Significance
Conventional models of cancer progression propose that single cells leave the primary tumor, enter the circulation, and seed clonal metastases. However, metastases can contain multiple clones, raising the question: How do polyclonal metastases form? We demonstrate that cancer cells seed distant organs as cohesive clusters, composed of two molecularly distinct subpopulations, whose proportions vary systematically during metastasis. We establish that collective dissemination is a frequent mechanism for metastasis and identify a molecular program in the most invasive, keratin 14+ (K14+) cancer cells, regulating cell–cell adhesion, cell–matrix adhesion, and immune evasion. We demonstrate that this metastatic phenotype is dependent upon K14 expression. Understanding the molecular basis of collective dissemination may therefore enable novel prognostics and therapies to improve patient outcomes.
Keywords: collective invasion, collective dissemination, polyclonal metastasis, breast cancer, keratin 14
Abstract
Recent genomic studies challenge the conventional model that each metastasis must arise from a single tumor cell and instead reveal that metastases can be composed of multiple genetically distinct clones. These intriguing observations raise the question: How do polyclonal metastases emerge from the primary tumor? In this study, we used multicolor lineage tracing to demonstrate that polyclonal seeding by cell clusters is a frequent mechanism in a common mouse model of breast cancer, accounting for >90% of metastases. We directly observed multicolored tumor cell clusters across major stages of metastasis, including collective invasion, local dissemination, intravascular emboli, circulating tumor cell clusters, and micrometastases. Experimentally aggregating tumor cells into clusters induced a >15-fold increase in colony formation ex vivo and a >100-fold increase in metastasis formation in vivo. Intriguingly, locally disseminated clusters, circulating tumor cell clusters, and lung micrometastases frequently expressed the epithelial cytoskeletal protein, keratin 14 (K14). RNA-seq analysis revealed that K14+ cells were enriched for desmosome and hemidesmosome adhesion complex genes, and were depleted for MHC class II genes. Depletion of K14 expression abrogated distant metastases and disrupted expression of multiple metastasis effectors, including Tenascin C (Tnc), Jagged1 (Jag1), and Epiregulin (Ereg). Taken together, our findings reveal K14 as a key regulator of metastasis and establish the concept that K14+ epithelial tumor cell clusters disseminate collectively to colonize distant organs.
During metastasis, cancer cells escape the primary tumor, travel through the circulation, and colonize distant organs. Conventional models of cancer progression propose that each metastasis arises from the clonal outgrowth of a single tumor cell and this conceptual framework is a foundation for models, such as epithelial-mesenchymal transition (EMT) and migratory cancer stem cells (1).
Challenging the generality of the single-cell/single-metastasis model are long-standing clinical observations that tumor cell clusters (also termed “tumor clumps”) are also observed across the stages of metastasis. Tumor cell clusters are detected in the bloodstream of cancer patients (2), clusters can efficiently seed metastases (3), and though rare, circulating tumor cell (CTC) clusters have prognostic significance (4, 5). Furthermore, metastases are composed of multiple genetically distinct tumor cell clones, in mouse models of breast, pancreas, and small cell carcinoma (5–7), and in human metastatic prostate cancer patients (8). Taken together, these observations provide accumulating evidence that tumor cell clusters contribute to metastasis. However, they leave unresolved two important questions: how do tumor cell clusters emerge from the primary tumor, and which molecular features identify cell clusters that metastasize?
An important clinical observation is that cancer cells invade the surrounding stroma as cohesive clusters in the majority of epithelial tumors, a process termed “collective invasion” (9, 10). In breast cancer, collective invasion is facilitated by invasive leader cells, a subpopulation of tumor cells that highly express keratin 14 (K14) and other basal epithelial markers (11). K14+ cells are migratory, protrusive, and lead trailing K14− cells, while maintaining cell–cell cohesion and E-cadherin–based cell contacts.
In this study, we sought to understand how these K14+ cells exit collective invasion strands in the primary tumor and travel to distant organs (12). One hypothesis is that collective invasion is an intermediate step toward eventual single-cell dissemination and monoclonal metastasis. However, tumor cell clusters are detected in circulation (5) and primary human breast tumors can disseminate collectively into the surrounding extracellular matrix in ex vivo assays (13–15). These data prompted an alternative hypothesis, that collectively invading K14+ cancer cells could initiate and complete the metastatic process as a cohesive multicellular unit. Here we define the clonal nature of metastases in a spontaneous mouse model of metastasis to the lungs (16, 17), in which the predominant invasive form in the primary tumor is collective invasion strands led by K14+ cells (11). We establish that the majority of metastases arise from polyclonal seeds, and show that disseminated tumor cell clusters are predominantly composed of K14+ cells. We propose a mechanism for polyclonal metastasis via the collective invasion, dissemination, and colonization of clusters of K14+ cancer cells.
Results
Multicolor Fluorescent Lineage Tracing Identifies Frequent Polyclonal Metastasis in a Mouse Model of Breast Cancer.
To define the clonal origin of metastasis in a model of collective invasion, we developed a multicolor lineage-tracing strategy in a commonly used mouse model of breast cancer, MMTV-PyMT, which has provided fundamental insights into diverse aspects of metastatic progression (16, 17). In this model, the MMTV promoter drives mammary epithelial expression of the polyoma virus middle T antigen (PyMT), a potent activator of PI3K signaling, a hallmark of human breast cancer. The mammary tumors formed in this model show gene expression most similar to luminal B, an aggressive subtype of human breast cancer prone to metastasis (18).
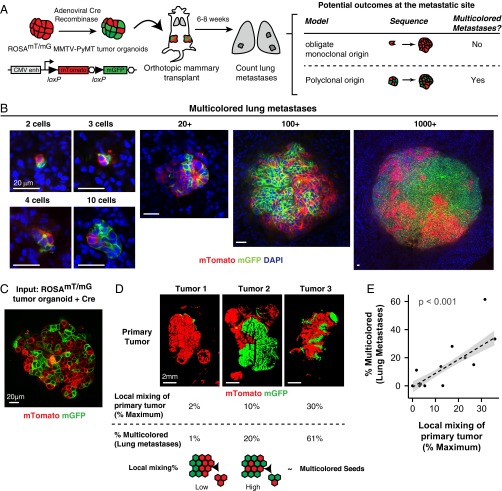
We generated a convertible double-fluorescent mammary tumor model, ROSAmT/mG;MMTV-PyMT, in which tumor cells express membrane-localized tdTomato (mTomato) and, upon treatment with adenoviral Cre recombinase (adeno-Cre), switch heritably to expression of membrane-localized enhanced GFP. ROSAmT/mG;MMTV-PyMT tumor organoids were treated with adeno-Cre to generate a mixture of red and green cells. Mosaic organoids were then injected orthotopically into the cleared mammary fat pad of nonfluorescent host mice. Because the host is nonfluorescent, this experimental system enabled us to unambiguously identify tumor metastases and their clonal origin (Fig. 1A). The principle of this strategy is that if there is an obligate single-cell intermediate during the metastatic process, we would expect to see a transition from a multicolored primary tumor (red and green) to singly red or green metastases.
Fig. 1.
Multicellular seeding is a frequent mechanism for distant metastasis. (A) Schema of multicolor lineage tracing assay. ROSAmT/mG;MMTV-PyMT tumor organoids were treated with adenoviral Cre to induce recombination from membrane tdTomato (mTomato) to membrane eGFP (mGFP). Mosaic tumor organoids were then transplanted into nonfluorescent NSG host mice. After 6–8 wk, lungs of these mice were harvested. If metastases arise exclusively from single-cell seeding, there should be only single color metastases. In contrast, multicellular seeding should produce metastases with both colors. (B) Representative micrographs of polyclonal lung metastases of different sizes. n = 355 polyclonal metastases, across 16 mice and 4 independent experiments. (C) Representative micrograph of a mosaic tumor organoid treated with adeno-Cre and grown in 3D Matrigel with intermixing of red and green tumor cell clones. (D and E) Representative micrographs of primary tumors arising from mosaic tumor organoids transplanted into NSG host mice. Primary tumors varied in their local mixing of red and green tumor cell clones (local mixing %). These differences correlated with the percentage of multicolored metastases detected in the lung (% multicolored). n = 12 mice, 4 independent experiments, 4,072 metastases. Correlation determined by Spearman rank test for samples with more than five lung metastases per mouse. (Scale bars, 20 μm in B and C, and 2 mm in D.)
In the lungs of these mice, we detected single disseminated tumor cells, micrometastases (2–50 cells), and macrometastases (>50 cells). Single disseminated tumor cells were 47-fold more frequent than metastases (range 10- to 150-fold, n = 10 mice). We reasoned that if lung metastases arose exclusively from seeding of single disseminated tumor cells, then each lung metastasis should express only one color. In contrast, multicellular seeding should be able to produce metastases with both colors. Interestingly, we observed lung metastases composed of both red and green tumor cells across a range of sizes, from 2 cells to >1,000 cells per metastasis (Fig. 1B) (n = 375 multicolored metastases). Multicolored metastases demonstrated significant intermixing of red and green tumor cells. Transplanted mice showed wide variation in the percentage of multicolored metastases from a minimum of 0% to a maximum of 61% (n = 158 multicolored out of 257 metastases). Taken together, our data show that multicolored metastases can occur frequently in the MMTV-PyMT model.
To understand our variable frequency of detection of multicolored metastases, we analyzed the degree of mixing of red and green tumor cell clones at each step in our experiments. Whereas red and green tumor cells were well mixed in the recombined tumor organoids used as input (Fig. 1C), the tumors that resulted from transplanting these organoids showed surprisingly wide variation in the mixing of red and green tumor cells. We observed tumors composed almost entirely of a single color to tumors with significant intermixing of red and green zones (Fig. 1D). To analyze this variation further, we quantified the local mixing of red and green cells within each tumor. Local mixing was assessed in 100 × 100-pixel (1.3 mm × 1.3 mm) regions tiled across the tumor by calculating the probability of selecting two colors when pixel values were chosen at random within each region. We observed a strong linear correlation between the extent of local mixing and the percentage of detected multicolored metastases (Fig. 1E). Thus, local proximity between clones of different colors in the primary tumor determines our ability to detect multicolored metastases. Based on our analysis of the relationship between local mixing and the detection of multicolored metastases (Fig. 1E), we estimate that >97% of metastases were formed from multicellular seeds (95% confidence interval 74–100%).
