Significance
How is chemical diversity generated by biological systems? Some biosynthetic pathways are constrained to produce one metabolite, but others are relatively relaxed and can produce a set of diverse compounds. Using the example of the tru pathway to cyanobactins, we propose a model of diversity-generating metabolism in which pathway flux and properties are strikingly different from the canonical design of conventional metabolic pathways.
Keywords: natural products, secondary metabolism, biosynthesis, RiPP, cyanobactin
Abstract
A conventional metabolic pathway leads to a specific product. In stark contrast, there are diversity-generating metabolic pathways that naturally produce different chemicals, sometimes of great diversity. We demonstrate that for one such pathway, tru, each ensuing metabolic step is slower, in parallel with the increasing potential chemical divergence generated as the pathway proceeds. Intermediates are long lived and accumulate progressively, in contrast with conventional metabolic pathways, in which the first step is rate-limiting and metabolic intermediates are short-lived. Understanding these fundamental differences enables several different practical applications, such as combinatorial biosynthesis, some of which we demonstrate here. We propose that these principles may provide a unifying framework underlying diversity-generating metabolism in many different biosynthetic pathways.
There are two fundamentally different types of metabolic pathways in living systems. The first are aimed to generate one or a few discrete chemicals; these comprise the majority of pathways. The second have evolved to produce large numbers of different metabolites (1, 2). Although perhaps fewer in number, this second class, which we will call “diversity generating” (DG), may be responsible for the majority of small molecules in living systems. A key difference is that the compounds produced in the latter generally have a more limited phylogenetic distribution.
The metabolic pathways first elucidated were for the synthesis of essential metabolites found in all cells, such as amino acids, purines, or pyrimidines. These conventional metabolic pathways typically comprise multiple metabolic steps, with the intermediates generated in each step converted only to the final product of the pathway. DG pathways, however, do not yield a single final product. Each enzyme in a DG pathway has relaxed substrate specificity and is able to handle a variety of compounds, carrying out the same chemical transformation on different substrates.
Previously, an evolutionary framework was developed to explain why some biosynthetic pathways produce many compounds (3–5). In this study we provide the first integrated overview, to our knowledge, of the multiple metabolic steps that comprise a DG biosynthetic pathway. We have uncovered striking differences in how this pathway differs from the canonical features of conventional pathways. Our results provide an initial framework for understanding how DG pathways are designed and how key features of such pathways diverge from the textbook model.
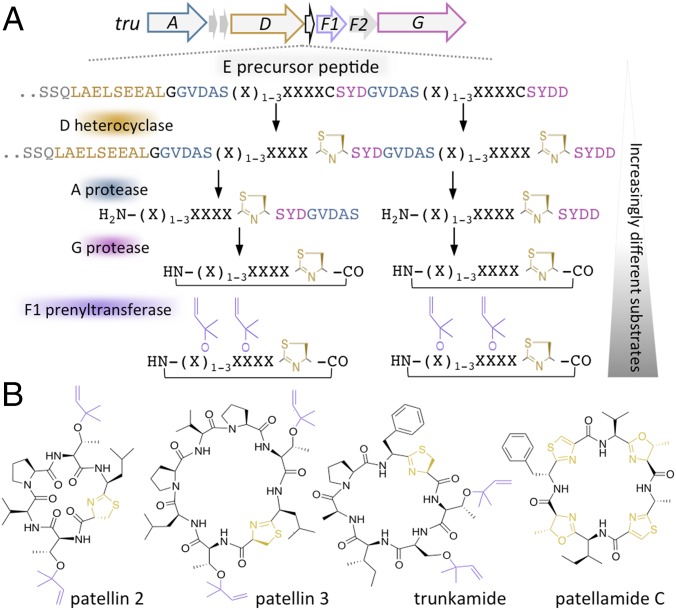
We specifically examine the tru and related pat cyanobactin pathways (1, 6). These ribosomally synthesized and posttranslationally modified (RiPP) secondary metabolite pathways were identified in cyanobacterial symbionts of coral reef animals. Their expression required transfer to a model host, Escherichia coli (Fig. 1) (7). In nature, tru and pat accept a wide variety of hypervariable substrates, but the enzymes are essentially identical in sequence. The tru pathway is capable of synthesizing potentially millions of compounds with highly diverse structures (8). In tru, the first step is the ribosomal synthesis of the precursor peptide, TruE, which encodes the production of two different patellin products (Fig. 1 and SI Appendix, Fig. S1) (7). Many different natural and artificial TruE variants exist, encoding different products (“X” in Fig. 1). TruE is the substrate for TruD, which inserts heterocycles (9), and TruA, a protease that removes the leader sequence (10). The product of TruA is the substrate of protease TruG, which macrocyclizes the N and C termini (10). Finally, TruF1 installs one or more isoprene units on the mature macrocycle (11).
Fig. 1.
