Significance
Cyclic di-GMP (cdiG) is an important bacterial signaling molecule because it regulates motility and affects surface colonization and biofilm formation. It has long been established that cdiG is made by GGDEF enzymes, which are named after five conserved amino acids in the catalytic domain. However, a major group of bacteria, proteobacteria, have a high abundance of these enzymes, which raises the possibility that some of these enzymes have alternate functions. This study details the discovery of GGDEF enzymes that can also make cyclic AMP-GMP (cAG), a recently identified signaling molecule that regulates surface sensing and attachment in pathogenic and environmental bacteria. This provides the first evidence to our knowledge that GGDEF enzymes can make alternative cyclic dinucleotides to cdiG and that cAG is more widespread in proteobacteria.
Keywords: bacterial signaling, surface sensing, second messengers, cyclic dinucleotides, fluorescent biosensor
Abstract
Over 30 years ago, GGDEF domain-containing enzymes were shown to be diguanylate cyclases that produce cyclic di-GMP (cdiG), a second messenger that modulates the key bacterial lifestyle transition from a motile to sessile biofilm-forming state. Since then, the ubiquity of genes encoding GGDEF proteins in bacterial genomes has established the dominance of cdiG signaling in bacteria. However, the observation that proteobacteria encode a large number of GGDEF proteins, nearing 1% of coding sequences in some cases, raises the question of why bacteria need so many GGDEF enzymes. In this study, we reveal that a subfamily of GGDEF enzymes synthesizes the asymmetric signaling molecule cyclic AMP-GMP (cAG or 3′, 3′-cGAMP). This discovery is unexpected because GGDEF enzymes function as symmetric homodimers, with each monomer binding to one substrate NTP. Detailed analysis of the enzyme from Geobacter sulfurreducens showed it is a dinucleotide cyclase capable of switching the major cyclic dinucleotide (CDN) produced based on ATP-to-GTP ratios. We then establish through bioinformatics and activity assays that hybrid CDN-producing and promiscuous substrate-binding (Hypr) GGDEF enzymes are found in other deltaproteobacteria. Finally, we validated the predictive power of our analysis by showing that cAG is present in surface-grown Myxococcus xanthus. This study reveals that GGDEF enzymes make alternative cyclic dinucleotides to cdiG and expands the role of this widely distributed enzyme family to include regulation of cAG signaling.
Four cyclic dinucleotides (CDNs) have been discovered to date and are found primarily in bacteria, although recent examples in eukaryotes have further raised the profile of this class of signaling molecules (1–5). Cyclic di-GMP (cdiG), which has been studied for over 30 years, is the most well-characterized CDN and is primarily associated with the transition from the motile planktonic to sessile biofilm-forming bacterial lifestyles (1, 2, 6). It was first discovered by Benziman and colleagues as a regulator of cellulose synthase in Gluconacetobacter xylinus (6). Synthesis of cdiG was later associated with the GGDEF domain (7, 8), which is found in 75% of bacterial species (9) and is named after five consecutive amino acids conserved in the catalytic domain. On the other hand, cyclic AMP-GMP (cAG or 3′, 3′-cGAMP) was first discovered as the product of the enzyme DncV in the El Tor strain of Vibrio cholerae (10). It is a regulator of V. cholerae motility and intestinal colonization in mammalian hosts. Other CDNs include cyclic di-AMP (cdiA), which is involved in bacterial cell wall homeostasis and sporulation (3, 11), and 2′, 3′-cyclic AMP-GMP, which is involved in the mammalian innate immune response (12–16).
We and others recently discovered that Geobacter, a genus of deltaproteobacteria, use cAG-sensing riboswitches to regulate genes associated with extracellular electron transfer (17, 18), an extraordinary activity that involves bacterial colonization on metal oxide surfaces (19). We further showed that Geobacter sulfurreducens produces cAG (17), but the synthase enzyme remained a mystery. Geobacter genomes have no homologs to the cAG synthases DncV or cGAS, which harbor oligoadenylate synthase (OAS)-like domains and produce structurally distinct isomers of cAG (3′, 3′-cGAMP and 2′, 3′-cGAMP, respectively) (12–14, 20). However, the cAG-sensing riboswitches in Geobacter gained function via adapting the ligand binding pocket of GEMM-I riboswitches, which typically bind cdiG (21). Thus, we considered that cAG signaling may have evolved in Geobacter by co-opting components from the cdiG signaling pathway.
The G. sulfurreducens genome encodes 29 GGDEF domain-containing enzymes that are assigned as diguanylate cyclases (DGCs) (SI Appendix, Fig. S1). These enzymes comprise nearly 1% of the coding sequences. In contrast, there are two predicted DAC domains, which are assigned as diadenylate cyclases. Whereas the existence of GGDEF domains in a genome is considered sufficient proof for cdiG signaling and has been used to establish the presence of cdiG signaling in the greater majority of sequenced bacterial species (9), the redundancy of GGDEF enzymes could have permitted at least one of these enzymes to evolve new functions. This led us to hypothesize that one or more GGDEF domains had gained cAG synthase activity.
