Abstract
Notch signaling largely determines intestinal epithelial cell fate. High Notch activity drives progenitors toward absorptive enterocytes by repressing secretory differentiation programs, whereas low Notch permits secretory cell assignment. Myeloid translocation gene-related 1 (MTGR1) is a transcriptional corepressor in the myeloid translocation gene/Eight-Twenty-One family. Given that Mtgr1−/− mice have a dramatic reduction of intestinal epithelial secretory cells, we hypothesized that MTGR1 is a key repressor of Notch signaling. In support of this, transcriptome analysis of laser capture microdissected Mtgr1−/− intestinal crypts revealed Notch activation, and secretory markers Mucin2, Chromogranin A, and Growth factor-independent 1 (Gfi1) were down-regulated in Mtgr1−/− whole intestines and Mtgr1−/− enteroids. We demonstrate that MTGR1 is in a complex with Suppressor of Hairless Homolog, a key Notch effector, and represses Notch-induced Hairy/Enhancer of Split 1 activity. Moreover, pharmacologic Notch inhibition using a γ-secretase inhibitor (GSI) rescued the hyperproliferative baseline phenotype in the Mtgr1−/− intestine and increased production of goblet and enteroendocrine lineages in Mtgr1−/− mice. GSI increased Paneth cell production in wild-type mice but failed to do so in Mtgr1−/− mice. We determined that MTGR1 can interact with GFI1, a transcriptional corepressor required for Paneth cell differentiation, and repress GFI1 targets. Overall, the data suggest that MTGR1, a transcriptional corepressor well characterized in hematopoiesis, plays a critical role in intestinal lineage allocation.—Parang, B., Rosenblatt, D., Williams, A. D., Washington, M. K., Revetta, F., Short, S. P., Reddy, V. K., Hunt, A., Shroyer, N. F., Engel, M. E., Hiebert, S. W., Williams, C. S. The transcriptional corepressors MTGR1 regulates intestinal secretory lineage allocation.
Keywords: Notchsignaling, Paneth cells, CBFA2T2
Intestinal differentiation programs are highly complex and dynamic. Stem cells reside at the crypt base intermingled with Wnt-agonist-secreting Paneth cells. Stem cell division gives rise to daughter cells that migrate along the crypt-villus axis and differentiate into secretory cells (goblet, enteroendocrine, and Paneth) or absorptive enterocytes (1, 2). The molecular mechanisms underlying how intestinal stem cells differentiate into secretory or absorptive cells are incompletely understood, but Notch signaling plays a critical role in lineage allocation (3).
The mammalian Notch pathway is activated by single transmembrane ligands [Delta-like (Dll) 1, 3, 4, and Jagged 1 and 2] binding to Notch receptors (Notch1–Notch4) expressed on adjacent cells (4). Dll1 and Dll4 in particular are indispensable mediators of Notch signaling and required for intestinal homeostasis (5, 6). Ligand binding to the Notch receptor induces intracellular proteolytic cleavage by γ-secretase releasing the Notch intracellular domain (N-ICD) to translocate to the nucleus, where it binds to the DNA binding transcription factor Suppressor of Hairless Homolog (CSL) (3). The N-ICD then cooperates with Mastermind-like Protein 1 to drive the expression of Hairy/Enhancer of Split 1 (HES1) and commits the intestinal progenitor to an absorptive fate (2, 7). Genetic experiments in mice show that ectopic expression of Notch receptors or the N-ICD results in villi populated predominantly by absorptive enterocytes at the expense of secretory lineage production and concomitant down-regulation of secretory transcript markers Mucin2 (Muc2) and Chromogranin A (Chga) (1, 2, 8, 9). Conversely, in the absence of Notch signaling, intestinal progenitor cells differentiate preferentially into secretory cells. For example, pharmacologic inhibition of Notch signaling using γ-secretase inhibitors (GSIs) or genetic ablation of Notch function causes villi to be populated by secretory cells at the expense of absorptive enterocytes (10–12).
Myeloid translocation gene-related 1 (MTGR1) is a member of the myeloid translocation gene (MTG) family of transcriptional corepressors. MTGs were originally discovered in acute myeloid leukemia and function as scaffolding proteins, orchestrating the formation of repression complexes (13). MTGR1 is widely expressed, including in the intestinal epithelium, and complexes with mSin3a, N-CoR, and histone deacetylases (14). Experiments with knockout mice for all 3 family members have identified MTGs as regulating developmental, differentiation, and cancer programs. Some mice lacking Mtg8, a family member of Mtgr1, show a deletion of the midgut, suggesting an important role for MTGs in gut development (15). Mtg16−/− mice revealed that Mtg16 was required for hematopoietic progenitor allocation and early progenitor differentiation (16). Finally, genetic ablation of Mtgr1 revealed an increase in intestinal proliferation and a dramatic reduction in intestinal secretory cells, indicating that Mtgr1 is required for appropriate lineage allocation in the gut (14, 17).
Given the importance of Notch signaling in determining intestinal differentiation, we hypothesized that unbridled Notch activation was driving the secretory lineage defect in Mtgr1−/− mice and that Notch inhibition could reverse the phenotype. RNA analysis of Mtgr1−/− intestinal crypts revealed increased Notch activity and down-regulation of secretory markers. Similarly, Muc2, Chga, and growth factor-independent 1 (Gfi1) were down-regulated in whole Mtgr1−/− intestines and Mtgr1−/− enteroids, consistent with Notch activation. We determined that MTGR1 interacts with CSL, a key Notch effector, and suppresses Notch-induced transcriptional activity in vitro. Pharmacologic Notch inhibition using the GSI dibenzazepine (DBZ) in Mtgr1−/− mice rescued the hyperproliferation observed in Mtgr1−/− intestine and increased production of the goblet and enteroendocrine lineages. GSI treatment did not increase Paneth cell number in Mtgr1−/− mice, however, suggesting that MTGR1 is required for GSI-dependent Paneth cell specification. Moreover, we show that MTGR1 interacts with GFI1, a transcriptional corepressor important in Paneth cell differentiation, and can repress GFI1 targets. In sum, our findings indicate that MTGR1, a previously identified transcriptional corepressor known to be critical in hematopoiesis, is an important regulator of intestinal lineage allocation.
MATERIALS AND METHODS
Mice and treatments
A total of 10 wild-type (WT) and 12 Mtgr1−/− mice were treated with vehicle or GSI at 10 μmol/kg twice daily for 5 consecutive days. GSI was prepared as previously described (18) and made fresh daily. GSI and vehicle were administered intraperitoneally. All in vivo procedures were carried out in accordance with protocols approved by the Vanderbilt Institutional Animal Care and Use Committee.
