Significance
Acyltransferases (ATs) are responsible for the selection and incorporation of acyl building blocks in the biosynthesis of various polyketide natural products. Proper protein–protein interactions between AT and cognate acyl carrier protein (ACP) are critical for the functional transfer of acyl groups. However, the ACP recognition mechanism has remained elusive because the structural determination of an AT–ACP complex is hampered by the weak and transient interactions between them. Herein, we describe the first crystal structure of the AT–ACP complex. To stabilize the weak protein–protein interaction sufficiently for analysis, we prepared a covalent AT–ACP complex using a cross-linking reagent for crystallization. The determined AT–ACP complex structure provides detailed mechanistic insights into ACP recognition by AT.
Keywords: polyketide, acyltransferase, acyl carrier protein, protein–protein interaction, cross-linking
Abstract
Acyltransferases (ATs) are key determinants of building block specificity in polyketide biosynthesis. Despite the importance of protein–protein interactions between AT and acyl carrier protein (ACP) during the acyltransfer reaction, the mechanism of ACP recognition by AT is not understood in detail. Herein, we report the crystal structure of AT VinK, which transfers a dipeptide group between two ACPs, VinL and VinP1LdACP, in vicenistatin biosynthesis. The isolated VinK structure showed a unique substrate-binding pocket for the dipeptide group linked to ACP. To gain greater insight into the mechanism of ACP recognition, we attempted to crystallize the VinK–ACP complexes. Because transient enzyme–ACP complexes are difficult to crystallize, we developed a covalent cross-linking strategy using a bifunctional maleimide reagent to trap the VinK–ACP complexes, allowing the determination of the crystal structure of the VinK–VinL complex. In the complex structure, Arg-153, Met-206, and Arg-299 of VinK interact with the negatively charged helix II region of VinL. The VinK–VinL complex structure allows, to our knowledge, the first visualization of the interaction between AT and ACP and provides detailed mechanistic insights into ACP recognition by AT.
Polyketide synthases (PKSs) are multifunctional enzymes responsible for the biosynthesis of various polyketide natural products (1). Bacterial modular PKSs comprise several catalytic modules that are each responsible for a single round of the polyketide chain elongation reaction. Each module minimally consists of a ketosynthase (KS) domain, an acyltransferase (AT) domain, and an acyl carrier protein (ACP) domain. The AT domain recognizes a specific acyl building block and catalyzes its transfer reaction onto the 4′-phosphopantetheine arm of the ACP. KS extends the polyketide chain by condensing the resulting ACP-bound building blocks with the elongated acyl–ACPs. Although standard modular PKSs contain AT domains in their modules, some modular PKSs lack AT domains in each module and instead receive their acyl building blocks by standalone trans-acting ATs (2).
The selection of the starter unit is generally governed by the substrate specificity of the AT domain in the loading module (1). In some modular PKS systems, a didomain-type loading module comprising a loading AT domain and an ACP domain selects an acyl starter building block such as an acetate unit to generate an acyl–ACP intermediate, which is transferred to the downstream extension module for polyketide chain elongation. Alternatively, in a KSQ-type loading module consisting of three domains, the AT domain selects an α-carboxyacyl substrate such as a malonyl group, and the KSQ domain subsequently catalyzes its decarboxylation to construct an acyl–ACP thioester. For polyketide chain elongation, the AT domain of the extension module generally recognizes a specific α-carboxyacyl–CoA as an extender building block (3). Malonyl– and methylmalonyl–CoA are commonly used as extender building blocks in biosynthetic pathways. Some ATs were reported to recognize ACP-bound substrates such as methoxymalonyl–, hydroxymalonyl–, and aminomalonyl–ACP (3, 4). Thus, ATs are key determinants of building block specificity in polyketide biosynthesis and attractive targets to change the substrate specificity to obtain biologically active unnatural polyketide products (5). However, the substitution of an AT domain by a homologous AT domain possessing different substrate specificity resulted in reduced or abolished production of polyketide analogs in many cases, probably because of disruption of proper protein–protein interactions or the inability of downstream modules to process polyketide analogs (5, 6).
The importance of protein–protein interaction between AT and ACP during the acyltransfer reaction was proposed in previous studies (7, 8). Wong et al. described that AT recognizes its cognate ACP from other ACPs through protein–protein interactions (7). Proper AT–ACP interactions are believed to be essential for kinetically efficient polyketide chain elongation. However, the mechanism of ACP recognition is not well understood because isolated AT structures provide no detailed information on the AT–ACP interactions (9). Structural determination of the AT–ACP complex is necessary for the complete understanding of the basis of ACP recognition for the acyltransfer reaction.
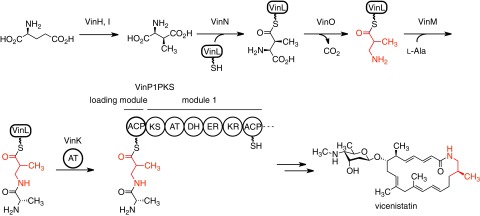
Macrolactam antibiotics are an important class of macrocyclic polyketides, and most contain a unique β-amino acid starter unit in their polyketide skeletons (10). Vicenistatin, produced by Streptomyces halstedii HC34, possesses a 3-aminoisobutyrate unit at the starter position of the polyketide backbone (11). This starter unit is biosynthesized from l-glutamate via (2S,3S)-3-methylaspartate, which is initially transferred onto the standalone ACP VinL by the adenylation enzyme VinN (12, 13). After decarboxylation, the resulting 3-aminoisobutyrate unit is aminoacylated with l-alanine to give dipeptidyl–VinL by another adenylation enzyme VinM. Then, the dipeptidyl moiety is transferred from VinL to the ACP domain (VinP1LdACP) of the VinP1PKS loading module by the trans-acting AT VinK (Fig. 1). These β-amino acid carrying enzymes are conserved in various macrolactam polyketide biosynthetic gene clusters, suggesting that β-amino acid starter units are loaded to PKS through the same mechanism in their biosynthesis (10).
Fig. 1.
Biosynthetic pathway of vicenistatin, including the VinK reaction. The 3-aminoisobutyrate unit is shown in red.
During the dipeptide transfer reaction from VinL to VinP1LdACP, VinK is supposed to recognize the VinL region as well as the dipeptidyl moiety to overcome the kinetic disadvantage of the diffusion-controlled limit. Additionally, VinK should distinguish VinP1LdACP as an acyl acceptor from other ACPs. Thus, we assume that the specific protein–protein interaction between VinK and two ACPs is important for the reaction. However, the origins of ACP selectivity cannot be predicted from the amino acid sequence of VinK. In this study, we carried out mutational and structural studies on VinK to clarify how VinK recognizes ACPs. The covalent VinK–VinL complex structure allows, to our knowledge, the first visualization of the interactions between AT and ACP and provides detailed mechanistic insights into ACP recognition by AT.
Results
Crystal Structure of VinK.
