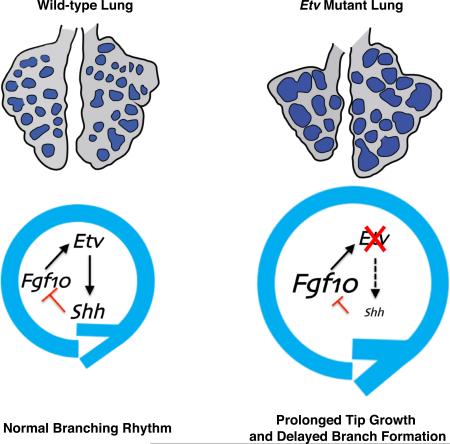
Summary
The mammalian lung forms its elaborate tree-like structure following a largely stereotypical branching sequence. While a number of genes have been identified to play essential roles in lung branching, what coordinates the choice between branch growth and new branch formation has not been elucidated. Here we show that loss of FGF-activated transcription factor genes, Etv4 and Etv5 (collectively Etv), led to prolonged branch tip growth and delayed new branch formation. Unexpectedly, this phenotype is more similar to mutants with increased, rather than decreased FGF activity. Indeed, an increased Fgf10 expression is observed and reducing Fgf10 dosage can attenuate the Etv mutant phenotype. Further evidence indicates that ETV inhibits Fgf10 via directly promoting Shh expression. SHH in turn inhibits local Fgf10 expression and redirects growth, thereby initiating new branches. Together, our findings establish ETV as a key node in the FGF-ETV-SHH inhibitory feedback loop that dictates the rhythm of branching.
Graphical Abstract
Introduction
In the mammalian lung, multiple rounds of branching morphogenesis are essential for generating the surface area required for efficient gas-exchange. In mice, lung development initiates at embryonic day (E) 9.5, with the outgrowth of a pair of epithelial buds from the embryonic foregut (Cardoso and Lü, 2006; Morrisey and Hogan, 2010). Subsequently, these buds elongate and begin the process of branching morphogenesis. During this process, new tips emerge through one of three defined modes of epithelial branching (Metzger et al., 2008; Short et al., 2013). The largely stereotypical nature of the branching sequence suggests strict genetic control of this process.
Many signaling pathways play critical roles in lung development (Warburton et al., 2005; Cardoso and Lü, 2006; Morrisey and Hogan, 2010). At the center of these is the fibroblast growth factor (hereafter FGF) signaling pathway. Multiple FGF ligands and all four FGF receptors are expressed in the lung at varying stages of development (Warburton et al., 2005; Cardoso and Lü, 2006; Morrisey and Hogan, 2010; Yu et al., 2010). The earliest requirement of FGF signaling is for FGF10 expressed in the mesoderm, which signals through the FGFR2IIIb isoform in the epithelium, and is essential for the formation of the initial lung buds (Min et al., 1998; Sekine et al., 1999; De Moerlooze et al., 2000). During branching morphogenesis, FGF10 acts as a chemoattractant for the distal epithelium (Bellusci et al., 1997b; Weaver et al., 2000), although whether localized Fgf10 expression is essential for branch formation is debated (Volckaert et al., 2013). This requirement for FGF10 in epithelial branch outgrowth is illustrated by Fgf10 hypomorphs and Fgf10 conditional knockout mice, which develop reduced lung epithelial branch number (Mailleux et al., 2005; Abler et al., 2009).
FGF has been shown to regulate the expression of multiple signaling pathway genes, the most prominent of which is sonic hedgehog (hereafter Shh) (Bellusci et al., 1997b; Pepicelli et al., 1998; Abler et al., 2009). In the lung, FGF10 and SHH form a feedback loop where FGF10 produced in the mesenchyme, signals to the distal epithelium to upregulate Shh expression. SHH then feeds back to inhibit Fgf10 expression in the adjacent mesenchyme, which in effect splits the Fgf10-expression domain in two. It is postulated that the new FGF10 signaling domains serve as two chemoattractant sources, which direct the bifurcation of the epithelial tip. This hypothesis is supported by mathematical modeling studies suggesting that the FGF10-SHH feedback loop is a central driver of the branching program (Hirashima et al., 2009; Menshykau et al., 2012; Iber and Menshykau, 2013). While each of the signaling components of the feedback loop has been shown to be essential for branching, how the balanced coordination of this feedback loop impacts branch growth and new branch formation has not been demonstrated.
In the FGF10-SHH feedback loop, FGF10 and SHH regulate each other's expression on the transcript level. Interestingly, while many studies have been devoted to examining the function of signaling pathway ligands and receptors during lung development, relatively few studies have examined how the downstream components, e.g. transcription factors, mediate this process. Consequently, the transcription factors involved in the FGF10-SHH feedback loop remain unknown. We have previously performed a genome scale transcription factor in situ hybridization screen to identify genes expressed in the branching lung (Herriges et al., 2012). From this screen, we identified the PEA3 group ETS domain transcription factor Etv5 as a gene expressed in the distal epithelium. Etv5 is one of three PEA3 group transcription factor genes, all of which are expressed in the developing lung. However, only Etv4 and Etv5 (hereafter collectively Etv) are strongly expressed in the lung epithelium (Chotteau-Lelièvre et al., 1997; Liu et al., 2003; Herriges et al., 2012). Previous studies have shown that exogenous FGF can induce Etv expression in the lung, making them attractive candidates for mediating FGF function during lung development (Liu et al., 2003; Lin et al., 2006). In addition, overexpression of a dominant-negative ETV5-Engrailed repressor fusion protein, has suggested that ETV transcription factors are necessary for proper epithelial patterning and cell differentiation (Liu et al., 2003). However, the potential role of ETV factors in mediating FGF function during branching morphogenesis remains unclear.
In this study we conditionally inactivated Etv genes in the lung epithelium and demonstrated that this leads to dilation of branch tips and reduced branch number. The epithelial Etv mutant lungs exhibit increased Fgf10 expression and decreased Shh expression. Our data suggest that ETV factors act as intermediates in the FGF-SHH negative feedback loop, by acting downstream of FGF signaling and promoting the activity of a long range Shh-enhancer. Loss of Etv thereby tipped the balance of the FGF-SHH feedback loop, leading to an altered branching rhythm resulting in increased tip size and decreased tip number.
Results
Inactivation of Etv4 and Etv5 led to dilated branch tips and reduced branch number
To investigate the requirement for Etv during development, we inactivated both Etv4 and Etv5 (hereafter collectively Etv) in the embryonic lung epithelium, a cell layer where they are co-expressed (Supplemental Fig.1A-D). This was achieved by combining an epithelial-specific Shhcre allele, with a conditional Etv5fl allele and a null Etv4− allele, which generated Shhcre/+;Etv4−/−;Etv5fl/fl (hereafter Etv mutant) embryos (Laing et al., 2000; Harfe et al., 2004; Zhang et al., 2009). We used Shhcre/+;Etv4+/−;Etv5fl/+ as controls unless otherwise stated. In the mutant, there was an ~85% reduction of the full-length Etv5 transcript by E11.5, demonstrating efficient inactivation by CRE (Fig. 1A). In addition to the Etv mutant, both Shhcre/+;Etv4+/+;Etv5fl/fl and Shhcre/+; Etv4+/−;Etv5fl/fl embryos exhibited lung branching phenotypes similar to albeit not as severe as the Etv mutant while other genotypes are normal (data not shown). In addition, further inactivating Etv1 in the Etv mutant background (generating Shhcre/+;Etv1fl/fl;Etv4−/−;Etv5fl/fl embryos) did not alter the phenotypes, consistent with the observation that Etv1 is primarily expressed in the lung mesenchyme (data not shown)(Chotteau-Lelièvre et al., 1997). Henceforth, we focused our analysis on the Etv mutant (Shhcre/+;Etv4−/−;Etv5fl/fl), as it exhibits the most severe lung phenotypes among the genotypes listed above.