Generated cell lines
Mouse small intestinal epithelial (MSIE) WT and Mtgr1−/− cell lines were generated and validated by the Vanderbilt Digestive Disease Research Center (DDRC) Novel Cell Line Development Subcore according to previously published protocols (19). MSIE cells and derivative lines have been used extensively to study intestinal biology in vitro (20–22). Briefly, Mtgr1−/− mice were crossed with the “Immorto Mouse,” which contains a transgene encoding a temperature-sensitive simian virus 40 large T tumor antigen (19). Intestinal epithelial cells were then isolated and cultured at permissive temperatures. The epithelial nature of both WT and Mtgr1−/− cell lines was confirmed by positive staining for epithelial markers LE61, keratin, and E-cadherin and negative staining for stromal marker SMA.
Laser capture microdissected array
Microarray analysis of WT and Mtgr1−/− crypts was conducted as previously described (17).
Small intestinal enteroid culturing
The enteroid culture method was modified from Sato et al. (23). A total of 8 cm of small intestine was dissected, flushed with ice-cold PBS, and opened lengthwise. Intestines were quickly rinsed in 0.01% bleach followed by PBS, dissected into 5 mm pieces, suspended in 5 ml ice-cold PBS, and incubated at 4°C for 15 min. Following incubation, fragments were vortexed for 3 s. PBS was aspirated, and samples were washed with an additional 10 ml PBS. Tissue was then transferred to 5 ml chelation buffer (2 mM EDTA, made fresh in Dulbecco’s PBS) and rocked for 30 min at 4°C prior to washing twice with 10 ml cold PBS. After washing, 5 ml cold shaking buffer (PBS with 43.3 mM sucrose and 54.9 mM sorbitol) was added, and tissues were gently shaken for 4 min to free intestinal crypts. Resulting crypts were filtered through a 70 μm filter into a prechilled 50 ml tube. The filter was rinsed with an additional 5 ml shaking buffer.
Complete crypts were counted, and enough volume for 1200 crypts was transferred to a prechilled 5 ml round-bottomed tube. Crypts were centrifuged at 150 × g for 10 min at 4°C. Shaking buffer was aspirated, and crypts were resuspended in 50 μl Matrigel (#356237; BD Biosciences, San Jose, CA, USA), per well, containing 50 ng/ml epidermal growth factor (#2028-EG-200; R&D Systems, Minneapolis, MN, USA), 100 ng/ml Noggin (#1967-NG-025/CF; R&D Systems), 500 ng/ml R-Spondin (#3474-RS-050; R&D Systems), and 50 μg/ml Wnt3a (#GF-160; Millipore, Billerica, MA, USA). Matrigel was overlaid with 500 μl Minigut culture media (Advanced DMEM/F12, #12634-010; Invitrogen, Carlsbad, CA, USA), l-glutamine (#25030; Invitrogen), Pen-Strep (#15140-148; Invitrogen), HEPES (#25-060-CI; Mediatech), N2 Supplement (#390155; R&D Systems), and B27 Supplement (#17504044; Invitrogen).
For RNA analysis, WT and Mtgr1−/− enteroids were harvested at 3 d postplating. Matrigel plugs were washed twice with ice-cold PBS, scraped, and centrifuged at 150 × g for 5 min at 4°C to pellet enteroids. The wash and centrifugation process was repeated twice more to remove remaining Matrigel. Enteroids were then resuspended in 1 ml TRIzol (Life Technologies, Grand Island, NY, USA) and homogenized by 3 passes through a 25-gauge needle prior to RNA extraction.
Immunohistochemistry
After the mice were sacrificed, the small intestine was harvested and irrigated with PBS. The intestine was cut transversely and divided into 3 equal sections: proximal, middle, and distal (termed “ileum”). The intestinal segments were then opened longitudinally and rolled orienting the most distal region of intestine such that it was located in the innermost part of the roll. The tissues were then fixed in formalin (1:10 dilution buffered) overnight. The solution was subsequently changed to 70% ethanol before standard paraffin embedding. Five-micron sections were cut and stained with hematoxylin and eosin (H&E) and periodic acid Schiff (PAS) by the Vanderbilt Tissue Core. Goblet cells were scored by PAS staining. Proliferation was measured by phospho-histone (pH3) staining using anti-pH3 at 1:150 (Upstate/Millipore, Billerica, MA, USA) and incubated overnight at 4°C. Enteroendocrine cells were assessed by chromogranin A (CgA) staining using anti-CgA at 1:1000 (ImmunoStar, Hudson, WI, USA) and incubated overnight at 4°C. Anti-lysozyme (Lys) antibody (Dako, Carpinteria, CA, USA) at 1:500 incubated at room temperature for 1 h was utilized to identify Paneth cells. VECTASTAIN ABC Kit (Vector Laboratories, Burlingame, CA, USA) was used to perform all immunohistochemistry reactions. Dewaxing and antigen retrieval processing of sections were conducted as previously described (24).
Quantitative RT-PCR
Intestinal tissue from mice was isolated and immediately placed into 350 μl RNALater (QIAGEN, Valencia, CA, USA) and stored at −80°C. RNA from tissue or cells was isolated using the RNAEasy kit (QIAGEN) according to the manufacturer’s “Animal Tissue” protocol. RNA was subsequently stored at −80°C. A total of 20 μl cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) from 1 μg RNA per sample. All RT-PCRs were carried out using SYBR Green reaction mix (Bio-Rad Laboratories) according to the manufacturer’s protocol.
The following primers were used to measure RNA expression: glyceraldehyde 3-phosphate dehydrogenase (#7317), Gfi1 (#2421), Notch2 (#4291), Notch3 (#4249), and Sox9 (#6220) (all from RealTimePrimers, Elkins Park, PA, USA).
The following sequences were used to measure expression levels of the listed genes: Muc2, forward 5′-TGCCCACCTCCTC2AAAGAC-3′ and reverse 5′-GTAGTTTCCGTTGGAACAGTGAA-3′; Chga, forward 5′-CCACTGCAGCATCCAGTT-3′ and reverse 5′-AGTCCGACTGACCATCATCTTTC-3′; Neuronal differentiation 1 (NeuroD1), forward 5′-ATGACCAAATCATACAGCGAGAG-3′ and reverse 5′-TCTGCCTCGTGTTCCTCGT-3′; SAM-pointed domain-containing ETS transcription factor (Spedf), forward 5′-AAGGCAGCATCAGGAGCAATG-3′ and reverse 5′-CTGTCAATGACGGGACACTG-3′; and Atonal homolog 1 (Atoh1), forward 5′-GAGTGGGCTGAGGTAAAAGAGT-3′ and reverse 5′-GGTCGGTGCTATCCAGGAG-3′.