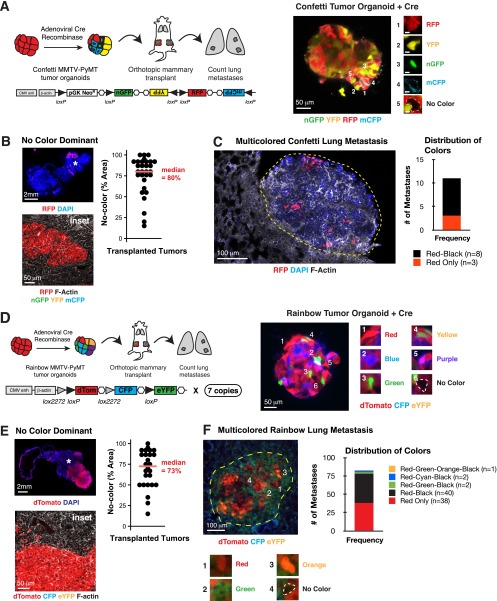
We also determined the frequency of multicolored metastases in two additional transplant models. In MMTV-PyMT tumors expressing the Confetti lineage reporter (Fig. S1A), tumor cells are induced by adeno-Cre to one of five distinct possible colors: cytoplasmic red fluorescent protein (RFP), cytoplasmic yellow fluorescent protein (YFP), nuclear GFP (nGFP), membrane cyan fluorescent protein (mCFP), or an unrecombined no-color (19). We observed zonal patches of a single color in the primary tumor, with a clear preference for the unrecombined no-color (Fig. S1B). Even with this limitation, we observed lung metastases composed of both no-color and RFP+ tumor cells (Fig. S1C). In MMTV-PyMT tumors expressing the Rainbow reporter (Fig. S1D), adeno-Cre induces recombination to one of 21 different color values (20). Despite the increased color diversity, we similarly observed zonal patches of a single color in the primary tumor, with preference for a no-color tumor cell (Fig. S1E). In the lungs of these mice, we observed multiple metastases composed of one to four different colors (Fig. S1F). Taken together, data from three distinct lineage analyses reveal polyclonal metastases in this genetically engineered mouse model of breast cancer metastasis.
Fig. S1.
Lineage tracing in Confetti and Rainbow mice identify multicolored lung metastases in the MMTV-PyMT model. (A, Left) Schema of multicolor lineage-tracing assay to determine clonality of metastases using the Confetti reporter. Confetti;MMTV-PyMT tumor organoids were treated with adeno-Cre to induce recombination to one of four colors: nuclear-GFP, membrane CFP, cytoplasmic YFP, or cytoplasmic RFP. Mosaic tumor organoids were then transplanted into nonfluorescent NSG host mice. After 6–8 wk, the lungs were harvested and the number of multicolored lung metastases was counted. (Right) Representative Adeno-Cre treated Confetti tumor organoid grown in 3D Matrigel. (B, Left) Representative micrograph of no-color dominance in a transplanted Confetti;MMTV-PyMT tumor stained with phalloidin (F-actin) and DAPI. (Lower) Inset of region denoted by star in Upper panel, showing solid boundary between RFP+ and no-colored cells. (Right) Median percentage area of the primary tumor occupied by no-color tumor cells presented as a dot plot (n = 30 tumors, 15 mice, 3 independent experiments). (C, Left) Representative multicolored lung metastasis in a transplanted Confetti;MMTV-PyMT mouse. Lung sections were stained with phalloidin (F-actin) and DAPI. (Right) Bar graph of color distribution of lung metastases (n = 11 metastases, 2 mice). (D, Left) Schema of multicolor lineage-tracing assay to determine clonality of metastases using the Rainbow reporter. Rainbow; MMTV-PyMT tumor organoids were treated with adeno-Cre to induce recombination to 1 of 21 potential colors. Mosaic tumor organoids were then transplanted into nonfluorescent NSG host mice. After 6–8 wk, the lungs were harvested and the number of multicolored lung metastases were counted. (Right) representative Adeno-Cre treated Rainbow tumor organoid grown in 3D Matrigel. (E, Left) Representative micrograph of no-color dominance in Rainbow;MMTV-PyMT transplanted tumor stained with phalloidin (F-actin) and DAPI. (Lower) Inset of region marked with star in Upper panel, showing solid boundary between RFP+ and no-colored cells. (Right) Median percentage area of the primary tumor occupied by no-color tumor cells presented as a dot plot (n = 26 tumors, 13 mice, 2 independent experiments). (F, Left) Representative multicolored lung metastasis identified in a Rainbow;MMTV-PyMT transplanted mouse. Lung sections were stained with phalloidin (F-actin) and DAPI. (Right) Bar graph of color distribution of lung metastases (n = 83 metastases, 6 mice, 2 independent experiments). [Scale bars, 50 μm (A, B, Lower, D, E, Lower), 2 mm (B, Upper, and E, Upper), and 100 μm (C and F).]
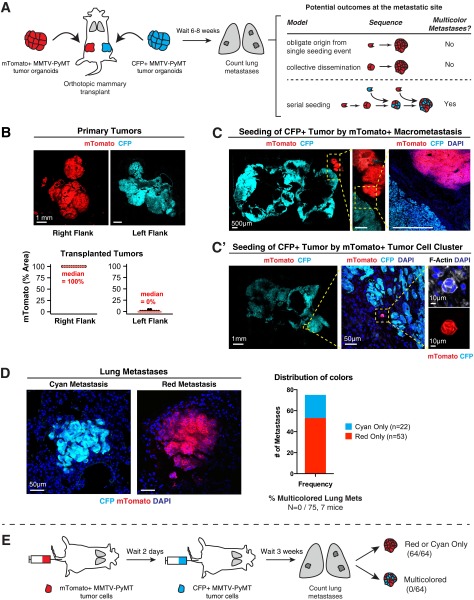
Polyclonal Lung Metastases Arise from Multicellular Seeds and Not by Serial Seeding of Single Tumor Cells.
The multicolored metastases that we observed could arise via the serial seeding of single tumor cells or via colonization by a multicellular cluster of tumor cells. We sought to distinguish these mechanisms in two different ways. To model stochastic serial seeding events integrated over time, we first isolated organoids from different MMTV-PyMT tumors that constitutively expressed either mTomato or CFP. We then transplanted mTomato+ tumor organoids to the right flank and CFP+ tumor organoids to the left flank of a nonfluorescent host (Fig. S2 A and B). Interestingly, we observed a small degree of colonization of CFP+ primary tumors by mTomato+ cancer cells, accounting for 1–5% of the total tumor area, determined macroscopically in 5 of 11 mice (Fig. S2B, Lower, and Fig. S2 C and C′). These data are consistent with the reported ability of metastases to reseed primary tumors, termed “tumor self-seeding” (21). Importantly, when we examined the clonal composition in the lungs, we only observed single-colored metastases (Fig. S2D). Taken together, these data establish that polyclonal metastases do not efficiently arise from serial seeding.
Fig. S2.
Polyclonal metastases arise from multicellular seeds not by serial seeding of single tumor cells. (A) Schema of transplant assay to model the contribution of serial seeding to polyclonal metastases. mTomato+ tumor organoids were isolated from ROSAmT/mG;MMTV-PyMT tumors and transplanted to the right flank of a nonfluorescent NSG mouse. Tumor organoids were isolated from β-actin–CFP;MMTV-PyMT tumors and transplanted to the left flank of the same mouse. After 6–8 wk, lungs of these mice were harvested and examined for multicolor metastases. There are two potential outcomes at the metastatic site. If metastases arise exclusively from a single seeding event, there should be only single-color metastases. In contrast, serial seeding by single cells should produce multicolored metastases. (B) Representative micrograph of transplanted tumors arising from either mTomato+ or CFP+ tumor organoids transplanted into NSG host mice (n = 22 tumors, 11 mice, 2 independent experiments). mTomato+ tumor cells were observed in 5 of 11 CFP+ tumors and accounted for 1–5% of the total tumor area. (C and C′) Representative micrographs of CFP+ tumor colonized by a large mTomato+ metastasis (C) and a small mTomato+ tumor cell cluster (C′). The tumor was stained with phalloidin (F-actin) and DAPI. (D) Representative micrographs of single colored CFP+ or mTomato+ lung metastases. Multicolored metastases were not observed (n = 0 of 75 metastases, 7 mice, 2 independent experiments). (E) Schema of tail vein assay that models serial waves of disseminated single tumor cells. 2 × 105 FACS-sorted mTomato+ single cells were injected into nonfluorescent NSG host mice and 2 d later, 2 × 105 FACS-sorted CFP+ single cells were injected into the same mice. Three weeks later, lungs were collected and analyzed. Multicolored metastases were not observed (n = 0 of 64 metastases, 4 mice). (Scale bars, 1 mm in B, 500 μm in C, 1 mm in C′, and 50 μm in D.)
To extend this finding, we modeled serial seeding by waves of disseminated tumor cells in the bloodstream. In this second experimental approach, we transplanted mTomato+ and CFP+ tumor cells serially via tail-vein injection (Fig. S2E). mTomato+ tumor cells were isolated by FACS and then injected as a single-cell suspension into a nonfluorescent host. Two days later, CFP+ tumor cells were isolated by FACS and injected as a single-cell suspension into the same mice. Three weeks later, lungs were collected and analyzed. In the lung, we observed exclusively single-colored metastases (Fig. S2E). Therefore, serial delivery of cancer cells to the lungs was not an efficient mechanism for generation of polyclonal metastases. In agreement with a recent study revealing metastases from oligoclonal clusters in breast cancer cell lines (5), our data are most consistent with a model in which polyclonal metastases occur via colonization by multicellular seeds rather than serial arrival and aggregation of single tumor cells.
Direct Observation of Polyclonal Collective Invasion, Polyclonal Disseminated Tumor Emboli, and Polyclonal CTC Clusters.