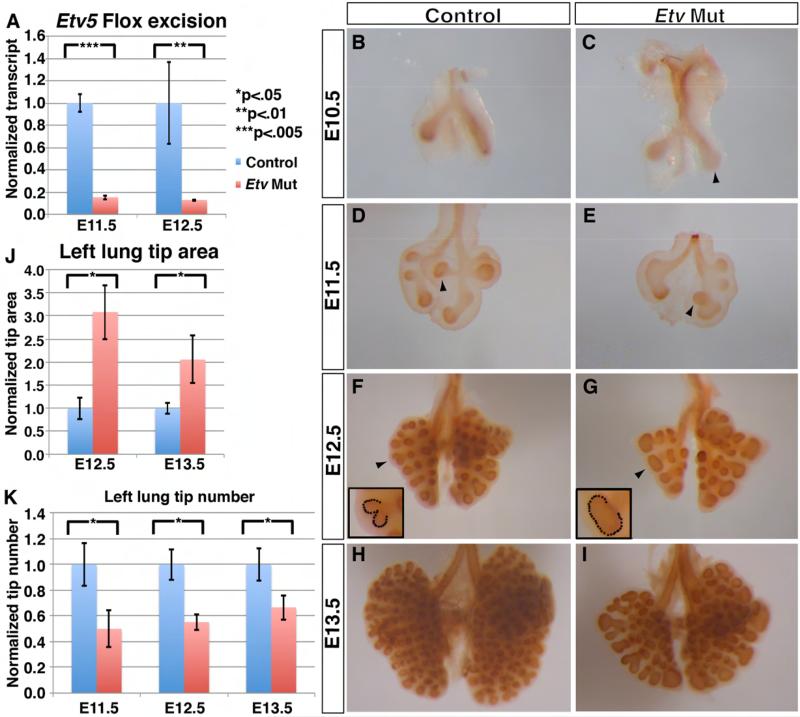
Figure 1. Inactivation of Etv in the lung epithelium led to epithelial branching defects.
(A) Etv5 inactivation was efficient as evidenced by the clear reduction of full-length transcripts in the Etv mutant lung (E11.5: 1.0 for controls, 0.14 for Etv mutants, p=0.005; E12.5: 1.0 for controls, 0.15 for Etv mutants, p=0.02, n=3 each group). (B-I) Representative control (B,D,F,H) and Etv mutant (C,E,G,I) whole lungs with epithelium outlined by anti-E-Cadherin immunohistochemical staining. In the mutant, the tip dilation phenotype was already apparent at E10.5 (arrowhead in C). The reduced tip number phenotype was apparent shortly after the initiation of secondary branching at E11.5; the position of the bud for the future accessory lobe was shifted more posteriorly compared to control (arrowheads in D and E). (J) Tip area is increased in the left lung at the indicated stages (E12.5: 1.0 for controls and 3.08 for Etv mutants, p=0.001; E13.5: 1.0 for controls and 2.05 for Etv mutants, p=0.018). (K) Tip number is decreased in the Etv mutant Left lung at the indicated stages (E11.5: 1.0 for controls, 0.5 for Etv mutants, p=.004; E12.5: 1.0 for controls, 0.55 for Etv mutants, p=0.011; E13.5: 1.0 for controls, 0.67 for Etv mutants, p=0.024. Actual tip numbers at the three stages shown are: E11.5: control 3.50 ± 0.57 versus mutant 1.75 ± 0.50; E12.5: control 9.67± 1.15 versus mutant 5.33 ± 0.58; E13.5: control 16.00 ± 2.00 versus mutant 10.67 ± 1.53). Examples of how the tip areas were defined were indicated by arrowheads and outlined in the insets in F-G. Quantification was carried out in n≥4 samples for each genotype and stage. Data are presented as standard error of the means (+SEM), as in graphs in all figures. See also Figure S1.
While Etv mutants did survive and appear to have a normal lifespan, they displayed smaller lungs at birth despite of a normal overall body size (Supplemental Fig. 1E,F). In tracing back the lung hypoplasia phenotype, we found that Etv mutant lungs exhibited morphological defects as early as E10.5, shortly after lung bud initiation (Fig. 1B,C). Anti-ECadherin antibody labeling of the epithelium revealed that the distal tips of nascent buds, especially the larger right lung bud, were consistently dilated in the mutant compared to control (Fig. 1C arrowhead). This tip dilation phenotype remained apparent throughout branching morphogenesis and the increase in tip size was statistically significant at both E12.5 and E13.5, as quantified either in two-dimension or in three-dimension (Fig. 1D-J, Supplemental Fig. 1G-I, movie). Branch number was reduced starting at E11.5 and persisted throughout the branching program (Fig. 1D-I,K).
A significant contributor to reduced branch number is a delay in branch formation. For example, at E11.5 in the left lobe, while two domain branches had formed in the control, none was observed in the mutant (Fig. 1D,E). However, by E12.5, it is apparent that the initial domain branches had formed, but the total branch number was still fewer compared to control (Fig. 1F,G). All three branch subroutines (domain branching, planar bifurcation and orthogonal bifurcation) were observed in the mutant, but all were reduced in number (Metzger et al., 2008; Short et al., 2013). These reductions resulted in mutant lungs that were roughly the same shape, but reduced in size compared to control (Supplemental Fig. 1E,F). No changes in cell proliferation or cell death was observed in either the lung epithelium or mesenchyme, suggesting that the morphological changes may be a combined result of cumulating subtle reduction in cell proliferation rate and altered cell morphogenesis (Supplemental Fig1J-M). Taken together, these results indicate that Etv genes are required in the epithelium for proper branching morphogenesis.
Cell differentiation is largely unperturbed in Etv mutant lungs
To determine if Etv genes play a role in lung patterning and/or cell differentiation, we first analyzed the expression of Sox2 and Sox9, markers for proximal versus distal epithelial cells, respectively. At E12.5 by quantitative RT-PCR (qRT-PCR) analysis, there is no difference in Sox9 expression but a trending increase in Sox2 expression. However, by RNA wholemount in situ hybridization, both genes are expressed in largely expected domains in the context of a dimorphic lung, suggesting no overt patterning defects (Supplemental Fig. 2A-E). We then analyzed the various cell types in the airway or alveolar epithelium. All analyzed cell types including club cells, pulmonary neuroendocrine cells, ciliated cells, basal cells, type I and type II cells appear present in normal proportion (Fig.2A-H, Supplemental Fig.2F-I). This result is unexpected as it is distinct from the reported airway and alveolar cell differentiation defects in transgenic lungs where an ETV5-engrailed transcriptional repressor domain fusion protein was overexpressed in the lung epithelium (Liu et al., 2003). This difference raises the possibility that the phenotypes in the transgenic line may be due to inhibitory effects of the ETV5-Engrailed fusion proteins on factors other than ETV4 and ETV5.
