Significance
Central biological processes in cells and tissues are intrinsically governed by the structural order of biomolecular assemblies. It is thus a key factor to decipher how these assemblies organize in complex molecular organizations, from the nanometric to the macroscopic scale. Polarized microscopy can access such information; however, signals are spatially averaged over the optical diffraction limit and are contaminated by the fluorophores’ orientational flexibility of their linker to the biomolecules. By bringing polarized fluorescence down to superresolution microscopy using single-molecule localization, we show that structural imaging can be scaled down to nanometric scales and is able to discriminate fluorophores’ flexibility from biomolecules’ orientational order. We demonstrate nanoscale structural imaging in fundamental biological filament organizations.
Keywords: single-molecule orientation, polarization, superresolution, actin order, dsDNA order
Abstract
Essential cellular functions as diverse as genome maintenance and tissue morphogenesis rely on the dynamic organization of filamentous assemblies. For example, the precise structural organization of DNA filaments has profound consequences on all DNA-mediated processes including gene expression, whereas control over the precise spatial arrangement of cytoskeletal protein filaments is key for mechanical force generation driving animal tissue morphogenesis. Polarized fluorescence is currently used to extract structural organization of fluorescently labeled biological filaments by determining the orientation of fluorescent labels, however with a strong drawback: polarized fluorescence imaging is indeed spatially limited by optical diffraction, and is thus unable to discriminate between the intrinsic orientational mobility of the fluorophore labels and the real structural disorder of the labeled biomolecules. Here, we demonstrate that quantitative single-molecule polarized detection in biological filament assemblies allows not only to correct for the rotational flexibility of the label but also to image orientational order of filaments at the nanoscale using superresolution capabilities. The method is based on polarized direct stochastic optical reconstruction microscopy, using dedicated optical scheme and image analysis to determine both molecular localization and orientation with high precision. We apply this method to double-stranded DNA in vitro and microtubules and actin stress fibers in whole cells.
Biological processes are inherently driven by the molecular-scale organization of biomolecular assemblies, which arrange in precise structures that are essential for biological functions in cells and tissues. The extent to which the biological function depends on the underlying molecular-scale organization is particularly evident in filamentous assemblies, such as DNA filaments and cytoskeletal protein filaments. Changes in the local higher-order organization of DNA filaments is tightly linked to essential DNA-mediated processes including control of gene expression, DNA replication, and DNA repair. However, how specific DNA-binding proteins affect DNA filament architecture and thus DNA-mediated functions is poorly understood (1). Similarly, the spatial organization of cytoskeletal filaments in cells and tissues is also weakly explored, despite their central role in generating forces and driving cell motility, cell division, and tissue morphogenesis (2). Electron microscopy has been widely used to provide molecular-scale images of the structure of such filament assemblies; however, it typically involves several daylong sample preparation and ultrathin sectioning of the biological material, thus limiting investigations in whole cells and tissues.
Polarized fluorescence imaging is a powerful approach for elucidating the structural organization of filament assemblies because it is compatible with a wide variety of microscopy techniques, thus enabling studies across multiple spatial and temporal scales. Polarized fluorescence imaging allows not only retrieval of structural information from molecular orientation measurements (3, 4) but also quantification of depolarization-induced processes such as in homo-fluorescence resonant energy transfer, which serve as indicators for biomolecular clustering or polymerization (5, 6). Molecules act as oriented absorption/emission dipoles, whose pointing direction can be monitored, exploiting the polarized nature of light. This property has been used to determine the organization of molecular assemblies whose orientation is constrained, in particular in the cell membrane (7–11). Recent works using polarized microscopy have also evidenced microscopic-scale organization of septin filaments in budding yeast (12, 13) and actin filaments in Drosophila tissues (14), opening a path for in vivo structural imaging. Mapping the local organization of complex filament assemblies using polarized fluorescence imaging thus appears as a key approach to understand fundamental biological functions as diverse as DNA-mediated processes and animal cell mechanics. Although significant advances have been made toward orientational order imaging using polarized fluorescence, investigations are still limited to the diffraction-scale resolution, which hampers single filament-scale observation that is required for a quantitative structural analysis of local disorder. Moreover, the retrieved information does not directly report the orientational organization of protein filaments, but rather the mixture between filament orientations and the intrinsic rotational mobility of the attached fluorescent probes (wobbling), which depends on the rigidity of their linker (10, 15).
It is possible to extract information on orientational mobility using single-molecule detection, which can reveal processes that are often missed in ensemble averaging. Seminal single-molecule studies have used light polarization to measure single-molecule orientations, using excitation polarization modulation (16), analyzed direction (17), or more refined schemes to access out-of-plane tilt information (for a review, see ref. 18). Another great advantage of single-molecule imaging is the possibility to achieve superresolution imaging, which relies on single-molecule localization to reconstruct images at nanometer-scale precision, providing that emitters emit temporally independently (19–21). Combining superresolution imaging with single-molecule orientation measurements would provide an ultimate way to image the structural organization of filamentous assemblies at high spatial resolution in vivo. Although a similar combination has been applied to probe the rotational mobility of single molecules in isotropic environments (22), its use to quantify orientations in ordered systems (e.g., filamentous structures) presents several challenges. First, molecular orientation can itself affect localization properties (23–25), and therefore the quality of image reconstruction. This effect has been shown, however, to be less dramatic when rotational mobility occurs in 2D in the sample plane, or in a large angular range (26). Second, the measurement of molecular orientation involves splitting the signal into polarized detection/excitation channels, which can decrease localization precision if not properly processed. At last, although in-plane components of single-molecule orientations are relatively simple to extract, the measurement of out-of-plane orientations of molecules requires more sophisticated experimental schemes and detection algorithms (18, 27). Several signal analysis solutions have been proposed for both orientation and localization monitoring (25, 28–30); however, there is no report yet on quantitative superresolution imaging of molecular orientations in ordered systems.
In this work, we present a quantitative method combining steady-state in-plane single-molecule orientation measurements and superresolution imaging, using polarization-resolved direct stochastic optical reconstruction microscopy (polar-dSTORM). We propose a simple experimental scheme, which is compatible with high signal-to-noise ratio conditions and which provides structural information in filamentous assemblies in 2D, which is sufficient for order interpretation. We present a dedicated algorithm for polarized single-molecule localization and quantification, and show that the imaging of orientational behaviors can be achieved with high accuracy, providing that stringent signal analysis is performed. We further show how to exploit the measured polarized signals to retrieve information on both the fluorescence label wobbling and the local disorder of biological filaments, two parameters that are not discernible in diffraction-limited ensemble methods. We illustrate the use of polar-dSTORM in microtubule networks in fixed cells and double-stranded DNA (dsDNA) in vitro, and evidence the effect of the fluorophore structure on its angular wobbling when linked to actin stress fibers in fixed cells via phalloidin conjugates. These results reveal that ensemble polarization-resolved methods generally overestimate molecular order, illustrating that single-molecule approaches are needed for determining the structural organization of biomolecular assemblies in an unbiased manner.
Results and Discussion
Model and Simulations for Disentangling Orientational Mobility of Fluorophores from Local Structural Disorder.