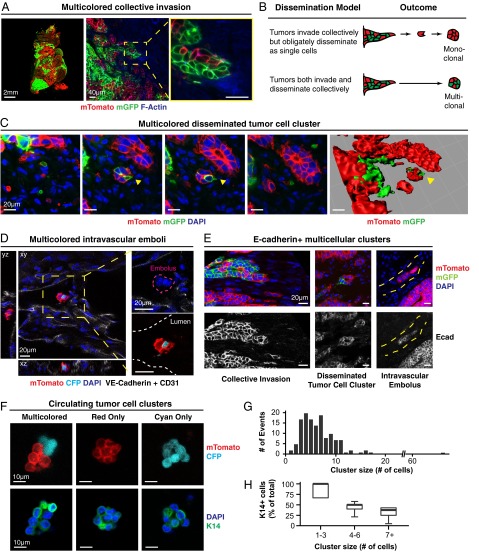
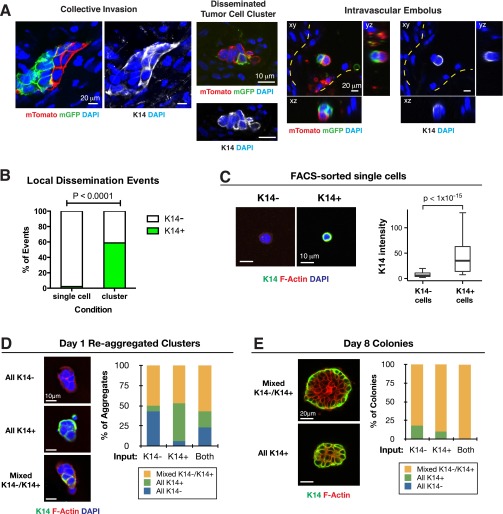
Having shown that polyclonal metastases arise from multicellular seeding, we next asked how multicellular seeds emerge from the primary tumor. To answer this question, we identified tumor cell clusters across stages of metastasis, starting from collective invasion at the primary tumor stromal interface. Consistent with our local mixing analysis (Fig. 1 D and E), when we imaged at single-cell resolution, transplanted tumors were composed of zonal patches and collective invasion strands were typically composed of a single color. Despite this technical barrier to detection of polyclonal groups, we observed multicolored collective invasion at interfaces between regions of red and green cancer cells (Fig. 2A). In these regions, we reasoned that there were two outcomes at the tumor–stromal interface corresponding to two distinct models for the collective invasive-disseminative transition (Fig. 2B). One possibility is that tumor cells invade collectively but that dissemination occurs obligately through single cells. In this case, because disseminated clusters should arise only from focal proliferation of single cells, disseminated clusters should be exclusively composed of singly red or green tumor cells. Alternatively, cancer cells could both invade and disseminate collectively as multicellular units. In this case, we would expect to identify multicolored disseminated clusters.
Fig. 2.
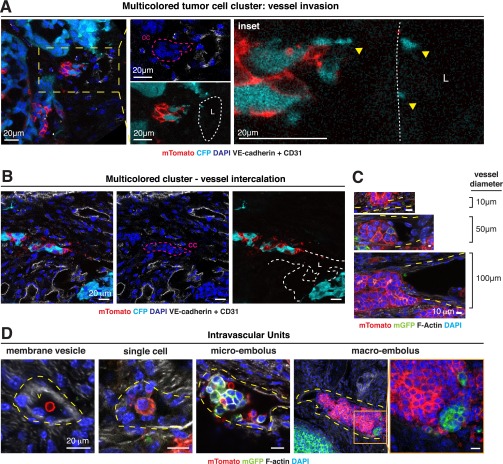
Direct observation of polyclonal collective invasion, polyclonal disseminated tumor emboli, and polyclonal CTC clusters. (A) Representative micrograph of polyclonal collective invasion arising from mosaic ROSAmT/mG;MMTV-PyMT transplanted tumors stained with phalloidin (F-actin) and DAPI (n = 75 units, 5 tumors). (B) Schema of two potential outcomes for disseminated tumor cell clusters at the tumor stromal interface. (C) Representative micrographs of a polyclonal disseminated tumor cluster (yellow arrow) in the x-y plane with successive images along the z axis (Left panels) and reconstructed 3D image (Right) (n = 25 units, 5 tumors). (D) Representative micrograph of a polyclonal disseminated tumor embolus contained within a vessel. The transplanted tumor is composed of mTomato+ and CFP+ tumor cells. Injection with VE-Cadherin and CD31 fluorescently labeled antibodies marked functional vasculature. (E) Representative micrographs demonstrating E-cadherin+ polyclonal collective invasion, dissemination, intravascular embolus (from left to right). Yellow hash marks: vessel lumen. (F) Representative micrographs of CTC clusters composed of mTomato+ and CFP+ tumor cells and stained for K14 and DAPI (n = 1 multicolored cluster, n = 13 mTomato+ clusters, n = 2 CFP+ cluster). (G) The number of events for each CTC cluster size is presented as a histogram (n = 134 events, 3 transplanted mice). (H) The median percentage of cells that are K14+ in CTCs of different cluster sizes are presented as a boxplot (n = 17 clusters). [Scale bars, 2 mm (A, Left), 40 μm (A, Right), 20 μm (C–E), and 10 μm (F).]
To distinguish these possibilities, we analyzed disseminated tumor cell clusters at three different stages of the metastatic sequence. First, we observed multicolored disseminated tumor cell clusters ≤200-μm away from the tumor–stromal interface, which we confirmed were fully disconnected from the primary tumor by 3D reconstruction (Fig. 2C). Next, we examined the adjacent vasculature at the tumor–stromal interface (Fig. S3 A–C). We observed both multicolored tumor microemboli and tumor macroemboli composed of >100 tumor cells within vessels (Fig. 2D and Fig. S3D). Consistent with their collective organization, we observed membrane-localized E-cadherin in collectively invading cancer cells, locally disseminated clusters, and tumor emboli (Fig. 2E). As a proof of concept, we next isolated CTCs from the systemic circulation of a mouse bearing fluorescently labeled tumors. Although this tumor had quite limited local mixing of blue and red clones, we identified both a multicolored and multiple singly colored CTC clusters (Fig. 2F). We analyzed CTC cluster size in three mice and found that they ranged from 2 to 66 cells per cluster, with a median of 6 cells (Fig. 2G). Taking these data together, our lineage-tracing studies enabled us to observe multicolored tumor cell clusters at five different stages of metastasis: collective invasion, locally disseminated clusters in the adjacent stroma, intravasated tumor emboli, CTC clusters, and distant micro- and macrometastases.
Fig. S3.
Multicolored disseminated tumor emboli are observed. (A) Representative micrograph of a multicolored disseminated tumor cell cluster invading into an adjacent vessel. The transplanted tumor is composed of mTomato+ and CFP+ tumor cells. Injection with fluorescently labeled VE-Cadherin and CD31 antibodies marked functional vasculature. Tumors were also stained with DAPI to mark nuclei. Leftmost panel demonstrates a collective cluster (cc) of tumor cells invading into a nearby vessel lumen (L). Inset shows CFP+ blebs in close proximity to the vessel (n = 12 units, 6 tumors). (B) Representative micrographs of multicolored disseminated tumor cell cluster intercalated into adjacent blood vessel wall. A mosaic mTomato+, CFP+ tumor was stained with DAPI. The mouse was injected with fluorescent VE-Cadherin + CD31 antibodies before sacrifice. White hash marks: vessel lumen. Magenta harsh marks: cell cluster. (C) Representative micrographs demonstrating intercalation of disseminated tumor cell clusters into vessels of varying caliber. Mosaic MMTV-PyMT;mT/mG ;MMTV-PyMT transplanted tumors were stained with phalloidin (F-actin) and DAPI. Yellow hash marks: vessel lumen. (D) Representative micrographs demonstrating intravascular dissemination units of different sizes in vessels adjacent to primary tumor. Mosaic MMTV-PyMT;mT/mG transplanted tumors were stained with phalloidin (F-actin) and DAPI. Yellow hash marks: vessel lumen. (Scale bars, 20 μm in A, B, and D, Left, and 10 μm in C.)
Aggregating Tumor Cells into Clusters Promotes Colony Formation ex Vivo and Lung Metastatic Colonization in Vivo.
An important question raised by these experiments is: Why are multicellular seeds advantageous for metastatic colonization? This question is interesting because tumor cells typically maintain extensive cell–cell contacts in vivo, and the organization of epithelial cells into cohesive clusters promotes tissue growth in a variety of normal and tumor contexts. For example, doublets of Paneth and Lgr5+ stem cells enhance the organoid-forming potential of single Lgr5+ stem cells (19), whereas the survival of colonic tumor organoids is dependent on retaining E-cadherin contacts (22).
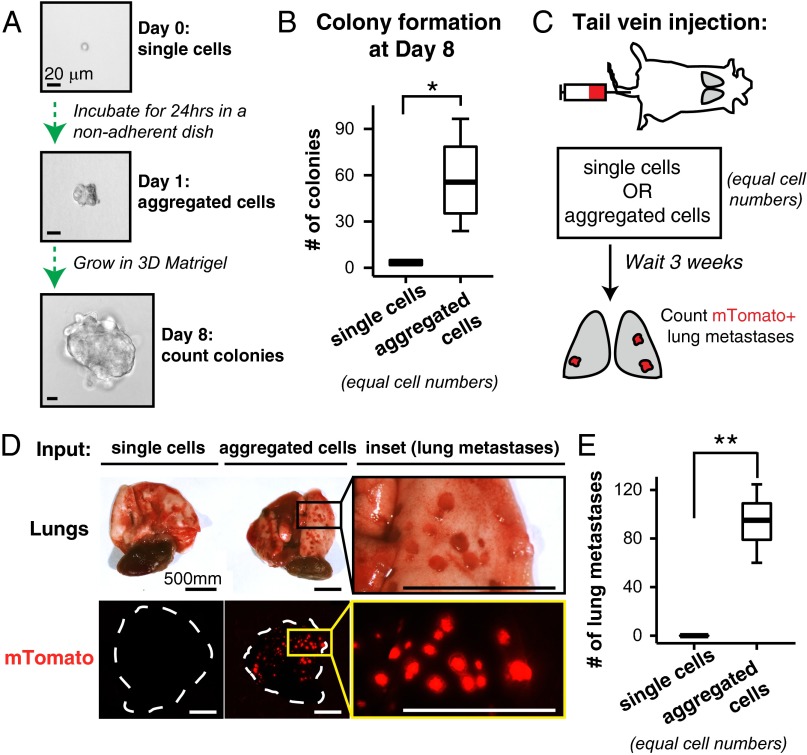
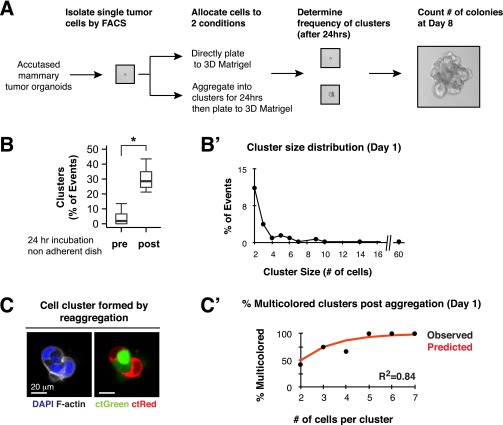
To model the colonization efficiency of multicellular seeding, we developed an experimental system to aggregate single tumor cells into cell clusters. Briefly, we isolated FACs-sorted single mTomato+ MMTV-PyMT tumor cells and incubated them overnight in a nonadherent dish to form cell clusters (Fig. 3A). Overnight incubation increased the median number of cell clusters by >15-fold relative to the input single cells (Fig. S4 A and B). The tumor cell clusters that resulted had a median size of 2 cells, and ranged from 2 to 60 cells in size (Fig. S4B′). Importantly, coincubation of red and green tumor cells confirmed that clusters were generated by aggregation rather than by clonal expansion of single tumor cells (Fig. S4C). Furthermore, the experimentally determined percentages of multicolored clusters as a function of cluster size matched the predicted percentages, assuming random assortment of red and green tumor cells into clusters (Fig. S4C′). Incubation in a nonadherent dish therefore induces the efficient aggregation of single cancer cells into clusters. We next used this approach to compare the efficiency of colony formation by single cells or aggregated clusters, while holding the total cell number constant in each condition at 10,000 cells per well. Because each cluster is formed from multiple cells, the total number of starting seeds is lower in the aggregated condition compared with the single cell condition. Despite this disadvantage, when cultured in 3D Matrigel, we observed that clusters were >15-fold more efficient at forming colonies compared with single tumor cells (Fig. 3 A and B). Thus, in an ex vivo setting, cluster organization strongly enhances colony formation.