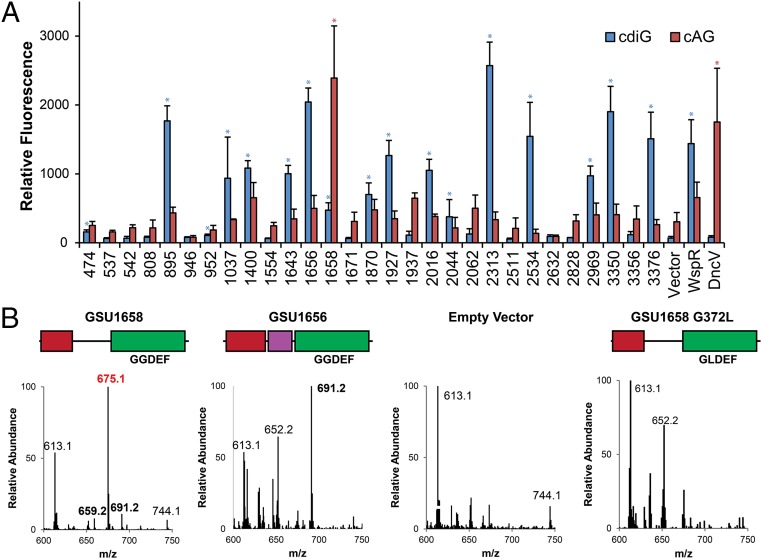
To test this hypothesis, an in vivo flow cytometry screen of all 29 GGDEF enzymes was performed using fluorescent riboswitch-based biosensors that respond selectively to cdiG or cAG (Fig. 1A) (17, 22). We posited that overexpression may drive dimerization of the GGDEF protein, thus enabling enzymatic activity to be assayed even in the absence of activating signal. Sixteen enzymes exhibited significant fluorescence turn-on with the cdiG biosensor and little to no turn-on with the cAG biosensor, in line with the results for WspR, a well-characterized diguanylate cyclase (23, 24). Twelve others exhibited little to no turn-on with both biosensors, indicating that these enzymes were not functional or were poorly expressed under the assay conditions in a heterologous host. However, the GGDEF enzyme encoded by the gene GSU1658 displayed a markedly different signal profile, namely significant fluorescence turn-on with the cAG biosensor and little turn-on with the cdiG biosensor. Similar results were observed for DncV, the cAG synthase from V. cholerae, although DncV and GSU1658 share no sequence homology.
Fig. 1.
In vivo fluorescent biosensor screen of 29 Geobacter GGDEF genes reveals a cAG synthase. (A) Average fluorescence measured by flow cytometry (n = 3; 10,000 cells per run) of E. coli BL21 (DE3) Star cells coexpressing the cdiG-selective biosensor Dp-Spinach2 (blue) or cAG-selective biosensor Gm0970-p1-4delA-Spinach (red) along with GGDEF domain proteins from G. sulfurreducens strain PCA, diguanylate cyclase WspR, cAG synthase DncV, or empty vector. Blue and red stars denote significant (P < 0.01) fluorescence turn-on by Student’s t test above control signal (i.e., significant signal above pCOLA background for the cdiG sensor; above WspR for the cAG sensor). (B) LC/MS analysis of E. coli cell extracts overexpressing constructs shown or empty vector; see SI Appendix, Fig. S1 for protein domain color scheme. Shown are the MS spectra from integrating the retention time region containing all three cyclic dinucleotides. Expected masses are for cdiG (m/z = 691), cAG (m/z = 675), and cdiA (m/z = 659).
To validate the results of the biosensor screen, we performed cell extract analysis of Escherichia coli expressing the candidate cAG synthase GSU1658, a candidate diguanylate cyclase GSU1656, or empty plasmid. LC-MS and MS/MS data showed that E. coli do not inherently produce cAG, but overexpression of GSU1658 leads to high cAG levels (Fig. 1B and SI Appendix, Fig. S2). Furthermore, this activity requires an active GGDEF domain, as no cAG was observed with the GLDEF mutant of GSU1658. In contrast, overexpression of GSU1656 leads to high cdiG levels, but no cAG. Together with the in vivo screening results, these data reveal GSU1658 as a GGDEF enzyme with unprecedented cAG synthase activity.
The cAG synthase DncV has a monomeric protein fold and a single active site with individual binding sites for ATP and GTP (25). Diguanylate cyclases, on the other hand, possess a C2-symmetric active site formed by the homodimeric association of two GGDEF domains, with one GTP bound per monomer (24, 26). To demonstrate that GSU1658 self-associates, we coexpressed C-terminal His- and HA-tagged monomers and showed that pulldowns with the His tag isolated the HA-tagged monomer (SI Appendix, Fig. S3). However, we could not measure a relevant dimerization constant because the wild-type protein is isolated as an auto-inhibited form with cyclic dinucleotide tightly bound to an inhibitory binding site (I-site) in the GGDEF domain (26, 27) (SI Appendix, Figs. S4 and S5). This form is inactive and appears to be mostly monomeric by size-exclusion chromatography (SI Appendix, Fig. S4), which is similar to observations for PleD, a classic diguanylate cyclase whose domain architecture resembles that of GSU1658 (26, 28). Both enzymes contain a response regulator receiver (Rec) domain before the GGDEF domain (29), as well as an I-site (26, 27). We identified and confirmed the I-site in GSU1658 by purifying the I-site mutant R393A, which did not coelute with bound cyclic dinucleotides (SI Appendix, Figs. S4 and S5). Unfortunately, the I-site mutant was largely insoluble, even with addition of a maltose-binding protein (MBP) tag, as it forms higher order aggregates at low micromolar concentrations in vitro, as observed by size exclusion chromatography and isothermal calorimetry experiments (SI Appendix, Figs. S3 and S4). This aggregation phenomenon may be due to misfolding of the R393A variant under in vitro conditions or may be naturally involved in synthase activity, as oligomerization of the diguanylate cyclase WspR has been observed in Pseudomonas aeruginosa to activate enzyme activity (30). In the latter case, WspR oligomerization is controlled by the phosphorylation status of the Rec domain, as opposed to changes in protein expression level. Prior transcriptional profiling of G. sulfurreducens grown under diverse conditions (biofilm, electrode, fumarate/acetate, fumarate/ferric citrate) showed that GSU1658 is constitutively expressed (31), so activation of enzyme dimerization or oligomerization likely occurs through the Rec domain as well.