In the filamentous biological structures studied here, fluorescent labels are likely to undergo fast wobbling (rotational diffusion at a timescale much smaller than the integration time) within an angular range that depends on the label linker rigidity. Instead of reporting the real structure that they are linked to, the monitored fluorophores’ orientation therefore includes both their averaged orientation and wobbling aperture angle, denoted and , respectively, in Fig. 1A. At a macroscopic scale in ensemble measurements, only an average of this information over many molecules can be measured, which obviously loses information on individual wobbling behaviors (Fig. 1A). Interestingly, if one would be able to extract averaged orientations from single molecules independently from their wobbling angle, a more quantitative picture of structural order in biological filaments could be afforded (Fig. 1B). Indeed, the angular extent of molecules over local filament regions is low in ordered structures and increases with disorder.
Fig. 1.
Molecular orientations and polarization factor P in a polar-dSTORM experiment. (A) Angular constraint of a label (here, Alexa Fluor 488; the red arrow representing its emission dipole direction) attached to a fiber. The schematic drawing defines a single-molecule wobbling aperture δ and its pointing direction , as well as the averaged cone aperture angle ψ and its average orientation (also corresponding to the average fiber direction). (B) Angular extent of single molecules’ average pointing direction in a portion of filaments organization, showing a high-order case (low ) and a low-order case (high ). (C) Experimental scheme defining the directions split by a polarizing beam splitter. (D) Calculated dependence for molecular angular distributions aligned in the sample plane (see SI Appendix, Note 1, for details on the sensitivity of this dependence to high NA collection and out-of-plane tilt angles). The averaged graph follows the same dependence because it mathematically preserves the same type of equations. (E) Simulated histograms of the polarization factor for single molecules in a global angular aperture ψ = 120°, for two different average directions = (90°, 45°) and several values of the wobbling aperture . Each molecule of this histogram has a wobbling pointing direction taken within the ψ aperture. These histograms are based on analytical expressions of taken every 0.3° for .
The goal of this work is to access structural information () at a nanoscale, and therefore to measure the average orientation of wobbling molecules, independently on their wobbling angle , in the same time as their location at a subdiffraction scale. Accessing and in 3D from single molecules is in principle possible by projecting their fluorescence signal over more than two incident/detected polarization directions (31, 32), or using defocused imaging schemes (27). However, because localization is required, high signal-to-noise ratios have to be ensured, which thus imposes minimal signal deformation or splitting. We thus choose to privilege a 2D readout of molecules’ orientations, by preferentially exciting in-plane molecules using normal incidence and circular excitation polarization, and projecting their signal over two orthogonal analyzing directions ∥ and (Fig. 1C). The incident circular polarization used here also ensures uniform excitation efficiency in the sample plane, in contrast with the more generally used linearly polarized excitation, which is appropriate only for isotropic samples (22). The measured polarized intensities and are combined into a polarization factor that is normalized to the total in-plane intensity :
In practice, one should include in this expression the diattenuation correction factor of the detection path optics (Materials and Methods) and correct it for the use of high numerical aperture (N.A.) collection that induces polarization mixtures (SI Appendix, Note 1 and Figs. S1 and S2). For a fixed emission dipole orientation, relates directly to the molecule’s orientation projected in the sample plane (SI Appendix, Note 1). Orientational wobbling of the fluorophore makes orientation retrieval more complex, and cannot be retrieved independently from . For instance, at low N.A., (SI Appendix, Note 1) (Fig. 1D).
To measure and independently and thus estimate the extent of filaments structural order (), we analyze local structures statistically. In filament structures, cylindrical symmetry is expected; thus, the average pointing direction of molecules lies along the local fiber direction in the sample plane (or its perpendicular, depending on the labeling type), which decreases the number of unknown parameters. Because is measurable from the image, the averaged orientational order (Fig. 1A) can be directly extracted from averaged values by the dependence , which follows the same characteristic shape as (Fig. 1D). At this stage, is an average and does not contain any information on individual wobbling. To extract this information, a closer inspection of the statistical distribution of is necessary using filament regions of known directions . Simulated distributions in Fig. 1E show in particular that, for a given average disorder , the histogram width decreases with an increasing extent of wobbling . More quantitatively, can be estimated in vertical or horizontal filaments for which . Indeed, in this case, is highly sensitive to and reaches its extreme value , allowing a determination of the upper limit of . With this estimation, single-molecule directions can then be directly extracted from (Fig. 1D), and thus structural-order information () can be given on filament regions. Note that, at this stage, an uncertainty remains between and . This can be circumvented using measurements on different fibers’ orientations. At in particular, one of the two solutions can be more easily ruled out, considering that all values stay preferentially in the proximity of the average fiber direction.
Ultimately, the angular extent relates to structural heterogeneities that are not detectable in nonpolarized superresolution images. Imaging single-molecule directions thus leads to superresolution structural imaging of filament organization, corrected from the label wobbling.
Experimental Validation.
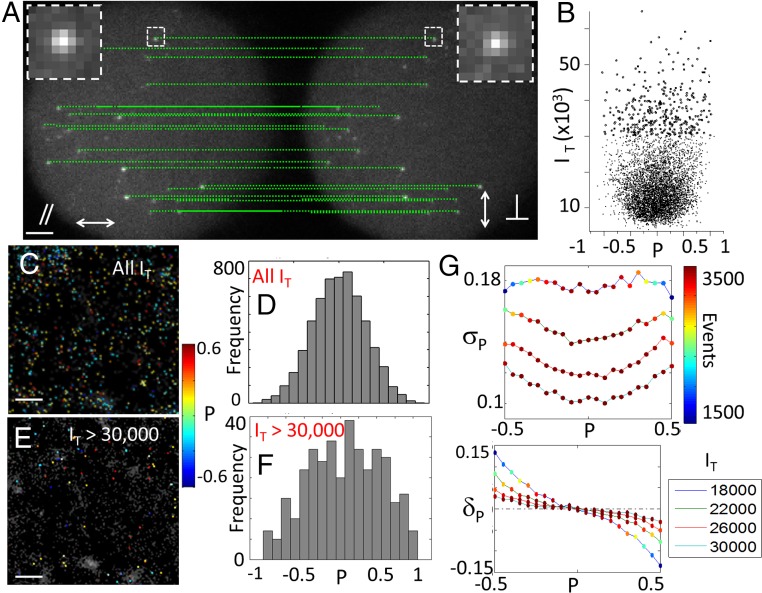
Single-molecule polar-dSTORM signals, generated from in-plane wide-field circular excitation, are recorded on two polarized images (Fig. 2A) using a 90× magnification and N.A. of 1.3, which permits consideration of negligible bias from out-of-plane tilts angles of molecules, in the frame of a 2D model where molecular wobbling lie preferentially in the sample plane (SI Appendix, Fig. S3). These images are formed by placing a polarization beam splitter in front of an EMCCD camera, with minimal intermediate optics to avoid image alterations. Each molecule is therefore visible as two separate polarized point spread function (PSF) images (Fig. 2A), with different intensities depending on its value.
Fig. 2.