Fig. 3.
Clustered organization significantly promotes colony formation in ex vivo culture and metastasis formation in vivo. (A) FACS-sorted single tumor cells from MMTV-PyMT tumor organoids were either (i) immediately plated into Matrigel or (ii) first incubated in a nonadherent dish for 24 h to promote cell aggregation and then subsequently plated into Matrigel. The same numbers of cells were used in each condition. Colony formation was assessed at day 8. (B) The median number of colonies per 10,000 input cells presented as boxplots (n = 4 independent experiments). *P < 0.05. (C) Schema of aggregation tail vein assay. mTomato+ tumor organoids were dissociated as in A to generate two input suspensions, single cells and aggregated cells, for tail-vein injection into NSG mice. (D) After 3 wk, lungs were visually inspected for mTomato+ metastases. (E) The median number of lung metastases per mouse presented as boxplots (two independent experiments, single cell suspension, n = 10 mice, aggregated cell suspension, n = 13 mice). **P < 0.001. (Scale bars, 20 μm in A and 500 μm in D.)
Fig. S4.
Single tumor cells aggregate into tumor cell clusters in a nonadherent dish. (A) Single tumor cells were isolated by FACS and either immediately plated into Matrigel or first incubated in a nonadherent dish for 24 h and then subsequently plated into Matrigel. The same numbers of cells were used in each condition. (B and B′) Single cells and clusters were counted and the percentage of events that were clusters was determined. (B) Twenty-four–hour incubation increases the percentage of cell clusters by >15-fold. n = 402 events, 5 independent experiments. P value determined by Wilcoxon rank sum test. *P < 0.05. (B′) Cluster size distribution of aggregated clusters, median: 2 cells, range: 2–60 cells (n = 80 clusters, 4 independent experiments). (C and C′) Single red or green tumor cells were isolated by FACS and either immediately plated into Matrigel or first incubated in a nonadherent dish for 24 h and then subsequently plated into Matrigel. The same numbers of cells were used in each condition. (C) Clusters resulting at day 1 consisted of both red and green cells, confirming that they were generated by aggregation, rather than by clonal expansion of a single tumor cell. (Scale bar, 20 μm.) (C′) The percentage of multicolored clusters formed postincubation stratified by cluster size. The predicted distribution was determined from binomal expectation. R2 was determined by Pearson correlation coefficient (n = 41 clusters, 2 independent experiments).
To assess metastatic efficiency in vivo, we next injected into the tail veins of nonfluorescent host mice either a single-cell suspension of mTomato+ tumor cells or an equal number of mTomato+ tumor cells that had been aggregated into clusters in vitro. We assessed lung metastatic burden at 3 wk (Fig. 3C). Single-cell suspensions generated zero to one metastases per mouse (Fig. 3 D and E). In contrast, aggregated clusters generated many large macroscopic mTomato+ metastases with >100-fold increase in metastatic efficiency relative to single cells (Fig. 3 D and E). Taken together, these data indicate that cluster organization is sufficient to significantly enhance tumor cell survival and promote metastasis formation.
Locally Disseminated Tumor Cell Clusters, CTC Clusters, and Micrometastases Are Enriched for K14+ Cells, Relative to Macrometastases.
Given the frequency of polyclonal metastases and the efficiency of metastasis formation by multicellular seeding, we next sought to determine the molecular characteristics of the multicellular seed. In MMTV-PyMT tumors, K14+ cells account for ∼2% of the total tumor cell population (11). These K14+ tumor cells lead >88% of collective invasion strands extending into the surrounding stroma (11). Consistent with the pioneer function of K14+ cells during metastasis, we observed K14+ cells in polyclonal collective invasion, disseminated tumor cell clusters, and intravascular emboli (Fig. S5A). We also observed significant differences in K14 expression between single cells and disseminated clusters, with 2% and 59% expressing K14, respectively (Fig. S5B). In addition, we observed that 94% of CTC clusters contained K14+ cells (n = 15 of 16 CTC clusters) (Fig. 2H). The fraction of cells within the cluster that were K14+ decreased as the size of the cluster increased (Fig. 2H). Our data therefore show that K14+ cells are enriched in the dissemination stages of metastasis.
Fig. S5.
Disseminated tumor cells are enriched for K14+ and FACS-sorted single K14+ and K14− tumor cells Interconvert to form uniformly K14+ clusters. (A) Representative micrographs demonstrating K14+ polyclonal collective invasion, dissemination, and intravascular embolus (from left to right). Mosaic ROSAmT/mG;MMTV-PyMT transplanted tumors were stained with DAPI and K14. Yellow hash marks: vessel lumen. (B) The percentage of disseminated single tumor cells and disseminated tumor cell clusters that are K14+ presented as a bar-graph (n = 45 single cells, n = 17 clusters, across 5 tumors). P value determined by Fisher’s exact test. (C) Single K14+ and K14− tumor cells were isolated by FACS from K14-actin-GFP;MMTV-PyMT tumors, and then subsequently stained for K14, phalloidin (F-Actin), and DAPI. (Right) The median intensity of K14 staining for each cell presented as a boxplot (n = 317 cells, 3 independent experiments). P value determined by Mann–Whitney test. (D, Left) Cell clusters formed at day 1 postaggregation were stained for K14, DAPI, and phalloidin (F-actin). (Right) Bar graph shows the percentage of clusters composed of only K14− cells, only K14+ cells, or both cell types (n = 119 clusters, 3 independent experiments). (E, Left) Colonies formed at day 8 were stained for K14 and phalloidin (F-actin). (Right) Bar graph shows the percentage of colonies composed of only K14− cells, only K14+ cells, or both cell types (>10 cells per colony, n = 55 colonies, 3 independent experiments). (Scale bars, 10 μm in A, Center, C, and D; 20 μm in A, Left and Right, and E.)
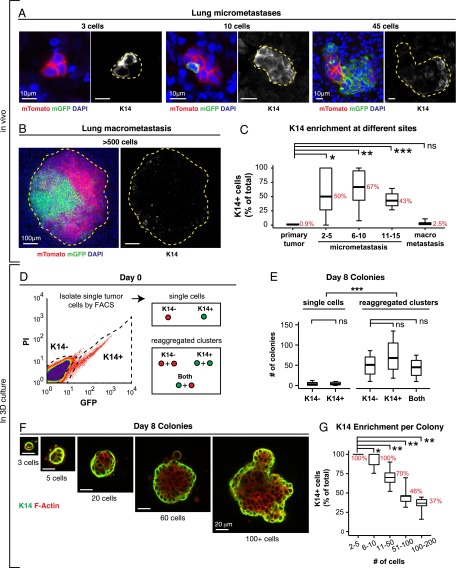
In lung metastases, the proportion of K14+ cells relative to the total cell number varied systematically with the size of the metastasis (Fig. 4 A and B). In the primary tumor, micrometastases, and macrometastases, K14+ cells accounted for 0.9%, >50%, and 2.5% of cells, respectively (Fig. 4C). Thus, K14+ cells are highly enriched in locally disseminated clusters, CTC clusters, and micrometastases relative to their frequency in the primary tumor, and the proportion of K14+ cells reverts to baseline in macrometastases.
Fig. 4.
Micrometastases are enriched for K14+ cells relative to macrometastases. (A–C) The percentage of cells that are K14+ varies inversely with the size of the metastasis. Representative micrographs of lung micrometastases of different sizes (A) and a lung macrometastasis (B) stained for K14 and DAPI. (C) The median percentage of cells that are K14+ at each stage of metastasis presented as a boxplot (n = 9 primary tumors, n = 49 micrometastases, n = 18 macrometastases). The median percentages for each condition are denoted in red. *P < 0.05, **P < 0.01, ***P < 0.001. (D) Schema of in vitro model of colonization. K14+ and K14− single-cell suspensions were isolated by FACS from K14-actin-GFP;MMTV-PyMT tumors. In addition, single-cell suspensions were aggregated into clusters, using as input either (i) K14+ cells, (ii) K14− cells, or (iii) a 50:50 ratio of both K14+ and K14− cells (both). (E) The median number of colonies formed at day 8 is presented as boxplots (n = the number of independent experiments. K14− single cells: n = 8, K14+ single cells: n = 8, K14− clusters: n = 3, K14+ clusters: n = 3, mixtures of both cell types: n = 3). ***P < 0.005. (F) K14+ and K14− cells and clusters generate colonies that are morphologically similar. Representative images of colonies of different sizes formed at day 8, stained for K14 and F-actin. (G) The median percentage of cells that are K14+ in different dissociated tumor cell clusters grown in Matrigel presented as a boxplot (n = 4,547 cells in 103 colonies). The median percentages for each condition are denoted in red. *P < 0.001. **P < 10−7. (Scale bars, 10 μm in A, 100 μm in B, and 20 μm in F.)
To model these transitions in 3D culture, we next isolated single K14− and K14+ cells by FACS from MMTV-PyMT tumors that genetically encode for a fluorescent biosensor for K14 expression (Fig. 4D and Fig. S5C) (11, 23). Accordingly, we observed that for individually purified K14− and K14+ populations, colony formation was >16-fold higher in aggregated clusters relative to single tumor cells (Fig. 4E). In contrast, colony formation was not significantly different for K14− single cells versus K14+ single cells, or for K14− aggregated cells versus K14+ aggregated clusters (Fig. 4E). Taken together, our data reveal that the difference in colony formation between K14− and K14+ cells is small relative to the difference between multicellular clusters and single cells.
One interpretation of these data could be that the interaction of K14− and K14+ tumor cells may synergistically enhance the efficiency of colony formation. Motivating this hypothesis, Wnt-driven murine mammary tumors contain two subclones that show interclonal cooperation during tumor progression (24). To test the synergy hypothesis, we used our aggregation assay to test the effect of aggregating equal mixtures of K14− and K14+ cells (Fig. 4D). However, mixing K14− and K14+ cells in aggregating clusters did not significantly enhance colony formation relative to either pure K14− or K14+ cell clusters (Fig. 4E).