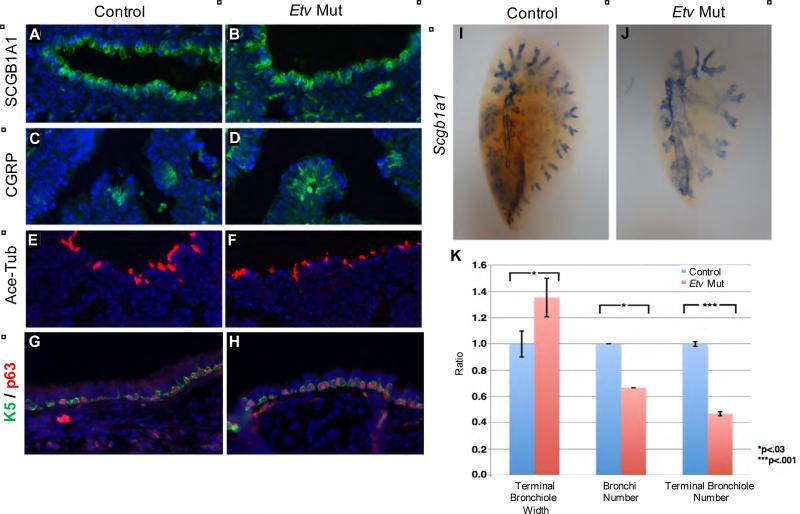
Figure 2. Cell differentiation is largely normal in Etv mutant lungs.
(A-H) Immunofluorescent labeling of airway cell types using indicated markers for club cells (A,B), pulmonary neuroendocrine cells (C,D), ciliated cells (E,F) and basal cells at E18.5 (A-F) or adult (G,H). (I,J) Wholemount RNA in situ hybridization of Scgb1a1 to outline the airway of E18.5 left lobe. (K) Quantification of E18.5 airway phenotype. The terminal bronchiole size was calculated by measuring the tips of Scgb1a1 outlined bronchiole alveolar junction (for ratio: 1.0 for controls and 1.35 for Etv mutants, p=0.011, n=3). The secondary bronchi number was calculated by counting all branches off the main left lung bronchus (for ratio: 1.0 for controls and 0.67 for mutants, p=0,016, n=3; actual average tip number 6±0 for controls and 4±0.8 for mutants). The terminal bronchiole number was calculated by counting the tips of Scgb1a1 outlined bronchiole alveolar junction (for ratio: 1.0 for controls and 0.47 for mutants, p=2.8×10−6, actual tip number 29±0.6 for controls and 14±0.5 for mutants). See also Figure S2.
While all epithelial cell types are present in normal proportion, wholemount staining of Scgb1a1, a club cell marker, outlines airways that are enlarged in diameter and reduced in number (Fig.2I-K). This pattern indicates that the earlier branching defect has a profound impact on later airway organization.
We also analyzed the mesenchymal cell types in the Etv mutants. We found that the smooth muscle cells and endothelial cells are normal (Supplemental Fig.2J-M). In contrast, there is a reduction and disorganization of cartilage cells (Supplemental Fig.2N,O), suggesting that there is a non-autonomous effect of loss of Etv in the epithelium on cartilage differentiation in the mesenchyme.
FGF signaling is increased, rather than decreased in Etv mutant lungs
As FGF promotes Etv expression, we postulated that Etv may serve as a positive mediator of FGF function. Surprisingly, the increase in tip size observed in Etv mutant lungs was more similar to mutants with increased, rather than decreased, FGF signaling (Abler et al., 2009; Tang et al., 2011; Volckaert et al., 2013). To determine if and how FGF activity was altered in the mutant, we examined the pattern of phosphorylated-ERK (hereafter pERK), a commonly assayed readout of FGF activity, using immunofluorescent staining. The pERK signal was elevated in intensity and expanded in domain in the mutant epithelium compared to the control (Fig. 3A-F, Supplemental Fig. 3A-F). Consistent with the expansion of the pERK domain, there is an increase in the expression of Spry2 and Bmp4, two genes known to be promoted by FGF activity by both RNA in situ hybridization and qRT-PCR (Supplemental Fig. 3G-K) (Abler et al., 2009).
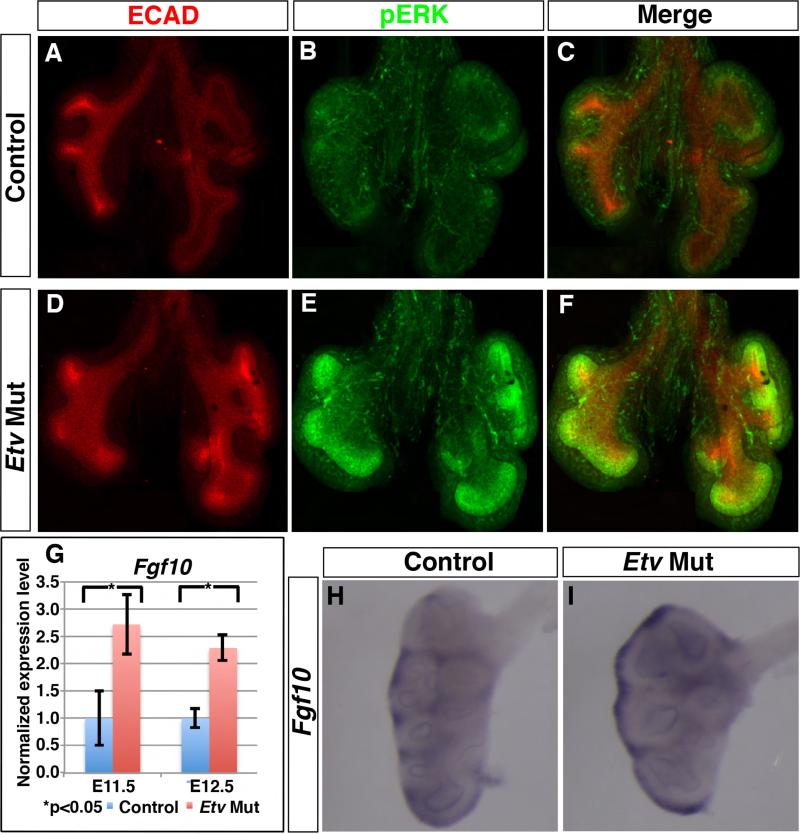
Figure 3. Etv mutant lungs exhibited an increase in FGF signaling activity.
(A-F) Representative E11.5 whole lung immunofluorescently stained for E-Cadherin (red) and pERK (green). pERK staining in the mutant lung was increased in level and expanded in domain compared to control. (G-I) Fgf10 expression was increased as shown by qRT-PCR at stages indicated (E11.5: 1.0 for controls, 2.72 for Etv mutants, p=0.03; E12.5: 1.0 for controls, 2.29 for Etv mutants, p=0.01, n=3 each group) (G) and RNA in situ hybridization at E12.5 (H,I). See also Figure S3.