Principle and interpretation of polar-dSTORM data, application to Atto633 molecules attached to PLL. (A) Polarized imaging, showing examples of detected single-molecule pairs. (B) Polarization factor versus total intensity (, in EMCCD counts) measured for the whole collection of detected molecules. (C) Polar-dSTORM image of values (each molecule spot is associated to a value (color scale), superimposed with the pure dSTORM image shown as gray background). (D) Corresponding histogram of values. Note that, in these histograms, events corresponding to very low or intensities (for which the detection algorithm fails) are not represented, even though they are detected from the polar-dSTORM algorithm. These events would indeed lead to with low precision because they correspond to very low intensity levels for one of the analyzed images. (E) Polar-dSTORM image with filtering total intensities above 30,000 counts. (F) Corresponding histogram. (G) Simulated intensity dependence of the SD (σP) and bias (δP) for expected between −0.5 and 0.5 (SI Appendix, Fig. S6). The scale represents the number of measured events after intensity filtering. The EMCCD gain used for simulations is 300, and background level is 1,000 counts. Both σP and bias δP increase are stronger at higher background level or higher gain level, which is expected from higher noise level. [Scale bars: 2 μm (A) and 500 nm (C and E).] [Gaussian blurring: 30 nm (C and E).]
Polar-dSTORM analysis requires that the measurement of is performed in conjunction with molecular localization, which requires a specific algorithm that can deal with low signal-to-noise conditions (due to the polarized splitting of the signal) and multiple subimages. Our polar-dSTORM algorithm accurately estimates both the polarization factor and the position for each molecule present in the dSTORM image stack (SI Appendix, Note 2). The detection method is optimal for low signal-to-noise ratio conditions, thanks to a generalized likelihood ratio test performed before subpixel PSF estimation (33). In the experimental setup used, the recorded frames include and polarized images, separated by a vector identical for all molecules of the image stack (Fig. 2A). Because the knowledge of single-molecule localizations is required at the subpixel level, this vector must also be determined with a high level of precision. Considering that a precalibration might depend on the optical conditions and on the sample used, we rather chose to measure this vector using detected molecules in the dSTORM image stack from a statistical analysis over many detected molecule pairs, guaranteeing its determination at a level of precision better than the localization precision (SI Appendix, Note 2 and Fig. S4). Once the vector is defined, single-molecule pairs are identified at 95% confidence level using a distance test based on their localization precision (SI Appendix, Note 2). Finally, and (signals integrated over the PSF size) are calculated for each detected molecule pair to estimate , and to reconstruct a polar-dSTORM image of in which the molecular positions are combinations of both positions weighted by their variances, which ensures optimal position estimation (SI Appendix, Note 2).
To validate the method, polar-dSTORM was first applied on a test sample where molecules are fixed and expected to be randomly oriented, using Atto 633 molecules deposited on a poly-l-lysine (PLL)-coated layer. First, recorded values shows a clear dependence on the intensity for low intensity counts (Fig. 2B), which thus affects superresolution images of and corresponding histograms (Fig. 2 C–F). Collecting all molecules whatever their intensity level leads to a histogram, which resembles a narrow peak centered on < > = 0, whereas for > 30,000 counts (“counts” denoting the recorded EMCCD signal), the statistics spreads over a larger ranges of values. This is due to the fact that low intensities induce a bias in the recorded values, which originates from noise and possibly out-of-plane molecular tilt angles (SI Appendix, Note 1 and Fig. S2). After intensity thresholding, the global histogram of resembles the expected stationary behavior (Fig. 2F), which was not visible before filtering (Fig. 2D). Signal level restriction is thus a determining step in the analysis of polar-dSTORM images.
To quantify the intensity effect on the quality of the polarization factor estimation, we ran the polar-dSTORM algorithm on simulated single-molecule images where the noise and background levels are similar to experimental conditions [the noise level of the EMCCD camera is included based on precalibration and noise analysis (SI Appendix, Fig. S5)]. We estimated the polarization factor for a collection of 4,000 single molecules of known and polarization factor (SI Appendix, Fig. S6). Its bias and SD are reported in Fig. 2G for a decreasing intensity level . Both quantities increase when the signal level decreases, with more pronounced effects away from . When the intensity falls below 10,000 counts (localization precision, >30 nm), the precision on drops down and single-molecule orientations cannot be measured with a precision better than 10° or more. However, these simulations show that a precision of below 0.05 (corresponding to a precision in single-molecule orientation of ∼3°) can be obtained providing that the total intensity is above 30,000 counts (which corresponds to a localization precision of 16 nm in the present experimental conditions). Ultimately, under high signal-to-noise conditions, a precision of ∼ 0.005 has been reached experimentally (SI Appendix, Fig. S7). In summary, even in conditions of signal-to-noise levels appropriate for dSTORM imaging, ensuring a high precision and low bias on requires more stringent algorithm detection conditions.
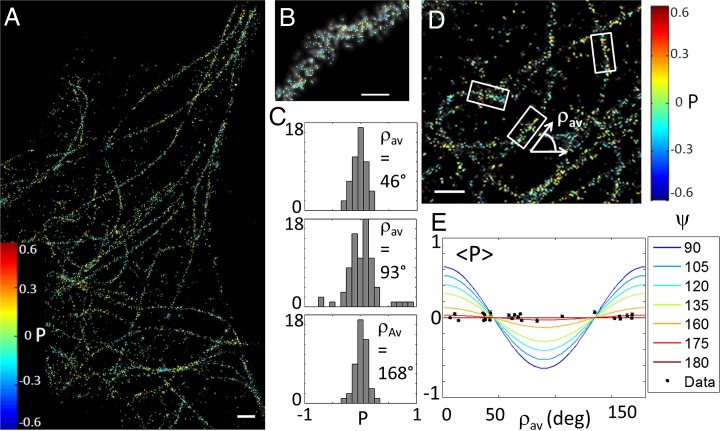
To further corroborate our method, we applied polar-dSTORM to biological filaments labeled with flexibly linked fluorophores, by indirect immunolabeling of microtubules using Alexa Fluor 647-conjugated secondary antibodies (Fig. 3). After intensity thresholding, histograms of individual fibers are seen to be centered on = 0 (Fig. 3 C and D), whatever the fiber orientation, showing a global range of explored angles of ψ ∼ 180° (Fig. 3E). Note that this thresholding leads to a reduction of the total number of measured localizations of about 70%. The SD of those histograms is essentially limited by noise, which is consistent with fast rotational motion (see corresponding theoretical case in Fig. 1E). The structural flexibility of antibody molecules together with the use of a primary–secondary antibody pair leaves a large orientational freedom to the fluorescent labels that explore random angles within the integration time of the EMCCD camera, making impossible any structural studies in this specific situation.
Fig. 3.
Polar-dSTORM imaging in microtubules. (A) Polar-dSTORM image of Alexa Fluor 647-immunolabeled microtubules in fixed cells (Intensity threshold of 12,000 counts, expected σP ∼ 0.12). (B) Zoom on a specific fiber (superimposed to a traditional dSTORM image, gray scale). (C) Individual histograms reported for individual portions of fibers exhibiting different mean orientations (). (D) Zoom on the global image showing typical selections for local fibers orientations, obtained from automatic detection of clusters of localizations (SI Appendix, Fig. S8). (E) Averaged , superimposed with experimental averaged values measured for several portions of fibers oriented with various directions (only portions for which the number of events is above 50 are kept; SEM is shown as an error bar). [Scale bars: 1 μm (A and D) and 150 nm (B).] [Gaussian blurring size: 30 nm (A, B, and D).]