Alternatively, phenotypic transitions between differentiation states could blunt our ability to detect differences in colony formation (25). We therefore examined K14 expression in aggregated tumor cells at day 0, day 1 postaggregation (Fig. S5D), and at day 8 (Fig. S5E). Interestingly, cell clusters at day 1 already contained mixtures of K14− and K14+ cells regardless of whether isolated K14− cells, K14+ cells, or mixtures of both cell types were used as input (Fig. S5D). Similarly, at day 8, >80% of colonies were composed of mixtures of K14− and K14+ cells in each condition (Fig. S5E). Morphologically, small colonies were primarily composed of K14+ cells and larger colonies were composed of an inner core of K14− cells and an outer layer of K14+ cells (Fig. 4F). Concordant with our observations of metastases in vivo, the proportion of K14+ cells varied with the size of clusters grown in 3D Matrigel. In small clusters, two to five cells in size, 100% of cells were K14+, and in larger clusters with ≥20 cells, all clusters contained multiple K14− cells (Fig. 4G). Thus, isolated K14− and K14+ cells converge toward cell colonies composed of both cell states, with K14+ cells predominant in small colonies in culture and in micrometastases. Conversely, K14− cells are predominant in large colonies and in macrometastases.
The Transcriptional Program of K14+ Tumor Cells Is Enriched for Desmosome and Hemidesmosome Adhesion Complex Genes, and Depleted for Genes Involved in MHC Class II Immunosurveillance.
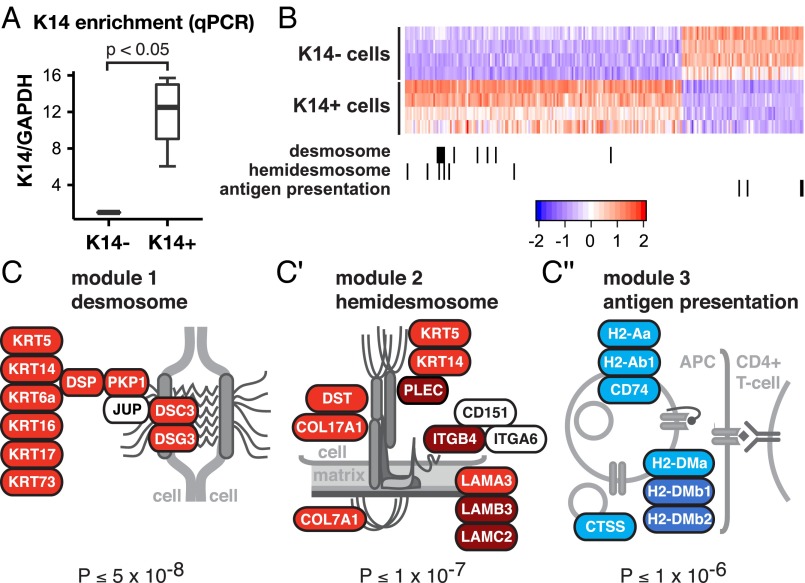
Given the enrichment for K14+ cells in disseminated tumor cell clusters and micrometastases, we next asked how the transcriptional profiles of K14+ and K14− cells differed. To answer this question, we compared the transcriptomes of K14+ and K14− cells from primary MMTV-PyMT tumors. To focus our attention on changes in gene expression between cancer cell subpopulations, we first isolated epithelial organoids from the tumor and then used differential centrifugation to deplete immune and fibroblastic stromal cells. We then processed these organoids to single cells and FACS-sorted them based on K14 status. As expected, quantitative RT-PCR revealed that the K14+ cell fraction exhibited >12-fold higher levels of K14 RNA expression (Fig. 5A). Our RNA-seq analysis revealed 239 genes differentially expressed between K14− and K14+ cells at a genome-wide significance level of P < 10−6 (mean-variance normalized heatmap in Fig. 5B). By DAVID gene ontology (GO) analysis (26), we identified significantly enriched GO categories in each cell subpopulation (Table S1). This analysis revealed significant enrichment for genes involved in extracellular matrix proteins, intermediate filament organization, and epithelial differentiation in K14+ cells. Interestingly, K14+ cells were enriched for transcripts encoding TNC, POSTN, and CTGF, proteins required in the metastatic niche (27–29). In addition, DAVID analysis revealed a number of genes involved in the regulation of the immune system that were significantly depleted in K14+ cells (Table S1). These GO categories included immune response, MHC class II antigen presentation, T-cell activation, and genes involved in chemotaxis. Genes involved in MHC class II presentation were among the most depleted in K14+ cells (Fig. 5C′′). Our data reveal that K14+ cells differ across multiple molecular programs that could be advantageous across distinct steps in metastasis, including immune evasion. Furthermore, two key features distinguishing K14+ cells from the bulk tumor cells are their increased expression of both cell–cell and cell–matrix adhesion genes.
Fig. 5.
The transcriptional program of K14+ tumor cells is enriched for desmosome and hemidesmosome adhesion complex genes, and depleted for genes involved in MHC class II immunosurveillance. (A) K14− and K14+ cells were isolated by FACS from K14-actin-GFP;MMTV-PyMT tumors and the relative mRNA expression of K14 relative to GAPDH was determined by quantitative RT-PCR. The relative mRNA expression is presented as a boxplot (n = 4 independent experiments). (B) Heatmap of the most differentially expressed genes determined by RNA sequencing of K14− and K14+ cells at a genome-wide significance of P < 10−6. (C–C′′) Genes that encode for desmosome, hemidesmosome, and MHC class II antigen presentation protein complexes are differentially expressed and coregulated in K14+ cells.
Table S1.
GO enrichment analysis in K14+ tumor cells
| GO term | P value | Genes |
| Functional categories enriched in K14+ cells | ||
| Proteinaceous extracellular matrix | 2.82E-10 | TNC, FBN1, HSPG2, POSTN, DCN, TIMP3, PRELP, COL17A1, TNFRSF11B, HMCN1, LAMA3, COL7A1, NAV2, CTGF, COL12A1, ADAMTS1, TNN, FBN2 |
| Epithelium development | 1.74E-08 | FGFR2, KRT6A, BMP2, PTGS2, TNC, GJA1, JAG1, GLI3, KRT17, CD44, IRF6, KRT14, TRP63, SEMA3C, ID3, BMP7 |
| Skeletal system development | 2.35E-07 | FGFR2, ZFAND5, BMP2, PTGS2, PDLIM7, TIPARP, HSPG2, IGF2, GLI3, IGSF10, CTGF, TRP63, RUNX1, AXIN2, BMP7 |
| Intermediate filament cytoskeleton organization | 1.20E-06 | TCHH, KRT6A, KRT17, KRT16, KRT14, DST |
| Cell adhesion | 2.25E-06 | MPDZ, TNC, HSPG2, ITGA2, POSTN, CXADR, LAMA3, CD44, COL7A1, PKP1, DSG3, CTGF, FAT2, DSC3, COL12A1, TNN, ANTXR1, DST, ABL2 |
| Negative regulation of cell proliferation | 4.98E-06 | CBLB, BMP2, CAV1, PTGS2, IRF6, BCL6, CXADR, BMP7, AXIN2, GLI3, KLF4, H19 |
| Functional categories depleted in K14+ cells | ||
| Immune response | 3.18E-12 | PTPRC, CCL3, LY86, CXCL2, H2-AB1, CLEC4N, CD74, FCGR3, C1QA, OSM, FCGR2B, CCR5, LILRB4, CX3CR1, FCER1G, IL1B, H2-AA, H2-DMA |
| Antigen processing and presentation of exogenous peptide antigen | 3.19E-12 | FCGR2B, CTSE, H2-AA, FCER1G, H2-AB1, H2-DMA, CD74, FCGR3 |
| Cell chemotaxis | 1.14E-11 | CORO1A, C5AR1, CXCL2, CX3CR1, FCER1G, IL1B, ITGAM, FCGR3 |
| External side of plasma membrane | 3.10E-08 | CD83, PTPRC, CCR5, ITGAX, EMR1, FCGR2B, H2-AA, FCER1G, H2-AB1, CD74, ITGAM |
| Positive regulation of T-cell activation | 2.79E-07 | CD83, PTPRC, CORO1A, IKZF1, H2-AA, H2-DMA, CD74 |
| Response to wounding | 1.30E-06 | C1QA, CCL3, CCR5, SAA2, GATM, SERPINA1B, SAA1, LY86, CXCL2, IL1B, FCGR3 |
In this table, RNA-seq data were analyzed as in Fig. 5. Functional enrichment for GO categories are shown that are significantly enriched or depleted in the transcriptional program of K14+ cells.
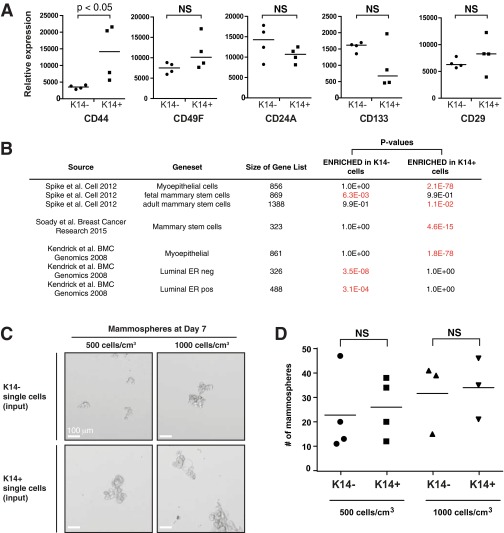
We also tested the extent to which the K14+ cell transcriptome correlated with “stemness” in three ways (30, 31). First, we looked at the expression of putative stem cell markers in K14− and K14+ cells. We observed that CD44 was differentially expressed between these cell populations, but CD49f, CD24a, CD133, and CD29 were not (Fig. S6A). Next, we tested for gene set enrichment in K14− and K14+ cells with published mouse mammary stem cell gene sets (Fig. S6B). Our gene set tests revealed that K14− cells were enriched for fetal mammary stem cell and luminal epithelial gene signatures. In contrast, K14+ cells were enriched for mammary stem cell, adult mammary stem cell, and myoepithelial gene signatures. A third approach we took to test stemness was to assay the ability of isolated single K14− and K14+ cells to form mammospheres, as in Spike et al. (31) (Fig. S6C). Our data show that both K14− and K14+ cells form mammospheres and that the mammosphere-forming efficiency was not significantly different between K14− and K14+ cells (Fig. S6D). Taken together, our findings are consistent with a model in which stemness features are observed in both K14− and K14+ cells, and that both cell types are capable of colony formation.
Fig. S6.
K14+ cells and K14− cells are enriched for different stemness genes and are not significantly different in their mammosphere forming ability. (A) Relative expression of different stem cell-associated genes presented as a dot-plot. P value determined by Mann–Whitney test. (B) Enrichment in K14+ and K14− cells was determined by Wilcoxon gene set testing using published stem cell gene sets identified by transcriptome profiling of mouse mammary gland. (C) Representative micrographs of mammospheres generated from K14− and K14+ single cells in suspension culture. (Scale bars, 100 μm.) (D) The median number of mammospheres is presented as a dot plot for K14− and K14+ single tumor cells isolated by FACS and plated at two different densities. P value determined by the Mann–Whitney test.