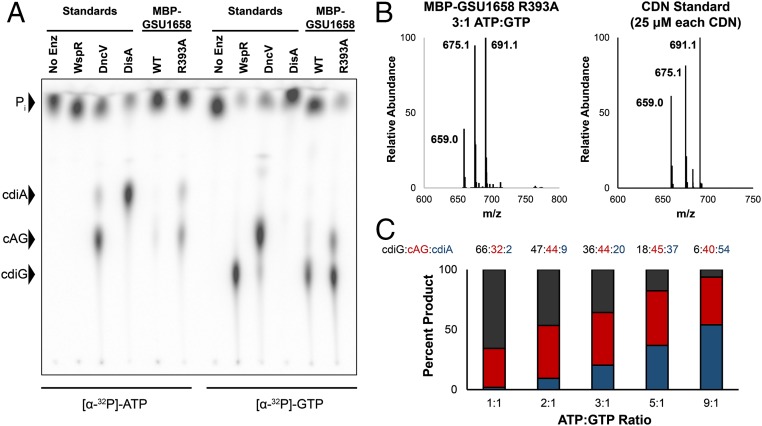
Absence of product bound at the I-site is expected to relieve autoinhibition of the enzyme. To assay GSU1658 activity in vitro, we incubated the enzyme with ATP and GTP doped with trace radiolabeled ATP or GTP and identified the CDNs produced via TLC. This assay allows newly synthesized CDNs to be distinguished from prebound ones. All GGDEF enzymes were analyzed at micromolar concentrations to favor active dimer or oligomer formation. As expected, the CDN-bound WT GSU1658 was less active in vitro than the R393A I-site mutant (Fig. 2A). Unexpectedly, however, both the WT and R393A mutant produced significant levels of cdiA and cdiG in addition to cAG. This finding contradicts the cell lysate data in which cAG was the predominant product. Closer inspection of the biosensor and cell lysate results for WT GSU1658, however, did reveal an increase in cdiG signal, albeit at lower levels than observed in vitro. We realized this product distribution can be explained by the homodimeric structure of the enzyme, if each of the two identical substrate-binding sites are able to recognize both ATP and GTP. Because R393A and WT GSU1658 had similar cell lysate profiles, we concluded that the I-site mutation has no effect on product ratios (SI Appendix, Fig. S6). Because we do not know the native oligomeric state of activated WT GSU1658, we cannot rule out that oligomerization affects product distribution.
Fig. 2.
GSU1658 produces different cyclic dinucleotides depending on ATP-to-GTP ratios. (A) Cellulose TLC analysis of radiolabeled products from enzymatic reactions with 1:1 ATP-to-GTP substrates in excess and doped with trace amounts of α-32P-labeled ATP or α-32P-labeled GTP. Before loading, reactions were quenched with alkaline phosphatase to digest unreacted nucleotides, resulting in production of inorganic phosphate (Pi). Residue R393 is located in the putative I-site. (B) Representative LC/MS analysis of an enzymatic assay performed with MBP-GSU1658 R393A at 3:1 ratio of ATP to GTP in comparison with a standard containing all three product CDNs at equal concentrations. Shown is the MS spectrum from integrating the retention time region containing all three cyclic dinucleotides. Expected masses are for cdiG (m/z = 691), cAG (m/z = 675), and cdiA (m/z = 659). (C) Analysis of product ratios for MBP-GSU1658 R393A at different ATP-to-GTP ratios based on LC/MS analysis and comparison with CDN standard to account for different ionization efficiencies. Average values of two replicate runs are shown.
The discrepancy between in vivo and in vitro results is likely due to two major factors. First, because cdiG is found naturally in E. coli, cdiG-specific phosphodiesterases exist for its breakdown (32). The presence of these phosphodiesterases, and the absence of cdiA- or cAG-specific phosphodiesterases, could have led to the lower observed levels of cdiG in the cell lysate. The enzyme TBD1265, for instance, is a canonical EAL domain-containing phosphodiesterase that is 33-fold more selective for cdiG over cAG, with no activity toward cdiA hydrolysis (33). Furthermore, of the 28 HD-GYP and EAL proteins in V. cholera El Tor, only three HD-GYP enzymes showed cleavage activity for cAG (34).
Second, the in vitro assays were carried out with 1:1 ATP to GTP, but in cells, ATP is usually found in excess relative to GTP. To examine this effect, we incubated R393A GSU1658 with different ratios of ATP to GTP. As we increased ATP relative to GTP, the product ratio skewed toward cAG and cdiA relative to cdiG (Fig. 2 B and C and SI Appendix, Fig. S7). The heterodimeric product, cAG, appeared to be the major product at ATP-to-GTP ratios between 3:1 and 5:1, in accordance with the measured physiological ranges for enterobacteria (between 2:1 and 3:1) (35, 36). Furthermore, it should be noted that published ratios are for total cellular ATP and GTP, whereas the pool of free GTP is likely lower. Taken together, these results reveal that we have discovered a GGDEF enzyme with dinucleotide synthase activity that produces different CDNs depending on the ratio of ATP to GTP.