To trace the cause of this increase in FGF activity, we examined the expression of upstream FGF pathway components. Using quantitative qRT-PCR, we found that the expression of Fgf10, a principle FGF ligand that is critical for branching, was significantly increased in the Etv mutant lung (Fig. 3G). Consistent with this Fgf10 increase, we found that there was an expansion of the Fgf10 expression domains in the mutant lung (Fig. 3H,I).
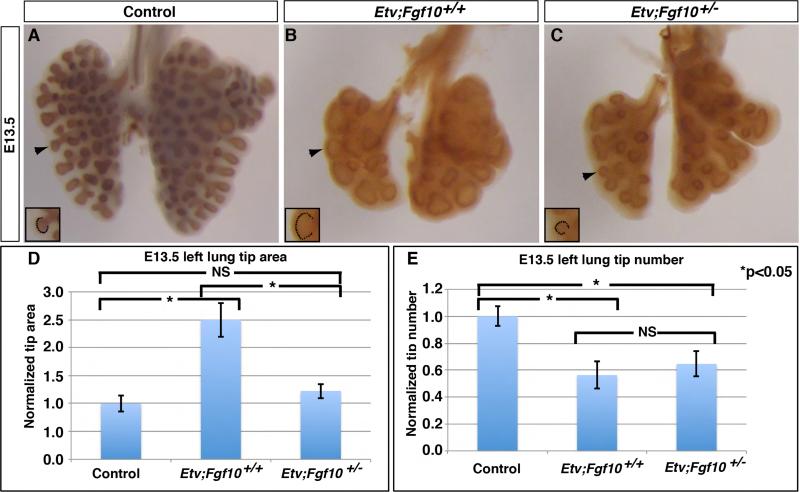
As increased FGF activity has been associated with increased lung epithelial lumen size (Tang et al., 2011), the above results raised the possibility that the increase in FGF activity could contribute to the branching defects observed in the Etv mutant lung. To address this possibility, we reduced FGF signaling in the mutant lung by inactivating one copy of Fgf10 in the Etv mutant background, generating Shhcre/+;Etv4−/−;Etv5fl/fl;Fgf10+/− (hereafter Etv;Fgf10) mutants (Sekine et al., 1999). Compared to Etv mutants, the Etv;Fgf10 mutants exhibited a mild but consistent attenuation of the distal dilation defect starting at E11.5 (Supplemental Fig.4). By E13.5, the epithelial tips of Etv;Fgf10 mutants were roughly 50% smaller than those of Etv mutants, and were not significantly different from Etv controls (tip area normalized to controls: 1.22 for Etv;Fgf10 mutants and 2.50 for Etv mutants, p=0.009 between Etv;Fgf10 and Etv mutants, p=0.075 between Etv;Fgf10 mutants and controls, n=3 each genotype) (Fig. 4A-D). While there was a slight attenuation of the reduced branch number phenotype, the difference between Etv;Fgf10 mutant and Etv mutant lungs was not significant (tip number normalized to controls: 0.64 for Etv;Fgf10 mutants and 0.56 for Etv mutants, respectively, p=0.32, n=3 each genotype), suggesting that in the Etv mutant background, the tip dilation phenotype responds more sensitively to Fgf10 dosage than the branch number phenotype (Fig. 4D,E).
Figure 4. Reducing Fgf10 gene dosage in the Etv mutant background led to attenuation of the tip dilation phenotype.
(A-C) Representative E13.5 lung of indicated genotype with the epithelium outlined by anti-E-cadherin immunohistochemical staining. (D) Introducing the Fgf10 mutant allele attenuated the branch tip area phenotype (area: 1.0 for controls, 2.5 for Etv mutants, 1.22 for Etv;Fgf10 mutants; p=0.009 for Etv mutants versus Etv;Fgf10 mutants, and p=0.075 for controls versus Etv;Fgf10 mutants). (E) Branch tip number was not attenuated by introducing the Fgf10 mutant allele (tip number: 1.0 for controls, 0.562 for Etv mutants, 0.654 for Etv;Fgf10 mutants, p=0.32). Quantification was carried out in n=3 samples for each genotype. See also Figure S4.
Etv mutant lungs exhibited a decrease in SHH signaling
The above data suggest that Etv and FGF engage in an inhibitory feedback loop, where FGF promotes the expression of Etv in the distal epithelium, and ETV inhibits the expression of Fgf10 in the distal mesenchyme. As Etv genes encode transcription factors that act cell-autonomously, their inhibition of Fgf10 expression in the mesenchyme is likely through ETV regulation of an intermediate cell surface/secreted factor that is expressed in the epithelium, which in turn impacts Fgf10 expression in the mesenchyme. One of the candidate mediators is FGF9, which is previously shown to promote Fgf10 expression in the mesenchyme (Colvin et al., 2001; del Moral et al., 2006). However, we found that Fgf9 expression was not altered in the Etv mutant lung (normalized to control at 1.0: Etv mutant 0.79, p=0.33, n=3 for each). A second candidate mediator is Shh. Like Etv, Shh is expressed in the epithelium (Bellusci et al., 1997a). Furthermore, the distal epithelial expression of Shh is positively regulated by FGF10 in the epithelium (Abler et al., 2009), and SHH in turn inhibits Fgf10 expression in the mesenchyme (Pepicelli et al., 1998). These data led to the hypothesis that ETV may mediate the FGF-SHH feedback loop by promoting Shh expression in the distal epithelium.
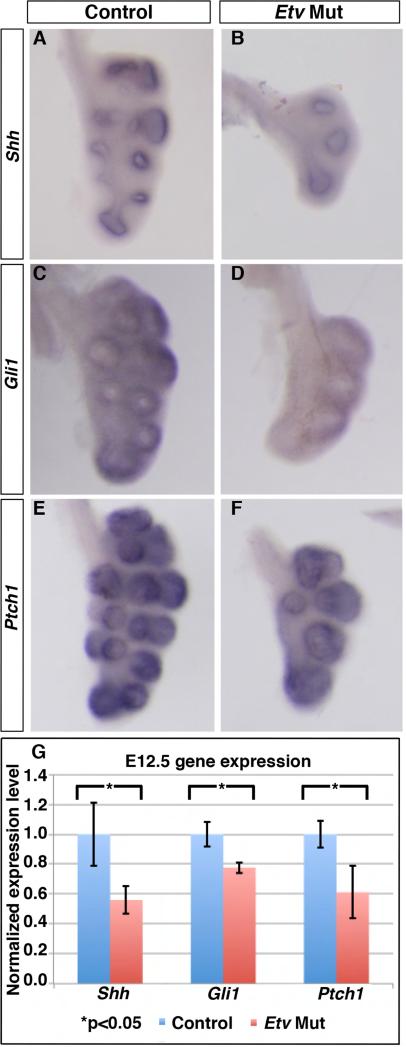
To test this hypothesis, we examined whether SHH signaling is altered in the Etv mutant lung. By in situ hybridization, we found that the staining intensity of Shh, as well as SHH pathway readouts, Gli1 and Ptch1, was decreased in Etv mutant lungs (Shhcre/+;Etv4−/−;Etv5fl/fl) compared to controls (Shhcre/+;Etv4+/−;Etv5fl/+) (Fig. 5A-F). This decrease was confirmed by qRT-PCR results (Fig. 5G), suggesting that Etv genes are required to promote Shh expression in the distal epithelial tips.
Figure 5. SHH signaling was decreased in the Etv mutant lung.