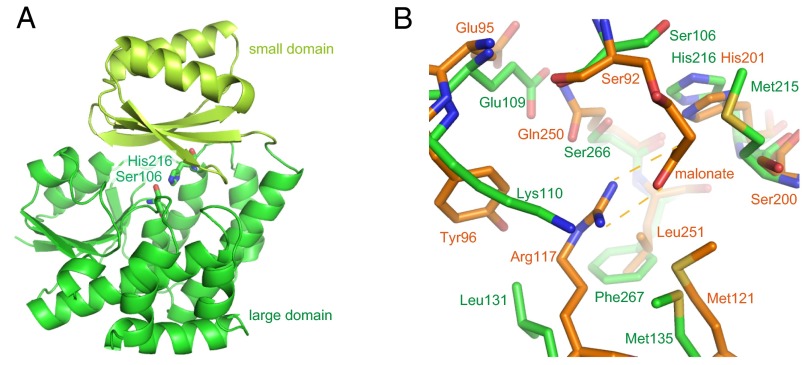
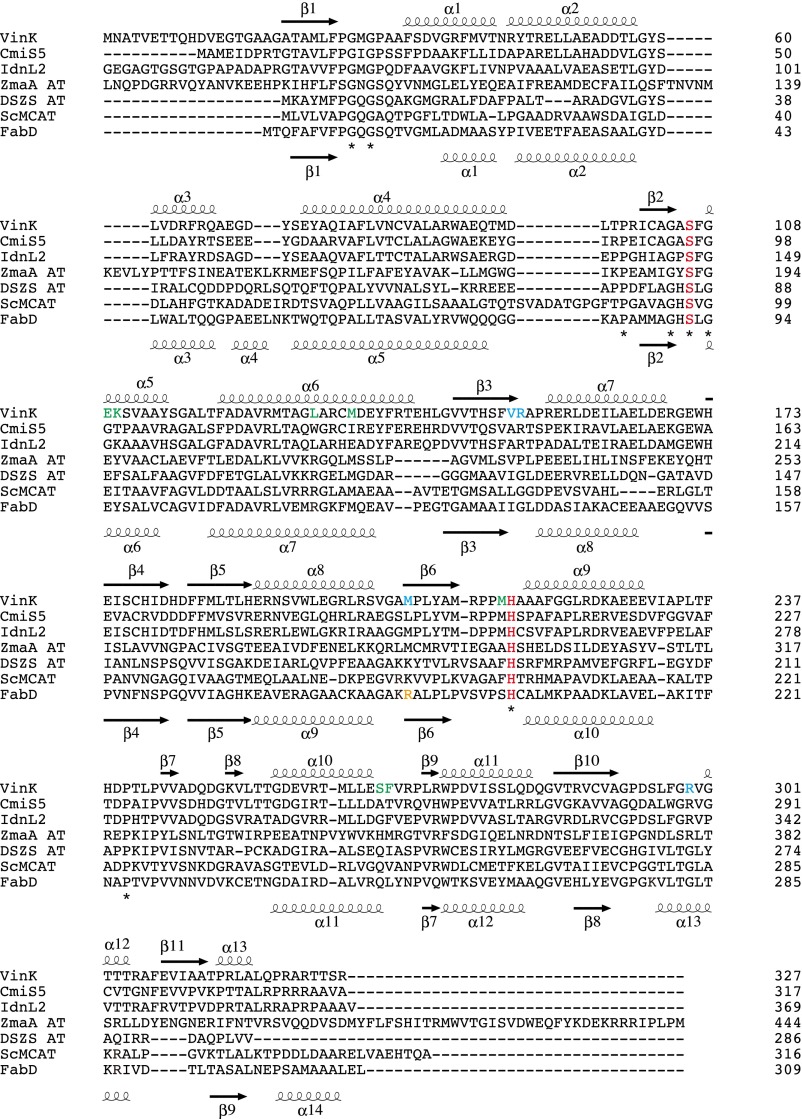
To obtain the structural basis of substrate and ACP recognition, we first determined the VinK crystal structure at 1.80-Å resolution (Table S1). VinK is organized into two domains, including a large domain (residues 19–143 and 214–319) and a small domain (residues 144–213) (Fig. 2A). The overall architecture of VinK is similar to those of other structurally characterized ATs including FabD [Protein Data Bank (PDB) ID code 2G2Z; 25% sequence identity; 3.0-Å root mean square deviation (rmsd) for Cα atoms], which is a discrete malonyl–CoA:ACP transacylase (MCAT) of Escherichia coli fatty acid synthase (FAS) (14), malonyl–CoA-specific trans-acting AT (PDB ID code 3RGI; 21% sequence identity; 2.6 Å rmsd for Cα atoms) from disorazole synthase (DSZS) (15), and ZmaA AT (PDB ID code 4QBU; 18% sequence identity; 3.6 Å rmsd for Cα atoms), which recognizes hydroxymalonyl–ACP as a substrate (4).
Table S1.
Data collection and refinement statistics
| Datasets | VinK apo | VinK–VinL complex | VinK SeMet* |
| PDB ID | 5CZC | 5CZD | |
| Data collection statistics | |||
| Beamline | PF BL-5A | PF AR-NE3A | PF AR-NW12A |
| Wavelength, Å | 1.00000 | 1.00000 | 0.97922 |
| Space group | P21 | P212121 | P21 |
| Unit-cell parameters | |||
| a, Å | 45.52 | 44.14 | 45.38 |
| b, Å | 74.81 | 68.23 | 75.27 |
| c, Å | 84.81 | 148.40 | 85.33 |
| α, deg | 90.00 | 90.00 | 90.00 |
| β, deg | 91.24 | 90.00 | 90.83 |
| γ, deg | 90.00 | 90.00 | 90.00 |
| Resolution, Å | 45.51–1.80 | 68.23–2.34 | 50.00–2.46 |
| (outer shell) | (1.90–1.80) | (2.46–2.34) | (2.50–2.46) |
| Unique reflections | 52,383 (7,647) | 19,598 (2,747) | 20,225 (1,005) |
| Redundancy | 3.3 (3.2) | 6.3 (5.4) | 3.6 (3.5) |
| Completeness, % | 99.3 (99.3) | 99.3 (97.4) | 96.5 (94.4) |
| Rmerge, % | 10.4 (49.0) | 14.0 (75.8) | 6.7 (11.2) |
| Mean <I/σ(I)> | 7.8 (2.2) | 9.1 (2.1) | 25.6 (9.9) |
| Refinement statistics | |||
| Rfac, % | 16.6 | 20.2 | — |
| Rfree, % | 21.6 | 27.3 | — |
| rmsd | |||
| Bond lengths, Å | 0.018 | 0.013 | — |
| Bond angles, deg | 1.827 | 1.627 | — |
| Average B factors, Å2 | |||
| VinK | 22.6 | 43.2 | — |
| VinL | — | 46.6 | — |
| 4′-phosphopantetheine | — | 54.7 | — |
| BMOE | — | 87.2 | — |
| Solvent | 30.9 | 40.1 | — |
| Ramachandran plot | |||
| Favored region, % | 97.8 | 96.0 | — |
| Allowed region, % | 2.0 | 4.0 | — |
| Outer region, % | 0.2 | 0.0 | — |
VinK SeMet derivative data were used only for initial phase determination.
Fig. 2.
Structure of VinK. (A) Overall structure of VinK. The large and small domains are shown in green and yellow-green, respectively. The catalytic residues are shown as sticks. (B) The structure of the substrate-binding pocket. The superimposed structures of VinK (green) and FabD (orange; PDB ID code 2G2Z) are shown. In the FabD structure, the malonyl moiety is covalently bound to the catalytic Ser-92.