Reaction conditions used were previously described (25).
Plasmids
The pCMV5 expression plasmids containing HA-tagged MTGR1 and HA-tagged MTG16 were previously described (26). The pCDNA3-myc-CSL, pCDNA3-N1-ICD, and the Hes1-luciferase reporter were previously described (27, 28). The GFI1-3X-Flag expression vector and p2XSATB-1-luciferase reporter were generous gifts to M.E.E. from Dr. Tarik Moroy and have been previously described (29).
Immunoprecipitation and Western blot
Immunoprecipitations and Western blot protocols were carried out as previously described (24). Briefly, expression vectors (HA-MTGR1, HA-MTG16, myc-CSL, Flag-GFI1, or Flag-N1-ICD) were transiently expressed in COS7L cells. Whole-cell lysates were subjected to immunoprecipitation with anti-myc monoclonal antibody 9E10 (Vanderbilt Monoclonal Antibody Core, Nashville, TN, USA) or anti-Flag monoclonal antibody (Sigma-Aldrich, St. Louis, MO, USA), and immune complexes were collected on Protein G Sepharose beads (Sigma-Aldrich). HA:MTGR1 or HA:MTG16 was immunoblotted for using anti-HA rabbit polyclonal antibody Y-11 (Santa Cruz Biotechnology, Dallas, TX, USA). Detection of tubulin using anti-tubulin (Sigma-Aldrich) served as a loading control.
Luciferase promoter assay
All luciferase promoter assays were conducted as previously described (27). Briefly, NIH 3T3 cells were grown on 6-well plates and transfected with Lipofectamine and lysed 48 h later according to the manufacturer’s protocol (Promega, Madison, WI, USA). Luciferase activity was quantified in whole-cell extracts by luminometry according to the manufacturer’s instructions. Results were normalized to an internal control constitutive reporter (pCMV-Renilla) and expressed as the mean ± sd.
Flow cytometry
Flow cytometry was conducted as previously described (30). Briefly, single-cell suspensions were formed after the thymus was minced into fragments and passed through a 70 μm filter. Cells were labeled with the appropriate combination of fluorochrome-conjugated anti-CD4 and anti-CD8 (eBioscience, San Diego, CA, USA). Analysis was performed with a 3-laser BD LSR II using FACSDiva software (BD Biosciences).
Statistical methods
Immunohistochemistry (number of positively stained cells) was analyzed by 1-way ANOVA. If only 2 groups were being compared, a Student’s t test was used. The observer was blinded to slide identity, and the slide was scored in an unbiased fashion.
RESULTS
Notch signaling is hyperactive in the setting of Mtgr1 loss
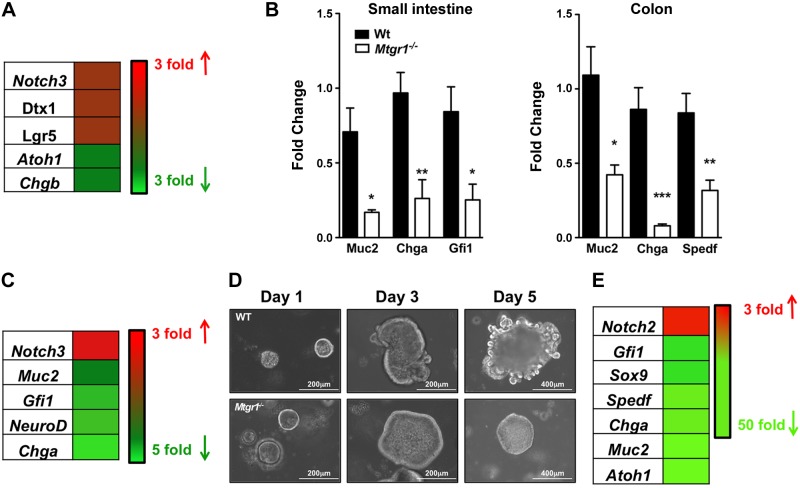
Histologic examination of Mtgr1−/− mice revealed reduced secretory cells, but the molecular mechanism underlying this defect was unclear (14). To identify whether hyperactive Notch was driving the reduction in secretory cells, we reviewed a previously reported transcriptome data set from laser capture microdissected small intestinal crypts of WT and Mtgr1−/− mice (17) and identified that Notch3 and Notch-effector Deltex homolog 1 (Dtx1) were up-regulated 2-fold, whereas secretory markers and targets of Notch repression Atoh1 and Chromogranin b (Chgb) were down-regulated 2-fold (Fig. 1A). Stem cell marker Leucine-rich repeat containing G protein–coupled receptor 5 (Lgr5) was also up-regulated 2-fold. Collectively, the data suggest a lack of intestinal differentiation (Fig. 1A). Transcriptome analysis of whole Mtgr1−/− small intestines by quantitative PCR (qPCR) confirmed a hyperactive Notch profile with goblet cell–associated Muc2, enteroendocrine-associated Chga, and Paneth/goblet-associated Gfi1 all down-regulated: Muc2, 3.5-fold (P < 0.05); Chga, 4-fold (P < 0.01); and Gfi1, 3.4-fold (P < 0.05) (Fig. 1B). To determine whether the changes in secretory markers were confined to the small intestine, we extended our analysis to the colon and observed similar decreases in secretory lineage markers such as Muc2, Chga, and Spedf: Muc2, 2-fold (P < 0.05); Chga, 10-fold (P < 0.001); and Spedf, 2.5-fold (P < 0.01) (Fig. 1B).
Figure 1.