Nanoscale Orientational Order Imaging in Biological Filaments.
We then applied polar-dSTORM to filaments where the orientational mobility of the fluorescent labels is expected to be constrained but not known. We first measured single dsDNA fibers in vitro on a coverslip surface, labeled with the intercalant YOYO-1, which has been previously used in dSTORM imaging (34) (Fig. 4). YOYO-1, whose planar structure has two conjugate moieties sandwiched in between consecutive base pairs (Fig. 4B), is known to be constrained and ordered in a direction roughly perpendicular to DNA at sufficiently low dye density (34, 35). The averaged dependence of the experimental data indeed shows that the mean pointing direction of the angular constraint is perpendicular to the fiber and on average contained within a cone aperture of ψ ∼ 90° (Fig. 4 C and D). Moreover, the histogram shapes show a degree of dye wobbling, their width being above noise, but still not as large as expected from a complete stationary situation within a 90° aperture (see theoretical histograms in Fig. 1E). Quantification of the wobbling angle δ for fibers oriented close to = 0° or 90° (Fig. 4E), as detailed above, gives an upper limit of δ ∼ 60°. This result is consistent with previous ensemble measurements that were performed using multiple polarization analysis (36). Polar-dSTORM, however, advantageously provides additional nanoscale spatial information on the structural disorder, compared with ensemble measurement. Assuming indeed that all molecules undergo wobbling to the same extent, their individual wobbling directions can be estimated from fibers oriented close to = 45° as mentioned above (Fig. 4F). We find that regions of typically a few hundreds of nanometer length show quite homogeneous distributions of for YOYO-1, over an angular extent of typically 20–30°, which represents a narrow angular freedom range (Fig. 4G). We could, however, observe locally dsDNA fiber regions where YOYO-1 orientations do not follow the fiber average direction, indicating local bending in DNA that is not detectable in the pure dSTORM image (Fig. 4G).
Fig. 4.
Polar-dSTORM imaging in DNA. (A) Polar-dSTORM images (superimposed to traditional dSTORM images) of local regions in dsDNA fibers labeled with YOYO-1, immobilized in vitro on a sample surface (Intensity threshold of 12,000 counts, expected σP ∼ 0.12). (B) Schematic representation of the behavior of YOYO-1 emission dipoles (red arrows) inside the dsDNA structure. (C) Histograms of polarization factors for three different fibers regions. (D) Averaged behaviors for several measured fibers, superimposed to the modeled dependence of . The determination of ψ is done by a fit of the data, supposing molecular distributions lying in the sample plane and including high N.A. corrections. Only portions for which the number of events is above 30 are kept; SEM is shown as an error bar. (E) Determination of an upper limit for the molecular wobbling angle δ using statistical analysis on a fiber oriented close to 0°. values are reported in correspondence to and values (both solutions are possible) for each measured single molecule. is calculated from the inversion of the analytical expression of for a known δ, here taken as 60°. The best-fitting solution δ is the one that encompasses all experimental points at the minimum of the dependence. For representation only, noise is added to the estimation of , accounting for the molecule total intensity (SI Appendix, Note 1). This example and other fibers show that δ lies in the range of 40–60°. (F) Determination of the extension of molecular average pointing directions , using statistical analysis on a fiber oriented close to 45°, using a similar representation as in E, and using δ = 60°. (G, Left) Superresolution image of the deduced values for this fiber, superimposed to the dSTORM image (gray scale), using sticks oriented by the angle in the plane relative to the horizontal (the color of the sticks follows the color bar in A). The arrow points to a location of the dsDNA fiber whose molecular order departs from the average behavior of the imaged region. (Right) Corresponding histograms of () for this molecular population are shown to quantify the extent of orientations within this fiber. [Scale bars: 100 nm (A and G).] [Gaussian blurring size: 30 nm (A and G).]
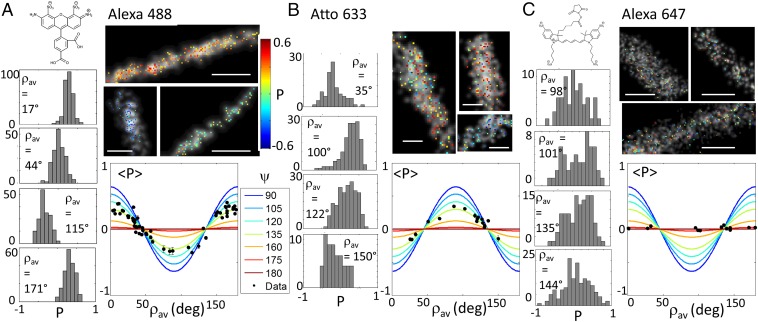
In a second example, we applied polar-dSTORM to actin stress fibers in fixed cells, with actin labeled using different phalloidin-conjugated fluorescent dyes (Fig. 5). Phalloidin itself provides a priori sufficient orientational rigidity with a very high affinity to actin filaments (37), but the conjugation chemistry for each fluorescent dye can lead to phalloidin–dye links of different flexibilities depending on the exact structure of the link. Polar-dSTORM imaging on Alexa Fluor 488–phalloidin for different fibers regions shows first that the averaged cone aperture of the angular constraint is ψ ∼ 130° (Fig. 5A). The histograms widths are slightly above the noise limit, indicating a degree of wobbling. The behavior of another fluorescent conjugate, Atto 633–phalloidin (Fig. 5B), in the same labeling conditions, shows a similar averaged cone aperture value except that histograms are slightly larger, illustrating a smaller wobbling angle than for Alexa Fluor 488. Interestingly, Atto 633 points perpendicularly to the fiber direction, whereas Alexa Fluor 488 points along it, which is a consequence of their specific molecular structure. At last, Alexa Fluor 647–phalloidin shows a completely isotropic (ψ ∼ 180°) and a quite stationary behavior (with a large histogram shape), most probably due to its larger and more extended molecular structure (Fig. 5C). This clearly makes this label not appropriate for imaging of actin fiber organization in fixed cells. Using a similar analysis as described above, Alexa Fluor 488 molecules are shown to exhibit a wobbling of higher limit δ ∼ 90°, whereas for Atto 633, δ ∼ 70°. Note that this slight difference in wobbling extent could be attributed to specific charge interactions of the fluorophore with the actin filaments (Alexa Fluor 488 being negatively charged, whereas Atto 633 is positively charged), and to the flexibility of the fluorophore molecular structure itself imposed by its own linker length. We have, however, observed that Atto 565 (a neutral molecule with similar linker length as Alexa Fluor 488) showed very little difference in wobbling behavior (SI Appendix, Fig. S9). This indicates that the dominant effect governing wobbling in phalloidin conjugates is rather the molecular linker length.
Fig. 5.