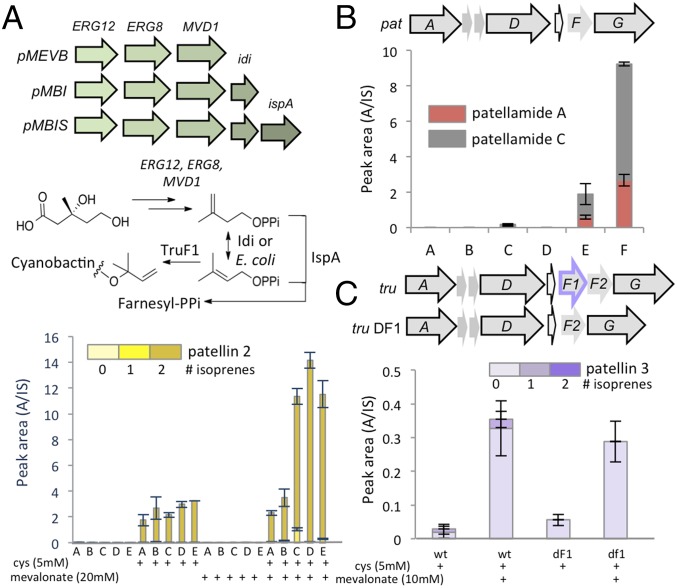
Generation of chemical diversity by cyanobactin pathways. (A) Biosynthesis of cyanobactins by the tru pathway. The relationship between the precursor peptide recognition sequence and the relevant enzyme is shown by color. For example, the TruA protease recognition sequence is shown in blue. “X” indicates regions that can be highly variable. Because these elements are progressively cleaved during biosynthesis, the substrate for each ensuing enzymatic step is progressively different in structure (SI Appendix, Fig. S1). (B) Representative natural coral reef compounds produced in this study.
Each enzyme has relaxed substrate specificity; thousands of derivatives have been synthesized (8, 12–14). At each ensuing biochemical step the substrates are increasingly different, because early substrates contain large conserved elements known as “recognition sequences,” which are progressively pared away in the course of biosynthesis and are not found in the products of the pathway (1, 15). Thus, late-stage enzymes encounter structurally divergent substrates, whereas the substrates of early-stage enzymes retain some conserved structural features.
Although tru has been expressed in E. coli to produce many compounds, a limitation was low and variable yield. Common methods afforded only modest yield improvements (16). Here, while improving yield by nearly 30,000-fold from an initial ∼1 μg/L, we discovered principles that may be fundamental in DG pathways.
Results
Optimization with Cysteine and Mevalonate Enables Determination of Biological Activity.
We first optimized production in E. coli by traditional methods using the vector pTru-SD encoding the tru pathway, resulting in a best yield of <10 μg/L over a 5-d fermentation. The low yield likely was caused by a well-known limitation: Although engineering pathways to produce active enzymes can be straightforward, this strategy often fails to provide the chemical products of those enzymes. Therefore, we sought to add compounds to modulate enzymatic efficiency in vivo, adopting a metabolite-directed approach.
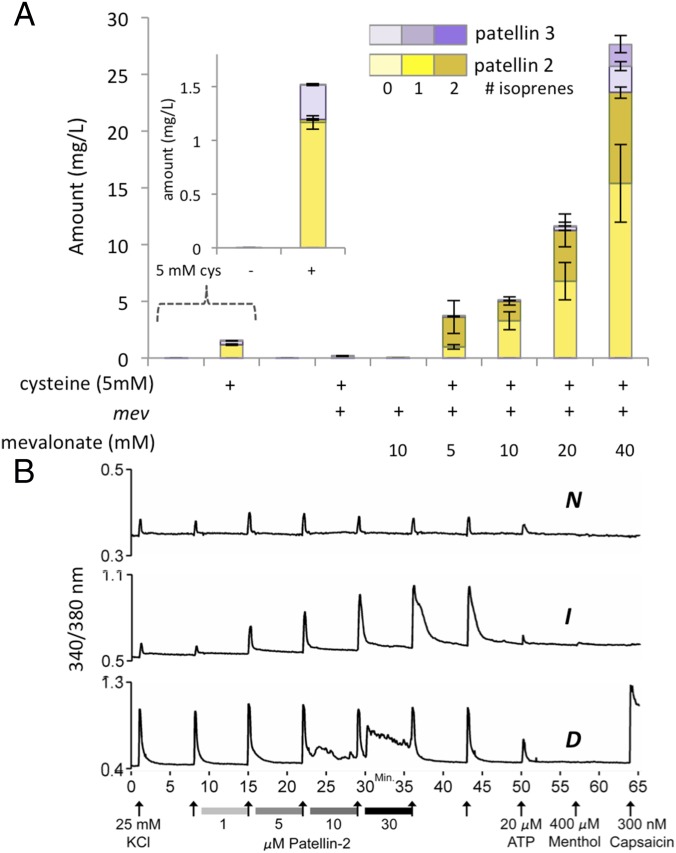
We found that the addition of cysteine (5–10 mM), along with minor process changes, reproducibly increased the yield of patellins by 150-fold, to ∼1.5 mg/L (Fig. 2A and SI Appendix, Fig. S2). A mixture of patellins with 0–2 units of isoprene was produced, and prenylation increased slowly over a series of days. Therefore, we increased production of the isoprene precursor, dimethylallylpyrophosphate (DMAPP), using the vector pMBI, which contains genes (mev) that convert mevalonic acid to DMAPP (17), which in turn is a substrate for prenyltransferase TruF1 (11). Coexpression of pTru-SD and pMBI led to a great increase in prenylation that depended on the dose of added mevalonate (Fig. 2A) and also, surprisingly, increased the total yield of patellins from cysteine alone by ∼18-fold, to 27 mg/L. This increase was dependent upon cysteine, without which mev/mevalonate did not increase the yield. We calculated that ∼250 mg/L of precursor peptide TruE-3-2 was synthesized, representing ∼25% of the dry weight of the cells. This striking effect may greatly exceed known yields of RiPPs by precursor weight.
Fig. 2.