(A-F) Representative whole mount RNA in situ hybridization of left lobes of E12.0-E12.5 lungs with each set of control and mutant as littermates. (G) Quantification of expression by qRT-PCR (Shh: 1.0 for controls, 0.56 for Etv mutants, p=0.027; Gli1: 1.0 for controls, 0.77 for Etv Mutants, p=0.013; Ptch1: 1.0 for controls, 0.61 for Etv mutants, p=0.022). Quantification was carried out in n≥3 samples for each.
Restoring SHH signaling activity attenuated the lumen size phenotype in Etv mutant lungs
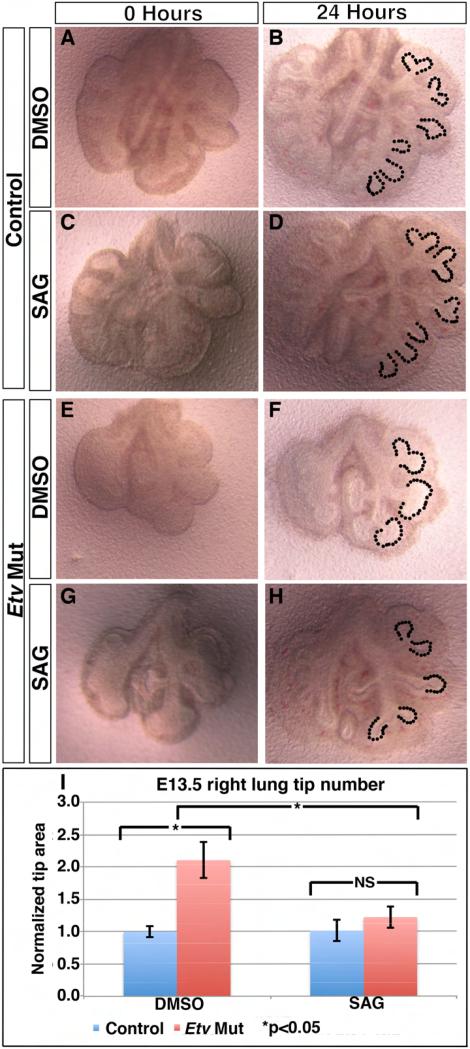
To determine if the decrease in SHH signaling in the Etv mutant contributes to the branching phenotypes, we enhanced SHH activity in the Etv mutant lung and assessed the epithelial branch structure. This enhancement was achieved by using a potent pharmacological activator of the SHH co-receptor Smoothened (Smoothened agonist, hereafter SAG) in an in vitro lung culture system (Chen et al., 2002; Liu et al., 2003; Warburton et al., 2010). Treatment of control lungs with SAG in culture was effective to induce the expression of the SHH pathway readout Ptch1 (Supplemental Fig. 5). Furthermore, Etv mutant lungs cultured in control media without SAG retained their dilated epithelial tip (tip area normalized to control genotype treated with DMSO: 2.10 for Etv mutants treated with DMSO, p=0.015, n=3 for each genotype), recapitulating the in vivo phenotype (Fig. 6A-B, E-F,I). Together these data demonstrate that this in vitro lung culture system would allow us to examine how SHH signaling is contributing to the epithelial phenotypes in the mutant lung.
Figure 6. SAG treatment of Etv mutant lungs led to attenuation of tip dilation phenotype.
(A-H) Representative images of E11.5 lungs cultured for 24 hours in either DMSO (A,B,E,F) or SAG (C,D,G,H). In the control lungs, SAG treatment did not affect tip size (areas outlined by dashes, 1.0 DMSO versus 1.01 SAG, p=0.89) (B,D). In the Etv mutant lungs, SAG treatment led to a reduction of tip size towards the size of the control (2.10 DMSO versus 1.22 SAG, p=0.015) (F,H). (I) Quantification of the right lung epithelial tip size (n=3 for each). See also Figure S5.
To test whether restoring SHH activity would attenuate Etv mutant epithelial phenotypes, we first did an SAG titration experiment and selected a concentration (7.26×10−6ug/ul) that does not affect epithelial tip size (Fig. 6A-D). In contrast, treatment of Etv mutant lungs with SAG at this concentration led to a significant decrease in the epithelial tip size compared to Etv mutant lungs treated with DMSO (tip size normalized to control genotype treated with DMSO: 1.22 for Etv mutants treated with SAG, and 2.10 for Etv mutants treated with DMSO, p=0.015 between mutants treated with SAG versus DMSO, n=3 each group) (Fig. 6E-I). Furthermore, in SAG treated Etv mutant lungs the size of the epithelial tip was not different from either DMSO or SAG treated control lungs (tip size normalized to control genotype treated with DMSO: 1.01 for control genotype treated with SAG, 1.22 for SAG Etv mutants treated with SAG, p=0.16 between the SAG treated controls versus mutants, n=3 each group) (Fig. 6I). These results suggested that the decrease in SHH signaling in the Etv mutant contributes to its phenotype, and that ETV restrict tip size partly through promoting Shh expression.
ETV factors regulate Shh expression through a long range Shh enhancer
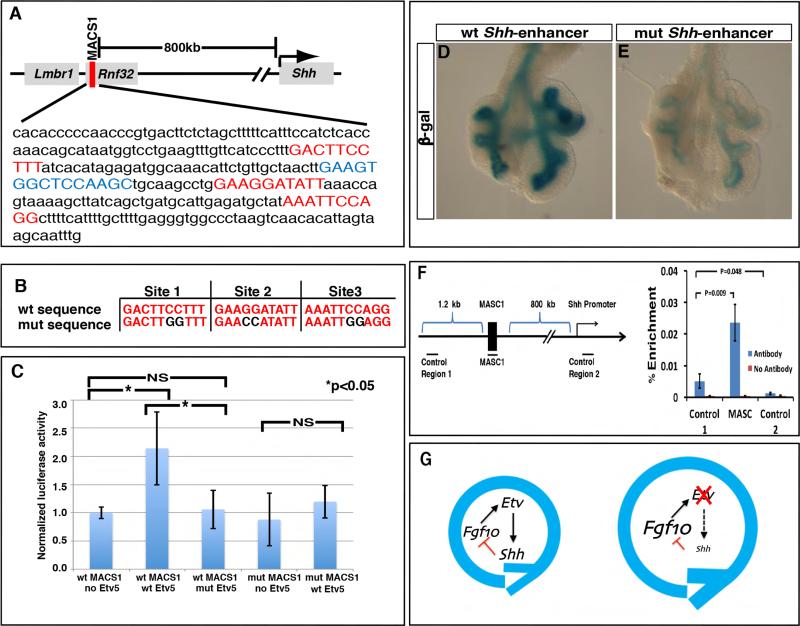
A previous study characterized a cluster of highly conserved long-range Shh enhancers that reside approximately 600 to 900kb upstream of the gene (Sagai et al., 2009). One of the enhancers, termed MACS1, was shown to drive expression activity in the lung epithelium in transient transgenic experiments. Using an established ETV DNA binding matrix (Wei et al., 2010), we identified three highly conserved ETV DNA binding sites in this Shh-enhancer (Fig. 7A,B). These binding sites are conserved across mammalian phyla, raising the possibility that they are functionally relevant for the activity of the Shh-enhancer.
