight at 4 °C in PBS supplemented with 1% BSA. After three washes, appropriate fluorochrome-conjugated secondary antibodies (Invitrogen, Life Technologies) were added for 90 min at RT, after which slides were washed and mounted in DAPI-containing medium (Vector Laboratories). X-gal stainings were performed as described in Supporting Information, X-Gal Staining.
Immunohistochemistry.
Six-micrometer tissue sections from frozen samples were placed on SuperFrost Ultra Plus slides (Histolab) and stored in –80 °C freezer until staining. Next, sections were air-dried 30 min and thereafter fixed in 4% (wt/vol) PFA (Sigma Aldrich) for 20 min at 4 °C. Sections were incubated with Bloxall (Vector Laboratories) for 10 min followed by blocking in serum-free protein block (Dako) for 30 min at RT. Samples were incubated with primary antibodies overnight at 4 °C. Subsequently, sections were incubated with ImmPRESS rabbit secondary antibodies (Vector Laboratories) for 30 min at RT, and specific staining was detected by incubation with DAB kit (Vector Laboratories). Finally, tissue sections were counterstained with Hematoxylin (Histolab), dehydrated, and mounted.
Processing of Paraffin-Embedded Sections.
Following deparafinization the tissue was quenched for endogenous peroxidase activity with 3% (vol/vol) H2O2 for 10 min at RT. Antigen retrieval was performed using high-pH (pH = 10) buffer solution in pressure cooker for 20 min. Blocking was performed in species-specific serum diluted in TNB. Primary antibodies were added overnight at 4 °C. Appropriate horseradish peroxidase-conjugated secondary antibodies were applied followed by amplification using the Vectastain ABC system (Vector Laboratories) and development with DAB kit (Vector Laboratories). Sections were further dehydrated, counterstained with hematoxylin, and mounted. For immunofluorescence after primary antibody incubation, fluorochrome-conjugated secondary antibodies were added to sections before mounting.
Cell Culture.
All cell lines were maintained at 37 °C in a humidified chamber with 5% CO2. Mouse pancreatic beta tumor cell lines (βTC) βTC3, βTCPO, βTC-99-3o, and βTC-1710-1; pancreatic beta hyperplastic cells, βHCII; and embryonic fibroblast cell line NIH/3T3 were maintained in DMEM supplemented with 10% (vol/vol) FCS and penicillin–streptomycin (all from Gibco, Life Technologies). Cells were grown in flasks for isolation of RNA and FACS analysis/sorting or in gelatin precoated coverslips or chamber slides (Lab-TekII; VWR) for immunostaining. When semiconfluent, cells were starved overnight before the addition of recombinant PDGF-DD (100–300 ng/mL; R&D) or PDGF-BB (100–300 ng/mL; PeproTech).
Spheroid assay.
βTC3 were cultured in six-well, ultralow-attachment plates (Corning Life Sciences) in serum-free DMEM:F12 medium (Gibco, Life Technologies) supplemented with B27 (1:50; Invitrogen, Life Technologies), 20 ng/mL Epidermal Growth Factor, 20 ng/mL basic Fibroblast Growth Factor (both from Prepotech), 5 μg/mL Insulin (Sigma-Aldrich), and 4μg/mL Heparin (STEMCELL Technologies). Spheres were counted manually under an inverted microscope.
Immunostaining of adherent cultured cells.
After stimulation with PDGF-DD or PDGF-BB, cells were washed with PBS and fixed with 4% PFA followed by permeabilization with PBS supplemented with 0.5% Tween20 and 1% BSA, 10 min in RT. Primary antibodies were incubated O/N at 4 °C. Slides were further processed as described in Supporting Information, Processing of Cryosections.
Isolation and Preparation of Tumors for Flow Cytometry.
Mice were anesthetized with avertin [2.5% (vol/vol), Sigma] and perfused with PBS, and tumors were excised from the pancreas. Tumors were digested at 37 °C for 15–20 min with a 20-mL collagenase mix containing 12,500 units Collagenase II (Worthington Biochemical Corp.), 12,500 units Collagenase IV (Gibco, Life Technologies), and 40 μL DNase I (Sigma). Cells from the digested tumors were passed through a 70-μm cell strainer and washed. Red blood cells were lysed with PharmLyse (BD Biosciences Pharmingen) for 10 min at RT.
Preparation of Cells from Adherent Cultures for Flow Cytometry.