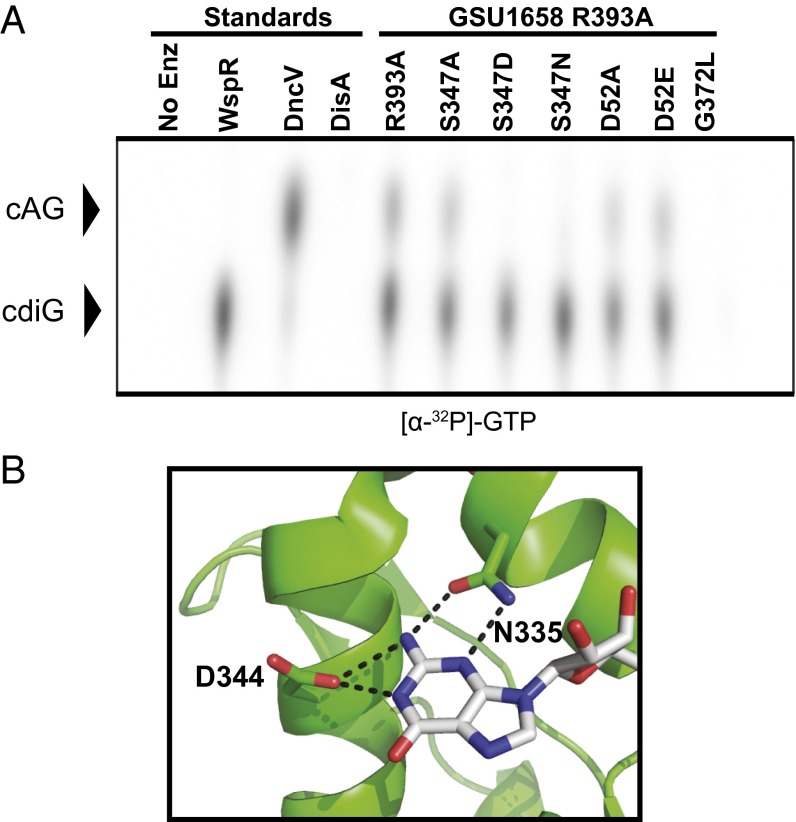
We analyzed several features of GSU1658 by mutational analysis. Phosphorylation of the Rec domain is known to activate homodimer formation of canonical GGDEF enzymes such as PleD (26, 28); however, treatment of GSU1658 with BeF3−, a known phosphomimic compound used to study Rec domains (37), resulted in protein precipitation. Phosphorylation site knock-out (D52A) and mimic (D52E) mutations of GSU1658 in the I-site mutant background had little to no effect on overall enzyme activity (Fig. 3A), and the D52E mutation had little to no effect on product ratios (SI Appendix, Fig. S8). These mutations may not adequately recapitulate the phosphorylated state of the Rec domain in GSU1658 nor the interactions with the cognate histidine kinase, which has not yet been identified.
Fig. 3.
Identification of specificity position in Hypr GGDEF active site. (A) Cellulose TLC of radiolabeled products from enzymatic reactions with 1:1 ATP-to-GTP substrates in excess and doped with trace amounts of α-32P-labeled GTP (full TLC and results for α-32P-labeled ATP are shown in SI Appendix, Fig. S10). Residue R393 is located in the putative I-site, S347 is located in the nucleotide binding site, and D52 is the putative phosphorylation site in the Rec domain. (B) Nucleotide binding region of PleD in complex with nonhydrolyzable GTP analog (Protein Data Bank 2V0N, ref. 26). Hydrogen bonding contacts between the guanine base and key protein residues are shown as dotted lines.
Sequence alignment and analysis of the X-ray crystal structure of PleD (26) revealed that GSU1658 may harbor a Ser residue (S347) in place of the Asp residue (D344) that interacts with the nucleobase of the GTP substrate (Fig. 3B and SI Appendix, Fig. S9). The ability of the side chain hydroxyl to serve as either hydrogen bond donor or acceptor could allow both GTP and ATP to serve as substrates (SI Appendix, Fig. S9). Thus, we analyzed mutations at this position in the substrate binding pocket of GSU1658 in the I-site mutant background.
Interestingly, the S347A mutant maintained dinucleotide cyclase activity (Fig. 3A and SI Appendix, Fig. S10), so the Ser side chain is not strictly necessary for recognition of ATP or GTP. However, the S347D mutant only makes cdiG, so the Asp side chain restores specificity for GTP. The majority (99%) of annotated GGDEF enzymes harbor an Asp at this position, as do the canonical diguanylate cyclases PleD and WspR.