Polar-dSTORM imaging in actin stress fibers. images (superimposed to traditional dSTORM images, gray scale) of local regions in phalloidin-labeled actin stress fibers in fixed cells, with corresponding histograms for different portions of fibers and averaged behaviors reported in the dependence. The determination of ψ is done by a fit of the data, supposing molecular distributions lying in the sample plane and including high N.A. corrections. Only portions for which the number of events is above 60 are kept; SEM is shown as an error bar. (A) Alexa Fluor 488. Intensity threshold: 50,000 counts; expected σP < 0.05. (B) Atto 633. Intensity threshold: 50,000 counts; expected σP < 0.05. (C) Alexa Fluor 647. Intensity threshold: 12,000 counts; expected σP ∼ 0.12. The known molecular structures are shown in Insets. [Scale bars: 100 nm (A and B) and 500 nm (C).] [Gaussian blurring size: 15 nm (A), 25 nm (B), and 40 nm (C).]
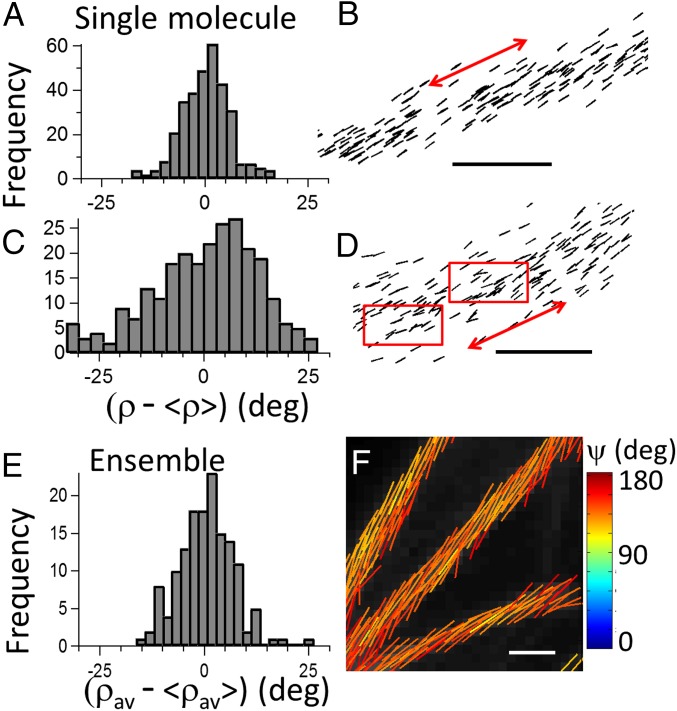
Overall, the wobbling angle values measured for these fluorophores are close to previously reported single-molecule measurements on F-actin Rhodamine labels in vitro (3). This experiment necessitated, however, recording individual single-molecule time traces analyzed over four excitation and four detected polarizations, which is incompatible with single-molecule localization imaging. Here, quantifying the fluorophores’ wobbling in conjunction with superresolution imaging allows accessing information on the local-scale morphology of filaments organization, both spatially and structurally. At this superresolution scale, 100-nm length fiber portions have been analyzed similarly as above for Alexa 488 (similar results are obtained for Atto 633). Pointing directions in actin stress fibers span on average ∼ 20–30° (Fig. 6 A and B), evidencing a high filament order. This shows the capacity of actin filaments to form straight structures of collective orientations that is likely to correlate to the mechanical role of stress fibers in cells. In the thickest measured fibers, however (above 50 nm diameter), this extent is seen to be heterogeneous, with increases that could reach up to ∼ 50°, which supports the formation of more complex filaments organizations (Fig. 6 C and D). This result shows that, at the nanometric scale of investigation of polar-dSTORM, the local disorder in the filament organization can be measurable, which is discernible by a local bending of molecule orientations that is not detectable in dSTORM images (Fig. 6D).
Fig. 6.
Comparison of nanoscale and diffraction-scale structural imaging in actin stress fibers (labeled with Alexa Fluor 488–phalloidin conjugate). (A) Reconstruction of single-molecule orientations using polar-dSTORM data, in an actin fiber portion oriented close to 45° and incorporating in the estimation the wobbling angle = 90°: histogram of molecular orientations with respect to the averaged population (), evidencing high degree of homogeneity. (B) Corresponding polar-dSTORM image where directions are represented by sticks oriented by the angle relative to the horizontal axis (the red arrow points the averaged fiber direction). (C) Same analysis in a heterogeneous actin filament, with (D) its polar-dSTORM image, where red rectangles point out regions of structural heterogeneity. (E and F) Ensemble confocal polarization-resolved fluorescence measurement (optical resolution, 200 nm) on actin stress fibers with the same label. The average fluorophores orientation and the angular aperture ψ of their distribution in each pixel are determined by fitting tunable polarization data (Materials and Methods). (E) Histograms of averaged orientations along a fiber portion of 2-μm length. (F) Each fitted pixel is represented by a stick encoded with its orientation angle and a color following the ψ color scale. The fluorescence intensity is in gray scale. [Scale bars: 100 nm (B and D) and 1.5 μm (F).]
It is finally interesting to compare the extent , e.g., the molecular orientational order corrected from fluorophore label mobility, with the averaged molecular order measured in ensemble measurements. Confocal polarization resolved images on actin-labeled cell samples using the same phalloidin conjugates as above are depicted in Fig. 6 E and F, following a method described in ref. 11, which relies on polarization incident tuning (Materials and Methods). In this measurement, the molecular order information is averaged over a spatial scale of about 200 nm, e.g., the diffraction limit of the used microscope. At this scale, the total angular distribution of labels is slightly higher than in single-molecule measurements, with ψ ∼ 140° (for both Alexa 488 and Atto 633) with homogeneous fibers pointing directions (Fig. 6E), which visibly hides any local heterogeneity. The fact that ψ surpasses the orientational disorder value measured from single-molecule averaged measurements shows that, at a 200-nm spatial scale, structural heterogeneities occur (note that single-molecule averages are typically given for fiber regions of 100-nm length). This value points out the incapacity of ensemble measurements to report real biomolecular order in filaments organizations. Indeed, as shown in single-molecule measurements, fluorophore label wobbling is often not negligible even in fixed cells, which clearly makes confocal polarized microscopy largely overestimating orientational information. The method developed here, by extracting information on the fluorophore rotational dynamics that is missed in ensemble measurements, provides thus access to resolved structures at the nanoscale, making it able to measure structural-order modifications that can occur at the single filament scale.
Conclusion
In this work, we have demonstrated polarized single-molecule–based superresolution imaging and its capacity to provide structural information that is not detectable in ensemble measurements or in pure superresolution imaging. A key element of the method is that the required precision for measuring single-molecule orientations is essentially governed by the recorded signal-to-noise level, which imposes careful signal analysis and intensity thresholding. This threshold decreases the number of reported molecules, compared with pure dSTORM imaging, which is based on a sole localization detection criterion. Nevertheless, standard dSTORM dyes and related fluorophores used for other superresolution techniques such as STORM and photoactivation light microscopy provide sufficient signal levels to reach a reasonable number of detected events in biomolecular structures. Applying this method to biological filaments, we have shown that statistical analysis allows retrieving wobbling and pointing directions independently, thus correcting the measured polarized information from fluorophores orientational flexibility, which is intrinsic to the label linker. In particular, we evidence residual wobbling in various systems, even in fixed cells and tightly packed DNA intercalants. We show, however, that, even though this wobbling is present, structural heterogeneity can be accessed at the nanoscale. dsDNA investigations evidence that this method can be applied to morphological studies that are particularly relevant to protein–DNA interaction studies, at a scale that could not be reachable with traditional polarized fluorescence microscopy (1, 36). Actin stress fibers studies show that particular care has to be brought on both fluorophore and labels, to access fine structural information.