βTCs were detached using Accutase (Gibco, Life Technologies) at 37 °C and washed in PBS. All cell suspensions were preblocked with a mouse Fc block (CD16/CD32, 1:100; BDBiosciences Pharmingen) for 10 min at 4 °C in the dark. Cells were then incubated with labeled primary antibodies against PDGFRβ (CD140b-APC, 1:40; eBioscience), PDGFRα (CD140a-PE, 1:40; eBioscience), and/or peptide Exendin 4-FAM or Exendin4-biotin (1:40; Phoenix Pharmaceuticals) for 30 min on ice in FACS buffer: DMEM supplemented with 0.2% BSA and 5% PBS-based dissociation buffer (Life Technologies). When necessary, cells were further incubated with Streptavidin-APC (Life Technologies) for 30 additional minutes before washing. All cells were washed, filtered, and labeled with 7AAD dye (eBioscience) or DAPI (Life Technologies) for exclusion of nonviable cells before acquisition.
Quantification of Metastasis.
The right lateral liver lobes from RIP1-TAg2 mice were paraffin embedded and sectioned entirely. Sections were deparaffinized and stained with H&E, and metastases were quantified under an observation microscope (Nikon Eclipse Ci).
X-Gal Staining.
Before X-gal staining, sections were washed in PBS and permeabilized in PBS, 4 mM MgCl2, 0.04% Nonidet P-40, and 0.02% Na-deoxycholate for at least 30 min at RT, with change of the solution two times. Staining was performed O/N at 37 °C in 1 mg/mL X-gal (Sigma-Aldrich) in PBS, 4 mM MgCl2, 0.04% Nonidet P-40, 0.02% Na-deoxycholate, 5 mM K4Fe(CN)6, and 5 mM K3Fe(CN)6. Sections were washed again in permeabilization solution before dehydration and mounting.
Antibodies for Immunostaining.
The following antibodies were used for immunostaining: Podocalyxin (AF1556, 1:100; R&D Systems), NG2 (AB5320, 1:400; Millipore), PDGFRβ (28E1, 1:100; Cell Signaling; and CD140b, 1:100; eBioscience), PDGFRα (D1E1E, 1:200; Cell Signaling and CD140a, 1:100; eBioscience), Desmin (1:400; Abcam), phospho-Histone H3 (Ser10, 1:200; Cell Signaling; or Ser-28, 1:400; Abcam), Cleaved-caspase 3 (1:100; Cell Signaling), BrdU (BU1/75, 1:100; Accurate Chemical), Insulin (1:300; Dako; or MAB1417, 1:600; R&D Technologies), α-Amylase (1:500; Sigma), F4/80 (1:200; Serotec), CD45 (30-F11, 1:200; BD Pharmingen), T-Antigen (1:10000, a kind gift from Douglas Hanahan, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), and Chromogranin A (DAK-A3, 1:200; Dako).
Microscope Image Acquisition and Analysis.
Images were acquired using either a Nikon Eclipse E800 microscope equipped with Nikon Plan Fluor objectives (10×, NA 0.30; 20×, NA 0.50; 40×, NA 0.75) and a SPOT RTKE camera using the SPOT advanced software or a Olympus BX63 microscope equipped with UPlanSApo objectives (10×/0.40; 20×/0.75; 40×/0.95) and DP80 camera using the CellSens Dimension imaging software. Positive structures were analyzed using Volocity software (Perkin–Elmer) and CellSens quantification software and quantified as number of positively stained pixels per area. For each analysis, more than seven images per mouse were used.
In Vivo Tumorigenesis Assays.
FACS sorted cells (PDGFRβ− βTC3 or PDGFRβ+ βTC3) were injected sc in a matrigel solution (BD Biosciences) into NOD-SCID mice that were monitored for the development of tumors.
Primers for Gene Expression Analysis.
Primers used for Pdgfd (QT00155799), Igf1 (QT02423379), Igf2 (QT00109879), Hgf (QT00158046), L19 (QT01779218), and Egf (QT00151018) were purchased from Qiagen. Primer sequences for other PDGF family members and reference gene were as follows:
Pdgfrα, forward TGGCATGATGGTCGATTCTA, reverse CGCTGAGGTGGTAGAAGGAG
Pdgfrß, forward CACCTTCTCCAGTGTGCTGA, reverse GGAGTCCATAGGGAGGAAGC
Pdgfa, forward GAGATACCCCGGGAGTTGAT, reverse TCTTGCAAACTGCAGGAATG
Pdgfb, forward CCTCGGCCTGTGGACTAGAAG, reverse CCTTGTCATGGGTGTGCTTA
Pdgfc, forward GTGGAGGAAATTGTGCCTGT, reverse TCCAGAGCCACATCAGTGAG
Rpl19, forward AGCCACGCTTTCATACTGCT, reverse TTCAGCTTGTGGATGTGCTC.
Assessment of Vascular Functionality.
Four minutes before sacrifice, mice were injected retroorbitally with 100 μL of 0.5 mg/mL of fluorescein-conjugated tomato lectin (Vector Laboratories). Pancreata and livers were collected for histology.