Optimization of patellin production in E. coli and discovery of neuroactivity. (A) Coral reef compounds patellins 2 and 3 were synthesized in E. coli using vector pTru-SD, with added agents shown on the x axis. Cultures were harvested on fermentation day 6. Yield was measured by comparison to an internal standard (SI Appendix, Fig. S2). Measurements were made from independent replicates performed in triplicate. Different degrees of prenylation are indicated by progressively darker color: yellow, patellin 2; purple, patellin 3. (B) Dorsal root ganglion assay using cultivated mouse neurons. Each trace represents the response of a single neuron in culture. KCl (arrows on the x axis) is added to the culture and removed, leading to depolarization and Ca2+ entry into the cell, measured using the fluorescence ratio of Ca2+-responsive Fura-2 dye (y axis). Between KCl additions, recombinant patellin 2 was added for 5 min and removed from cells at increasing doses, leading to response by neurons. About 800 individual neurons were examined in this study (SI Appendix, Fig. S4). Shown are typical nonresponsive neurons (N, top trace). In comparison, a neuron is shown that increases (I) the KCl-responsive Ca2+ flux after patellin 2 is added in some cells (middle trace). In some neurons, a direct depolarization (D) is seen when patellin 2 is added (bottom trace).
Patellin 2 is a seldom-found natural product from coral reef animals with unknown bioactivity (7, 18). With robust expression, we synthesized wild-type patellin 2 in E. coli (SI Appendix, Fig. S3) and screened it in a variety of assays such as constellation pharmacology, which is not traditionally used in natural products research. Constellation pharmacology with mouse dorsal root ganglion neurons (19–21) showed that patellin 2 exhibits an activity profile that may indicate potassium channel inhibition (Fig. 2B and SI Appendix, Fig. S4 A and B).
Cysteine Functions by Liberation of Hydrogen Sulfide.
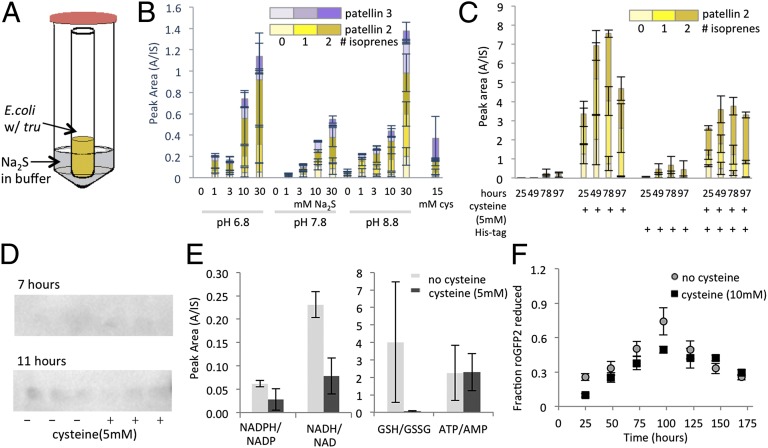
Cultures with cysteine had unusual characteristics, leading us to speculate that cysteine may not be the direct molecular initiator of increased yield. Cystine could replace cysteine (SI Appendix, Fig. S5A), and yield depended upon vessel size and shape. Serendipitously, we used cysteine in an experiment in which the cover of a multiwell plate was not pierced to allow gas exchange. In that event, control wells lacking cysteine exhibited a great increase in patellin production. Therefore, we suspected that the active reagent was volatile. Hydrogen sulfide is produced when E. coli degrades extracellular cysteine to sulfide (22–24). Indeed, added cysteine and cystine were completely degraded during E. coli growth (see SI Appendix, Fig. S11A). We liberated H2S gas into cultures at varying concentrations (Fig. 3A and SI Appendix, Fig. S5B); H2S alone was sufficient to recapitulate the increased patellin yields found with cysteine (Fig. 3B).
Fig. 3.
Sulfide liberated from cysteine increases compound production. (A) Experimental setup for generating H2S gas in E. coli cultures. By adding sodium sulfide externally to growing cultures at varying pH, consistent low doses of H2S are generated. (B) With this setup, the addition of sulfide leads to cyanobactin production. Cultures were grown for 3 d, and 2-mL cultures were extracted and analyzed as described in Methods. (C) Production of patellin 2 in vectors with and without a His-tag shows that production is cysteine responsive and increases even after cysteine is depleted from culture (SI Appendix, Fig. S11A) and for days after TruE-2-2 disappears. Measurements were performed in triplicate from independent cultures. (D) TruE is expressed early in the log phase, and its synthesis or degradation does not differ with or without cysteine. The Western blot against His-tagged TruE-2-2 shows that the protein is visible only at 7 and 11 h but is not visible at later time points, independent of cysteine addition. Additional points and conditions are shown in SI Appendix, Fig. S7D. (E) The addition of cysteine (5 mM) leads to the oxidation of redox-sensitive metabolites but has little effect on the ATP/AMP ratio in E. coli. Measurements were performed in four independent cultures using 100-mL–scale cultures harvested after 24 h. (F) Adding 10 mM cysteine has little effect on the growth of E. coli but leads to oxidation of the cytoplasm, as measured by the redox state of redox-responsive roGFP2. Each data point is the average of at least six independent cultures. Spectra of reduced and oxidized roGFP in E. coli are shown in SI Appendix, Fig. S5E.
Molecular Mechanism of H2S.
Four major possibilities were tested in which H2S might impact (i) the abundance of cyanobactin proteins; (ii) the availability of metabolites or cofactors; (iii) cellular redox; and (iv) the activity of proteins.