K14 Expression Is Required for Distant Metastasis and Regulates Gene Expression of Multiple Metastasis Effectors.
Our gene-expression study revealed that K14+ cells displayed coordinated up-regulation of most desmosome (10 of 11) and hemidesmosome (10 of 12) complex genes (Fig. 5 C and C′) (32). These observations were intriguing to us because: (i) K14 is the major intermediate filament in hemidesmosomes; (ii) leader cells require expression of basal epithelial genes, including K14, for their collective invasive behavior (11); and (iii) a recent study has implicated desmosomal function in CTC clusters from human cancer patients (5).
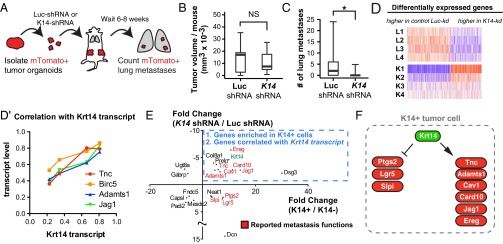
To test the hypothesis that K14 expression is required for distant metastasis, we transplanted mTomato+ tumor organoids, either expressing control Luciferase or K14 shRNA, into host mice (Fig. 6A). Primary tumors derived from Luciferase control knockdown organoids did not show statistically significant differences in size compared with primary tumors derived from K14 knockdown tumor organoids (Fig. 6B). In contrast, K14 knockdown tumors had a sevenfold reduction in the mean number of metastases relative to control knockdown tumors (Fig. 6C).
Fig. 6.
K14 is required for distant metastasis and regulates the expression of multiple metastasis effectors. (A) Schema to test the in vivo requirement for K14 in metastasis. Fluorescent mTomato+ tumor organoids were transduced with either lentiviral Luc-shRNA or K14-shRNA, selected with puromycin, and transplanted into the cleared mammary fat pads of nonfluorescent congenic hosts. Mice were harvested from 6 to 8 wk and lungs were sectioned and counted for mTomato+ lung metastases. (B and C) Median tumor size in cubic millimeters and number of lung metastases for Luc-shRNA and K14-shRNA tumors, with data presented as boxplots (n = 11 mice for Luc-shRNA, n = 10 mice for K14-shRNA). *P < 0.05. (D) Heatmap of the most differentially expressed genes determined by RNA sequencing of luciferase control knockdown and K14 knockdown tumor organoids at an FDR < 0.05. (D′) Scatterplot of correlation of transcript expression of top ranked genes versus Krt14. (E) Scatterplot of the most differentially expressed genes with twofold or greater significant enrichment in K14+ cells or K14− cells and whose gene expression was significantly associated with K14 transcript levels (FDR < 0.05 for both datasets). Krt14 is highlighted in green. Published metastasis genes reported in red. (F) Schema of metastasis genes enriched in K14+ cells and regulated by Krt14 expression.
Having shown that K14 expression is required for distant metastasis, we next asked whether knockdown of K14 altered gene transcription. We determined the transcriptomes of luciferase control knockdown and K14 knockdown tumor organoids by RNA-Seq and identified 1,584 genes differentially expressed between conditions (mean-variance normalized heatmap in Fig. 6D). To dissect this list further, we next ranked genes by their association with Krt14 transcript levels [487 genes at a false-discovery rate (FDR) < 0.05]. The top four genes most correlated with Krt14 transcript expression were enriched for multiple major metastasis effector genes that promote metastatic niche remodeling (Tnc, AdamTs1, Jag1) and metastasis survival (AdamTs1, Birc5) (Fig. 6D′) (27, 29, 33, 34). Thus, disruption of K14 expression abrogates the expression of multiple metastasis effectors.
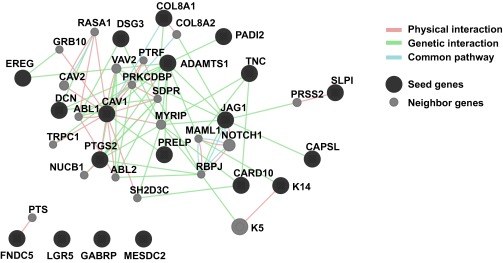
To identify the core molecular program expressed in K14+ cells and regulated by K14 expression, we then integrated this transcriptome data with our RNA-seq enrichment data for K14+ cells. We identified 14 genes with twofold or greater significant enrichment in K14+ cells and whose gene expression was significantly associated with Krt14 transcript levels (Fig. 6E). Consistent with our observations in the K14 knockdown dataset, we observed that nine of these genes were previously reported in metastasis regulation, including in metastatic niche, vascular remodeling, and immunosurveillance, including Tnc, AdamTs1, Jag1, as well as Card10, Cav1, Ereg, Lgr5, Slpi, and Ptgs2 (29, 35–38). Core genes showed evidence of multiple physical and genetic interactions and occurrence along common pathways (Fig. S7). Interestingly, the gene most highly enriched in K14+ cells and positively regulated by Krt14 transcript levels was Dsg3, which encodes for the major desmoglein in desmosomes. Thus, K14 expression also regulates the expression of a major desmosome component. Taken together, our data establish that K14 expression is required for distant metastasis and that K14 is a regulator of multiple genes that function at distinct steps of the metastatic cascade (Fig. 6F).
Fig. S7.
Gene network of K14+ metastasis genes. The network of coregulated genes seeded using the most differentially expressed genes in Fig. 6E. Seed genes in black, neighbor genes in gray. Physical interactions, genetic interactions, and common pathway interactions were included in the network.
Discussion
A major goal of our study was to determine how breast cancer cells seed distant metastases. Our lineage analysis enabled us to identify polyclonal collective invasion strands, polyclonal disseminated tumor cell clusters in the stroma, polyclonal CTC clusters, and polyclonal micro- and macrometastases. Our findings, in conjunction with recent reports in CTC clusters (5), and genomic sequencing of metastases (7, 8), provide strong evidence for polyclonal seeding as a major mechanism for metastatic spread.
We next focused on the molecular features of the tumor cell clusters that give rise to polyclonal metastases. We have previously established that the cells leading collective invasion near uniformly express K14 (11). We reveal profound and systematic changes in the relative proportions of K14+ and K14− cells from micrometastatic to macrometastatic stages. We detect K14+ cells in disseminated tumor cell clusters, in intravascular tumor emboli, CTC clusters, and in micro- and macrometastases. Importantly, both CTC clusters and micrometastases are >20-fold enriched for K14+ cells relative to primary tumors or macrometastases. Thus, K14+ cells are significantly enriched in breast cancer cells during the phases of metastasis most associated with systemic spread. Conversely, K14− cells are significantly enriched in the phases of metastasis most associated with proliferation. Our in vivo data cannot distinguish between conversion of cells from a K14+ to a K14− state after arrival in the lung versus expansion of K14− cancer cells that arrived as a component of a mixed K14+/K14− cluster. However, our ex vivo assays provide clear evidence for interconversion between epithelial states in both directions. The molecular mechanisms that drive transitions between K14+ and K14− states are an important area for further study.
In our study, we also revealed transcriptomic differences between K14− and K14+ tumor cells and identified genes coding for protein complexes distinct to each compartment. Concordant with a recent report on plakoglobin, our data implicate the desmosome in polyclonal seeding (5). Intriguingly, K14+ cells are also enriched for hemidesmosome complex genes and genes encoding for proteinaceous extracellular matrix, and K14 expression is required for gene expression of multiple metastasis effectors involved in niche remodeling (29, 37). It is likely that polyclonal tumor cell clusters provide an efficient nidus for interactions with collagen I, fibroblasts, macrophages, endothelial cells, and other tumor microenvironmental components in the multicellular seeding process (39–44).
Our data do not exclude the possibility of single-cell seeding events by either EMT or migratory stem cells. However, our lineage analysis does put an upper limit on the relative contribution of these events to metastasis in the MMTV-PyMT model. In agreement with our data, a recent study used lineage analysis in spontaneous mouse models of breast cancer to reveal that EMT does not significantly contribute to the formation of lung metastases in MMTV-PyMT mice, and that inhibition of EMT through miR-200 overexpression does not impair the ability of MMTV-PyMT tumor cells to form metastases (45). However, it remains technically prohibitive to image the transit of a cancer cell continuously from escape out of the primary tumor through to establishment of a distant site, and so some inference remains necessary to construct models of the process. Accordingly, it is important to acknowledge contrary observations. We observe K14− single cells in the mammary stroma and real-time intravital imaging was recently used to observe single MMTV-PyMT cancer cells intravasating at sites of macrophage–endothelial cell connections (44). Our data are compatible with a model in which K14 is required for collective invasion and dissemination, but not single-cell dissemination. Furthermore, our data do not exclude the possibility of transitions between single cell and multicellular organization in the mammary stroma before intravasation. Future studies are needed to map the relative frequency of different single-cell and multicellular configurations and their molecular features across the entire mammary fat pad and in different cancer models.
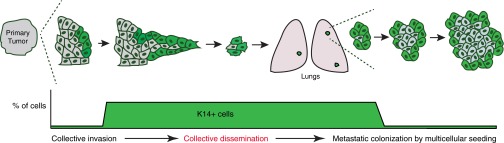
The simplest interpretation of our data is that breast cancer metastases form via the local dissemination, entry into circulation, and distal seeding of a multicellular cluster containing K14+ cancer cells (Fig. S8). Within this framework, the survival advantage of multicellular organization provides a mechanism by which genetically distinct clones can jointly found a micrometastasis. Eradicating polyclonal micrometastases in the adjuvant setting will be particularly challenging when these founders have intrinsic differences in treatment sensitivity. Our findings also support the concept that micrometastatic and macrometastatic disease may require distinct therapeutic strategies (46). Finally, it will be important in future studies to determine how tumor cell clusters gain access to the lymphatic and venous circulation.
Fig. S8.
Polyclonal metastasis occurs through collective dissemination of K14+ tumor cell clusters. Primary tumors invade and disseminate collectively into surrounding tissue. This disseminated unit efficiently colonizes distant sites to form polyclonal metastasis. In MMTV-PyMT tumors, the traveling unit is composed of multiple K14+ epithelial tumor cells. Our model is supported by multiple lines of experiments in our study: the high frequency of multicolored metastases arising from well mixed multicolored primary tumors in which clonality can most readily be assessed (Fig. 1 and Fig. S1), the lack of multicolored metastases when the different colored cells are deployed at different locations or at different times (Fig. S2), the observation of polyclonal clusters across major stages of metastasis (Fig. 2), the large survival advantage conferred by multicellular organization both in culture and in vivo (Fig. 3), the enrichment for K14 expression in disseminated tumor cells and micrometastases (Fig. 4), the expression of epithelial desmosome and hemidesmosome gene programs in K14+ cells (Fig. 5), and the requirement for K14 expression for both distant metastasis and the expression of multiple metastasis effectors, crucial for remodeling of the metastatic niche, including Tnc, Adamts1, Jag1, and Ereg (Fig. 6).