Figure 7. ETV controls Shh expression through putative binding sites in a long-range enhancer.
(A) The MACS1 lung epithelium enhancer lies approximately 800kb upstream of the Shh transcriptional start site, and contains three highly conserved ETV binding sites (red) and one highly conserved NKX2-1 binding site (blue). (B) The three ETV binding sites were each mutated to nucleotides previously shown to abolish binding (de Launoit et al., 1998). (C) Relative luciferase activity from MLE12 cells transfected with either wild-type (wt) or mutant (mut) MACS1 enhancer; together with either Etv5 empty vector, wild-type (wt) Etv5 vector, or mutant (mut) Etv5 with disrupted DNA binding domain vector (1.0 for WT MACS1+no Etv5, 2.14 for WT MACS1+wt Etv5, 1.06 for wt MACS1+mut Etv5, 0.88 for mut MACS1+no Etv5, and 1.19 mut MACS1+wt Etv5, n=3 for each group). (D,E) Representative β-gal staining of transgenic lungs carrying lacZ reporter driven by either wt or mut Shh enhancer. (F) Percent recovery compared to inputs of either MACS1, a control fragment approximately 1.2 kb upstream of MASC1 (control region 1), or a control fragment approximately 800kb downstream of MACS1 near the Shh gene (control region 2), by anti-Flag antibody against ETV5-Flag, or no antibody control. The extract was prepared from lung epithelial MLE12 cells with overexpression of Etv5-Flag and Nkx2-1 plasmids. (G) A model of Etv regulation of lung epithelial branching. The outside circle represents the growth and bifurcation that constitute each reiterated cycle of branching. In the Etv mutant, decrease in Shh and increase in Fgf10 leads to prolonged growth and delayed branching. See also Figure S6.
To assess whether ETV factors regulate Shh expression via these predicted sites in the long-range enhancer, we first performed luciferase reporter assays in cultured MLE12 cells. In these cells, overexpression of Etv5 promoted the activity of the Shh-enhancer (Fig. 7C). This promotion is dependent on intact ETV5 DNA binding domain, as deletion of the N-terminal portion of this domain abolished the increase in enhancer activity (Fig. 7C). To determine whether the three conserved ETV binding sites are required for the enhancer response to ETV5, we mutated the core ETV binding sequence from GGAA/T (CCTT/A) to CCAA/T (GGTT/A) (Fig. 7B). A previous study showed that similar mutations in other conserved ETV sites abolished the ability of ETV5 to activate transcription via these sites (de Launoit et al., 1998). We found that the mutated Shh enhancer no longer responded to full-length ETV5 (Fig. 7C). Furthermore, we mutated each of the putative ETV binding sites individually, and found that mutating sites 2 and 3, but not site 1, affected responsiveness to ETV (Supplemental Fig.6A). Together, these data suggest that ETV5 is able to promote the activity of the Shh-enhancer, and both an intact DNA binding domain on the protein and the highly conserved ETV DNA binding sites on the enhancer are required for this interaction.
A previous study showed that ETV5 can interact with NKX2-1 at the protein level to promote the expression of Sftpc in a lung epithelial cell line (Lin et al., 2006). Within the Shh-enhancer, we identified a highly conserved NKX2-1 binding site near one of the ETV sites (Fig. 7A). In the MLE12 cell luciferase assay, we found that ETV5 can act synergistically with NKX2-1 to induce the activity of the Shh lung enhancer (Supplemental Fig. 6A).
To assess the role of the ETV binding sites for Shh enhancer activity in an in vivo context, we generated transgenic embryos that carry lacZ transgenes expressed under the control of either the wild-type or mutant Shh lung enhancer. We analyzed enhancer activity by staining for β-galactosidase (hereafter β-gal) enzyme activity in the lungs dissected from these embryos. By genotyping, the injection yielded 20 embryos carrying the wild-type Shh enhancer transgene, and 23 embryos carrying the mutant Shh enhancer transgene. As expected, in transgenics carrying either construct, β-gal activity ranged from no expression to specific expression in lung epithelium, presumably due to variations in insertion sites and transgene copy number. Nearly 50% (9/20) of the wild-type enhancer transgenic embryos displayed only epithelium-specific expression, in agreement with a previous study (Sagai et al., 2009), whereas such expression was observed in only 26% (6/23) of mutant Shh enhancer transgenics (Fig. 7D,E, and Supplemental Fig. 6A). Moreover, β-gal activity was strong in 4 of these 9 control enhancer transgenics whereas such strong activity was not observed in any of the mutant enhancer transgenics (Supplemental Fig. 6B). Together these data suggest that the conserved ETV binding sites are necessary for normal and robust activity of the Shh-enhancer in an in vivo context.
We then addressed if ETV may promote Shh expression via direct binding to its enhancer. Chromatin immunoprecipitation experiment performed in MLE-12 cells show that immunoprecipitation of ETV5 led to increased pull-down of the Shh MACS1 enhancer compared to control DNA fragment ~1 kb away from the enhancer (Fig. 7F). This preferential pull-down of the enhancer suggests that ETV5 promotes Shh expression by directly binding to the MACS1 enhancer.
Discussion
While it is well established that multiple signals essential for normal lung branching regulate each other's expression on the transcript level, relatively little is known about which transcription factors mediate these interactions, and their role in the branching process. In this study, we initially set out to address if Etv genes may serve as a mediator of FGF10 function in promoting branch growth. Instead, we found that the Etv mutant phenotype is distinct from that of Fgf loss of function mutants. Our evidence supports that Etv genes feed back to inhibit Fgf10 expression. And they do so via regulating the expression of Shh, thereby acting as a key component of the FGF-SHH signaling loop during lung branching.
It has been shown that lung branching morphogenesis follows a largely stereotypical spatial and temporal program (Metzger et al., 2008; Short et al., 2013). Three subroutines, planar bifurcation, orthogonal bifurcation and domain branching have been identified to function as building blocks of the branching sequence (Metzger et al., 2008; Short et al., 2013). Execution of the subroutines relies on a balanced choice between branch growth and new branch formation. For example, for planar bifurcation, each tip grows along one direction for a measurable extent before splitting to follow two new directions, and the cycle repeats. It has been postulated that the decision between growth and bifurcation is driven by an FGF-SHH feedback loop, where FGF stimulates growth, and SHH locally inhibits FGF, leading to bifurcation (Hirashima et al., 2009; Menshykau et al., 2012; Iber and Menshykau, 2013). Our data indicate that Etv serves as a unique node of the feedback loop by controlling the balance of these signals (Fig. 7G). In the absence of Etv function, Shh expression is reduced while Fgf10 expression is increased. Thereby, the Etv mutant epithelium show prolonged growth and delayed branch formation, resulting in fewer and larger branch tips.
In the kidney, Etv genes have also been shown to play a role in branching (Kuure et al., 2010). Instead of Fgf10, Etv genes are regulated by GDNF signaling via RET, also a tyrosine kinase receptor, similar to FGF receptors. Compound Etv mutants exhibit lack or greatly reduce ureteric bud branching, mimicking the Gdnf/Ret mutant phenotypes. Furthermore, in a chimeric kidney, defective behavior of the Etv mutant cells mimic that of Ret mutant cells (Kuure et al., 2010). Thus, in the kidney, Etv genes appear to be a more mainstream mediator of receptor tyrosine kinase signaling, distinct from our findings here in the lung.