Notch signaling is hyperactive, and secretory differentiation markers are down-regulated in the setting of Mtgr1 loss. A) Heatmap of dysregulated Notch-associated genes in microarray from laser-captured small intestinal crypts from WT and Mtgr1−/− mice (17). B) qPCR of the indicated lineage markers in small intestines (left) and colons (right) of WT (n = 4) and Mtgr1−/− (n = 4) mice. C) Heatmap of secretory-associated transcripts in WT and Mtgr1−/− MSIE cells. D) Representative images of enteroid cultures from WT and Mtgr1−/− mice 1, 3, and 5 d after plating. E) Heatmap of Notch-associated genes in WT and Mtgr1−/− small intestinal-derived enteroids. Red denotes increased fold expression relative to WT, and green denotes decreased fold expression relative to WT. The statistical test used was the Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
An ex vivo approach to test whether loss of epithelial Mtgr1 resulted in Notch activation was next employed. First, we generated a conditionally immortalized Mtgr1−/− intestinal epithelial cell line by harvesting intestinal epithelial cells from Mtgr1−/− mice crossed with the Immorto Mouse (19). RNA analysis of this line identified transcriptome changes consistent with increased Notch signaling and a lack of differentiation. Notch3 was increased 2.8-fold, whereas differentiation markers Muc2, Gfi1, NeuroD, and Chga were down-regulated 1.5-, 2-, 2.2-, and 5-fold, respectively (Fig. 1C). Second, we generated Mtgr1−/− intestinal enteroids to assess the effect of Mtgr1 loss in a 3-dimensional intestinal culture model (Fig. 1D). We were unable to culture Mtgr1−/− enteroids longer than 7 d. However, during this time frame, we were able to observe that Mtgr1−/− enteroids remained in a stem-spheroid morphology. The persistence of stem-spheroids suggested that the Mtgr1−/− enteroids failed to differentiate (Fig. 1D). In support of this, we observed RNA changes indicating hyperactive Notch signaling. Notch2 was elevated 3-fold, whereas secretory markers ChgA, Muc2, and Atoh1 were down-regulated 40-fold (Fig. 1E). Thus, our ex vivo data closely paralleled our in vivo observations. Overall, the data indicate that Mtgr1 loss results in Notch activation and preferential shunting to absorptive intestinal lineages.
MTGR1 suppresses CSL-dependent transcription
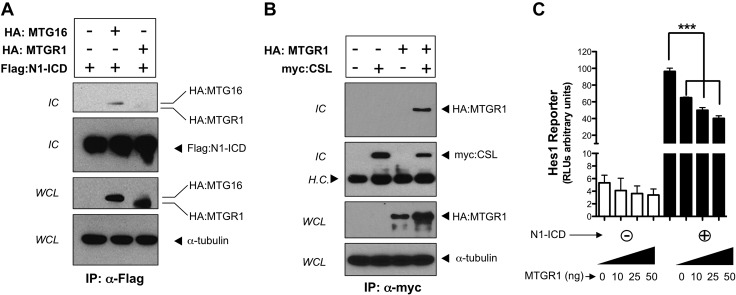
Given that ectopic expression of Notch receptors or the N-ICD resulted in a phenotype similar to deletion of Mtgr1 (1, 8, 9), we hypothesized that MTGR1 could negatively regulate Notch-induced transcription. It has previously been reported that MTG family members can interact with the Notch1 intracellular domain (N1-ICD) and the key Notch effector CSL (27), but the functional consequence of this is unclear. To identify a molecular mechanism by which MTGR1 could regulate Notch activity in vitro, we tested whether MTGR1 could interact with the N1-ICD or CSL by coimmunoprecipitation. We did not detect an interaction between MTGR1 and the N1-ICD (Fig. 2A). We did, however, identify that MTGR1 interacts with CSL (Fig. 2B). Because the prototypic function of MTGR1 is transcriptional repression, we next used a luciferase reporter assay to examine the impact of MTGR1 on the Notch target gene Hes1. MTGR1 transfection into NIH 3T3 cells with or without N1-ICD coexpression resulted in dose-dependent repression of a Hes1 luciferase reporter plasmid (Fig. 2C). Overall, the data suggest that MTGR1 acts as an upstream regulator of Notch-dependent transcriptional activity by interacting with CSL.
Figure 2.
MTGR1 interacts with CSL and suppresses Notch-induced transcriptional activity. A) HA-epitope–tagged MTGR1 and Flag-epitope–tagged N1-ICD were transiently expressed in COS7L cells. HA:MTGR1 was blotted for in immune complexes (IC) (lane 3). HA-MTG16 and Flag-N1-ICD were also transiently transfected and immunoprecipitated as a positive control (lane 2). B) HA:MTGR1 and myc-epitope–tagged CSL were transiently expressed in COS7L cells in the combinations shown. The presence of myc:CSL and HA:MTGR1 was determined in immune complexes. C) MTGR1, in the quantities shown, was transiently expressed in NIH 3T3 cells with or without N1-ICD coexpression, along with the transcriptional reporter Hes1-Lux. Luciferase activity was quantified in whole-cell extracts by luminometry. Results were normalized to an internal Renilla constitutive reporter. The statistical test used was the Student’s t test. H.C., heavy chain; WCL, whole-cell lysate. ***P < 0.001.
GSI rescues the hyperproliferative phenotype in Mtgr1−/− intestine
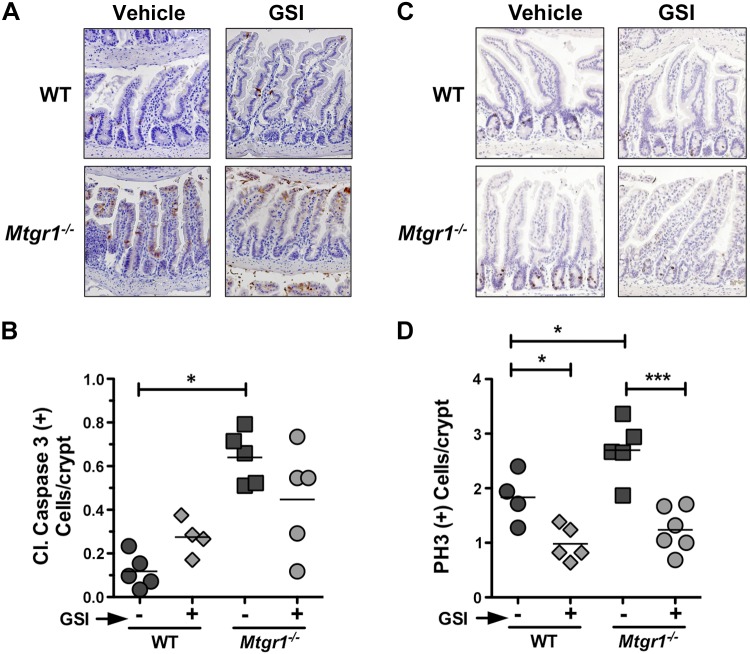
To test whether Notch inhibition was sufficient to rescue loss of secretory cells in the Mtgr1−/− mice, we treated 8- to 12-wk-old Mtgr1−/− mice with DBZ, a Notch inhibitor/GSI. GSI treatment effectively blocked Notch activation because we observed an increase in CD4−/CD8−, CD4+/CD8−, and CD4−/CD8+ T cells and a decrease in CD4+/CD8+ T cells (Supplemental Fig. 1). Mtgr1−/− mice demonstrate increased intestinal proliferation and apoptosis at baseline as previously reported (17), and we observed similar increases in vehicle-treated Mtgr1−/− mice compared to WT (Fig. 3). Apoptosis, as determined by cleaved caspase-3, was not significantly affected by GSI treatment (Fig. 3B). Immunohistochemical analysis for pH3 (Ser10), however, showed that GSI reduced the number of proliferating cells per crypt in both WT and Mtgr1−/− small intestine (Fig. 3C, D), indicating that the previously reported basal increase in proliferation in Mtgr1−/− intestine (17) is at least partially due to Notch activation.