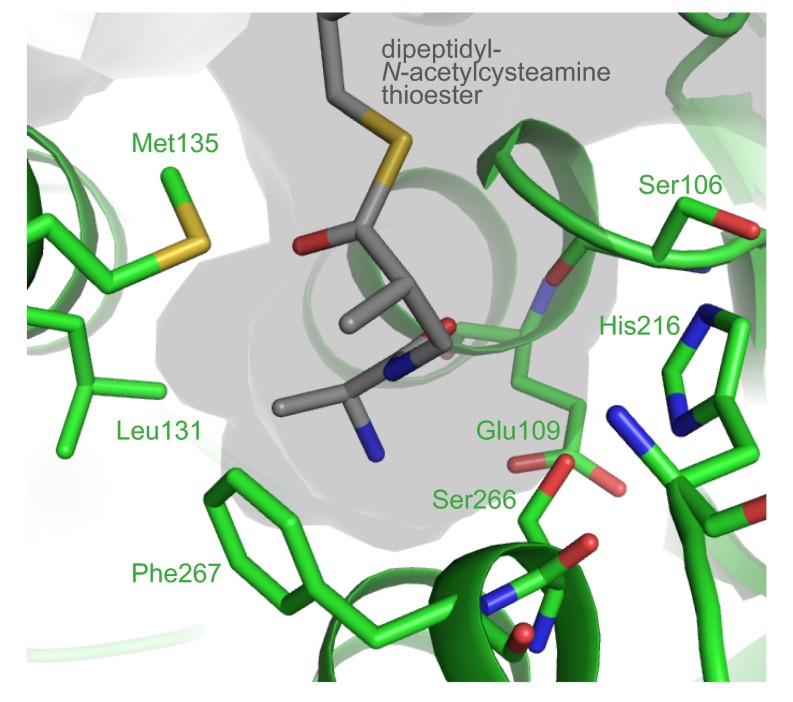
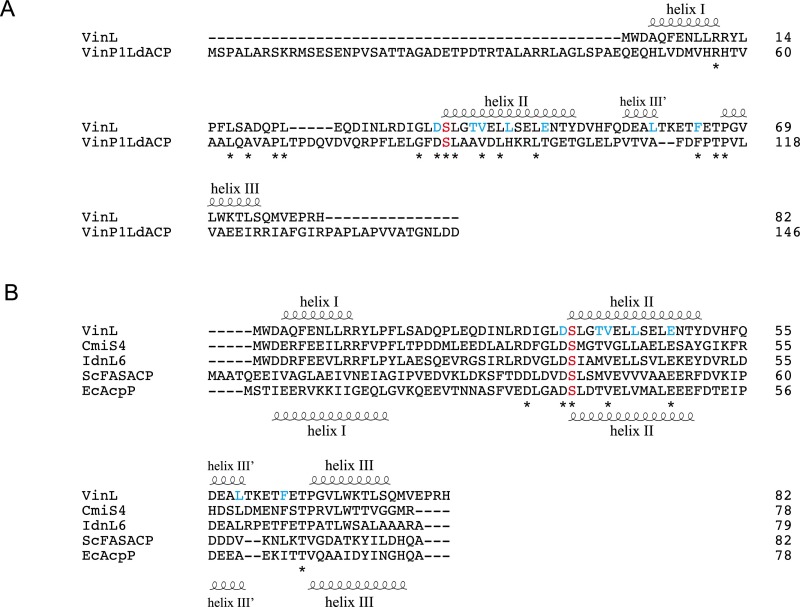
VinK has two conserved catalytic residues, Ser-106 and His-216, at the interface between the large domain and small domain as observed in other ATs (Fig. 2A and Fig. S1). The substrate-binding cavity of VinK is constructed by residues Glu-109, Lys-110, Leu-131, Met-135, Met-215, Ser-266, and Phe-267, which are located in the large domain (Fig. 2B and Fig. S2). FabD has an Arg-117 at the substrate-binding cavity for the recognition of the carboxyl group of the malonate substrate (14), whereas VinK has a Leu-131 at the corresponding position (Fig. 2B and Fig. S1). This Leu-131 residue swings away from the catalytic Ser-106 and appears to provide the space for the bulky dipeptidyl moiety. Thus, VinK seems to have a larger substrate-binding tunnel than that of malonyl–CoA-specific ATs.
Fig. S1.
Amino acid sequence alignment of VinK with other VinK-like proteins involved in the macrolactam biosynthesis (CmiS5 involved in the biosynthesis of cremimycin from Streptomyces sp. MJ635-86F5 and IdnL2 involved in the biosynthesis of incednine from Streptomyces sp. ML694-90F3) and other related AT proteins (ZmaA AT involved in the biosynthesis of zwittermicin A from Bacillus cereus UW85, DSZS AT involved in the biosynthesis of disorazole from Sorangium cellulosum, ScMCAT involved in the biosyntheses of actinorhodin and fatty acid from Streptomyces coelicolor, and FabD involved in the biosynthesis of fatty acid from E. coli). The secondary structural elements of VinK and FabD are indicated above and below the sequences, respectively. Catalytic residues are shown in red. The residues of substrate-binding pocket in VinK are shown in green. Arg-153, Met-206, and Arg-299 of VinK are shown in cyan. FabD Arg-190 for the interaction with the phosphate group of CoA ribose moiety is shown in orange.
Fig. S2.
The structure of substrate-binding pocket of VinK. The docked dipeptidyl-N-acetylcysteamine thioester molecule is shown in gray.
Cross-Linking Reaction to Obtain VinK–ACP Complexes.
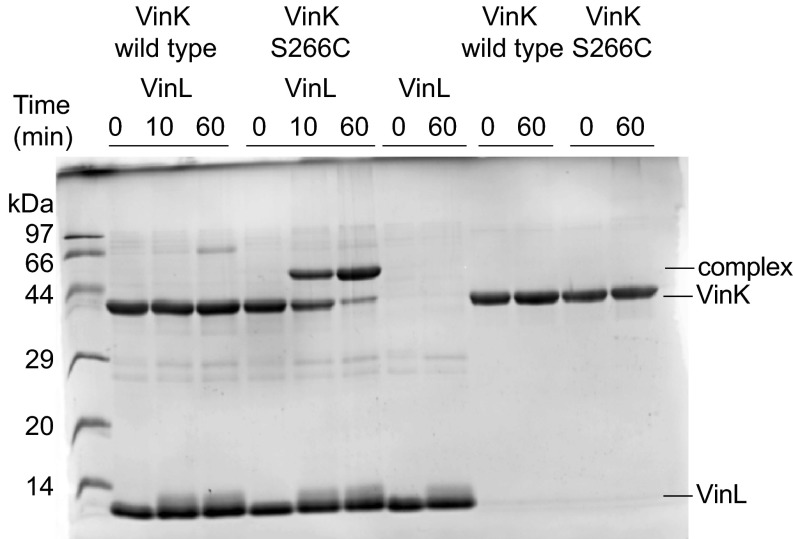
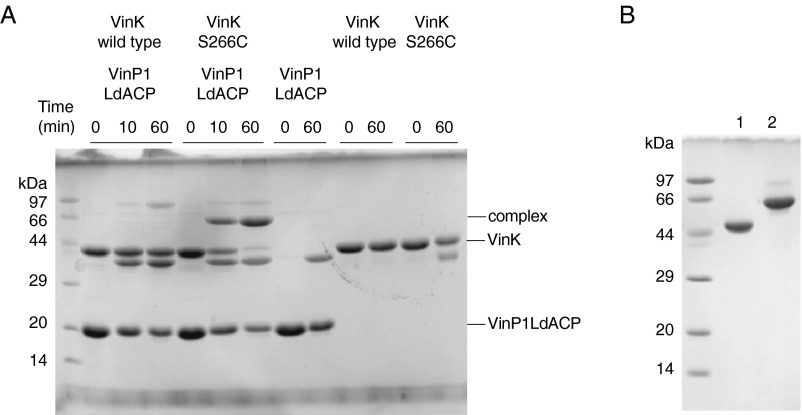
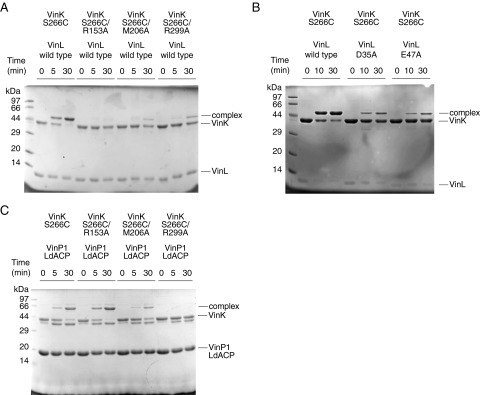
To gain greater insight into how VinK recognizes ACPs during the reaction, we attempted to crystallize VinK–VinL and –VinP1LdACP complexes. Because transient enzyme–ACP complexes are generally difficult to crystallize, we designed a site-specific covalent cross-linking method using bifunctional maleimide reagents for trapping the VinK–ACP complexes. We introduced the Cys mutation at the position of Ser-266, which is a noncatalytic residue at the base of the substrate-binding tunnel in VinK (Fig. 2B) and then used this S266C mutant for cross-linking to the 4′-phosphopantetheine arm of ACPs. The cross-linking reaction between the VinK S266C mutant and VinL in the presence of 1,2-bismeleimidoethane (BMOE) gave a covalent complex in sufficient yield (Fig. 3). In contrast, no apparent cross-linking was observed between the VinK wild type and VinL. This result suggested that the specific cross-link formation occurred at the substrate-binding pocket of the VinK S266C mutant. Similarly, the cross-linking reaction between the VinK S266C mutant and VinP1LdACP in the presence of BMOE gave a covalent complex (Fig. S3A). Next, we purified these covalent VinK–VinL and –VinP1LdACP complexes for crystallization (Fig. S3B). We obtained reproducible VinK–VinL complex crystals, although we failed to obtain a VinK–VinP1LdACP crystal. The mixture of the VinK S266C mutant with VinL in the absence of BMOE did not give any complex crystals, indicating the necessity of using BMOE to capture VinL.