Other natural variant residues identified by bioinformatics analysis were analyzed by TLC or cell lysate experiments (Fig. 3A and SI Appendix, Fig. S10). In support of our model for side chain–nucleobase interactions, the S347N mutant makes cdiG and the S347T mutant makes all three bacterial CDNs (Fig. 3A and SI Appendix, Figs. S10 and S11). These data confirm that this position strongly influences recognition of ATP or GTP. However, the protein background also appears important, as the inverse D344S mutant of PleD was inactive (SI Appendix, Fig. S12), rather than making all three CDNs as predicted. Likewise, the inverse D-to-S mutations of several Geobacter diguanylate cyclases (GSU1400, GSU2313, and GSU2534) resulted in enzyme inactivation, whereas the GSU3350 mutant retained diguanylate cyclase activity (SI Appendix, Fig. S13). The inability of these inverse mutations to generate a cAG synthase may be due to Ser having a shorter side chain than Asp, so these diguanylate cyclases may not bring the residue in close enough proximity to engage with the substrate. The binding pocket of GSU1658 appears to be more plastic, as it remains active while accommodating both larger (S347D) and smaller (S347A) residues.
We further found that replacing the full GGDEF domain of PleD with the one from GSU1658 generates a functional dinucleotide cyclase (SI Appendix, Fig. S12). However, the ratio of cdiG to cAG produced by the protein chimera is higher than for GSU1658, which suggests that the difference in conformation between the chimeric and natural homodimers may influence product ratios. Thus, we have identified that a natural amino acid variation in GGDEF domains can change the specificity of the enzyme and showed that features outside of the GGDEF domain also affect product ratios.
Whereas previous GGDEF enzymes were uniformly assigned as DGCs if they have the active motif [G/A/S]G[D/E]E[F/Y] (2), our results reveal that GGDEF enzymes are a family of dinucleotide cyclases, in which DGCs are the major subfamily. GSU1658 is the founding member of a distinct subfamily of GGDEF enzymes that make hybrid CDNs and are promiscuous for ATP and GTP substrates. Thus, we propose that this newfound subfamily be called hybrid, promiscuous (Hypr) GGDEF enzymes, and that GSU1658 be renamed HyprA.
To survey this newfound Hypr subfamily, we performed a bioinformatics analysis of 32,587 predicted active GGDEF enzymes to identify sequences that harbor the D-to-S or D-to-T variation at the specificity position (SI Appendix, Table S1). These two variants, which we predict give rise to Hypr activity, are rare and comprise only 0.17% of all GGDEF domains. All sequenced Geobacter and Pelobacter species have at least one Hypr enzyme (SI Appendix, Fig. S9) and have riboswitch effectors that regulate genes in response to cAG (17, 18). A second Hypr is predicted in G. sulfurreducens, but GSU1937 was not active in the biosensor-based screen (Fig. 1A). This protein has a predicted transmembrane HAMP domain, so it likely requires membrane insertion for activity and was inactive when expressed heterologously.
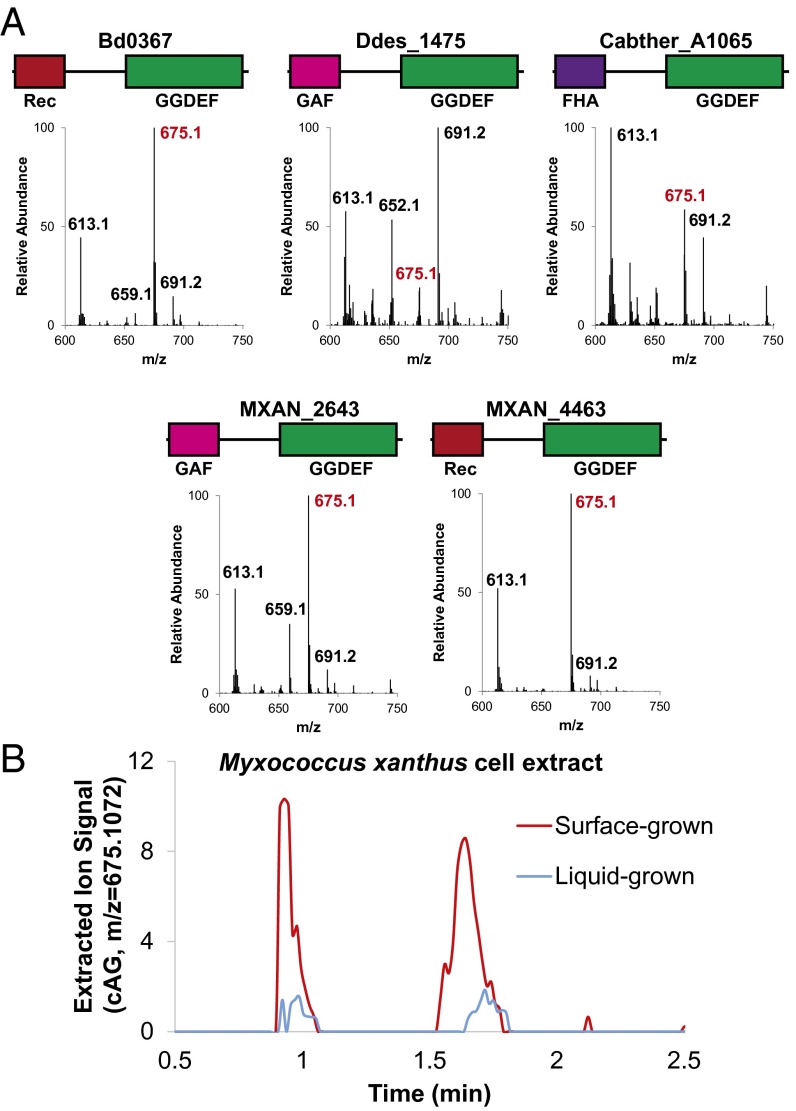
Intriguingly, bacteria that do not harbor cAG-selective riboswitches also appear to encode candidate Hypr enzymes in their genomes (SI Appendix, Table S1). For example, Myxococcus xanthus, a social bacterium that serves as a model for studying group behavior (38) and cell–cell communication (39), harbors one protein homologous to HyprA (MXAN_4463) and another protein with a GAF sensory domain preceding the Hypr GGDEF catalytic domain (MXAN_2643, renamed HyprB). Bd0367 (also called DgcA) from Bdellovibrio bacteriovorus, an intracellular pathogen of other bacteria described as a “living antibiotic,” has a similar Rec-GGDEF architecture to HyprA and has been shown to regulate gliding motility and prey escape, a phenotype distinct from those controlled by the other three GGDEF enzymes (40).