Figure 3.
GSI reduces proliferation but has no effect on apoptosis in the Mtgr1−/− intestine. Representative sections are shown of distal intestine (ileum) from vehicle or GSI-treated WT and Mtgr1−/− mice stained for (A) apoptosis marker cleaved caspase-3 or (C) proliferation marker pH3. Positive staining cells for (B) cleaved caspase-3 and (D) pH3 were quantified per crypt-villus unit. A minimum of 25 crypt-villus units was counted per mouse. The statistical test used was 1-way ANOVA. *P < 0.05; ***P < 0.001. Images were captured using a ×20 objective.
GSI increases production of goblet and enteroendocrine cells in Mtgr1−/− intestine but fails to augment Paneth cell production
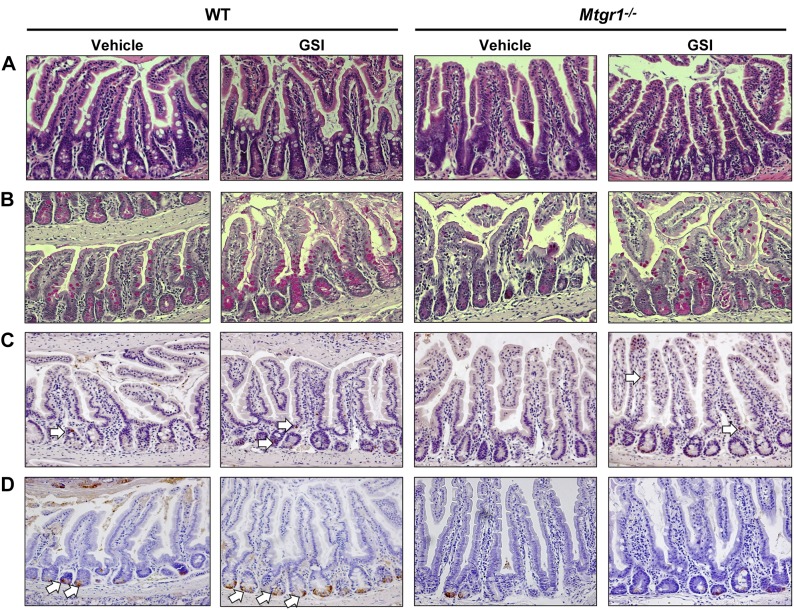
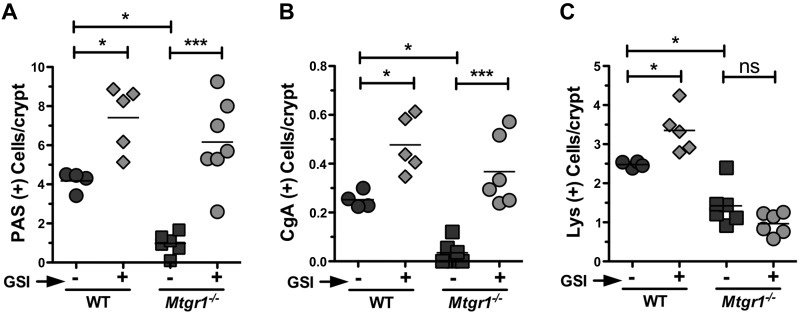
Histologic architecture of both WT and Mtgr1−/− small intestine was unchanged by GSI treatment (Fig. 4A). GSI treatment increased WT and Mtgr1−/− goblet cell indices in comparison to vehicle-treated mice (Fig. 4B), with the most pronounced effects seen in the ileum: Vehicle 0.97 vs. GSI 6.1 PAS+ cells/villus-crypt unit in Mtgr1−/− (P < 0.001) (Fig. 5A and Supplemental Fig. 2). To identify changes in enteroendocrine cell number, we stained for CgA (Fig. 4C). Similarly, GSI treatment significantly augmented the number of enteroendocrine cells compared to vehicle controls: Vehicle 0.035 vs. GSI 0.36 CgA+ cell/villus-crypt unit in Mtgr1−/− (P < 0.001) (Fig. 5B and Supplemental Fig. 2). Interestingly, whereas anti-Lys staining for Paneth cells (Fig. 4D) revealed that GSI treatment increased the number of Paneth cells in WT mice, GSI treatment had no effect on Paneth cells in any segment of the small intestine in Mtgr1−/− mice [Vehicle 0.96 vs. GSI 1.4 Lys+ cell/villus-crypt unit in Mtgr1−/− (P < 1.0)] (Fig. 5C and Supplemental Fig. 2), revealing a previously unrecognized role for MTGR1 in Paneth cell differentiation. Taken together, our findings indicate that MTGR1 is a key regulator of intestinal lineage allocation.
Figure 4.
Notch inhibition increases goblet and enteroendocrine, but not Paneth, cells in Mtgr1−/− mice. Vehicle and GSI (DBZ, 10 μM/kg twice daily for 5 d)-treated WT and Mtgr1−/− mice are shown. A) H&E staining of distal intestine sections. PAS staining for goblet cells (B), CgA for enteroendocrine cells (C), and Lys staining for Paneth cells (D) in the distal intestine. White arrows indicate positive-staining cells. Images were captured using a ×20 objective.
Figure 5.
Mtgr1 is required for Paneth lineage production after Notch inhibition. Quantification is shown of (A) PAS+, (B) CgA+, and (C) Lys+ cells per intestinal crypt/villus structure in WT and Mtgr1−/− mice treated with vehicle or GSI. A minimum of 25 crypt-villus units was counted per mouse. ns, not significant. The statistical test used was 1-way ANOVA. *P < 0.05; ***P < 0.001.