Mechanism 1: Protein Abundance Is Not Altered.
In pTru-derived vectors, the tru operon is under control of the lac promoter. Using a lac promoter-GFP fusion, we showed that GFP is modestly decreased when cysteine is introduced (SI Appendix, Fig. S6A). Similarly, when gfp was fused directly to the last gene in tru (16), fluorescence was modestly diminished with cysteine in comparison with cysteine-free controls (SI Appendix, Fig. S6B). We examined the production and stability of all tru enzymes under control of individual T7-lac promoters and observed no significant effect.
We investigated whether the concentration of the substrate TruE increased upon the addition of cysteine in the native tru context. We modified the truE-2-2 gene (which encodes patellin 2 in both cassettes) by inserting a 6xHis tag into the leader sequence (Fig. 3 C and D). In Western blots, TruE-2-2 was observed after 7 h of fermentation (early log phase) and was optimal around 11 h (midlog) but was not detectable by late-log phase (17 h and later). No difference was observed in the presence or absence of cysteine (SI Appendix, Fig. S7D). In contrast, detection of cyclic products was greatly increased when cysteine was added. Thus, the effects of cysteine are not explained by increased TruE production or stability.
Mechanism 2: Metabolism Is Not Altered Significantly by Cysteine.
ATP is used in heterocyclization (9, 25), so it might affect the pathway directly, whereas other metabolites might act indirectly. Two types of metabolomics experiments were used. The first examined a panel of primary metabolites using authentic standards, such as ATP and NADH (Fig. 3E and SI Appendix, Fig. S8A). No significant differences were observed except with redox-responsive metabolites, which were greatly altered. We also performed untargeted LC-MS and observed only minor differences (SI Appendix, Fig. S8B) (26, 27).
Mechanism 3: Cellular Redox Is Not Responsible for the Effect.
Because cysteine changed the cellular redox state, leading to greatly increased glutathione disulfide, it was possible that this change also increased the yield of products. Cysteine recently was shown to oxidize the E. coli cytoplasm (28). Using metabolomics and redox-sensitive GFP, we confirmed that cysteine greatly oxidizes E. coli (Fig. 3F and SI Appendix, Fig. S8A). However, H2S was equally effective in producing patellins, but instead of oxidizing cells it mildly reduced them (SI Appendix, Fig. S5C). Moreover, H2S and cysteine were equally effective in producing patellins in aerobic and anaerobic fermentation. Therefore, a change in cellular redox state did not cause increased patellin production.
Mechanism 4: Modulation of Protein Activity Underlies the Cysteine Effect.
Although gross cellular redox does not correlate with increased patellin production, the redox state of individual thiols in proteins changes with increased intracellular H2S (29, 30). Therefore, it was possible that the addition of sulfide would alter protein activity. Critically, we noticed that, the higher the yield of patellin 2 in a culture, the more cysteine-dependent that yield became. One example can be seen in Fig. 3C, in which introduction of a His-tag to TruE-2-2 decreases the yield of patellin 2 in comparison with the unmodified vector; more patellin 2 is produced without cysteine with this His-tagged vector. Therefore, we proposed that a pTru plasmid-encoded protein acts as a negative regulator of compound synthesis that is relieved by the addition of sulfide.
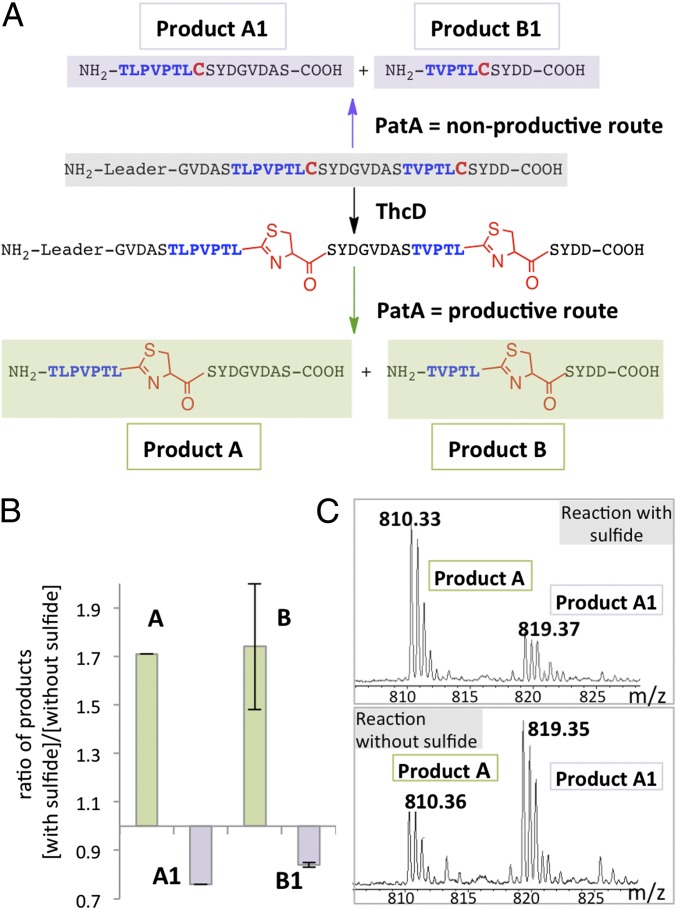
One possible negative regulator is TruA, which is redox sensitive and is the only enzyme in the pathway that is capable of derailing the biosynthesis by making nonproductive intermediates. This hypothesis was tested by combining the relevant proteins in vitro and determining the effect of sulfide on product formation. We treated TruE-3-2 with the TruD homologs ThcD and PatA (98% identical to TruA) in the presence or absence of 500 μM sulfide. Absent sulfide, we observed mainly the incorrect products that are made when PatA acts before the action of ThcD (Fig. 4 and SI Appendix, Fig. S9). With sulfide, in contrast, we saw mostly the correct products made when ThcD acted before PatA. When PatA or ThcD was used singly with its authentic substrate, it performed without significant differences with or without the addition of 500 μM sulfide, indicating that this effect was not caused by gross inhibition of enzymes or a by change in the fundamental properties of enzymes or substrates. We propose that sulfide specifically modulates the substrate preference of mature enzymes in the cyanobactin pathway, enabling posttranslational control of product formation in vivo.