These three and other candidate Hypr enzymes were cloned into E. coli and analyzed by cell extraction followed by LC-MS. In all tested cases that expressed well (SI Appendix, Fig. S14), we observed the production of cAG, although to varying levels (Fig. 4). In contrast, overexpression of PleD or WspR, two well-characterized diguanylate cyclase GGDEFs, led to production of cdiG only. The cAG-to-cdiG ratios are different between Hypr enzymes, which reinforces that the Ser specificity residue is not the sole determinant of product ratios. The majority of the Hypr enzymes we tested preferentially make cAG under in vivo conditions in which ATP is in excess. Furthermore, we recapitulated that I-site mutants of the M. xanthus and B. bacteriovorus HyprA enzymes produce cAG in vitro in a similar fashion to GSU1658 at 1:1 ATP-to-GTP ratios (SI Appendix, Fig. S14).
Fig. 4.
Validation of Hypr activity in select Deltaproteobacteria and Acidobacteria. (A) LC/MS analysis of E. coli cell extracts overexpressing candidate Hypr enzymes; see SI Appendix, Fig. S1 for protein domain color scheme and SI Appendix, Fig. S12 for corresponding protein gel. Bd, Bdellovibrio bacteriovorus; Ddes, Desulfovibrio desulfuricans; Mxan, Myxococcus xanthus; Cabther, Candidatus Chloracidobacterium thermophilum. Shown are the MS spectra from integrating the retention time region containing all three cyclic dinucleotides. Expected masses are for cdiG (m/z = 691), cAG (m/z = 675), and cdiA (m/z = 659). (B) LC/MS analysis of M. xanthus cell extracts from surface- or liquid-grown samples. Shown is the extracted ion trace for cAG (m/z = 675.1072; ppm < 10 cutoff) normalized to the weight of extracted cells. A second biological replicate is shown in SI Appendix, Fig. S15.
Our biochemical validation of cAG synthase activity for diverse Hypr GGDEFs strongly predicted that cAG was an endogenous signaling molecule in these other organisms. We initially found cdiG but not cAG in cell extracts of wild-type M. xanthus cultured in solution. However, based on the involvement of cAG signaling in processes related to surface sensing (intestinal colonization for V. cholerae and extracellular electron transfer for Geobacter), we hypothesized that cAG was similarly associated with M. xanthus growth on solid surfaces. Lysis conditions first required optimization to account for higher exopolysaccharide (EPS) content in surface-grown cells, which otherwise impeded successful extraction of all cyclic dinucleotides. This method was used to analyze cyclic dinucleotide content of M. xanthus grown in solution versus on 1.5% agar and revealed that cAG is produced at higher levels upon surface growth (Fig. 4 and SI Appendix, Fig. S15). Positive identification of cAG was confirmed by multiple methods, including comparison of HRMS and tandem MS/MS spectra and S1 nuclease digestion profile to synthetic standards (SI Appendix, Figs. S15–S17).
These results provide the first evidence to our knowledge for cAG signaling in myxobacteria and are in line with the conservation of HyprA and HyprB across all Myxococcales species. In particular, we showed that cAG levels are modulated by solution versus solid growth conditions. This correlates with a proposed role in surface sensing, which provides an advanced starting point for future phenotypic analysis and efforts to discover other components in the putative cAG signaling pathway. Taken together, our results reveal that Hypr activity is more widespread in bacteria than the distribution of cAG riboswitches, expanding the potential scope of cAG signaling (Fig. 5).
Fig. 5.
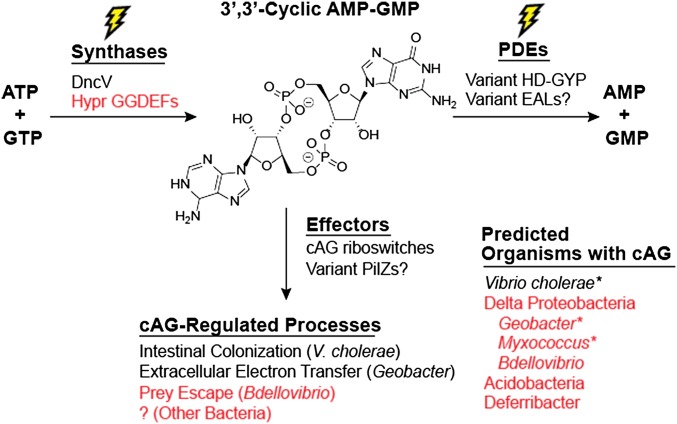
Expanded understanding of the function and evolution of cAG signaling. Levels of the second messenger cAG are regulated by the activity of cAG synthases and phosphodiesterases (PDEs) in response to primary environmental signals. Effectors that bind cAG then propagate downstream effects on bacterial physiology. In V. cholerae, DncV serves as the synthase and variant HD-GYP domains as the phosphodiesterases, but no effectors are known. We have shown that diverse Deltaproteobacteria and Acidobacteria contain Hypr GGDEF enzymes that can act as cAG synthases and control various processes through cAG riboswitches (Geobacter) and other unidentified effectors. An asterisk indicates organisms confirmed to have endogenous cAG. Text in red indicates new information put forth in this paper.