MTGR1 interacts with GFI1 and can repress GFI1 targets
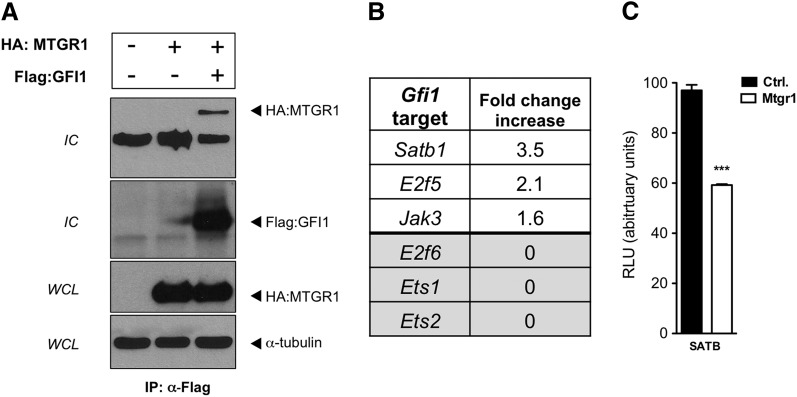
GSI treatment of Mtgr1−/− intestine increased Atoh1 and decreased Hes1 mRNA (Supplemental Fig. 3A, B) and induced greater goblet and enteroendocrine cell production (Figs. 4 and 5). Paneth lineages in Mtgr1−/− intestines, however, were not increased after GSI treatment. GFI1 is a key transcriptional corepressor in Paneth cell allocation (31). Similar to GSI’s effect on Atoh1 transcription, GSI treatment increased Gfi1 expression in Mtgr1−/− intestines (Supplemental Fig. 3C), suggesting that MTGR1 does not directly regulate Gfi1 transcription. Given that MTGR1 orchestrates the assembly of repression complexes containing histone deacetylases and transcriptional corepressors such as HDAC3, mSin3A, and N-CoR (14, 17), we postulated that MTGR1 could interact with GFI1 and may contribute to repression of a subset of GFI1 targets. In support of this, MTGR1 coimmunopurified with GFI1 (Fig. 6A). Analysis of a laser capture microdissection expression data set derived from WT and Mtgr1−/− intestinal crypts (17) indicated that Satb1, E2f5, and Jak3, targets of GFI1 repression (32), were up-regulated in Mtgr1−/− intestine (Fig. 6B). Other GFI1 targets such as E2f6, Ets1, and Ets2 were unchanged (Fig. 6B), suggesting that MTGR1 may be required to repress a subset of GFI1 targets. Furthermore, MTGR1 was capable of repressing a Satb-luciferase reporter activity by 50% (Fig. 6C). Our data indicate that MTGR1 can interact with GFI1 and cooperate in repressing a subset of GFI1 targets.
Figure 6.
MTGR1 interacts with transcriptional corepressor GFI1 and can repress GFI1 targets. A) HA:MTGR1 and Flag:GFI1 were transiently expressed in COS7L cells. Flag:GFI1 was immunoprecipitated, and HA:MTGR1 was blotted for in immune complexes (IC). B) GFI1 repression targets Satb1, E2f5, and Jak3 were up-regulated in laser-captured crypts from WT and Mtgr1−/− mice (17). C) HA:MTGR1 was transfected into NIH 3T3 cells with the transcriptional reporter Satb1-Lux. Luciferase activity was quantified in whole-cell extracts by luminometry. Results were normalized to an internal Renilla constitutive reporter. Ctrl., control. The statistical test used was the Student’s t test. ***P < 0.001.
Discussion
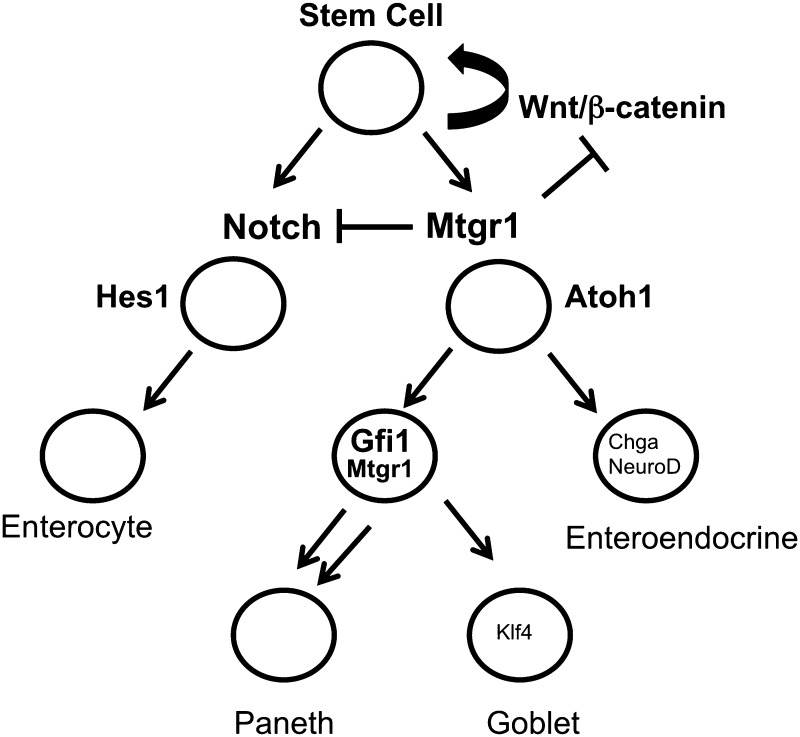
Notch signaling plays a critical yet incompletely understood role in determining intestinal cell fate (1, 2). When Mtgr1−/− mice were generated and examined for intestinal phenotypes, they had unexpected characteristics: 1) increased proliferation in the small intestine and colon, and 2) the progressive depletion of the secretory lineage in the small intestine (14, 17, 33), suggesting increased Notch tone and identifying MTGR1 as an important regulator of fate determination in the intestine. Here, we show that Notch inhibition by GSI treatment increased production of the goblet and enteroendocrine lineages in the Mtgtr1−/− intestine but failed to increase the production of the Paneth lineage. The results suggest a model in which MTGR1 represses Notch activation through an interaction with CSL (Fig. 7), and in its absence, unbridled Notch signaling drives the intestinal progenitors to a predominantly absorptive epithelium. Inhibiting Notch, as was achieved with a GSI, increased the production of secretory cells, with the exception of the Paneth lineage, suggesting that in addition to acting at the Notch/CSL interface, MTGR1 also regulates downstream lineage allocation. Because we identified that MTGR1 interacts with GFI1, a key transcriptional corepressor in Paneth cell differentiation, and can repress GFI1 targets, MTGR1 could be a requirement for GFI1-mediated Paneth cell differentiation.