Fig. 4.
Hydrogen sulfide modulates the substrate preference of early cyanobactin enzymes. The purified enzymes ThcD and PatA were combined with the substrate TruE-3-2 in vitro, and reactions were run for 6 h in triplicate. More of the correct products were obtained after 6 h when Na2S (500 μM) was added to the reaction medium than without sulfide (see SI Appendix, Fig. S9 for a full set of conditions and reactions). (A) Schematic of productive and nonproductive reaction pathways. (B) More of the correct products A and B than incorrect products A1 and B1 are synthesized when sulfide is added to cultures. (C) Mass spectra are shown for product A and product A1 with and without the addition of sulfide to the medium.
DMAPP from the mev Pathway Is Incorporated into Patellins.
E. coli normally contains the deoxyxylulose pathway to isoprenoids and therefore cannot use mevalonate (17). When 1-13C–labeled mevalonate was added to the culture, MS analysis showed that the major ion of the doubly prenylated peak was +2 Da in comparison with unlabeled material. MS/MS experiments localized this mass difference to DMAPP (SI Appendix, Fig. S10).
The Effect of Mevalonate Is Independent of Prenylation.
Previously, we showed that the rates of purified enzymes alone or in combination were not affected by DMAPP concentration (11). We used a series of vectors leading to different isoprene products, demonstrating that the increased yield could be observed no matter whether DMAPP, isopentenyl pyrophosphate, or farnesyl pyrophosphate was the primary product of the pathway (Fig. 5A). We also expressed the pat pathway, which is closely related to tru except that it lacks a functional prenyltransferase. Nonetheless, pat still was responsive to both cysteine and mevalonate/mev, requiring both to be detected at reasonable levels (Fig. 5B). Finally, the tru prenyltransferase gene truF1 and its nonprenylating homolog truF2 were knocked out individually or in tandem from the tru pathway (Fig. 5C). In all cases, production was still responsive to both cysteine and mevalonate. In sum, although this study did not determine the mechanism behind the increase in the patellin yield with mev/mevalonate, it was clear that direct prenylation of cyanobactins was not responsible. We also show that TruF2 is not a prenyltransferase and that TruF1 is solely responsible for prenylation in the pathway.
Fig. 5.
Mevalonate increases cyanobactin synthesis independently of prenylation. (A) Pathways for the conversion of mevalonate to DMAPP, the precursor for prenylation of cyanobactins. (Top) The diagram shows which genes are present in plasmids containing the mevalonate pathway used in this study. (Middle) The biochemical process catalyzed by the enzymes that convert mevalonate to DMAPP and other products. (Bottom) Cyanobactin production is greatest in the presence of mevalonate. This effect requires cysteine. Cultures were grown as described and were harvested after 5 d. Each set represents triplicate independent replicates. Expression conditions are shown as A, tru; B, tru/pBBR; C, tru/mevb; D, tru/mbi; E tru/mbis. (B) Expression from the pat pathway is increased with pMBI and mevalonate, even though patellamides A and C are not prenylated. Expression conditions are shown as A, pat; B, pat + 5 mM cysteine; C, pat/mbi + 5 mM cysteine; D, pat/mbi + 10 mM mevalonate; E, pat/mbi + 5 mM cysteine + 5 mM mevalonate; F, pat/mbi + 5 mM cysteine + 10 mM mevalonate. (C) Expression from the tru pathway is increased with pMBI and mevalonate, even in the absence of prenylation. pTru-b and pTru-DF1 were expressed as described. Cultures were harvested after 5 d and were analyzed for patellin production. df1, TruF1 knockout; wt, wild type.TruF2 and TruF1-TruF2 were deleted also, providing a similar effect. Note the absence of prenylated products when truF1 is removed from the tru pathway.
Each Ensuing Metabolic Step Is Increasingly Slower in Vivo.