Fig. 3.
Cross-linking reaction of VinK with VinL in the presence of BMOE. SDS/PAGE after the reaction of VinK with VinL for 10 and 60 min is shown. The bands corresponding to the complex formation are evident in the reaction mixture of the VinK S266C mutant with VinL.
Fig. S3.
SDS/PAGE analysis of VinK–ACPs cross-linking products. (A) Cross-linking reaction of VinK with VinP1LdACP in the presence of BMOE. SDS/PAGE after the reaction of VinK with VinP1LdACP for 10 and 60 min is shown. The bands corresponding to the complex formation were apparent from the reaction mixture of the VinK S266C mutant with VinP1LdACP. (B) Purified covalent VinK–VinL (lane 1) and VinK–VinP1LdACP (lane 2) complexes.
Crystal Structure of the VinK–VinL Complex.
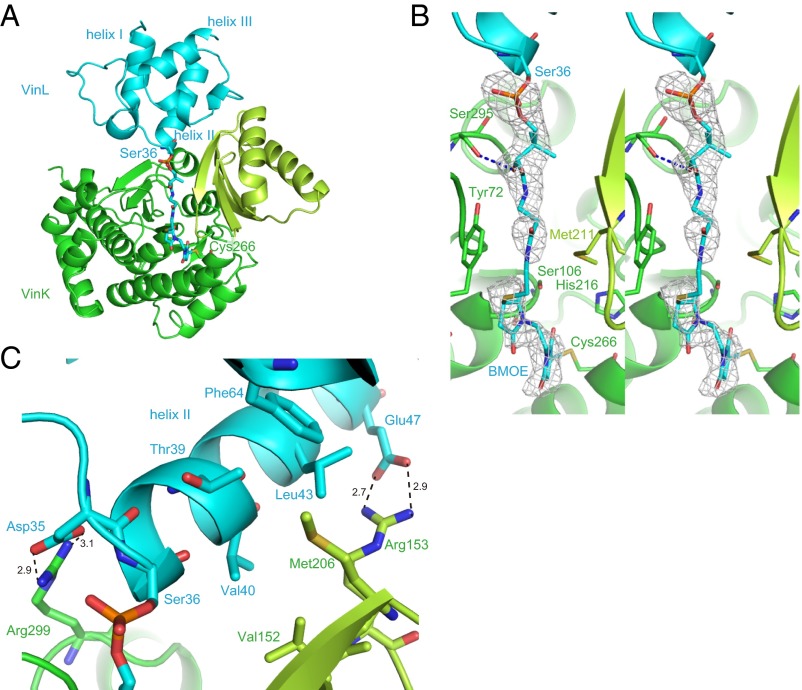
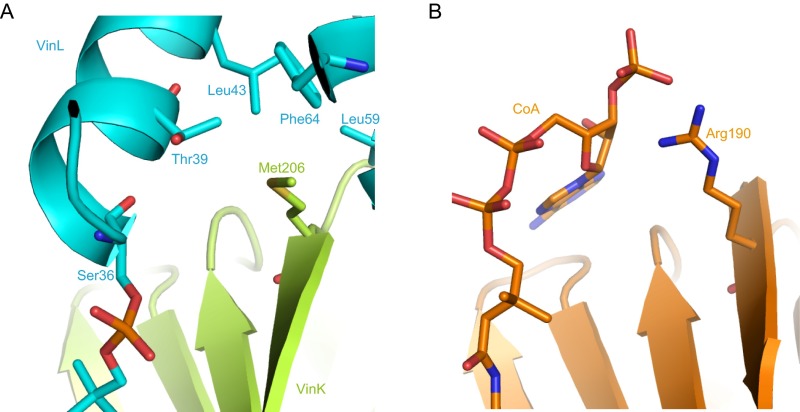
We determined the crystal structure of the VinK–VinL complex at 2.34-Å resolution (Table S1). In the complex structure, one VinK molecule and one VinL molecule are present in the crystallographic asymmetric unit (Fig. 4A). The overall architecture of VinL is similar to those of other structurally characterized carrier proteins (CPs) including E. coli FAS ACP (AcpP) (PDB ID code 2FAE; 21% sequence identity; 2.0 Å rmsd for Cα atoms; ref. 16). VinL has a helix bundle fold with three major helices (helices I, II, and III) and one minor distorted helix III′, as observed in most of the known ACP structures (9). A 4′-phosphopantetheine arm is covalently attached to Ser-36, which is located at the beginning of helix II (Fig. 4 B and C). The 4′-phosphopantetheine arm is orientated into the VinK active site through the binding tunnel. Thus, this VinK–VinL complex structure is mechanistically reasonable for the reaction. The maleimide groups were observed to be covalently attached to both the side-chain sulfhydryl group of the mutated Cys-266 residue of VinK and the terminal sulfhydryl group of the 4′-phosphopantetheine arm of VinL, although its electron density was not clearly defined probably because of its flexibility (Fig. 4B). The 4′-phosphopantetheine linker region exhibits only a few interactions with VinK. The β-alanine moiety of the 4′-phosphopantetheine linker is stacked between Tyr-72 and Met-211. The hydroxy group of the 4′-phosphopantetheine linker forms a hydrogen bond with Ser-295 of VinK.
Fig. 4.
Structure of the VinK–VinL complex. The VinK large domain, VinK small domain, and VinL are shown in green, yellow-green, and cyan, respectively. (A) Overall structure of the VinK–VinL complex. Ser-36 of VinL, 4′-phosphopantetheine arm, BMOE, and Cys-266 of VinK are shown as sticks. (B) Stereoview of substrate-binding tunnel region in the VinK–VinL complex. An Fo − Fc electron density map contoured at 2.5σ was constructed before incorporation of the BMOE and 4′-phosphopantetheine moieties. (C) The binding interface between VinK and VinL. Salt-bridge interactions are shown as broken lines.
Interactions Between VinK and VinL.
In the complex structure, VinL primarily contacts the small domain of VinK (Fig. 4A). The interface between VinK and VinL comprises ∼650 Å2, representing 12.6% of the surface area of VinL and 5.1% of the surface area of VinK. This small contact area, which is similar to those of other enzyme–CP complex structures such as the FabA–AcpP structure (17, 18), is consistent with the transient nature of the enzyme–ACP interactions. Arg-153 and -299 of VinK form salt bridges with Glu-47 and Asp-35 of VinL, respectively (Fig. 4C). Met-206 of VinK extends into a hydrophobic pocket formed by Thr-39, Leu-43, Leu-59, and Phe-64 of VinL. Thus, VinK seems to recognize the helix II region of VinL through salt bridges and hydrophobic contacts.