Acknowledgments
K.P. is the Göran and Birgitta Grosskopf Professor at Lund University. The research presented herein is supported by a Consolidator Grant from the European Research Council (the TUMORGAN project), the Swedish Research Council, the Swedish Cancer Society, the STARGET consortium (a Swedish Research Council Linnaeus network), BioCARE, and Lund University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509384113/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Cortez E, Roswall P, Pietras K. Functional subsets of mesenchymal cell types in the tumor microenvironment. Semin Cancer Biol. 2014;25:3–9. doi: 10.1016/j.semcancer.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heldin C-H, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79(4):1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 5.Pietras K, Sjöblom T, Rubin K, Heldin CH, Ostman A. PDGF receptors as cancer drug targets. Cancer Cell. 2003;3(5):439–443. doi: 10.1016/s1535-6108(03)00089-8. [DOI] [PubMed] [Google Scholar]
- 6.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 7.Druker BJ, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344(14):1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 8.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22(10):1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergsten E, et al. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat Cell Biol. 2001;3(5):512–516. doi: 10.1038/35074588. [DOI] [PubMed] [Google Scholar]
- 10.LaRochelle WJ, et al. PDGF-D, a new protease-activated growth factor. Nat Cell Biol. 2001;3(5):517–521. doi: 10.1038/35074593. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Heritable formation of pancreatic β-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315(6015):115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 12.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest. 2003;111(9):1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderberg C, et al. Deficiency for endoglin in tumor vasculature weakens the endothelial barrier to metastatic dissemination. J Exp Med. 2013;210(3):563–579. doi: 10.1084/jem.20120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, et al. Emerging roles of PDGF-D in EMT progression during tumorigenesis. Cancer Treat Rev. 2013;39(6):640–646. doi: 10.1016/j.ctrv.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindblom P, et al. Endothelial PDGF-B retention is required for proper investment of pericytes in the microvessel wall. Genes Dev. 2003;17(15):1835–1840. doi: 10.1101/gad.266803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xian X, et al. Pericytes limit tumor cell metastasis. J Clin Invest. 2006;116(3):642–651. doi: 10.1172/JCI25705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song S, Ewald AJ, Stallcup W, Werb Z, Bergers G. PDGFRbeta+ perivascular progenitor cells in tumours regulate pericyte differentiation and vascular survival. Nat Cell Biol. 2005;7(9):870–879. doi: 10.1038/ncb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franco M, Pàez-Ribes M, Cortez E, Casanovas O, Pietras K. Use of a mouse model of pancreatic neuroendocrine tumors to find pericyte biomarkers of resistance to anti-angiogenic therapy. Horm Metab Res. 2011;43(12):884–889. doi: 10.1055/s-0031-1284381. [DOI] [PubMed] [Google Scholar]
- 19.Uutela M, et al. PDGF-D induces macrophage recruitment, increased interstitial pressure, and blood vessel maturation during angiogenesis. Blood. 2004;104(10):3198–3204. doi: 10.1182/blood-2004-04-1485. [DOI] [PubMed] [Google Scholar]
- 20.Nozawa H, Chiu C, Hanahan D. Infiltrating neutrophils mediate the initial angiogenic switch in a mouse model of multistage carcinogenesis. Proc Natl Acad Sci USA. 2006;103(33):12493–12498. doi: 10.1073/pnas.0601807103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerr EM, et al. Defining molecular classifications and targets in gastroenteropancreatic neuroendocrine tumors through DNA microarray analysis. Endocr Relat Cancer. 2008;15(1):243–256. doi: 10.1677/ERC-07-0194. [DOI] [PubMed] [Google Scholar]
- 22.Tornehave D, Kristensen P, Rømer J, Knudsen LB, Heller RS. Expression of the GLP-1 receptor in mouse, rat, and human pancreas. J Histochem Cytochem. 2008;56(9):841–851. doi: 10.1369/jhc.2008.951319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 24.Efrat S, et al. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc Natl Acad Sci USA. 1988;85(23):9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connor DT, Deftos LJ. Secretion of chromogranin A by peptide-producing endocrine neoplasms. N Engl J Med. 1986;314(18):1145–1151. doi: 10.1056/NEJM198605013141803. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Tong R, Cochran DM, Jain RK. Blocking platelet-derived growth factor-D/platelet-derived growth factor receptor beta signaling inhibits human renal cell carcinoma progression in an orthotopic mouse model. Cancer Res. 2005;65(13):5711–5719. doi: 10.1158/0008-5472.CAN-04-4313. [DOI] [PubMed] [Google Scholar]