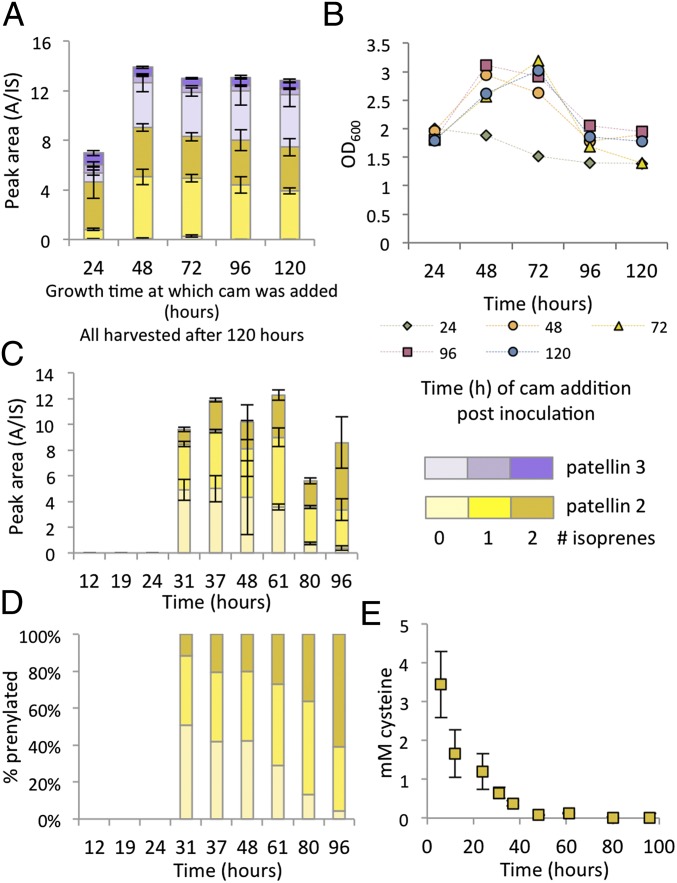
The first step in the synthesis of patellins is translation of the substrate (TruE) and required enzymes. We added the translation inhibitor chloramphenicol to E. coli cultures every 24 h beginning at the 24-h time point, with and without cysteine (Fig. 6 A and B). E. coli growth ceased upon the addition of chloramphenicol, but compound production did not, indicating that transcription and translation of tru proteins is completed during cell growth. This conclusion also is supported by the experiments with GFP described above, in which maximum fluorescence is achieved during log phase. In contrast, accumulation of cyclic and prenylated products continues for days after growth ceases.
Fig. 6.
Enzymatic activity continues for days after translation stops. (A) Chloramphenicol (cam) was added every 24 h to cultures containing cysteine, and cells were harvested from all wells at 120 h. Triplicate independent measurements are shown. The resulting production of patellins after 120 h is shown. (B) Growth curves are shown in which OD600 was measured every 24 h. Growth ceases upon addition of chloramphenicol. (C and D) Expressions using TruE-2-2 showing the onset of macrocyclization (C) corresponding with the depletion of cysteine (D). (E) When yields are normalized, it is clear that prenylation by TruF1 continues for days and is very slow. Expressions using TruE-T-T are shown in SI Appendix, Fig. S11B.
Following translation, TruE is heterocyclized by TruD. In Western blots following TruE, TruE-2-2 was barely visible at any time point (Fig. 3D and SI Appendix, Fig. S7D), even though His-tagged TruE derivatives under the control of the much stronger T7 promoter were readily visualized when T7 RNA polymerase expression was coinduced (SI Appendix, Fig. S7A). Because heterocyclization must precede the degradation of TruE, TruE-2-2 was being consumed over the course of its synthesis, demonstrating that the first enzymatic step (heterocyclization) is fast in vivo.
In contrast, macrocyclization was much slower. With the His-tagged TruE vector, cyclic peptide was observed at 24 h, with production increasing for a further 2 d (Fig. 3C). We examined the timing of production from the vectors containing TruE-2-2 and TruE-T-T (containing the trunkamide sequence in both cassettes) (Fig. 6 C and D and SI Appendix, Fig. S11B). Notably, the onset of cyclic peptide was observed in tandem with the timing of cysteine depletion and the onset of the stationary phase. Cyclic peptides continued to accumulate for approximately 24–72 h after cysteine depletion. Similar time courses were observed without cysteine, although much less macrocycle was produced. Thus in vivo macrocyclization is slower than the preceding steps in E. coli. Under all conditions, prenylation was even slower, continuing after the termination of the macrocyclization reaction (Fig. 6 D and E).
Discussion
We show that the DG tru pathway exhibits a surprising property: Each metabolic step in the pathway is progressively slower as biosynthesis proceeds in living cells. All metabolic steps except for precise timing of TruA cleavage were observed by quantifying pathway intermediates and products, leading to a metabolic flux model (SI Appendix, Fig. S12). (i) TruE precursor peptide accumulates in early log phase. (ii) TruD acts on TruE essentially immediately, before any proteolysis of TruE. (iii) TruE is rapidly proteolyzed, completely disappearing as cysteine is depleted and log phase ends. The timing of this disappearance is identical with and without cysteine. (iv) As log phase ends and cysteine is depleted, cyclic peptides accumulate slowly, requiring 24–48 h for completion. (v) Prenylated intermediates are made even more slowly, continuing to accumulate for >72 h and not being complete even at that time. Thus, in vivo each metabolic step requires a substantially longer period than the preceding one. The increasing sluggishness parallels the increasing differentiation of chemical substrates at each ensuing biochemical step and is in striking contrast to that in conventional pathways, in which the first metabolic step is limiting (and usually regulated with the pathway), designed to prevent the accumulation of intermediates or byproducts.
A caveat is that these results were generated using a heterologous host and thus may not reflect nature. The most striking discovery is the effect of cysteine on the cyanobactin pathway. Recently, it has been established that disulfide bonds are reduced across the proteome as cyanobacteria transition from dark to light (31) and that these changes are coupled to secondary metabolism (32). Reduction inhibits TruA, whereas only reduced TruE reacts with TruD. These results suggest that the effects of sulfide on cyanobactin biosynthesis in E. coli reflect natural cycles in cyanobacteria, providing a biochemical link to the disulfide cycle.