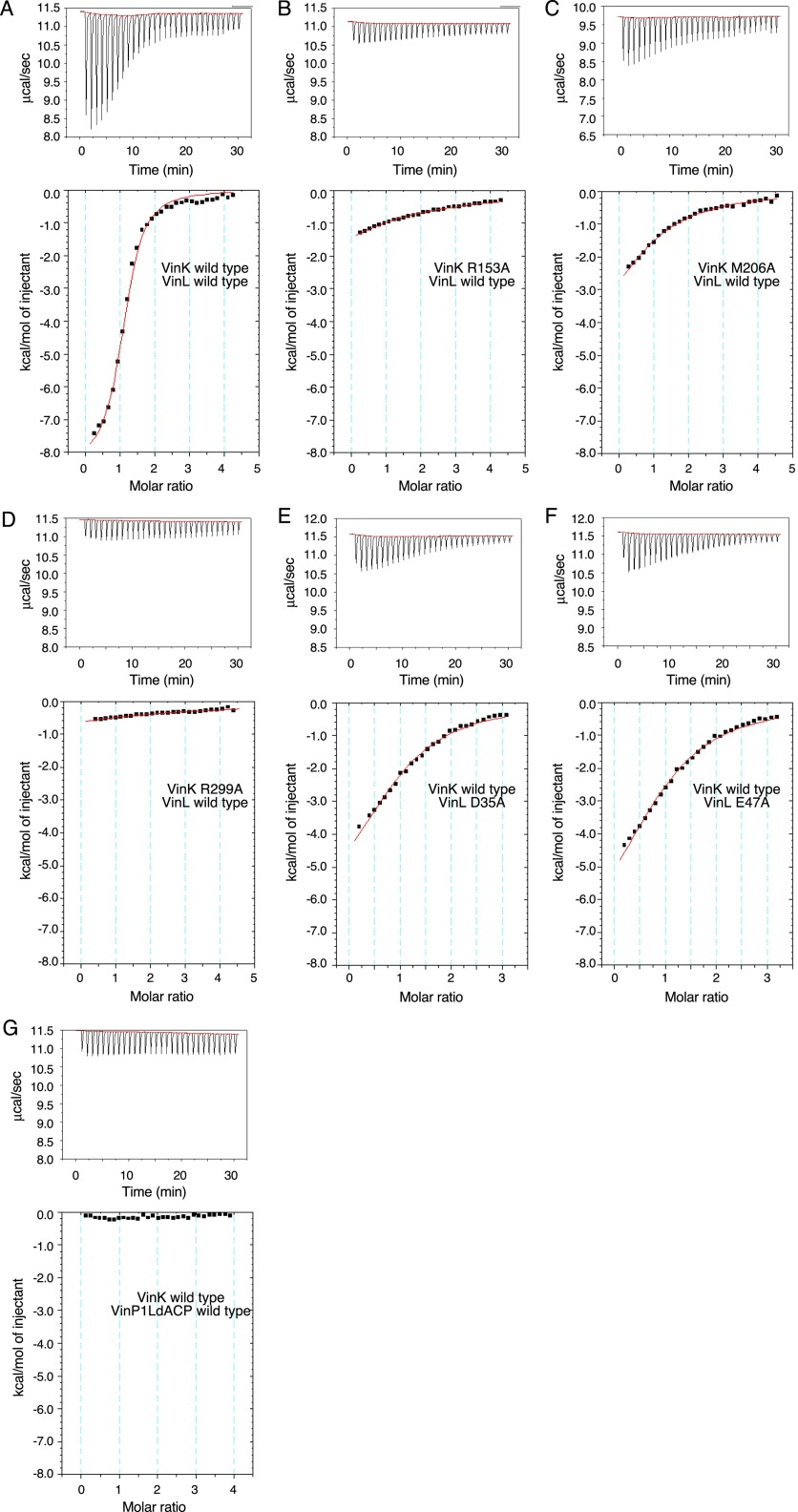
To confirm that these salt bridges and hydrophobic contacts are functional interactions with VinL, we constructed VinK R153A, M206A, and R299A mutants and then analyzed their abilities to interact with VinL by isothermal titration calorimetry (ITC). The VinK wild type was shown to bind VinL with a Ka value of 8.2 ± 0.8 × 104 M−1 (Table 1 and Fig. S4A), which is similar to the affinities of other MCATs for ACPs (19, 20). The R153A, M206A, and R299A mutants showed significantly reduced affinity for VinL (Fig. S4 B–D). We also carried out cross-linking assays to evaluate the interaction of each VinK mutant with VinL. The S266C/R153A, S266C/M206A, and S266C/R299A mutants all had significantly reduced cross-linking efficiency (Fig. S5A), consistent with the ITC results. Furthermore, we mutated VinL counterpart residues such as Asp-35 and Glu-47 and analyzed by ITC and cross-linking experiments. VinL D35A and E47A mutants showed weaker affinity (Table 1 and Fig. S4 E and F) and reduced cross-linking efficiency (Fig. S5B), as in the cases of VinK mutants. Thus, these mutational results showed that the presence of Arg-153, Met-206, and Arg-299 of VinK is crucial for the interaction with VinL, supporting the structural observation of the VinK–VinL complex.
Table 1.
Affinity of VinK to VinL determined by ITC
| VinK | VinL | Ka × 103,* M−1 | ΔG, kcal/mol | ΔH, kcal/mol* | −TΔS, kcal/mol | n |
| Wild type | Wild type | 82.1 ± 7.7 | −6.7 | −8.4 ± 0.2 | 1.7 | 1.11 ± 0.02* |
| R153A | Wild type | 1.9 ± 0.0 | −4.4 | −6.3 ± 0.1 | 1.9 | 1.0† |
| M206A | Wild type | 6.2 ± 0.2 | −5.1 | −5.5 ± 0.1 | 0.4 | 1.0† |
| R299A | Wild type | 0.8 ± 0.1 | −4.0 | −5.6 ± 0.3 | 1.6 | 1.0† |
| Wild type | D35A | 12.0 ± 0.6 | −5.6 | −6.7 ± 0.1 | 1.1 | 1.0† |
| Wild type | E47A | 10.9 ± 0.5 | −5.5 | −7.9 ± 0.1 | 2.4 | 1.0† |
Values are ± SD from the fit.
n was fixed at 1 for the purposes of fitting the data because the affinity was below the level for accurate determination of binding stoichiometry.
Fig. S4.
ITC data in the interaction between VinK and ACPs. The ITC data obtained for VinL to VinK (A–F) and the ITC data obtained for VinP1LdACP to VinK (G) are shown. Upper shows raw binding heat, and Lower shows the integrated binding heat minus the dilution control heat.
Fig. S5.
SDS/PAGE analysis of cross-linking assays. (A) Cross-linking reaction of VinK mutants with VinL in the presence of BMOE. SDS/PAGE after the reaction of VinK with VinL for 5 and 30 min is shown. (B) Cross-linking reaction of VinK with VinL mutants in the presence of BMOE. SDS/PAGE after the reaction of VinK with VinL for 10 and 30 min is shown. (C) Cross-linking reaction of VinK mutants with VinP1LdACP in the presence of BMOE. SDS/PAGE after the reaction of VinK with VinP1LdACP for 5 and 30 min is shown.
Interactions Between VinK and VinP1LdACP.
We next investigated the interactions between VinK and VinP1LdACP by ITC. However, the VinK wild type showed a significantly low affinity for VinP1LdACP (Fig. S4G), which made it difficult to calculate accurate thermodynamic parameters. Therefore, we decided to evaluate the interactions of the VinK mutants with VinP1LdACP by cross-linking assays. Among the VinK mutants, the S266C/R299A mutant had significantly decreased cross-linking efficiency compared with the S266C mutant (Fig. S5C), suggesting that Arg-299 of VinK is important for the interaction with VinP1LdACP. Given that Asp-35 of VinL is equivalent to Asp-86 of VinP1LdACP (Fig. S6A), Arg-299 might interact with Asp-86 of VinP1LdACP. In contrast, the VinK S266C/R153A and S266C/M206A mutants showed similar levels of efficiency to the S266C mutant, suggesting that Arg-153 and Met-206 do not largely interact with VinP1LdACP. This result may be explained by the fact that most of the VinL residues important for the interaction with Arg-153 and Met-206 of VinK are not conserved in VinP1LdACP. Thr-39, Leu-43, and Glu-47 of VinL are substituted with Ala-90, His-94, and Thr-98 of VinP1LdACP, respectively (Fig. S6A). Thus, VinP1LdACP appears to have a decreased number of salt bridges and hydrophobic interactions with VinK, likely leading to its lower affinity to VinK.
Fig. S6.
Amino acid sequence alignment of VinL with other ACPs. The secondary structural elements of VinL and EcAcpP are indicated above and below the sequences, respectively. A conserved Ser residue for the attachment of 4′-phosphopantetheine arm is shown as red. VinL residues important for the interaction with VinK are shown as cyan. (A) Amino acid sequence comparison of VinL with VinP1LdACP. (B) Amino acid sequence alignment of VinL with other VinL-like proteins involved in the macrolactam biosynthesis (CmiS4 from Streptomyces sp. MJ635-86F5 and IdnL6 from Streptomyces sp. ML694-90F3) and other related ACP proteins (ScFASACP from S. coelicolor and EcAcpP from E. coli).