Experimentally, dividing the signal in two polarization states, as we propose here, is a minimal scheme of which the important advantage is to preserve high signal-to-noise ratio levels, compatible with localization precisions of 20–30 nm. This simple experimental scheme is extendable to multiple configurations such as multicolor for detecting orientational correlations between interacting biomolecules, or live imaging to access dynamics information, if combined to single-molecule tracking. It is also extendable to 3D orientations measurements, in a version that would require more than two emission polarization states (31, 32), at the expense, however, of lower signal-to-noise ratio conditions. Ultimately, this technique can serve as a quantitative indicator for local structural properties at the nanoscale in more complex filament organizations, such as the hierarchical assembly of amyloid protofilaments (38) or the dynamic actin filaments reorganization involved in cell shape changes (39).
Materials and Methods
Polar-dSTORM.
Measurements are carried out on a custom epifluorescence microscope, adapted to retrieve the perpendicular (“⊥” corresponding to the vertical axis of the sample plane) and parallel (“∥,” corresponding to the horizontal axis of the sample plane) polarization states of the fluorescence images. The excitation light sources are polarized continuous wave lasers emitting at 639 or 488 nm (Coherent), with typical averaged power density of about 5–8 kW/cm2 before the objective. For both laser lines, the laser beam is expanded by a telescope and circularly polarized by quarter wave plates (WPQ10M; Thorlabs). A mirror reflects the beam toward the microscope, followed by a large focal length lens (f = 400 mm) to focus the beam in the backfocal plane of the objective. This provides a wide-field illumination of diameter of about 100 μm in the sample plane. After the reflection on a dichroic mirror (FF 500/646; Semrock), the excitation light is focused onto the sample by an oil-immersion objective lens (Plan Apo 60×; N.A., 1.3; Nikon). The emitted fluorescence is collected by the same objective lens in an epigeometry, passes through the dichroic mirror and a band pass emission filter (FF01-535/675; Semrock). The imaging lens provides a total magnification of 90× or 150×. At the microscope exit, a Wollaston prism (separation angle, 5°; CVI Laser Optics) is placed just before the EMCCD camera (Quantem 512sc; Photometrics), such as to fill the CCD chip with two perpendicularly polarized images. The size of the images is set by a diaphragm placed in the first image plane at the exit of the microscope. The polarization factor quality is seen to be homogeneous over the whole image field of view.
In addition to fluorescence excitation, another weak beam of 980 nm (about 1 mW from a laser diode coupled into a single mode fiber) is used as a source for a home-built autofocus system. This beam is brought off axis with a diameter smaller than the entrance pupil of the objective, retro-reflected, and its specular reflection by the sample coverslip surface is directed by a 50/50 beam splitter toward a camera that records the position of the reflected beam. The center of this position is compared with a reference calibration curve, which allows to correct back on the sample axial Z position, which is ensured by the control of the piezoelectric holder (MCL Nano-T Series) on which the sample is mounted. The acquisition and camera control are realized by Micromanager.
For dSTORM imaging, a first fluorescence image is recorded with low intensity (∼500 W/cm2, below STORM blinking conditions), ensuring the identification of relevant parts of the sample. The intensity is then raised to 5 kW/cm2, which is a typical level to provide a good compromise between signal level and blinking rate. The images are acquired at a rate of 30 ms per image, with a total of 4,000–80,000 images depending on the molecular density.
For polarization factor superresolution imaging, a correction factor has to be accounted for, which includes the diattenuation factor introduced by the different optics present in the detection path, making the efficiency of the ⊥ and ∥ channels be slightly different. The determination of this factor (denoted G, which has a value between 1 and 1.6, depending on the wavelengths detected) is made by imaging a solution of fluorophores freely diffusing/rotating in water, and systematically accounted for as a factor in the estimation of P: .
Polarized Confocal Microscopy.
The polarized confocal setup is based on the principle of linear dichroism, recording unanalyzed fluorescence signals for a tunable incident polarization (11) (SI Appendix, Fig. S10). A custom confocal epi microscope system is used with continuous wave lasers at 491 nm (Calypso; Cobolt) and 633 nm (Obis; Coherent). After recombination of the two lasers by a dichroic mirror (HC Beamsplitter BS 506; Semrock) and reflection on a two color dichroic mirror (HC Dual Line Notch Beamsplitter 500/646; Semrock), the incident angle of linear polarization is sequentially rotated by an achromatic half-wave plate (AHWP10M-600; Thorlabs) mounted on a motorized rotation stage (PR50CC; Newport), typically between 0° and 180° in 10° steps. The excitation light is focused onto the sample by a water-immersion objective lens (C-Apochromat 40× UV-VIS-NIR; N.A., 1.2; Carl Zeiss). Imaging is performed by scanning a set of two galvanometric mirrors placed between the dichroic mirror and the objective lens. The fluorescence is splitted by a dichroic mirror (HC Beamsplitter BS 605; Semrock) detected by two single-photon counting modules (SPCM-AQR-14; Perkin-Elmer), using emission filters (HQ540/80M-2P and 675/100; Chroma). The major part of polarization distortions are corrected for by placing the polarization rotation unit after the excitation dichroic mirror. Residual distortions are measured and included in the model used to retrieve information on the orientational order (11). The achromaticity of polarization distortions in the detection path is ascertained by a dedicated measurement at different wavelengths, following the procedure described in ref. 11. To quantify molecular orientational order, we model the angular distribution function either by a cone with a total aperture angle ψ and oriented in the sample plane with an angle , or by a torus shape of width ψ with molecules pointing in the direction. (ψ, ) are determined from the analysis of the Fourier series coefficients of the fluorescence modulation obtained by rotating the incident polarization (programmed under MATLAB; MathWorks), following a methodology detailed in ref. 11. The minimum level of signal is set at a total intensity of 5,000 photons, ensuring a precision of 2° on both ψ and (as estimated by Monte Carlo simulation under Poisson noise conditions; see ref. 9). ψ values are moreover selected only when passing a fit quality check (selecting data not affected by sample motion or bleaching during the polarization tuning measurements, as detailed in ref. 11).
Fluorophores.
The used fluorophores are Alexa Fluor 488 (absorption/emission maximum wavelengths: 495 nm/519 nm; Life Technologies), YOYO-1 (491 nm/509 nm; Life Technologies), Alexa Fluor 647 (650 nm/668 nm; Life Technologies), and Atto 633 (629 nm/657 nm; Atto-Tec).
Imaging Buffer.
The imaging buffer includes mercaptoethylamine (MEA) (Fluka) to achieve a final thiol concentration of 100 mM. MEA was stored as a solid at 4 °C and prepared fresh as a 1 M stock solution in water with pH adjusted to ∼8 with 1 M aqueous KOH. Additionally, an enzymatic oxygen-scavenging system was added to the switching buffer to oxygen removal: 0.5 mg/mL glucose oxidase (Sigma-Aldrich), 40 μg mL/L catalase (Sigma-Aldrich), and 10% (wt/vol) glucose (Fisher Scientific).
Fluorophores Attached on PLL-Coated Cover Glasses.