Our data indicate that in the lung epithelium, Etv genes promote Shh expression. This is interesting as ETV factors have been previously shown to inhibit Shh expression in the limb bud, opposite to its role in the lung (Mao et al., 2009; Zhang et al., 2009). We speculate that this difference may be due to differences in binding partners. In the limb bud, evidence suggests that ETV inhibits Shh expression indirectly through regulating the homo or heterodimerization of two E-box transcription factors TWIST1 and HAND2, which in turn control Shh expression (Firulli et al., 2007; Zhang et al., 2010). In the lung, our data suggest that ETV may directly promote Shh expression through highly conserved ETV binding sites in the MACS1 Shh lung enhancer. MACS1 may not be the only enhancer through which ETV regulates Shh expression. A recent paper reported the identification of an additional Shh enhancer, SLGE, that displays activity in the lung epithelium (Tsukiji et al., 2014). SLGE also contains a putative ETV binding site. Although this enhancer, different from MACS1, is not conserved, it is possible that ETV may regulate Shh expression through SLGE in addition to MACS1 in mice.
In addition to Etv, several other genes, Sproutys, Dusp6 and Sef, are also commonly used as FGF signaling readouts, because their expression is positively regulated by FGF activity (Tsang and Dawid, 2004). Both Sproutys and Dusp6 are known to act in the cytoplasm as direct negative regulators of the FGF pathway. Among the FGF readouts, Etv are the only genes that encode transcription factors. Thus we had speculated that the phenotype of Etv mutant lungs would mimic that of loss-of-FGF function mutants. Instead, like Sproutys and Dusp6, Etv genes function in lung as inhibitors of FGF activity. However, unlike Sproutys and Dusp6, Etv genes act indirectly through regulating Shh. It is intriguing that there are multiple inhibitors of FGF activity built in downstream of the ligands and receptors, suggesting that keeping FGF activity in check is of cardinal importance to tissue development.
Inhibitory feedback mechanisms are often revealed as drivers for reiterative biological processes. For example, in somite formation, the transcription factors Hairy/HES-based inhibitory feedback loop couples with signals such as Notch and FGF to drive the segmentation of the presomitic mesoderm into a sequence of organized blocks (Aulehla and Pourquié, 2008). In circadian rhythm generation, transcription factors Clock and BMAL1 are at the core of a complex feedback machinery that repeats the daily cycles of sleep, metabolism, and other circadian physiological functions (Takahashi et al., 2008). Our data presented here support the conclusion that Etv genes serve as a critical component of an inhibitory feedback loop that control the rhythm of the reiterative lung branching process.
Experimental Procedures
Mice
Mice were housed and all experimental procedures were performed in an American Association for Accreditation of Laboratory Animal Care-accredited laboratory animal facility at the University of Wisconsin. This study was approved by the University of Wisconsin Animal Care and Use Committee and conformed to the Guide for the Care and Use of Laboratory Animals. The embryos used in these experiments were harvested from time-mated females, with noon the day when the vaginal plug was observed counted as E0.5. The mutant alleles used in this study have been described previously: Etv5fl (Zhang et al., 2009), Etv4− (Laing et al., 2000), Fgf10− (Sekine et al., 1999), and Shhcre (Harfe et al., 2004). Shhcre/+;Etv4+/−;Etv5fl/+ littermate embryos were used as controls unless otherwise indicated.
Antibody staining
Antibody staining was performed following a previously published protocol. The primary antibodies used were rabbit anti-E-cadherin (Cell Signaling, 1:200 dilution), rat anti-E-cadherin (DECMA Sigma, 1:100 dilution), rabbit anti-phosphorylated-Erk (Cell Signaling, 1:200 dilution). The secondary antibodies used were Cy3-conjugated goat anti-mouse, FITC-conjugated goat anti-rabbit, HRP-conjugated goat anti-rabbit, and Biotin-conjugated goat anti-rabbit (Jackson ImmunoResearch, 1:200 dilution). The tertiary antibody used to detect pERK in whole mount lungs was FITC conjugated-Streptavidin (Jackson ImmunoResearch, 1:200 dilution).
RNA in situ Hybridization
RNA in situ hybridization was performed following a previously published protocol (Herriges et al., 2012).
Quantitative analysis of lung phenotypes
Two methods were used for quantification of embryonic lung tip areas. First, following wholemount immunohistochemical anti-E-Cadherin staining, lungs were imaged under brightfield on a Zeiss Axioscope Imager A2. ImageJ software was used to draw a freeform trace around each lung bud tip, and area within the trace was measured. Second, following wholemount immuofluorescent anti-E-Cadherin staining, lungs were imaged on a Zeiss 510 confocal laser scanning microscope. Z-stacks of single slice images were used to generate whole lobe three-dimensional surface projections using Bitplane Imaris software. Distal tip dimensions were measured in ImageJ and the tip volume was calculated from the formula V=4/3πabc where V is the volume of an ellipsoid and a, b, and c are radius of the tip in each of the three dimensions. For quantifying tip number, we manually counted anti-E-Cadherin antibody outlined epithelial tips at the indicated stages.
Quantitative RT-PCR
Total RNA was extracted from tissue using Trizol and an RNeasy-micro Qiagen RNA extraction kit (Invitrogen, Qiagen). RNA was reverse-transcribed using Superscript-III first-strand synthesis system (Invitrogen). Quantitative PCR was performed using SYBRgreen (Applied Biosystems) and in an ABI PRISM 7000 sequence detection system. The primer pairs used are listed in Supplemental Table 1. Three biological replicates and two technical replicates were performed for all qPCR. Expression values were normalized using β-actin and results were compared using the Student's t-test.
Luciferase reporter assay
To generate the wild-type enhancer luciferase reporter, the mouse MACS1 Shh enhancer was amplified using primers found in Supplemental table 1 and was cloned into the Pgl3 vector (Promega). To generate the mutated enhancer luciferase reporter, the GGAA/T core binding sequence was mutated to CCAA/T using the primers found in Supplemental table 1. The constructs containing the wild-type Etv5 open reading frame and a truncated Etv5 open reading frame were previously described (Zhang et al., 2009).
MLE-12 cells were transfected with either of the reporter constructs (200ng), and either an empty vector, wild-type Etv5 construct, or DNA-binding domain truncated mutant Etv5 construct (600ng) using lipofectamine 2000 (Invitrogen). A Renilla luciferase plasmid was used as a transfection control. Luciferase activity was analyzed using the Dual-Luciferase reporter system (Promega). Three biological replicates and two technical replicates were performed and results were compared using the Student's t-test.
In vitro Lung culture
Lungs from control and Etv mutant embryos were harvested at E11.5. Lungs were placed on a Nucleopore Trak-Etch membrane (Whatman, 8um), and cultured at the air/liquid interface with DMEM-F12 and 10% FBS. To activate SHH signaling, smoothened agonist (SAG) was added at a final concentration of 7.26×10−6ug/ul. DMSO was used as a diluent control. Lungs were cultured at 37°C in 5% CO2 for 24 hours before harvest and analysis.