One-Pot Enzymatic Reaction of VinK Wild Type and Mutants.
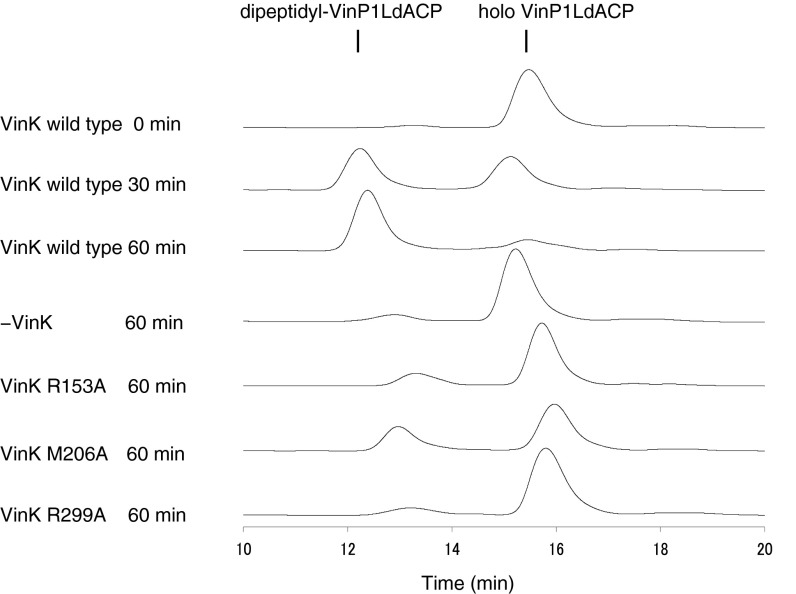
To compare activities of VinK wild type and mutants, we assayed their transacylation activities by a one-pot enzymatic reaction. Incubation of VinK wild type with VinN, VinO, VinM, VinL, VinP1LdACP, and associated substrates gave dipeptidyl–VinP1LdACP efficiently (Fig. S7). In contrast, all three VinK mutants showed significantly reduced formation of dipeptidyl–VinP1LdACP. Thus, disruption of interaction between VinK and VinL resulted in the decreased transacylation activity of VinK.
Fig. S7.
Comparison of dipeptide transfer activity of VinK wild type and mutants. HPLC traces (detection at 210 nm) of one-pot enzymatic reaction mixtures are shown.
Discussion
The proper protein–protein interactions between CP and its partner enzyme are essential for efficient enzyme turnover in the biosynthesis of fatty acids, polyketides, and nonribosomal peptides. The binding affinities of CP for its partner enzymes are weak, enabling it to interact reversibly with each enzyme in the biosynthetic pathway (9). Because of these weak and transient interactions, structural characterization of the enzyme–CP complex is difficult. Recently, several methods using synthetic probes to capture CP in functional association with a partner enzyme were developed, allowing the structural characterization of these complexes to visualize the protein–protein interactions (17, 21–23). However, no crystal structure of an AT–ACP complex was reported to date. Recently, Whicher et al. described the electron cryomicroscopy structure of modular PKS PikAIII that the ACP domain is positioned near the AT domain (24). Although the phosphopantetheinylated Ser residue of ACP domain is directed toward the AT active site in this structure, the distance between them is 35 Å, which is not close enough for the acyltransfer reaction. Thus, the exact AT–ACP contact interface remains elusive.
Wong et al. described the preparation of a covalent DSZS AT–ACP complex using cross-linkers such as 1,3-dibromopropanone and BMOE (15). Their method seems to be useful to capture ACP for the crystallization of the AT–ACP complex. However, only a modest fraction of DSZS AT was cross-linked to ACP. In this study, we attempted to obtain covalent VinK–ACP complexes using BMOE as a cross-linker. Although our strategy is similar to the previous DSZS AT cross-linking experiment (15), we changed the Cys mutation position for the cross-linking reaction. The catalytic Ser-86 of DSZS AT, which corresponds to Ser-106 in VinK, was mutated to Cys in the previous cross-linking experiment, whereas we selected the noncatalytic Ser-266 located at the base of the substrate-binding tunnel. In the VinK structure, Ser-266 is placed 5 Å deeper from the entrance of the tunnel compared with the catalytic Ser-106 (Fig. 2B). Therefore, we expected the S266C mutant to accommodate the BMOE molecule between the mutated Cys-266 residue and the 4′-phosphopantetheine arm of VinL. We obtained the covalent VinK–ACP complexes using BMOE in sufficient yield for crystallization (Fig. 3). We also succeeded in the determination of the VinK–VinL complex structure, which, to our knowledge, is the first crystal structure of an AT–ACP complex (Fig. 4). Thus, bifunctional maleimide reagents such as BMOE are useful tools to trap the transient AT–ACP complex for structural characterization. This cross-linking reaction requires only the 4′-phosphopantetheine arm of ACP and a Cys residue at the appropriate position of the partner enzyme. Therefore, this cross-linking strategy using bifunctional maleimide reagents would be a versatile method for trapping other enzyme–ACP complexes.
Most ATs accept acyl–CoAs as substrates and transfer the acyl group to the partner ACPs. In the complex structure of E. coli FabD with malonyl–CoA, Arg-190, which is located at the positively charged surface near the entrance of the substrate-binding tunnel, interacts with the phosphate group of the ribose moiety of CoA (14). The acyl–CoA-specific ATs generally have an Arg or Lys residue at this position (Fig. S1). At the corresponding position, VinK has Met-206, which interacts with the hydrophobic residues of VinL (Fig. S8). VinK homolog enzymes involved in the biosynthesis of other macrolactam antibiotics also have Met or Leu residues (Fig. S1). Furthermore, acyl–ACP-specific ATs such as ZmaA and ZmaF contain a Met residue at this position (4). Thus, the presence of a hydrophobic residue at this position might be a conserved feature in acyl–ACP-specific ATs.
Fig. S8.
Structural comparison of VinK with FabD. (A) The binding interface between VinK and VinL in the VinK–VinL complex structure. (B) The structure of FabD complexed with CoA (PDB ID code 2G2Z).
Helix II of several ACPs was observed to be involved in an interaction with their partner enzymes in the complex structures (9, 17, 18, 25–28). In terms of the interaction of AT with ACP, Streptomyces coelicolor MCAT was suggested to recognize the helix II of FAS ACP, based on the previous docking and mutational studies (19). However, the details of the binding and binding interface remain elusive. In this study, the VinK–VinL complex structure clearly shows the interaction of VinK with the negatively charged helix II region of VinL (Fig. 4B), which is, to our knowledge, the first direct evidence for the helix II recognition of ACP by AT. Compared with VinL, VinP1LdACP has a less acidic helix II and seems to make fewer interactions with VinK. Because VinK showed a much weaker affinity for VinP1LdACP than VinL, VinP1LdACP itself might not be enough to establish the interaction with VinK. The presence of other domains of modular VinP1PKS might affect the binding of VinK to the VinP1LdACP region in the actual vicenistatin biosynthetic pathway. Although it was suggested from the VinK–VinL structure that the negatively charged helix II region of ACP is used for the recognition by AT, it is still unclear whether this binding model can be applied to all cases of AT–ACP interaction. Some PKS ATs are predicted to bind ACP in a different manner based on docking analysis. For example, S. coelicolor MCAT was suggested to interact with the loop region between helix I and II of actinorhodin PKS ACP (19, 29). Alternatively, the presence of an additional domain such as the KS-AT linker domain in modular PKS might be involved in the binding mode of ACP, as proposed in the interaction between ATDYN10 and ACPDYN involved in dynemicin biosynthesis (30). For complete understanding of ACP recognition by AT, it is necessary to obtain other AT–ACP complex structures.