Figure 7.
Schematic of MTGR1 regulating multiple branch points in intestinal lineage differentiation. We propose that MTGR1 acts at multiple levels in regulating intestinal epithelial differentiation. MTGR1 negatively regulates Wnt signaling by competing with β-catenin for TCF4 occupancy. As the stem cell divides and moves along the crypt-villus axis, MTGR1 acts as a negative regulator of Notch signaling by interacting with CSL and repressing Notch-induced transcriptional activity. As cells differentiate into either HES1+ enterocytes or ATOH1+ secretory progenitors, MTGR1 is also required downstream for Paneth cell differentiation potentially through its interaction with GFI1.
Although the role of MTGs in hematopoiesis is well characterized, studies using knockout animal models revealed that MTGs have unanticipated roles in the gut. MTGs regulate intestinal development and lineage allocation programs, but each family member appears to regulate unique differentiation programs. A small percentage of Mtg8−/− mice show a deletion of their midgut, and those that survive postnatally demonstrate severe intestinal architectural disruption (15). Mtg16−/− mice show normal gut architecture, but they have decreased goblet cells (unpublished observations). Mtgr1−/− mice, on the other hand, show a marked reduction in all secretory indices (14). Given the importance of Notch signaling in the intestine, the phenotypes strongly suggested that MTGs modulate Notch. Components of the Notch pathway can interact with MTG16 in vitro (27), but no in vivo Notch studies have been conducted until now. Our results are the first to show that inhibition of Notch signaling can partially generate secretory lineages in Mtgr1−/− intestine.
The balance between absorptive and secretory cell fate in the intestine is largely governed in a reciprocal fashion by Notch and Atoh1 signaling pathways. Notch ligands activate Notch receptors that are subsequently cleaved by γ secretase to produce the N-ICD, resulting in N-ICD translocation to the nucleus where it increases the expression of Hes1, a repressor of the prosecretory cell transcription factor Atoh1. It is interesting to note that Mtgr1−/− mice closely phenocopy the pan-secretory-deficient Atoh1 mutants, suggesting a potential interplay between MTGR1 and ATOH1. How ATOH1 and MTGR1 cooperate in intestinal fate decisions is not known.
The inability of Notch inhibition to induce Paneth cell differentiation in the Mtgr1−/− mice identifies MTGR1 as a new Paneth cell allocation constituent. The mechanism of Paneth cell differentiation is controversial, but it is believed that a goblet/Paneth cell progenitor is marked by Gfi1 expression (31, 34) (Fig. 7). Gfi1-deficient mice have increased enteroendocrine cells, suggesting that loss of GFI1 drove supernumerary enteroendocrine cells because of the inability of the progenitor to differentiate into mucous and Paneth cells (31). It has also been proposed that GFI1 represses Neurog3 and, by doing so, “stabilizes” the Paneth and mucous cell lineages (34). In Gfi1-deficient mice, unchecked Neurog3 expression drives progenitor cells toward enteroendocrine cells. How MTGR1 is positioned in this model is unclear, but Gfi1 transcripts were increased after GSI administration in Mtgr1−/− intestine (Supplemental Fig. 3C), suggesting that MTGR1 is not involved in regulating Gfi1 expression. Our data suggest that GFI1 requires MTGR1 to repress a subset of targets and therefore initiate programming to properly form Paneth cells in the setting of Notch inhibition. Indeed, we determined that MTGR1 and GFI1 can complex and that MTGR1 was capable of repressing a GFI1 target reporter (Fig. 6A, C), suggesting that MTGR1 could interact with and enable GFI1-mediated repression. In support of this, we found a subset of GFI1 targets that was up-regulated in Mtgr1−/−crypts (Fig. 6B), suggesting that GFI1 may require MTGR1 to initiate some differentiation programs.
Paneth cell function is an intense area of investigation because of its potential supporting role in crypt stem cell biology (35–37). Understanding transcriptional programs regulating Paneth allocation is critical to defining their function in the intestinal stem cell niche. More broadly, intestinal secretory cell differentiation is still incompletely understood. MTGR1, a transcriptional corepressor originally discovered to be critical in hematopoiesis, appears to be an important and heretofore unrecognized contributor to intestinal cell lineage allocation.
Supplementary Material
Acknowledgments
The authors would like to thank the members of the C.S.W. lab for helpful discussions and proofing of the manuscript. The authors would also like to thank Robert Whitehead and the Vanderbilt DDRC Novel Cell Line Development Subcore for their assistance in generating the Mtgr1−/− MSIE line, and Dr. Tarik Moroy for graciously providing reagents to M.E.E. This work was supported by the U.S. National Institutes of Health (NIH) Grants K08DK080221 (to C.S.W.), R01DK092306 (to N.F.S.), R01CA1428260 (to N.F.S.), K08DK080190 (to M.E.E.) R01CA178030 (to S.W.H), P30DK058404 (Vanderbilt Digestive Disease Research Center), P50CA095103 (to M.K.W.), F30DK096718-01 (to B.P.), and T32 GM07347 (NIH/National Institute of General Medical Sciences) (to B.P.), Merit Review Grants from the Office of Medical Research, U.S. Department of Veterans Affairs 1I01BX001426 (to C.S.W.), and ACS-RSG 116552 (to C.S.W.). This publication was also supported in part by the National Cancer Institute Cancer Center Support Grant P30CA068485. This publication was also supported by the St. Baldrick's Foundation (to M.E.E.). This publication was also supported by CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH.