Materials and Methods
Mouse Lines and Breeding.
Animal protocols were approved by The Johns Hopkins University Institutional Animal Care and Use Committee. FVB/N-Tg(MMTV-PyVT)634Mul/J (MMTV-PyMT) (17), B6(D2)-Tg(CAG-Brainbow1.0)2Eggn/J (Rainbow) (20), Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J (Confetti) (19), B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (ROSAmT/mG) (47), and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from the Jackson Lab. K14-actin-GFP mice (23) were a gift from Elaine Fuchs (Rockefeller University, New York, NY). B6.129(ICR)-Tg(CAG-ECFP)CK6Nagy/J (β-actin-CFP) mice (48) were a gift from Mikala Egeblad (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). For lineage-tracing experiments, MMTV-PyMT mice were crossed with ROSA mT/mG mice, Confetti mice, Rainbow mice, or β-actin–CFP mice and transplanted into NSG host mice. For sorting of K14+ cells, MMTV-PyMT mice were crossed with K14-GFP-actin mice.
Statistics.
All analyses were conducted using the program R or Graphpad Prism. For all boxplots, the whiskers represent the 5th and 95th percentiles. P value determined by Mann–Whitney test unless otherwise noted. P < 0.05 was considered significant.
See SI Materials and Methods for a complete description of protocols for organoid isolation, orthotopic transplantation, lentiviral transduction, tail-vein assays, FACs sorting of K14+ cells, RNA-seq, colony-forming assays, mammosphere assays, determination of local mixing, and isolation of CTCs.
SI Materials and Methods
Isolation of Primary Mammary Tumor Organoids.
Primary tumor organoids were isolated from mammary tumors by step-wise mechanical disruption, enzymatic digestion, and differential centrifugation according to our published protocols (11, 14). Tumors were harvested from 8- to 10-wk-old mice, minced with a scalpel, and digested for 1 h at 37 °C in collagenase solution: (DMEM (10565–018; Gibco Life Technologies) with 2 mg/mL collagenase (C2139; Sigma-Aldrich), 2 mg/mL trypsin (27250–018; Gibco Life Technologies), 5% (vol/vol) FBS (F0926; Sigma-Aldrich), 5 μg/mL insulin (I9278; Sigma-Aldrich), and 50 μg/mL gentamicin (15750; Gibco Life Technologies). The suspension was centrifuged at 422 × g to remove cellular debris, and the pellet was treated with 2 U/μL DNase (D4263; Sigma-Aldrich) to separate out organoids. Tumor organoids were separated from single cells by differential centrifugation and counted under a microscope.
Mammary Fat Pad Transplantation.
Tumor organoids from MMTV-PyMT;ROSAmT/mG, MMTV-PyMT;Confetti, or MMTV-PyMT;Rainbow were incubated with 1:50 adeno-CMV-Cre (1045; Vector BioLabs)/DMEM overnight in a nonadherent 96-well plate. Cre expression was induced successfully in more than 75% of the organoids. To wash out adeno-Cre, the samples were collected in a BSA-coated microcentrifuge tube and centrifuged at 422 × g for 10 min. For intermediate mosaicism, tumor organoids were mixed 1:1 with unrecombined tumor organoids. Tumor organoids were resuspended in a 50% (vol/vol) DMEM/50% (vol/vol) Matrigel (354230; Corning) solution at a density of 25–40 organoids per microliter and kept at 4 °C during transplantation. We conducted orthotopic transplantations into 3- to 4-wk-old NSG mice in a sterile hood. The mice were anesthetized with 2.5% isoflurane, immobilized and the surgical site was made sterile using ethanol. A 1-cm midline incision was made, the skin was retracted, and the no. 4 mammary gland was exposed. The fat pad proximal to the lymph node was removed. Using a syringe, 40 μL of the tumor organoid suspension was injected into the mammary fat pad. The skin was then locally infiltrated with 5–10 μL of 0.25% bupivacaine. The same procedure was repeated for the contralateral mammary gland. The surgical wounds were closed using 9-mm autoclips and tissue glue. Triple antibiotic ointment was applied to the incision. The mice were closely monitored, and autoclips were removed 10 d postsurgery. Between 6 and 10 wk, tumors and lungs from these mice were harvested.
Differencial Interference Contrast Imaging and Confocal Microscopy.
Differential interference contrast imaging of tumor organoids was conducted using a Zeiss Cell Observer system with a Zeiss AxioObserver Z1 and an AxioCam MRM camera (Carl Zeiss). Confocal imaging was conducted with a Zeiss 780 laser scanning confocal microscope (Carl Zeiss).
Immunofluorescence Staining.
Tumors or lungs harvested from transplant mice were fixed in 4% or 1% paraformaldehyde, respectively, for 4 h. They were then incubated in a 25% sucrose solution overnight, embedded in Tissue Tek Optimal Cutting Temperature compound (OCT, Sakura) and frozen at −80 °C. OCT blocks were sectioned at 50-μm thickness using a Leica cryostat (Leica Biosystems) set to −27 °C. For antibody staining, the OCT was removed by rinsing with PBS for 45 min. Samples were blocked for 2 h with 10% FBS/1% BSA/PBS (vol/vol) solution, incubated with the conjugated primary antibodies overnight at 4 °C in 1% FBS/1% BSA/PBS solution, and rinsed two times in PBS for 10–20 min. Slides were optically cleared, mounted, and sealed with coverslips. Primary antibodies used were: Alexa Fluor Phalloidin (Life Technologies) anti-K14 (PRB-155P; Covance), anti–E-Cad (24E10; Cell Signaling Technologies), and DAPI (D3571; Life Technologies). To label functional vasculature, 10 μg of anti-CD31 (102416; BioLegend) and anti-CD144 (138108; BioLegend) antibodies were injected into the mouse by tail vein (49). After 20 min, mice were killed and tumors and lungs were collected.
Isolation of CTCs.
Approximately 500 μL of blood was harvested from each mouse via the right atrium using a 20-G needle (305176; BD Biosciences) and 1-mL syringe (309659; BD Biosciences). Before cardiac puncture, tumors were mechanically palpated to liberate CTCs into the systemic circulation, as in ref. 3. Blood was transferred to 1 mL K2EDTA tubes (365974; BD Biosciences) and placed on an inverter to prevent clotting. Blood was mixed 1:2 with ACK Lysis Buffer (118-156-721; Quality Biological) and centrifuged at 500 × g for 5 min. The pellet was washed with 500 μL PBS without Mg+2/Ca+2 (10010-023; Life Technologies) and centrifuged at 500 × g for an additional 5 min. The pellet was resuspended in 450 μL total volume with PBS wthout Mg+2/Ca+2 and spread onto slides with a proprietary adhesion coating (0906000, Marienfeld). Cells were left to settle on the slide for 5 min. The slide was then incubated at 37 °C for 60 min. Slide supernatant was discarded and adherent cells were fixed with 4% PFA at room temperature for 30 min. Slides were then washed with PBS without Mg+2/Ca+2 and permeabilized with 0.1% Triton-X at room temperature for 10 min. Slides were again washed with PBS without Mg+2/Ca+2 and then blocked at 37 °C for 20 min with an Fc receptor-blocking antibody diluted 1:20 (101320, BioLegend) in 10% Normal Goat Serum (50062Z, Life Technologies). Slides were incubated with conjugated primary antibodies diluted in PBS without Mg+2/Ca+2 at 37 °C for 1 h. Antibodies used were pancytokeratin-AlexaFluor488 (1:50; 4523S, Cell Signaling), Keratin 14-AlexaFluor488 (1:250; PRB-155P, BioLegend), and CD45-AlexaFluor647(1:50; 103124, BioLegend). Slides were washed with PBS without Mg+2/Ca+2, mounted and coverslipped using ProLong Diamond Anti-Fade with DAPI (P36971, Life Technologies) and 24 × 60 coverglass (12-545-M, Fisher). Slides were left to dry overnight in a dark chamber and imaged on a Zeiss Axio Imager Z2 (Carl Zeiss Microscopy) using Metafer 5 software (v3.11.3) (MetaSystems). High-resolution 3D images were collected on a Zeiss 780 laser-scanning confocal microscope (Carl Zeiss Microscopy).
FACS of Single K14− and K14+ Tumor Cells.
MMTV-PyMT; K14-actin-GFP were harvested for tumor organoids. Organoids were subsequently dissociated to single cell suspensions in StemPro Accutase Cell Dissociation Reagent (A11105-01; Life Technologies) for 20 min. Cells were then resuspended in HBSS/1%BSA. The resulting cell suspensions were filtered through 40-μm cell strainers and analyzed on a Beckman-Coulter MoFlo Cytometer by gating cellular distributions according to the eGFP reporter. Single cells and clusters were gated by FSLin vs. Pulse Width. The nozzle size chosen for sorting was 100+ μm. Dead cells were excluded by propidium iodide fluorescence. For each condition, cells were sorted into 200 μL Matrigel and cultured in 24-well plates with DMEM media supplemented with the growth factor basic human FGF (bFGF) (F0291; Sigma-Aldrich) at 37 °C, 5% CO2. For the aggregation experiments, 10,000 single cells were sorted into DMEM plus bFGF media and allowed to aggregate overnight in nonadherent-well dishes at 37 °C, 5% CO2. The samples were then resuspended into 200 μL Matrigel and plated as mentioned above. The samples were fixed at day 8 for counting of colonies. To confirm aggregation, fractions of single-cell suspensions were incubated for 1 h with 10 µM Cell Tracker Green and Cell Tracker Red (C7025 and C34552, Life Technologies), respectively, before sorting. An equal number of red and green cells where plated for a total of 10,000 single cells per well. Cell aggregates were analyzed the following day.
Tail Vein Injections.
NSG mice were injected with 200,000 single cells or an equivalent number of aggregated cells obtained from ROSAmT/mG;MMTV-PyMT or β-actin-CFP;MMTV-PyMT tumor cells isolated by FACS. Lungs from these mice were harvested 3 wk later, and lung metastases were counted under the dissection scope or by counting metastases from 50-μm lung sections.
Lentiviral Transduction of Tumor Organoids for Tumor Transplantation.