In searching for the enzyme responsible for cAG synthesis in Geobacter, we discovered a subfamily of homodimeric GGDEF proteins that have the capacity to produce all three bacterial cyclic dinucleotides, which we term Hypr GGDEFs. An efficient and selective in vivo fluorescent biosensor assay enabled the discovery of this previously unidentified activity, highlighting RNA-based biosensors as an attractive technology platform for enzyme screening. Alternative screening strategies such as fractionation (15, 16), in vitro enzyme screening (41), or a phenotype-based assay in the native organism (42) are relatively more time and resource intensive. The in vivo biosensor assay is particularly useful for signal transduction enzymes that are activated by dimerization, because this can be mimicked by overexpression in a heterologous host.
Unlike the other cAG synthases, DncV and cGAS, which have individual binding sites for ATP and GTP, Hypr GGDEFs have two symmetrical binding sites that accommodate either ATP or GTP as substrates. Notably, both DncV and cGAS display weak activity for synthesizing cdiG or cdiA in vitro as well, especially in the presence of only one type of substrate (10, 13, 25). However, these enzymes preferentially make cAG (3′,3′ or 2′,3′, respectively) at equimolar concentrations of NTPs, whereas the ability of HyprA to make cAG is tuned by the relative excess of ATP to GTP under physiological conditions.
One outstanding question is whether Hypr GGDEFs are responsible for cAG signaling only or if the ability of these enzymes to produce all three cyclic dinucleotides could be modulated and exploited by bacteria that use them as endogenous second messengers for distinct signaling pathways. Whereas we currently cannot exclude the latter possibility, the former possibility is more straightforward and consistent with current models for cdiG and cdiA signaling pathways, which are composed of separate sets of enzymes and effectors. In Geobacter, we propose that HyprA (GSU1658) and perhaps GSU1937, which also harbors the D-to-S variation, act as cAG synthases in vivo, and activation of these signaling enzymes turn on genes involved in extracellular electron transfer through cAG-specific riboswitches (Fig. 5) (17). The discovery of HyprA brings us closer to elucidating a primary signal that regulates this unique metabolic activity, as this enzyme harbors a Rec domain and thus is presumably part of a two-component system. In general, further work is needed to elucidate the activation mechanisms for Hypr GGDEFs and to address whether Hypr GGDEFs have promiscuous functions.
Through analysis of sequence determinants for HyprA activity and validation of additional Hypr GGDEFs, we predicted that these enzymes are more broadly distributed than cAG riboswitches, which have been the only identified sensors for cAG to date (Fig. 5). Hypr GGDEFs appear highly conserved in the bacterial orders Desulfuromonadales (includes Geobacter), Myxococcales, and Bdellovibrionales. Additionally, a number of bacteria in the Acidobacteria and Deferribacter phyla also contain candidate Hypr GGDEFs. Based on our demonstration of endogenous cAG production in wild-type M. xanthus, we expect that cAG signaling also is present in these other bacteria due to Hypr GGDEF activity, although other components remain to be identified. Another Hypr GGDEF that we showed in vitro is capable of producing cAG is Bd0367 from B. bacteriovorus (Fig. 4). Prior studies highlighted the conundrum for how Bd0367, which was called DgcA, could give rise to a distinct phenotype from the other three GGDEF enzymes, given that the small size of the bacteria “make c-di-GMP spillover unavoidable” (40). Based on our discovery of Hypr activity, one plausible explanation is that this enzyme is a HyprA and synthesizes a different signal than the other GGDEFs. Interestingly, gliding motility in B. bacteriovorus is associated with type IV pili, whose assembly is predicted to be controlled by cAG riboswitches in Geobacter and that is known to play a role in surface-associated twitching motility in M. xanthus (38), suggesting a unified connection between cAG and type IV pili-mediated processes.
Following our original hypothesis that components of the cdiG signaling pathway were co-opted for cAG signaling, we predict that variant PilZ domains and potentially other effectors exist in these cAG-producing organisms. Our results reveal that the large abundance and redundancy of GGDEF genes in bacterial genomes have allowed this enzyme family to diverge and evolve toward new synthase activity; the distribution of HD-GYP and EAL phosphodiesterase domains also are expanded in Deltaproteobacteria (9), and at least one variant HD-GYP domain has been shown to degrade cAG (34). Besides synthesizing cAG, we also showed that GGDEF domains can make cdiA, an activity that had been speculated (43), but only now proven. We conceive that larger distortions of the substrate-binding pocket, including in the signature GGDEF motif, could accommodate synthesis of pyrimidine-containing CDNs, which would expand the palette of CDN signaling molecules in nature.
Materials and Methods
Fluorescent Biosensor Screening Assay.