Glossary
- Atoh1
Atonal homolog 1
- CgA
chromogranin A
- Chga
Chromogranin A
- Chgb
Chromogranin b
- CSL
suppressor of hairless homolog
- DBZ
dibenzazepine
- DDRC
Digestive Disease Research Center
- Dll
Delta-like
- Dtx1
Deltex homolog 1
- Gfi1
growth factor-independent 1
- GSI
γ-secretase inhibitor
- H&E
hematoxylin and eosin
- HES1
Hairy/Enhancer of Split 1
- Lgr5
Leucine-rich repeat containing G protein–coupled receptor 5
- Lys
lysozyme
- MSIE
mouse small intestinal epithelial
- MTG
myeloid translocation gene
- Mtgr1
myeloid translocation gene-related 1
- Muc2
Mucin2
- N1-ICD
Notch1 intracellular domain
- NeuroD1
Neuronal differentiation 1
- N-ICD
Notch intracellular domain
- PAS
periodic acid Schiff
- pH3
phospho-histone H3
- qPCR
quantitative PCR
- Spedf
SAM-pointed domain-containing ETS transcription factor
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. (2005) Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964–968 [DOI] [PubMed] [Google Scholar]
- 2.Noah T. K., Donahue B., Shroyer N. F. (2011) Intestinal development and differentiation. Exp. Cell Res. 317, 2702–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray S. J. (2006) Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 4.Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellegrinet L., Rodilla V., Liu Z., Chen S., Koch U., Espinosa L., Kaestner K. H., Kopan R., Lewis J., Radtke F. (2011) Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240, e1–e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stamataki D., Holder M., Hodgetts C., Jeffery R., Nye E., Spencer-Dene B., Winton D. J., Lewis J. (2011) Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS One 6, e24484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh M., Katoh M. (2007) Notch signaling in gastrointestinal tract (review). [review] Int. J. Oncol. 30, 247–251 [PubMed] [Google Scholar]
- 8.Stanger B. Z., Datar R., Murtaugh L. C., Melton D. A. (2005) Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA 102, 12443–12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zecchini V., Domaschenz R., Winton D., Jones P. (2005) Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 19, 1686–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Es J. H., van Gijn M. E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D. J., Radtke F., Clevers H. (2005) Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 [DOI] [PubMed] [Google Scholar]
- 11.Riccio O., van Gijn M. E., Bezdek A. C., Pellegrinet L., van Es J. H., Zimber-Strobl U., Strobl L. J., Honjo T., Clevers H., Radtke F. (2008) Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen J., Pedersen E. E., Galante P., Hald J., Heller R. S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O. D. (2000) Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44 [DOI] [PubMed] [Google Scholar]
- 13.Davis J. N., McGhee L., Meyers S. (2003) The ETO (MTG8) gene family. Gene 303, 1–10 [DOI] [PubMed] [Google Scholar]
- 14.Amann J. M., Chyla B. J. I., Ellis T. C., Martinez A., Moore A. C., Franklin J. L., McGhee L., Meyers S., Ohm J. E., Luce K. S., Oulette A. J., Washington M. K., Thompson M. A., King D., Guataum S., Coffey R. J., Whitehead R. H., Hiebert S. W. (2005) Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol. Cell. Biol. 25, 9576–9585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calabi F., Pannell R., Pavloska G. (2001) Gene targeting reveals a crucial role for MTG8 in the gut. Mol. Cell. Biol. 21, 5658–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chyla B. J., Moreno-Miralles I., Steapleton M. A., Thompson M. A., Bhaskara S., Engel M., Hiebert S. W. (2008) Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol. Cell. Biol. 28, 6234–6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore A. C., Amann J. M., Williams C. S., Tahinci E., Farmer T. E., Martinez J. A., Yang G., Luce K. S., Lee E., Hiebert S. W. (2008) Myeloid translocation gene family members associate with T-cell factors (TCFs) and influence TCF-dependent transcription. Mol. Cell. Biol. 28, 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Es J. H., de Geest N., van de Born M., Clevers H., Hassan B. A. (2010) Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat. Commun. 1, 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehead R. H., Robinson P. S. (2009) Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G455–G460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead R. H., VanEeden P. E., Noble M. D., Ataliotis P., Jat P. S. (1993) Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc. Natl. Acad. Sci. USA 90, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musch M. W., Petrof E. O., Kojima K., Ren H., McKay D. M., Chang E. B. (2004) Bacterial superantigen-treated intestinal epithelial cells upregulate heat shock proteins 25 and 72 and are resistant to oxidant cytotoxicity. Infect. Immun. 72, 3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard J. K., McCann S. P., Bhardwaj V., Washington M. K., Frey M. R. (2012) Neuregulin-4 is a survival factor for colon epithelial cells both in culture and in vivo. J. Biol. Chem. 287, 39850–39858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., Clevers H. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 [DOI] [PubMed] [Google Scholar]
- 24.Barrett C. W., Fingleton B., Williams A., Ning W., Fischer M. A., Washington M. K., Chaturvedi R., Wilson K. T., Hiebert S. W., Williams C. S. (2011) MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res. 71, 1302–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazanjian A., Noah T., Brown D., Burkart J., Shroyer N. F. (2010) Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology 139, 918–928, e1–e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amann J. M., Nip J., Strom D. K., Lutterbach B., Harada H., Lenny N., Downing J. R., Meyers S., Hiebert S. W. (2001) ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21, 6470–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel M. E., Nguyen H. N., Mariotti J., Hunt A., Hiebert S. W. (2010) Myeloid translocation gene 16 (MTG16) interacts with Notch transcription complex components to integrate Notch signaling in hematopoietic cell fate specification. Mol. Cell. Biol. 30, 1852–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Signalling downstream of activated mammalian Notch. Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 29.Vassen L., Fiolka K., Möröy T. (2006) Gfi1b alters histone methylation at target gene promoters and sites of gamma-satellite containing heterochromatin. EMBO J. 25, 2409–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt A., Fischer M., Engel M. E., Hiebert S. W. (2011) Mtg16/Eto2 contributes to murine T-cell development. Mol. Cell. Biol. 31, 2544–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shroyer N. F., Wallis D., Venken K. J. T., Bellen H. J., Zoghbi H. Y. (2005) Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 19, 2412–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Z., Horwitz M. (2003) Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl. Acad. Sci. USA 100, 5932–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez J. A., Williams C. S., Amann J. M., Ellis T. C., Moreno-Miralles I., Washington M. K., Gregoli P., Hiebert S. W. (2006) Deletion of Mtgr1 sensitizes the colonic epithelium to dextran sodium sulfate-induced colitis. Gastroenterology 131, 579–588 [DOI] [PubMed] [Google Scholar]
- 34.Bjerknes M., Cheng H. (2010) Cell Lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev. Biol. 345, 49–63 [DOI] [PubMed] [Google Scholar]
- 35.Durand A., Donahue B., Peignon G., Letourneur F., Cagnard N., Slomianny C., Perret C., Shroyer N. F., Romagnolo B. (2012) Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl. Acad. Sci. USA 109, 8965–8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim T. H., Escudero S., Shivdasani R. A. (2012) Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. USA 109, 3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker N., van Oudenaarden A., Clevers H. (2012) Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell 11, 452–460 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.