Cover glasses and slides were cleaned with acetone–ddH2O–methanol–ddH2O (five times for 3 min each time) under running flow, and then sonicated in 1 M KOH for 30 min. They were rinsed thoroughly with water under running flow, followed by immersing in acetone for 10 min. Before use, they were extensively rinsed under water, and blown dry. Cleaned cover glasses were coated with PLL (Mr, 70,000–150,000; Sigma-Aldrich) by adsorption from a filtered 0.1 g/L solution for 1 h, incubated for 3 h at 37 °C, and afterward washed three times in ddH2O and blown dry. A drop of Atto 633 (30 µL; 1 mM) was deposited on the PLL-coated cover glasses, and then let dry for 30 min, and afterward washed three times in ddH2O, and blown dry. The cover glass with Atto 633 was then attached to a Cover-Well imaging chamber (Grace Bio Labs), which contained the switching buffer.
Cell Culture.
All experiments were carried out on fibroblast-like COS 7 cells (African green monkey kidney; SV40 transformed; ATCC CRL 1651). COS 7 cells were grown in an incubator at 37 °C and 5% CO2 in Lab-Tek chambers (Nunc) in DMEM (Gibco), pH 7.25, plus 10% (vol/vol) FCS, 1% sodium pyruvate, and 4 mM l-glutamine, for 12–24 h before the experiment.
Direct Labeling of Actin in Fixed Cells with Phalloidin–Alexa Fluor 488, Atto 633, and Alexa Fluor 647.
For polar-dSTORM measurements, COS 7 cells were washed briefly with 1 mL of prewarmed (37 °C) PBS and followed by fixation with 4% (vol/vol) PFA in PBS for 10 min at room temperature (20 °C). After washing the cells with PBS (three times, 5 min per each), cells were permeabilized with 0.5% (vol/vol) Triton X-100 in PBS for 10 min. They were washed again with PBS twice, 5 min per each, and then saturated with 5% (wt/vol) BSA in PBS for 30–60 min at 4 °C. Actin filaments were labeled by adding phalloidin–dye, diluted in PBS to a final concentration of 0.5–0.9 nM. The sample was wrapped with aluminum foil to protect from light. Incubation was done overnight at 4 °C. Phalloidin solution was then exchanged by 0.1% (vol/vol) Tween 20 in PBS in each chamber and incubated for 5 min at room temperature (20 °C), two times. Samples were stored at 4 °C in PBS until imaging. For ensemble confocal measurements, COS 7 cells were cultured under standard conditions. Cells were fixed [4% (vol/vol) paraformaldehyde, 0.5% Triton X-100, and 2% (wt/vol) BSA, 1 h at 4 °C], stained with Alexa Fluor 488/647 or Atto 633 phalloidin (1 nM for 20,000 cells, 30 min at 4 °C), and washed with 0.1% Tween 20. Measurements were performed at room temperature in PBS.
Immunofluorescent Labeling of Microtubules in Fixed Cells.
Cells were washed gently with prewarmed (37 °C) PBS, and followed by fixation with 4% (vol/vol) PFA in PBS for 10 min at room temperature (20 °C). The fixed cells were washed in PBS (5 min, three times). They were permeabilized with 0.1% Triton in PBS for 10 min. They were saturated with 5% (wt/vol) BSA in PBS (blocking buffer, 1 h at 4 °C). They were then stained with the primary antibody against tubulin (2.5 μg/mL mouse anti–β-tubulin; Sigma-Aldrich) for 60 min in blocking buffer. The sample was then rinsed with washing buffer (0.2% BSA, 0.1% Triton X-100 in PBS) three times. Goat anti-mouse Alexa Fluor 647 antibody was diluted in the same blocking buffer to 2.5 μg/mL and was added to the sample for 60 min at 4 °C. The antibody solution was changed in each chamber with 0.1% (vol/vol) Tween 20 in PBS for 5 min at room temperature (20 °C), twice. A second fixation was done with 4% (vol/vol) PFA in PBS for 5 min. Finally, cells were rinsed three times with PBS for 5 min and stored at 4 °C in PBS until imaging.
In Vitro λ-DNA.
Coverslips were treated using acetone–methanol–ddH2O five times (5 min each), and then sonicated in 1 M KOH for 30 min, and then rinsed thoroughly with running ddH2O, acetone, and again ddH2O for 5 min each, then dried by blow drying. Cleaned coverslips were coated with PLL (Mr, 70,000–150,000; Sigma-Aldrich) by adsorption from a filtered 0.1 g/L solution for 1 h, carefully rinsed with water, and blown dry. Samples were prepared by mixing λ-DNA (500 μg/mL; New England Biolabs) with YOYO-1 (10 μM stock solutions in DMSO; Life Technologies) to obtain 1:20 dye/bp in total volume 150 mL with TE50 buffer [50 mM Tris⋅HCl, 1 mM EDTA, pH 7.5 (with KaOH)]. The mixture was incubated for 1 h at room temperature. DNA–YOYO-1 mixture was purified by centrifugation in MicroSpin S-400 HR Columns (GE Healthcare; Life Technologies). The DNA was thawed and spin-coated onto a PLL-coated coverslip by dropping 40–80 μL, waiting for 30 s, and then rotating at low speed. The coverslip was then rinsed with 4 mL of ddH2O at a rotation speed of 2,000 × g for 1 min to stretch the DNA and to remove unbound material as an extra purification. The coverslips with labeled DNA–YOYO-1 were then closed to a Cover-Well imaging chamber (Grace Bio Labs) that contained the imaging buffer.
Supporting Information.
SI Appendix contains details on the experimental setups, the polar-dSTORM algorithm, calibrations, models, and simulations.
Supplementary Material
Acknowledgments
We thank M. Modesti for assistance with the dsDNA samples, N. Kumar Balla for assistance in the optical setup, as well as D. Marguet and G. Baffou for helpful comments and discussions. This work has been supported by Contracts ANR-10-INBS-04-01 (France-BioImaging Infrastructure Network), ANR-11-INSB-0006 (France Life Imaging Infrastructure Network), the A*MIDEX Project (ANR-11-IDEX-0001-02) funded by the “Investissements d’Avenir” French Government Program, managed by the French National Research Agency, and finally Conseil Régional Provence Alpes Côte d’Azur. C.A.V.C. and H.A.S. received a scholarship from the Erasmus Mundus Doctorate Program Europhotonics (Grant 159224-1-2009-1-FR-ERA MUNDUS-EMJD).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516811113/-/DCSupplemental.
References
- 1.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark AG, Wartlick O, Salbreux G, Paluch EK. Stresses at the cell surface during animal cell morphogenesis. Curr Biol. 2014;24(10):R484–R494. doi: 10.1016/j.cub.2014.03.059. [DOI] [PubMed] [Google Scholar]
- 3.Forkey JN, Quinlan ME, Goldman YE. Protein structural dynamics by single-molecule fluorescence polarization. Prog Biophys Mol Biol. 2000;74(1-2):1–35. doi: 10.1016/s0079-6107(00)00015-8. [DOI] [PubMed] [Google Scholar]
- 4.Brasselet S, et al. 2013. Imaging molecular order in cell membranes by polarization-resolved fluorescence microscopy. Fluorescent Methods to Study Biological Membranes, Springer Series on Fluorescence, eds Mély Y, Duportail G (Springer, Heidelberg), Vol 13, pp 311–337.