Chemically competent E. coli BL21 (DE3) Star cells (Life Technologies) were cotransformed with different combinations of biosensor plasmid (pET31b with GM0970 P1-4delA-Spinach or Dp-Spinach2) and enzyme construct plasmid (pCOLA-Duet1, various enzymes). Single colonies from LB/Carb/Kan plates were picked for overnight starter cultures, which were used to inoculate fresh liquid cultures. Cells were grown aerobically to an OD600 ∼0.3, then biosensor and enzyme expression was induced with 1 mM IPTG at 37 °C for 6 h. A total of 2 µL of each culture was diluted into 60 µL of 1× PBS containing 50 µM 3,5-difluoro-4-hydroxybenzylidine imidazolinone (DFHBI). Cellular fluorescence was measured for at least 10,000 cells using a BD Fortessa ×20 flow cytometer with BD FACSDiva software (version 1.0.0.650) located in the Flow Cytometry Core Facility at the University of California at Berkeley. Flow cytometry data were then analyzed by FlowJo (version 10.0.7).
Cell Extraction of E. coli.
Cyclic dinucleotides were extracted as described previously (44), with slight modifications. For full procedure, see SI Appendix.
Cell Extraction of M. xanthus.
The frozen cell pellet was weighed, thawed, resuspended in 2 mL of water and incubated with DNase I (10 mg/mL, Sigma) and lysozyme (10 mg/mL, Sigma) at 37 °C with agitation for 6 h to fragment the exopolysaccharide matrix, resulting in a less viscous cell suspension. Cells were lysed and extracted by addition of acetonitrile (4 mL) and methanol (4 mL). After centrifugation for 45 min at 13,855 × g, the supernatant was carefully removed and stored at −20 °C. Supernatant was evaporated to dryness by rotary evaporation, and the dried material was suspended in 400 µL ddH2O. The extract was filtered through a 3-kDa MWCO Amicon Ultra-4 Protein Concentrator (Millipore) and used immediately or stored at −20 °C.
In Vitro Activity Assay of Dinucleotide Cyclases Using Radiolabeled NTPs.
In vitro activity assays were performed as previously described by Kranzusch et al., with slight modifications (20). For full procedure, see SI Appendix.
Additional data and supplementary methods are available in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge Anthony T. Iavarone from the QB3/Chemistry Mass Spectrometry Facility for assistance with mass spectrometry experiments and Yichi Su for helpful discussion of the manuscript. This work was supported in part by NIH Grant DP2 OD008677 (to M.C.H.) and a National Science Foundation graduate fellowship (to Z.F.H.). M.C.H. holds a Career Award at the Scientific Interface (CASI) from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515287113/-/DCSupplemental.
References
- 1.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corrigan RM, Gründling A. Cyclic di-AMP: Another second messenger enters the fray. Nat Rev Microbiol. 2013;11(8):513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 4.Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154(5):962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488(7413):680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325(6101):279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 7.Ausmees N, et al. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett. 2001;204(1):163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 8.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187(5):1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seshasayee ASN, Fraser GM, Luscombe NM. Comparative genomics of cyclic-di-GMP signalling in bacteria: Post-translational regulation and catalytic activity. Nucleic Acids Res. 2010;38(18):5970–5981. doi: 10.1093/nar/gkq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witte G, Hartung S, Büttner K, Hopfner K-P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30(2):167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Reports. 2013;3(5):1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellenberger CA, et al. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci USA. 2015;112(17):5383–5388. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson JW, et al. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci USA. 2015;112(17):5389–5394. doi: 10.1073/pnas.1419264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435(7045):1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 20.Kranzusch PJ, Lee AS-Y, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Reports. 2013;3(5):1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren A, et al. Structural basis for molecular discrimination by a 3′,3′-cGAMP sensing riboswitch. Cell Reports. 2015;11(1):1–12. doi: 10.1016/j.celrep.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J Am Chem Soc. 2013;135(13):4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102(40):14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De N, et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6(3):0601–0617. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kranzusch PJ, et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell. 2014;158(5):1011–1021. doi: 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan C, et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101(49):17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christen B, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281(42):32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 28.Wassmann P, et al. Structure of BeF3- -modified response regulator PleD: Implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15(8):915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 29.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47(6):1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 30.Huangyutitham V, Güvener ZT, Harwood CS. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio. 2013;4(3):e00242–e13. doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu Y, et al. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res. 2010;20(9):1304–1311. doi: 10.1101/gr.107540.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38(1):128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanahan CA, Gaffney BL, Jones RA, Strobel SA. Identification of c-di-GMP derivatives resistant to an EAL domain phosphodiesterase. Biochemistry. 2013;52(2):365–377. doi: 10.1021/bi301510v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J, et al. Identification and characterization of phosphodiesterases that specifically degrade 3′3′-cyclic GMP-AMP. Cell Res. 2015;25(5):539–550. doi: 10.1038/cr.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257(16):9759–9769. [PubMed] [Google Scholar]
- 36.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190(2):718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan D, et al. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc Natl Acad Sci USA. 1999;96(26):14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet. 2011;45(1):21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassallo C, et al. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci USA. 2015;112(22):E2939–E2946. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobley L, et al. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 2012;8(2):e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110(22):9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lori C, et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523(7559):236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 43.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9(12):834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spangler C, Böhm A, Jenal U, Seifert R, Kaever V. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods. 2010;81(3):226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.