In conclusion, we succeeded in obtaining, to our knowledge, the first crystal structure of an AT–ACP complex. This complex structure should be useful for the prediction of a model for other AT–ACP complex interactions. This study could provide useful clues for the engineering of ATs to optimize protein–protein interactions with ACP for production of unnatural polyketide products.
Materials and Methods
VinK, VinL, and VinP1LdACP proteins were expressed and purified as described (12). VinK mutants were prepared by site-directed mutagenesis. The cross-linking reactions were performed in the presence of BMOE (Thermo Fisher Scientific). VinK protein and the VinK–VinL complex were crystallized by using the vapor-diffusion method. The VinK structure was solved by the single-wavelength anomalous dispersion method. ITC experiments were carried out at 25 °C by using the MicroCal iTC200 (GE Healthcare). The details for all of the experimental protocols are described in SI Materials and Methods.
SI Materials and Methods
Preparation of Purified VinK, VinL, and VinP1LdACP Proteins.
For the expression of VinK wild-type and mutant proteins, E. coli BL21(DE3) cells harboring pCold I-vinK were grown at 37 °C in Luria Bertani broth containing 50 mg/L ampicillin. After the optical density at 600 nm reached 0.6, protein expression was induced by the addition of 0.2 mM isopropyl β-d-1-thiogalactopyranoside, and the cells were then cultured for an additional 16 h at 15 °C. For the expression of selenomethionine-substituted VinK, E. coli B834(DE3) cells harboring pCold I-vinK were similarly grown in M9 medium supplemented with 50 mg/L l-selenomethionine and 50 mg/L ampicillin. VinL and VinP1LdACP proteins were coexpressed with the sfp gene to obtain holo-ACPs, as described (12). The construct of VinP1LdACP protein spans residues 1–146, comprising the N-terminal linker, the loading ACP domain, and downstream ACP-KS linker regions (Fig. S6A) of VinP1PKS (5,826 residues). VinN, VinO, and VinM proteins were expressed as described (12, 13). The recombinant proteins, which were collected from cell-free extracts prepared by sonication, were purified on a TALON affinity column (Clontech). The protein was then desalted and concentrated by using a PD-10 column (GE Healthcare) and an Amicon Ultra centrifugal filter (Merck Millipore), respectively. For crystallization, VinK was further treated with Factor Xa protease to remove His-tag and subjected to another round of TALON affinity column. For preparation of cross-linked VinK–VinL complex on a large scale, the N-terminal His-tag of VinL was removed by the treatment with thrombin protease.
Site-Directed Mutagenesis.
For the construction of the VinK R153A, M206A, S266C, and R299A mutants, the pCold I-vinK plasmid was used for the site-directed mutagenesis. Site-directed mutagenesis was performed with a QuikChange site-directed mutagenesis kit (Stratagene) by using the following oligonucleotides: R153A, 5′-CACTCCTTCGTCGCCGCCCCTCGGGAGCGG-3′; M206A, 5′-CGGTCGGTGGGCGCCGCGCCGCTGTACGCC-3′; S266C, 5′-CCATGCTCCTGGAGTGCTTCGTCCGCCC-3′; R299A, 5′-GACAGCCTCTTCGGGGCGGTCGGCACCACC-3′; and their complementary oligonucleotides. For the construction of the VinL D35A and E47A mutants, the pET28–vinL plasmid was used for the site-directed mutagenesis. Site-directed mutagenesis was performed by using the following oligonucleotides: D35A, 5′-GACATCGGCCTCGCCTCGCTCGGCACCGTC-3′; E47A, 5′-CTCTCCGAGCTGGCGAACACCTACGACGTC-3′; and their complementary oligonucleotides. The mutations were confirmed by determining the nucleotide sequences. These plasmids were transformed into E. coli BL21(DE3) cells, and the mutated enzymes were prepared as described above. We confirmed that VinL mutants were obtained as holoform by liquid chromatography-electrospray ionization-MS analysis as described (12).
Cross-Linking Reaction.
For the VinK–VinL cross-linking assays, 15 μM VinK protein was mixed with 50 or 150 μM VinL protein and 0.3 mM BMOE (Thermo Fisher Scientific) in a buffer containing 40 mM NaH2PO4/K2HPO4 (pH 7.0) and 5% (vol/vol) glycerol and incubated on ice. For VinK–VinP1LdACP cross-linking assays, 25 μM VinK protein was mixed with 50 μM VinP1LdACP protein and 0.1 mM BMOE in a buffer containing 40 mM NaH2PO4/K2HPO4 (pH 7.0) and 5% (vol/vol) glycerol and incubated at 23 °C. At the time points of 5, 10, 30, and 60 min, 15 μL of the reaction mixture was taken, and the reaction was quenched by the addition of SDS/PAGE loading buffer.
Preparation and Purification of the Cross-Linked VinK–ACP Complexes.
In a large-scale cross-linking reaction for the VinK–VinL complex, 50 μM His-tagged VinK S266C mutant protein was mixed with 350 μM His-tag free VinL protein in a buffer containing 40 mM NaH2PO4/K2HPO4 (pH 7.0) and 5% (vol/vol) glycerol (total volume 4.5 mL). The cross-link reaction was initiated by the addition of 0.3 mM BMOE. The reaction was carried out on ice for 4 h. After the reaction, BMOE was removed by using a PD-10 column. The obtained VinK–VinL complex protein was then purified through a TALON affinity column. In a large-scale cross-linking reaction for the VinK–VinP1LdACP complex, 60 μM VinK S266C mutant protein was mixed with 50 μM VinP1LdACP protein in a buffer containing 40 mM NaH2PO4/K2HPO4 (pH 7.0) and 5% glycerol (total volume 4.5 mL). The cross-link reaction was initiated by the addition of 0.13 mM BMOE. The reaction was carried out at 23 °C for 2 h. After the reaction, BMOE was removed by using a PD-10 column. The obtained VinK–VinP1LdACP complex was then purified through Superdex 200 and Resource Q columns (GE Healthcare).
Crystallization, Data Collection, and Structure Determination.
Crystals of native VinK and selenomethionine-substituted VinK were grown from a 1:1 mixture of a protein solution [15 mg/mL in 5 mM Tris⋅HCl (pH 7.5)] and a reservoir solution [0.1 M calcium acetate and 20% (vol/vol) polyethylene glycol 3350, pH 7.0] by using the sitting- or hanging-drop vapor-diffusion method at 26 °C. Crystals of VinK–VinL cross-linked complex were grown from a 1:1 mixture of a protein solution [12 mg/mL in 5 mM Tris⋅HCl (pH 7.5)] and a reservoir solution [0.05 M magnesium chloride, 20% (vol/vol) polyethylene glycol 8000, and 0.1 M Tris⋅HCl (pH 8.5)] by using the sitting-drop vapor-diffusion method at 5 °C. Before X-ray data collection, crystals were transferred into the reservoir solution supplemented with 25% (vol/vol) glycerol as a cryoprotectant and flash-frozen in the liquid nitrogen stream. X-ray data were collected by using beamlines BL-5A, AR-NW12A, and AR-NE3A (Photon Factory, Tsukuba, Japan). All diffraction data were indexed, integrated, and scaled by using the program iMosflm (31). Selenomethionine-substituted VinK crystal was used for the single-wavelength anomalous dispersion phasing method. Selenium position and initial phases were calculated by using the program AutoSol (32). After density modification, the program ARP/wARP (33) was used for automatic initial protein model building and refinement. The initial model of VinK–VinL complex was determined by the molecular replacement method using the program MOLREP (34) with the coordinates of the refined solo VinK structure as a template. Coot (35) was used for visual inspection and manual rebuilding of the model. Refmac (36) was used for refinement. The figures were prepared by using PyMOL (https://www.pymol.org). Interface area between VinK and VinL was calculated by the PISA server (37). The geometries of the final structures were evaluated by using the program Rampage (38).
Docking Analysis.