The in vitro enzymology of each cyanobactin pathway enzyme has been investigated (9–11, 33, 34). The individual enzymes are slow, with the in vitro trends paralleling the in vivo observations in this study. When all enzymes are combined in vitro, intermediates accumulate, as found in E. coli (11). The best investigation of individual DG metabolic enzymes involved a comparison of broad and narrow substrate-tolerant LanM family lanthionine synthases (2, 35, 36). The narrow substrate-tolerant enzyme HalM2 produces a single product very efficiently. However, the broad substrate-tolerant ProcM accepts widely different substrates with completely different skeletal features. In comparison with the substrate-restricted HalM2, the substrate-tolerant ProcM is slower and does not perform proofreading. Additionally, the first step is the fastest with ProcM but is the slowest (rate-limiting) with HalM2. Extrapolating these single-enzyme data to whole pathways, we propose that DG metabolic pathways may follow common principles.
In addition to the conceptual framework they provide, these principles have implications in biotechnology. Typically, heterologous expression is accomplished by modulating regulatory elements. However, this strategy provided only modest improvements with tru in our hands (16). Here, we report a metabolite-directed approach. Although sulfide does not increase the abundance or lifetime of tru proteins, it modulates the relative efficiency of the steps. In the case of conventional pathways, this strategy may not be necessary, because the gate from common biological substrates is at an early step, and all ensuing steps are relatively rapid. Perhaps such switches represent an alternative to the “committed step” of conventional pathways.
With sufficient patellins in hand, we discovered potentially useful neuroactivity. Patellins are rare (7, 18, 37). Indeed, most coral reef biodiversity comprises small animals that are not amenable to drug-discovery programs (16). By producing practical yields of five different coral reef animal products, we show that the supply problem can be circumvented, indicating a way that these small and rare samples can be practically coupled to drug discovery. In addition, we previously showed that the tru pathway can synthesize millions of derivatives (8, 12, 33). The ability to scale production thus is game-changing in terms of developing combinatorial biosynthetic strategies.
We propose general principles underlying DG metabolism. It would be expected that DG metabolism would not use the same substrate fidelity mechanisms as conventional metabolism. We show that for tru these mechanisms are, in fact, nearly the opposite. In tru, later metabolic steps are increasingly slow, and isolable intermediates accumulate, features that are likely hallmarks of DG metabolism. The advantage of using the tru pathway is not that it is unique but rather that its wide substrate tolerance has been established experimentally in both laboratory and natural conditions. However, many pathways are believed to exhibit relaxed substrate tolerance (38–40) and may share unifying features of DG metabolism.
In addition to sharing common mechanisms, DG pathways likely share a common purpose: generating metabolites aimed externally at other organisms rather than at the producing organisms themselves (38–40). The ability to diversify chemistry enables the producer to adapt as predators, prey, and competitors change. DG metabolism is not the only way that chemistry can adapt rapidly. Many metabolic pathways are relatively fixed and ancient, dedicated to producing specific compounds. In these cases, lateral pathway transfer is one mechanism that enables organisms to obtain new chemistry. In contrast, DG provides a way for organisms to diversify rapidly around an active pharmacophore. The resulting diversification is reminiscent of a synthetic medicinal chemistry approach in which a lead compound is diversified to produce useful drugs. DG metabolism thus is a widespread natural method of chemical optimization in response to challenge.
Methods
E. coli tru Expression Vectors.
In most experiments, the tru pathway was expressed from pTru-SD1, in which the genes truA, -B, -C, -D, -E, -F1, -F2, and -G were codon optimized for E. coli, and the intergenic sequence was wild type. In addition, the start of the operon (truA) was fused to the E. coli lac promoter, and the end of truG was fused to a terminator. In most vectors, the precursor peptide was TruE-3-2, where patellin 3 is encoded in the first cassette, and patellin 2 is in the second. In some vectors, TruE-2-2 (with two copies of the patellin 2 cassette) or TruE-T-T (with two copies of the trunkamide cassette) was used. A full set of vectors used is given in SI Appendix, SI Methods.
Production of Patellins in E. coli.
Six colonies of transformed E. coli DH10B cultures were picked from plates and grown in seed cultures overnight. The resulting cultures were combined and used to seed deep-well 24-well plates containing 2×YT (growcells.com) broth (6 mL per well) and ampicillin (50 μg/mL). Other additives, such as varying amounts of cysteine and/or mevalonate, were placed in individual wells. Plates were sealed, and holes were pierced above each well with a toothpick. Plates were incubated with shaking (100 rpm) for 5–7 d. Where indicated, cultures were harvested at several time points. Harvesting consisted of centrifugation followed by immediate analysis or storage at −80 °C until analysis.
Analytical Methods.
All experiments reported were performed a minimum of three times, in at least triplicate replicate samples. Cells were prepared by centrifugation to remove medium before analysis. To analyze fluorescence or cell growth, whole cells were used. To analyze patellin production, cells were extracted with acetone (2 mL), and the resulting acetone extract was analyzed by mass spectrometry in comparison with authentic standards and a standard concentration curve.
Complete details are provided in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01 GM102602 and NIH Postdoctoral Fellowship GM103219 (to E.P.). The purchase of the Agilent 6550 mass spectrometer was funded by NIH Grant OD016232 (J.E.C., Principal Investigator).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525438113/-/DCSupplemental.