- 27.Kong D, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27(8):1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong D, et al. Platelet-derived growth factor-D overexpression contributes to epithelial-mesenchymal transition of PC3 prostate cancer cells. Stem Cells. 2008;26(6):1425–1435. doi: 10.1634/stemcells.2007-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, et al. PDGF-D improves drug delivery and efficacy via vascular normalization, but promotes lymphatic metastasis by activating CXCR4 in breast cancer. Clin Cancer Res. 2011;17(11):3638–3648. doi: 10.1158/1078-0432.CCR-10-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Fredriksson L, Li X, Eriksson U. PDGF-D is a potent transforming and angiogenic growth factor. Oncogene. 2003;22(10):1501–1510. doi: 10.1038/sj.onc.1206223. [DOI] [PubMed] [Google Scholar]
- 31.Ustach CV, et al. A potential oncogenic activity of platelet-derived growth factor d in prostate cancer progression. Cancer Res. 2004;64(5):1722–1729. doi: 10.1158/0008-5472.can-03-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim Biophys Acta. 2010;1806(1):122–130. doi: 10.1016/j.bbcan.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramsson A, Lindblom P, Betsholtz C. Endothelial and nonendothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J Clin Invest. 2003;112(8):1142–1151. doi: 10.1172/JCI18549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franco M, Roswall P, Cortez E, Hanahan D, Pietras K. Pericytes promote endothelial cell survival through induction of autocrine VEGF-A signaling and Bcl-w expression. Blood. 2011;118(10):2906–2917. doi: 10.1182/blood-2011-01-331694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10(2):138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Z, et al. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell. 2014;25(3):350–365. doi: 10.1016/j.ccr.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seton-Rogers S. Microenvironment: Endothelial cells create a niche. Nat Rev Cancer. 2014;14(5):298. doi: 10.1038/nrc3730. [DOI] [PubMed] [Google Scholar]
- 38.Uutela M, et al. Chromosomal location, exon structure, and vascular expression patterns of the human PDGFC and PDGFD genes. Circulation. 2001;103(18):2242–2247. doi: 10.1161/01.cir.103.18.2242. [DOI] [PubMed] [Google Scholar]
- 39.Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: Evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62(13):3729–3735. [PubMed] [Google Scholar]
- 40.Pietras K, Ostman A. Hallmarks of cancer: Interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 41.Levéen P, et al. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 1994;8(16):1875–1887. doi: 10.1101/gad.8.16.1875. [DOI] [PubMed] [Google Scholar]
- 42.Soriano P. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 1994;8(16):1888–1896. doi: 10.1101/gad.8.16.1888. [DOI] [PubMed] [Google Scholar]
- 43.Ehnman M, Li H, Fredriksson L, Pietras K, Eriksson U. The uPA/uPAR system regulates the bioavailability of PDGF-DD: Implications for tumour growth. Oncogene. 2009;28(4):534–544. doi: 10.1038/onc.2008.410. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, et al. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 2011;478(7369):349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46(3):225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inda MM, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514(7520):54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Archetti M, Ferraro DA, Christofori G. Heterogeneity for IGF-II production maintained by public goods dynamics in neuroendocrine pancreatic cancer. Proc Natl Acad Sci USA. 2015;112(6):1833–1838. doi: 10.1073/pnas.1414653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 51.Fjällskog ML, Lejonklou MH, Oberg KE, Eriksson BK, Janson ET. Expression of molecular targets for tyrosine kinase receptor antagonists in malignant endocrine pancreatic tumors. Clin Cancer Res. 2003;9(4):1469–1473. [PubMed] [Google Scholar]
- 52.Fjällskog ML, Hessman O, Eriksson B, Janson ET. Upregulated expression of PDGF receptor beta in endocrine pancreatic tumors and metastases compared to normal endocrine pancreas. Acta Oncol. 2007;46(6):741–746. doi: 10.1080/02841860601048388. [DOI] [PubMed] [Google Scholar]
- 53.Banck MS, et al. The genomic landscape of small intestine neuroendocrine tumors. J Clin Invest. 2013;123(6):2502–2508. doi: 10.1172/JCI67963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raymond E, et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N Engl J Med. 2011;364(6):501–513. doi: 10.1056/NEJMoa1003825. [DOI] [PubMed] [Google Scholar]
- 55.Pietras K, Hanahan D. A multitargeted, metronomic, and maximum-tolerated dose “chemo-switch” regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer. J Clin Oncol. 2005;23(5):939–952. doi: 10.1200/JCO.2005.07.093. [DOI] [PubMed] [Google Scholar]
- 56.Cunha SI, et al. Genetic and pharmacological targeting of activin receptor-like kinase 1 impairs tumor growth and angiogenesis. J Exp Med. 2010;207(1):85–100. doi: 10.1084/jem.20091309. [DOI] [PMC free article] [PubMed] [Google Scholar]