The docking study was carried out by using the AutoDock program (Version 4.2; ref. 39). The dipeptidyl-N-acetylcysteamine thioester molecule was generated by using the PRODRG2 Server (40). Chain A of the crystal structure of VinK was used for the docking study. By using AutoDockTools, polar hydrogen atoms were added to amino acid residues, and Gasteiger charges were assigned to all atoms of the protein. All rotatable bonds of the ligand molecule were set to be flexible for the calculation, whereas all of the protein residues were kept rigid. The cubic energy grid was centered at the substrate binding pocket and had an extension of 50 Å in each direction.
ITC Analysis of the Interaction Between VinK and ACPs.
ITC experiments were carried out at 25 °C by means of standard procedures using the MicroCal iTC200 (GE Healthcare). A concentration of 0.15 mM VinK protein and 4.0 mM ACPs in 50 mM Tris⋅HCl (pH 8.0) were prepared for the 200-μL reaction cell and 40-μL microsyringe, respectively. Each 1-μL aliquot of ACPs was successively injected at 1-min intervals for 30 min into VinK solution stirred at 1,000 rpm. The integrated heat effect was analyzed by using a single binding site model implemented in the ORIGIN software. The fitted data yielded the binding stoichiometry (n), the association constants (Ka), and the enthalpy changes (ΔH). Gibbs free energy (ΔG) and entropy changes (ΔS) were calculated by using the standard thermodynamic equation (−RTLnKa = ΔG = ΔH − TΔS).
One-Pot Enzymatic Reaction.
The activities of VinK wild type and mutants were assayed by using a one-pot enzymatic reaction. The standard mixture contained 0.05 μM VinK wild type or mutants, 5 μM VinN, 5 μM VinO, 5 μM VinM, 2.5 μM VinL, 60 μM VinP1LdACP, 2 mM ATP, 1 mM dl-threo-β-methylaspartate, 1 mM l-alanine, 1 mM MgCl2, 50 mM Tris⋅HCl (pH 8.0), and 10% (vol/vol) glycerol in a volume of 20 μL. After incubation at 23 °C for 30 or 60 min, the reaction was quenched by the addition of acetonitrile (20% volume), and the aliquot (1.5 μL) of the supernatant was analyzed by HPLC. HPLC was carried out by using a Hitachi instrument (Chromaster 5110 Pump and UV Detector l-7405; Hitachi) equipped with a Protein-R C18 column (300 Å, 4.6 × 150 mm; Nacalai Tesque). The flow rate was 0.8 mL/min, and the detection wavelength was 210 nm. The elution condition was a linear gradient of 38–41% (vol/vol) acetonitrile in water containing 0.05% (vol/vol) trifluoroacetic acid over a period of 20 min.
Acknowledgments
This work was performed with the approval of the Photon Factory Program Advisory Committee (Proposals 2012G508 and 2014G530) and was supported in part by Japan Society for the Promotion of Science Grants-in-Aid for Young Scientists (B) 15K18679 (to A.M.); and Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research in Innovative Areas 22108003 (to T.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.K. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5CZC and 5CZD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520042113/-/DCSupplemental.
References
- 1.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48(26):4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 2.Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2010;27(7):996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 3.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26(1):90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park H, Kevany BM, Dyer DH, Thomas MG, Forest KT. A polyketide synthase acyltransferase domain structure suggests a recognition mechanism for its hydroxymalonyl-acyl carrier protein substrate. PLoS One. 2014;9(10):e110965. doi: 10.1371/journal.pone.0110965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn BJ, Khosla C. Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J R Soc Interface. 2013;10(85):20130297. doi: 10.1098/rsif.2013.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liou GF, Khosla C. Building-block selectivity of polyketide synthases. Curr Opin Chem Biol. 2003;7(2):279–284. doi: 10.1016/s1367-5931(03)00016-4. [DOI] [PubMed] [Google Scholar]
- 7.Wong FT, Chen AY, Cane DE, Khosla C. Protein-protein recognition between acyltransferases and acyl carrier proteins in multimodular polyketide synthases. Biochemistry. 2010;49(1):95–102. doi: 10.1021/bi901826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Z, Musiol EM, Weber T, Williams GJ. Reprogramming acyl carrier protein interactions of an Acyl-CoA promiscuous trans-acyltransferase. Chem Biol. 2014;21(5):636–646. doi: 10.1016/j.chembiol.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosby J, Crump MP. The structural role of the carrier protein--active controller or passive carrier. Nat Prod Rep. 2012;29(10):1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 10.Kudo F, Miyanaga A, Eguchi T. Biosynthesis of natural products containing β-amino acids. Nat Prod Rep. 2014;31(8):1056–1073. doi: 10.1039/c4np00007b. [DOI] [PubMed] [Google Scholar]
- 11.Shindo K, Kamishohara M, Odagawa A, Matsuoka M, Kawai H. Vicenistatin, a novel 20-membered macrocyclic lactam antitumor antibiotic. J Antibiot (Tokyo) 1993;46(7):1076–1081. doi: 10.7164/antibiotics.46.1076. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara Y, Kudo F, Eguchi T. A natural protecting group strategy to carry an amino acid starter unit in the biosynthesis of macrolactam polyketide antibiotics. J Am Chem Soc. 2011;133(45):18134–18137. doi: 10.1021/ja208927r. [DOI] [PubMed] [Google Scholar]
- 13.Miyanaga A, Cieślak J, Shinohara Y, Kudo F, Eguchi T. The crystal structure of the adenylation enzyme VinN reveals a unique β-amino acid recognition mechanism. J Biol Chem. 2014;289(45):31448–31457. doi: 10.1074/jbc.M114.602326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oefner C, Schulz H, D’Arcy A, Dale GE. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 6):613–618. doi: 10.1107/S0907444906009474. [DOI] [PubMed] [Google Scholar]
- 15.Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Structure and mechanism of the trans-acting acyltransferase from the disorazole synthase. Biochemistry. 2011;50(30):6539–6548. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roujeinikova A, et al. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365(1):135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen C, et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature. 2014;505(7483):427–431. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohman JR, et al. The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins. Proteins. 2014;82(7):1210–1218. doi: 10.1002/prot.24485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arthur CJ, et al. Structure and malonyl CoA-ACP transacylase binding of streptomyces coelicolor fatty acid synthase acyl carrier protein. ACS Chem Biol. 2009;4(8):625–636. doi: 10.1021/cb900099e. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, et al. Malonyl-CoA: Acyl carrier protein transacylase from Helicobacter pylori: Crystal structure and its interaction with acyl carrier protein. Protein Sci. 2007;16(6):1184–1192. doi: 10.1110/ps.072757307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem Biol. 2012;19(2):188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haslinger K, et al. The structure of a transient complex of a nonribosomal peptide synthetase and a cytochrome P450 monooxygenase. Angew Chem Int Ed Engl. 2014;53(32):8518–8522. doi: 10.1002/anie.201404977. [DOI] [PubMed] [Google Scholar]
- 23.Finzel K, Lee DJ, Burkart MD. Using modern tools to probe the structure-function relationship of fatty acid synthases. ChemBioChem. 2015;16(4):528–547. doi: 10.1002/cbic.201402578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whicher JR, et al. Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature. 2014;510(7506):560–564. doi: 10.1038/nature13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cryle MJ, Schlichting I. Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc Natl Acad Sci USA. 2008;105(41):15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parris KD, et al. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure. 2000;8(8):883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 27.Babu M, et al. Structure of a SLC26 anion transporter STAS domain in complex with acyl carrier protein: Implications for E. coli YchM in fatty acid metabolism. Structure. 2010;18(11):1450–1462. doi: 10.1016/j.str.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Guy JE, et al. Remote control of regioselectivity in acyl-acyl carrier protein-desaturases. Proc Natl Acad Sci USA. 2011;108(40):16594–16599. doi: 10.1073/pnas.1110221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keatinge-Clay AT, et al. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure. 2003;11(2):147–154. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 30.Liew CW, et al. Crystal structure of the acyltransferase domain of the iterative polyketide synthase in enediyne biosynthesis. J Biol Chem. 2012;287(27):23203–23215. doi: 10.1074/jbc.M112.362210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Lovell SC, et al. Structure validation by Calpha geometry: ϕ,ψ and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 39.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]