- 5.Ghosh S, Saha S, Goswami D, Bilgrami S, Mayor S. Dynamic imaging of homo-FRET in live cells by fluorescence anisotropy microscopy. Methods Enzymol. 2012;505:291–327. doi: 10.1016/B978-0-12-388448-0.00024-3. [DOI] [PubMed] [Google Scholar]
- 6.Vishwasrao HD, Trifilieff P, Kandel ER. In vivo imaging of the actin polymerization state with two-photon fluorescence anisotropy. Biophys J. 2012;102(5):1204–1214. doi: 10.1016/j.bpj.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axelrod D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys J. 1979;26(3):557–573. doi: 10.1016/S0006-3495(79)85271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benninger RKP, et al. Live cell linear dichroism imaging reveals extensive membrane ruffling within the docking structure of natural killer cell immune synapses. Biophys J. 2009;96(2):L13–L15. doi: 10.1016/j.bpj.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kress A, et al. Probing orientational behavior of MHC class I protein and lipid probes in cell membranes by fluorescence polarization-resolved imaging. Biophys J. 2011;101(2):468–476. doi: 10.1016/j.bpj.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazar J, Bondar A, Timr S, Firestein SJ. Two-photon polarization microscopy reveals protein structure and function. Nat Methods. 2011;8(8):684–690. doi: 10.1038/nmeth.1643. [DOI] [PubMed] [Google Scholar]
- 11.Kress A, et al. Mapping the local organization of cell membranes using excitation-polarization-resolved confocal fluorescence microscopy. Biophys J. 2013;105(1):127–136. doi: 10.1016/j.bpj.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443(7110):466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- 13.DeMay BS, Noda N, Gladfelter AS, Oldenbourg R. Rapid and quantitative imaging of excitation polarized fluorescence reveals ordered septin dynamics in live yeast. Biophys J. 2011;101(4):985–994. doi: 10.1016/j.bpj.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavrakis M, et al. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat Cell Biol. 2014;16(4):322–334. doi: 10.1038/ncb2921. [DOI] [PubMed] [Google Scholar]
- 15.Sosa H, Peterman EJ, Moerner WE, Goldstein LS. ADP-induced rocking of the kinesin motor domain revealed by single-molecule fluorescence polarization microscopy. Nat Struct Biol. 2001;8(6):540–544. doi: 10.1038/88611. [DOI] [PubMed] [Google Scholar]
- 16.Ha T, Laurence TA, Chemla DS, Weiss S. Polarization spectroscopy of single fluorescent molecules. J Phys Chem B. 1999;103(33):6839–6850. [Google Scholar]
- 17.Kinosita K, Jr, et al. Dual-view microscopy with a single camera: Real-time imaging of molecular orientations and calcium. J Cell Biol. 1991;115(1):67–73. doi: 10.1083/jcb.115.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backlund MP, Lew MD, Backer AS, Sahl SJ, Moerner WE. The role of molecular dipole orientation in single-molecule fluorescence microscopy and implications for super-resolution imaging. ChemPhysChem. 2014;15(4):587–599. doi: 10.1002/cphc.201300880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 20.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 22.Gould TJ, et al. Nanoscale imaging of molecular positions and anisotropies. Nat Methods. 2008;5(12):1027–1030. doi: 10.1038/nmeth.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enderlein J, Toprak E, Selvin PR. Polarization effect on position accuracy of fluorophore localization. Opt Express. 2006;14(18):8111–8120. doi: 10.1364/oe.14.008111. [DOI] [PubMed] [Google Scholar]
- 24.Engelhardt J, et al. Molecular orientation affects localization accuracy in superresolution far-field fluorescence microscopy. Nano Lett. 2011;11(1):209–213. doi: 10.1021/nl103472b. [DOI] [PubMed] [Google Scholar]
- 25.Backlund MP, et al. Simultaneous, accurate measurement of the 3D position and orientation of single molecules. Proc Natl Acad Sci USA. 2012;109(47):19087–19092. doi: 10.1073/pnas.1216687109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lew MD, Backlund MP, Moerner WE. Rotational mobility of single molecules affects localization accuracy in super-resolution fluorescence microscopy. Nano Lett. 2013;13(9):3967–3972. doi: 10.1021/nl304359p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backer AS, Moerner WE. Determining the rotational mobility of a single molecule from a single image: A numerical study. Opt Express. 2015;23(4):4255–4276. doi: 10.1364/OE.23.004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toprak E, et al. Defocused orientation and position imaging (DOPI) of myosin V. Proc Natl Acad Sci USA. 2006;103(17):6495–6499. doi: 10.1073/pnas.0507134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguet F, Geissbühler S, Märki I, Lasser T, Unser M. Super-resolution orientation estimation and localization of fluorescent dipoles using 3-D steerable filters. Opt Express. 2009;17(8):6829–6848. doi: 10.1364/oe.17.006829. [DOI] [PubMed] [Google Scholar]
- 30.Mortensen KI, Churchman LS, Spudich JA, Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat Methods. 2010;7(5):377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg SA, Quinlan ME, Forkey JN, Goldman YE. Rotational motions of macro-molecules by single-molecule fluorescence microscopy. Acc Chem Res. 2005;38(7):583–593. doi: 10.1021/ar040137k. [DOI] [PubMed] [Google Scholar]
- 32.Ohmachi M, et al. Fluorescence microscopy for simultaneous observation of 3D orientation and movement and its application to quantum rod-tagged myosin V. Proc Natl Acad Sci USA. 2012;109(14):5294–5298. doi: 10.1073/pnas.1118472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sergé A, Bertaux N, Rigneault H, Marguet D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods. 2008;5(8):687–694. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 34.Flors C, Ravarani CNJ, Dryden DTF. Super-resolution imaging of DNA labelled with intercalating dyes. ChemPhysChem. 2009;10(13):2201–2204. doi: 10.1002/cphc.200900384. [DOI] [PubMed] [Google Scholar]
- 35.Larsson A, Carlsson C, Jonsson M, Albinssont B. Characterization of the binding of the fluorescent dyes YO and YOYO to DNA by polarized light spectroscopy. J Am Chem Soc. 1994;116(19):8459–8465. [Google Scholar]
- 36.Mameren-Schotvanger J. 2008 Integrating single-molecule visualization and DNA micromanipulation. Available at dare.ubvu.vu.nl/handle/1871/15645. Accessed January 5, 2015.
- 37.Oda T, Namba K, Maéda Y. Position and orientation of phalloidin in F-actin determined by X-ray fiber diffraction analysis. Biophys J. 2005;88(4):2727–2736. doi: 10.1529/biophysj.104.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminski Schierle GS, et al. In situ measurements of the formation and morphology of intracellular β-amyloid fibrils by super-resolution fluorescence imaging. J Am Chem Soc. 2011;133(33):12902–12905. doi: 10.1021/ja201651w. [DOI] [PubMed] [Google Scholar]
- 39.Dolat L, et al. Septins promote stress fiber-mediated maturation of focal adhesions and renal epithelial motility. J Cell Biol. 2014;207(2):225–235. doi: 10.1083/jcb.201405050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.