References
- 1.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2(12):729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 2.Li B, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci USA. 2010;107(23):10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams DH, Stone MJ, Hauck PR, Rahman SK. Why are secondary metabolites (natural products) biosynthesized? J Nat Prod. 1989;52(6):1189–1208. doi: 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- 4.Jones CG, Firn RD. On the evolution of plant secondary chemical diversity. Phil Trans. 1991;333(1267):273–280. [Google Scholar]
- 5.Fischbach MA, Clardy J. One pathway, many products. Nat Chem Biol. 2007;3(7):353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102(20):7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4(6):341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruffner DE, Schmidt EW, Heemstra JR. Assessing the combinatorial potential of the RiPP cyanobactin tru pathway. ACS Synth Biol. 2015;4(4):482–492. doi: 10.1021/sb500267d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McIntosh JA, Donia MS, Schmidt EW. Insights into heterocyclization from two highly similar enzymes. J Am Chem Soc. 2010;132(12):4089–4091. doi: 10.1021/ja9107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131(6):2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sardar D, Lin Z, Schmidt EW. Modularity of RiPP enzymes enables designed synthesis of decorated peptides. Chem Biol. 2015;22(7):907–916. doi: 10.1016/j.chembiol.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134(1):418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto Y, Ito Y, Kato Y, Tsunoda S, Suga H. One-pot synthesis of azoline-containing peptides in a cell-free translation system integrated with a posttranslational cyclodehydratase. Chem Biol. 2014;21(6):766–774. doi: 10.1016/j.chembiol.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Houssen WE, et al. An efficient method for the in vitro production of azol(in)e-based cyclic peptides. Angew Chem Int Ed Engl. 2014;53(51):14171–14174. doi: 10.1002/anie.201408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sardar D, Pierce E, McIntosh JA, Schmidt EW. Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth Biol. 2015;4(2):167–176. doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donia MS, Ruffner DE, Cao S, Schmidt EW. Accessing the hidden majority of marine natural products through metagenomics. ChemBioChem. 2011;12(8):1230–1236. doi: 10.1002/cbic.201000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 18.Carroll AR, et al. Patellins 1-6 and trunkamide A: Novel cyclic hexa-, hepta- and octa-peptides from colonial ascidians, Lissoclinum sp. Aust J Chem. 1996;49(6):659–667. [Google Scholar]
- 19.Teichert RW, et al. Characterization of two neuronal subclasses through constellation pharmacology. Proc Natl Acad Sci USA. 2012;109(31):12758–12763. doi: 10.1073/pnas.1209759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teichert RW, Memon T, Aman JW, Olivera BM. Using constellation pharmacology to define comprehensively a somatosensory neuronal subclass. Proc Natl Acad Sci USA. 2014;111(6):2319–2324. doi: 10.1073/pnas.1324019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichert RW, Schmidt EW, Olivera BM. Constellation pharmacology: A new paradigm for drug discovery. Annu Rev Pharmacol Toxicol. 2015;55:573–589. doi: 10.1146/annurev-pharmtox-010814-124551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berglin EH, Carlsson J. Potentiation by sulfide of hydrogen peroxide-induced killing of Escherichia coli. Infect Immun. 1985;49(3):538–543. doi: 10.1128/iai.49.3.538-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awano N, Wada M, Mori H, Nakamori S, Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl Environ Microbiol. 2005;71(7):4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton WA, Snell EE. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci USA. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: Microcin B17 synthase. Science. 1996;274(5290):1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 26.Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci USA. 2013;110(6):2336–2341. doi: 10.1073/pnas.1218524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodwin CR, et al. Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Anal Chem. 2014;86(13):6563–6571. doi: 10.1021/ac5010794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtsu I, et al. The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J Biol Chem. 2010;285(23):17479–17487. doi: 10.1074/jbc.M109.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mustafa AK, et al. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2(96):ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul BD, Snyder SH. Protein sulfhydration. Methods Enzymol. 2015;555:79–90. doi: 10.1016/bs.mie.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Guo J, et al. Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol Cell Proteomics. 2014;13(12):3270–3285. doi: 10.1074/mcp.M114.041160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diamond S, Jun D, Rubin BE, Golden SS. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci USA. 2015;112(15):E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIntosh JA, et al. Circular logic: Nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J Am Chem Soc. 2010;132(44):15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntosh JA, Donia MS, Nair SK, Schmidt EW. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J Am Chem Soc. 2011;133(34):13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thibodeaux CJ, Ha T, van der Donk WA. A price to pay for relaxed substrate specificity: A comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J Am Chem Soc. 2014;136(50):17513–17529. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y, Mukherjee S, van der Donk WA. Product formation by the promiscuous lanthipeptide synthetase ProcM is under kinetic control. J Am Chem Soc. 2015;137(15):5140–5148. doi: 10.1021/jacs.5b01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zabriskie TM, Foster MP, Stout TJ, Clardy J, Ireland CM. Studies on the solution- and solid-state structure of patellin 2. J Am Chem Soc. 1990;112(22):8080–8084. [Google Scholar]
- 38.Firn RD, Jones CG. A Darwinian view of metabolism: Molecular properties determine fitness. J Exp Bot. 2009;60(3):719–726. doi: 10.1093/jxb/erp002. [DOI] [PubMed] [Google Scholar]
- 39.Weng JK, Noel JP. The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb Symp Quant Biol. 2012;77:309–320. doi: 10.1101/sqb.2012.77.014787. [DOI] [PubMed] [Google Scholar]
- 40.Olivera BM. Conus peptides: Biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281(42):31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.