ChIP
MLE-12 cells were transfected with 3X FLAG tagged full-length mouse Etv5 cDNA. Approximately 1×107 cells per sample were cross-linked with 1% formaldehyde for 20 min at 37°C. The samples were sonicated with the Covaris E210 sonicator to obtain chromatin fragment lengths of 200–1500 bp. Fragmented chromatin was incubated overnight at 4°C with anti-FLAG antibody conjugated to magnetic beads (Sigma, Slbl1128v). Immunoprecipitates were washed sequentially with low salt, high salt and LiCl wash buffers. The protein and DNA complex was eluted and the DNA was purified by phenol/chloroform extraction and ethanol precipitation. The DNA was used as template for qPCR.
Transgenic
The transgenic construct was produced by PCR amplification of the MACS1 enhancer and cloning it into the Hsp68-LacZ vector (Addgene). The primers used to clone and amplify up the MACS1 enhancer can be found in supplemental table 1. Production of transgenic mice was performed as previously reported (Anderson et al., 2013).
β-gal staining
Lungs from control and mutant Shh transgenic lungs were harvested at E11.5. Lungs were fixed in 4% PFA at 4°C for 10 min, and put through a standard β-gal staining protocol. All lungs were exposed to the β-gal stain for 2 hours. After two hours the lungs were removed from the stain, put in a 4% PFA postfix, and imaged.
Supplementary Material
Acknowledgements
We would like to thank members of the Sun lab for insightful discussions and critical readings of the manuscript, Dr. Kevin Eliceri and Dr. Ruth Sullivan for the use of imaging instrumentation and Imaris software at UW Laboratory for Optical and Computational Instrumentation. We would like to thank Dr. Xuehua Zhong and Dr. Shuimin Qian for sharing expertise on chromatin immunoprecipitation. This work was supported by NHLBI HL113870, 119946, 122406, Wisconsin Partnership Program Collaborative Research Award #2900 (to X.S.); and American Heart predoctoral fellowship # 11PRE5540006 and NIH predoctoral training grant to University of Wisconsin Genetics program T32 GM007133 (to J.C.H) and intramural support by the National Institutes of Health, National Cancer Institute and Center for Cancer Research (to M.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
John Herriges and Jamie Verheyden performed the majority of the Etv mutant analysis. Zhen Zhang generated the Etv lung mutants and carried out the initial analysis. Pengfei Sui and Yan Zhang carried out the ChIP analysis, Ying Zhang, Matthew Anders, Deborah Swing and Mark Lewandoski carried out the enhancer transgenic analysis. Xin Sun led the study.
References
- Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn. 2009;238:1999–2013. doi: 10.1002/dvdy.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Naiche LA, Wilson CP, Elder C, Swing DA, Lewandoski M. TCreERT2, a transgenic mouse line for temporal control of Cre-mediated recombination in lineages emerging from the primitive streak or tail bud. PLoS One. 2013;8:e62479. doi: 10.1371/journal.pone.0062479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulehla A, Pourquié O. Oscillating signaling pathways during embryonic development. Curr Opin Cell Biol. 2008;20:632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, Hogan BL. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997a;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997b;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotteau-Lelièvre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene. 1997;15:937–952. doi: 10.1038/sj.onc.1201261. [DOI] [PubMed] [Google Scholar]
- Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- de Launoit Y, Audette M, Pelczar H, Plaza S, Baert JL. The transcription of the intercellular adhesion molecule-1 is regulated by Ets transcription factors. Oncogene. 1998;16:2065–2073. doi: 10.1038/sj.onc.1201726. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- del Moral PM, De Langhe SP, Sala FG, Veltmaat JM, Tefft D, Wang K, Warburton D, Bellusci S. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol. 2006;293:77–89. doi: 10.1016/j.ydbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Redick BA, Conway SJ, Firulli AB. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Herriges JC, Yi L, Hines EA, Harvey JF, Xu G, Gray PA, Ma Q, Sun X. Genome-scale study of transcription factor expression in the branching mouse lung. Dev Dyn. 2012;241:1432–1453. doi: 10.1002/dvdy.23823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima T, Iwasa Y, Morishita Y. Mechanisms for split localization of Fgf10 expression in early lung development. Dev Dyn. 2009;238:2813–2822. doi: 10.1002/dvdy.22108. [DOI] [PubMed] [Google Scholar]
- Iber D, Menshykau D. The control of branching morphogenesis. Open Biol. 2013;3:130088. doi: 10.1098/rsob.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuure S, Chi X, Lu B, Costantini F. The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development. Development. 2010;137:1975–1979. doi: 10.1242/dev.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing MA, Coonrod S, Hinton BT, Downie JW, Tozer R, Rudnicki MA, Hassell JA. Male sexual dysfunction in mice bearing targeted mutant alleles of the PEA3 ets gene. Mol Cell Biol. 2000;20:9337–9345. doi: 10.1128/mcb.20.24.9337-9345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Perl AK, Shannon JM. Erm/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. J Biol Chem. 2006;281:16716–16726. doi: 10.1074/jbc.M602221200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jiang H, Crawford HC, Hogan BL. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev Biol. 2003;261:10–24. doi: 10.1016/s0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- Mailleux AA, Kelly R, Veltmaat JM, De Langhe SP, Zaffran S, Thiery JP, Bellusci S. Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development. 2005;132:2157–2166. doi: 10.1242/dev.01795. [DOI] [PubMed] [Google Scholar]
- Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menshykau D, Kraemer C, Iber D. Branch mode selection during early lung development. PLoS Comput Biol. 2012;8:e1002377. doi: 10.1371/journal.pcbi.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- Sagai T, Amano T, Tamura M, Mizushina Y, Sumiyama K, Shiroishi T. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development. 2009;136:1665–1674. doi: 10.1242/dev.032714. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Short K, Hodson M, Smyth I. Spatial mapping and quantification of developmental branching morphogenesis. Development. 2013;140:471–478. doi: 10.1242/dev.088500. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Marshall WF, McMahon M, Metzger RJ, Martin GR. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science. 2011;333:342–345. doi: 10.1126/science.1204831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 20042004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Tsukiji N, Amano T, Shiroishi T. A novel regulatory element for Shh expression in the lung and gut of mouse embryos. Mech Dev. 2014;131:127–136. doi: 10.1016/j.mod.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Bellusci S, De Langhe S, Del Moral PM, Fleury V, Mailleux A, Tefft D, Unbekandt M, Wang K, Shi W. Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr Res. 2005;57:26R–37R. doi: 10.1203/01.PDR.0000159570.01327.ED. [DOI] [PubMed] [Google Scholar]
- Warburton D, El-Hashash A, Carraro G, Tiozzo C, Sala F, Rogers O, De Langhe S, Kemp PJ, Riccardi D, Torday J, Bellusci S, Shi W, Lubkin SR, Jesudason E. Lung organogenesis. Curr Top Dev Biol. 2010;90:73–158. doi: 10.1016/S0070-2153(10)90003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Poe B, Schwarz M, Elliot SA, Albertine KH, Fenton S, Garg V, Moon AM. Fetal and postnatal lung defects reveal a novel and required role for Fgf8 in lung development. Dev Biol. 2010;347:92–108. doi: 10.1016/j.ydbio.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, Behringer RR, Sun X. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development. 2010;137:3417–3426. doi: 10.1242/dev.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev Cell. 2009;16:607–613. doi: 10.1016/j.devcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.