Lentiviral transduction of tumor organoids was conducted according to our published protocols (11). Approximately 800 ROSAmT/mG;MMTV-PyMT organoids were resuspended in 100 μL of DMEM/F12, and allowed to settle in a 96-well nonadherent dish for 1 h at 37 °C. The media was removed, and the organoids were infected with 50 μL of lentivirus (Mission shRNA; Sigma-Aldrich) and 3 μL of ViroMag beads (VM4100; OZ Biosciences). The 96-well plate was incubated on top of a magnetic plate (MF10000; OZ Biosciences) at 37 °C for 1.5 h, then the magnetic plate was taken off and the 96-well plate was incubated overnight at 37 °C. On day 2, 75 μL of media was removed and replaced with 200 μL of fresh FGF2 organoid media. On day 3, 175 μL of media was removed, and 175 μL of FGF2 organoid media supplemented with 2.5 nM bFGF and 4 μg/mL puromycin was added. After 3 d, the surviving organoids were collected for orthotopic transplantation into the no. 4 mammary fat pads of FVB host mice. Tumor dimensions were measured at regular intervals (one to two times a week) using digital vernier calipers. For analysis of disseminated tumor cell clusters, tumor–stroma border of primary tumor sections were scanned for dissemination events and confirmed to be completely detached from the primary tumor by sequential z-stacks. Dissemination events were scored as single cells or clusters (two cells or more).
Estimation of Local Mixing in the Primary Tumor.
We took low-power montages of primary tumors in transplanted mice on a Zeiss 780 confocal microscope. Fiji custom scripts were written to binarize these images and extract the percentage of pixels that were green in color (%Green). We then divided images into 100 × 100-pixel grids (1.3 mm × 1.3 mm) and determined the probability of choosing two pixels of different colors in each grid. We defined “% local mixing” as two times the average probability of choosing two pixels of different colors averaged over all grids. Using robust linear regression, we then estimated the percentage of multicolored metastases at maximal local mixing for data in Fig. 1E.
High-Throughput RNA Sequencing of K14− and K14+ Tumor Cells.
For each replicate, K14-actin-GFP;MMTV-PyMT tumor organoids were first dissociated into a single-cell suspension, and then single K14− and K14+ cells were isolated by FACS. Four independent experiments were conducted. For each cell population, total RNA was extracted using TRizol (15596-026; Life Technologies) and RNeasy (Qiagen). With 10–100 ng of RNA collected per sample, we generated barcoded NuGen RNA-seq v2 libraries and ran paired-end, 100-bp, 50-cycle sequencing on a HiSEq. 2000 (The Johns Hopkins Medical Institutions Deep Sequencing and Microarray Core Facility). Paired-end, nonstrand-specific RNA-seq reads were mapped to the mouse reference genome (Genomic Reference Consortium build 38) using RNA-STAR v2.3.0 (50). We achieved 80–85% read pair alignment to the reference genome and estimated the number of reads mapped to each gene using HTSeq v0.6.1p1 (51), with gene coordinates from the reference genome Generic Feature Format file. Raw counts were normalized and P values were calculated for paired sample differential expression using EdgeR (51). Genes were excluded that did not have at least one read per million in four of eight samples. This procedure yielded 12,190 mapped genes, of which 11,968 had gene annotation. Sequence data will be uploaded to the Sequence Read Archive. GO category enrichment was determined by DAVID Gene Set Analysis, v6.7, using the functional annotation chart algorithm, mouse genome as background, and goterm_bp_fat, goterm_cc_fat, and goterm_mf_fat gene lists (26). P values for module enrichment were calculated using a Fisher exact test (1,561 of 11,968 genes with FDR < 0.05).
High-Throughput RNA Sequencing of Luc-shRNA and K14-shRNA Transduced Tumor Organoids.
For each replicate, MMTV-PyMT tumor organoids were transduced with luciferase control shRNA or K14 shRNA, and selected with puromycin as we have done previously (11). Transduced organoids were embedded in 3D Matrigel and after 2 d in culture, harvested for RNA. Four independent experiments were conducted. For each condition, total RNA was extracted using RNeasy (Qiagen). With 10–100 ng of RNA collected per sample, we generated cDNA using the SMART-Seq v4 Ultra Low Input RNA Kit for sequencing (Clontech), barcoded libraries with the Nextera XT DNA Library Preparation kit (Illumina), and ran paired end, 100-bp, 50-cycle sequencing on a HiSEq. 2500 (GE Healthcare Seqwright). Paired-end, nonstrand-specific RNA-seq reads were mapped to the mouse reference genome (Genomic Reference Consortium build 38) using RNA-STAR (50). We achieved 73–86% read pair alignment to the reference genome and estimated the number of reads mapped to each gene using HTSEq. (51), with gene coordinates from the reference genome Generic Feature Format file. Raw counts were normalized and P values were calculated for paired sample differential expression using the limma package (52). Genes were excluded that did not have at least one read per million in four of the eight samples. This procedure yielded 13,332 mapped genes, of which 12,919 had gene annotation. Sequence data will be uploaded to the Sequence Read Archive. For each replicate (replicates one to four), the knockdown efficiency was determined by dividing the Krt14 transcript level in the K14-shRNA condition by the Krt14 transcript level in the Luc-shRNA condition. A linear model was then fitted to the voomed expression data using a design matrix parameterizing batch effects and differences in knockdown efficiency between replicates. With this fitted model, genes were then ranked by P value and FDR. The functional association network (Fig. S7) was determined using the GeneMANIA web interface (53).
Mammosphere Assay.
For suspension mammosphere culture, MMTV-PyMT; K14-actin-GFP tumors were first dissociated into a single-cell suspension, and then single K14+ and K14− cells were isolated by FACS. Cells were plated on ultralow-adherence plates (Thermo Scientific) at 500 cells/cm2 and 1,000 cells/cm2, respectively. Cells were culture in Basal Epicult-B mouse media (Stem Cell Technologies) containing B-supplement, 10 ng/mL rhEGF, 10 ng/mL rhbFGF, 4 μg/mL heparin, and 1% penicillin/streptomycin. The number of spheres for each well was counted under the microscope on day 7. The average number of spheres was compared across cell types and densities using the Mann–Whitney test.
Acknowledgments
We thank members of the A.J.E. laboratory for comments on the manuscript; B. Stanger and R. Maddipati for sharing unpublished data; Hao Zhang of The Johns Hopkins School of Public Health Flow Cytometry Core Facility for assistance with FACS; and Haiping Hao of The Johns Hopkins Medical Institutions Deep Sequencing and Microarray Core Facility for assistance with RNA-seq. K.J.C. is supported by a Postdoctoral Fellowship from the US Department of Defense (W81XWH-12-1-0018) and a Burroughs Wellcome Fund Career Award for Medical Scientists. A.J.E. is supported by a Research Scholar Grant (RSG-12-141-01-CSM) from the American Cancer Society, by funds from the National Institutes of Health/National Cancer Institute (P30 CA006973), by a grant from the Mary Kay Ash Foundation (036-13), by funds from the Cindy Rosencrans Fund for Triple Negative Breast Cancer Research, by a Research Leadership Award from the Metastatic Breast Cancer Network, and by an award from The Pink Agenda and The Breast Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq data have been uploaded to the NCBI Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession nos. SRP066319 and SRP066316).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508541113/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Moore GE, Sandberg AA, Watne AL. The comparative size and structure of tumor cells and clumps in the blood, bone marrow, and tumor imprints. Cancer. 1960;13:111–117. doi: 10.1002/1097-0142(196001/02)13:1<111::aid-cncr2820130121>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 3.Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 1976;36(3):889–894. [PubMed] [Google Scholar]
- 4.Hou JM, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30(5):525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 5.Aceto N, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maddipati R, Stanger BZ. Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov. 2015;5(10):1086–1097. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McFadden DG, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156(6):1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gundem G, et al. ICGC Prostate UK Group The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronsert P, et al. Cancer cell invasion and EMT marker expression: A three-dimensional study of the human cancer-host interface. J Pathol. 2014;234(3):410–422. doi: 10.1002/path.4416. [DOI] [PubMed] [Google Scholar]
- 10.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 11.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155(7):1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung KJ, Ewald AJ. Illuminating breast cancer invasion: Diverse roles for cell-cell interactions. Curr Opin Cell Biol. 2014;30:99–111. doi: 10.1016/j.ceb.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedl P, Zänker KS, Bröcker EB. Cell migration strategies in 3-D extracellular matrix: Differences in morphology, cell matrix interactions, and integrin function. Microsc Res Tech. 1998;43(5):369–378. doi: 10.1002/(SICI)1097-0029(19981201)43:5<369::AID-JEMT3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen-Ngoc KV, et al. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci USA. 2012;109(39):E2595–E2604. doi: 10.1073/pnas.1212834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedl P, et al. Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res. 1995;55(20):4557–4560. [PubMed] [Google Scholar]
- 16.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163(5):2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10(5):R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143(1):134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Tabansky I, et al. Developmental bias in cleavage-stage mouse blastomeres. Curr Biol. 2013;23(1):21–31. doi: 10.1016/j.cub.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo J, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci USA. 2011;108(15):6235–6240. doi: 10.1073/pnas.1015938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell. 2002;3(3):367–381. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 24.Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508(7494):113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 26.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 28.Malanchi I, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481(7379):85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 29.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17(7):867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soady KJ, et al. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res. 2015;17:31. doi: 10.1186/s13058-015-0539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spike BT, et al. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10(2):183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broussard JA, Getsios S, Green KJ. Desmosome regulation and signaling in disease. Cell Tissue Res. 2015;360(3):501–512. doi: 10.1007/s00441-015-2136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19(2):192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka K, et al. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer res. 2000;6(1):127–134. [PubMed] [Google Scholar]
- 35.Devoogdt N, et al. Secretory leukocyte protease inhibitor promotes the tumorigenic and metastatic potential of cancer cells. Proc Natl Acad Sci USA. 2003;100(10):5778–5782. doi: 10.1073/pnas.1037154100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goetz JG, et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146(1):148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta GP, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446(7137):765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 38.Pan D, et al. 2015. The CBM complex underwrites NF-kappaB activation to promote HER2-associated tumor malignancy. Mol Cancer Res, 10.1158/1541-7786.MCR-15-0229-T.
- 39.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Condeelis J, Pollard JW. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Calvo F, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15(6):637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22(5):697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stacker SA, Achen MG, Jussila L, Baldwin ME, Alitalo K. Lymphangiogenesis and cancer metastasis. Nat Rev Cancer. 2002;2(8):573–583. doi: 10.1038/nrc863. [DOI] [PubMed] [Google Scholar]
- 44.Harney AS, et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov. 2015;5(9):932–943. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer KR, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mina LA, Sledge GW., Jr Rethinking the metastatic cascade as a therapeutic target. Nat Rev Clin Oncol. 2011;8(6):325–332. doi: 10.1038/nrclinonc.2011.59. [DOI] [PubMed] [Google Scholar]
- 47.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 48.Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruns I, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20(11):1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 52.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 53.Warde-Farley D, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]