Abstract
Circular RNAs (circRNAs) are now recognized as large species of transcripts in eukaryotic cells. From model organisms such as C. elegans, Drosophila, mice to human beings, thousands of circRNAs formed from back-splicing of exons have been identified. The known complexity of transcriptome has been greatly expanded upon the discovery of these RNAs. Studies about the biogenesis and physiological functions have yielded substantial knowledge for the circRNAs, and they are now more likely to be viewed as regulatory elements coded by the genome rather than unavoidable noise of gene expression. Certain human diseases may also relate to circRNAs. These circRNAs show diversifications in features such as sequence composition and cellular localization, and thus we propose that they may be divided into subtypes such as cytoplasmic circRNAs, nuclear circRNAs, and exon-intron circRNAs (EIciRNAs). Here we summarize and discuss knowns and unknowns for these RNAs, and we need to keep in mind that the whole field is still at the beginning of exciting explorations.
Keywords: Biogenesis, circRNA, EIciRNA, microRNA, microRNA sponge, ncRNA, Transcription.
INTRODUCTION
Early days of molecular biology guided under the so called central dogma had led to the identification of RNA molecules such as rRNA, mRNA, and tRNA between 1950s and 1960s [1, 2]. These RNAs are essential to all the cells in fulfilling the task of expressing information coded by genomic DNA into functional proteins. They also had set up one of the general features of RNA as linear molecule with a 5’ end and a 3’ end. As a special form of RNA comparing to the “common” linear form, circular RNAs (circRNAs) were firstly described in 1976 that several plant viroids were covalently closed and single-stranded circular molecules without any end [3].
The decade following the discovery of plant viroids as circRNAs had witnessed the identification of RNA genomes of several plant and then animal viruses as circular molecules [4, 5]. The possible existence of circRNAs with covalent linkages in the cytoplasm of mammalian cells was indicated by electron microscopy in 1979 [6]. It was also reported that some yeast mitochondrial RNAs might be circular in 1980 [7]. The decade between late 1970s and 1980s with research about RNA splicing had demonstrated that introns were spliced out as another nonlinear form of “lariat” [8]. An important discovery coming out later was that an RNA exonuclease RNase R could digest all linear RNAs in isolated total RNA and leave only the loop portion of the intronic lariat and circRNAs [9, 10]. Mammalian cells might also have endogenous intronic circRNAs presumably coming from the loop of intronic lariat [10, 11]. The resistance to RNase R digestion has now become one of the golden markers of circRNA.
It has taken more than two decades before the realization that a large number of circRNAs actually exist in mammalian cells. In 1991, Nigro et al. identified several “abnormally” spliced transcripts corresponding to a tumor suppressor gene DCC, in which exons were joined accurately at consensus splice sites, but in an order different from that present in the primary transcript [12]. The authors called these transcripts “scrambled exons”. One year later, another example of scrambled exons was reported in which a human c-ets-1 transcript demonstrated a scrambled order of exons different from that in genomic DNA [13]. The authors also showed that the scrambled transcript was non-polyadenylated, and was not a product either from genomic rearrangement or from any ets-1 pseudogene. Finally in 1993, two reports came out about the formation of circRNAs from exons. In the first report, the previously identified scrambled transcripts of human ets-1 were demonstrated to be circRNA molecules [14]. In the second report, the important testis-determining gene sex determining region Y (Sry) was shown to express high abundant circular transcripts in testis of adult mice [15]. In the following years, examples of circRNAs formed by exonic sequences were discovered for genes such as rat P450 2C24, rat androgen binding protein gene, mouse Formin, human P450 2C18, human MLL, human dystrophin, mammalian sodium-calcium exchanger, and Drosophila muscleblind [16-24]. Examples of circRNAs corresponding to linear noncoding RNA (ncRNA) as well as antisense RNA were seen in 2010 and 2011 [25, 26]. It was in 2012 and 2013, with the development of high-throughput RNA sequencing (RNA-seq) and bioinformatic analysis, that circRNAs have begun to be recognized as large species of RNAs with thousands of members in mammalian cells [27-29].
Salzman J et al. reported in the 2012 paper, that a significant portion of the spliced transcripts from hundreds of human genes were circRNAs based on deep sequencing data from multiple normal and abnormal cell types [27]. In a 2013 paper, Jeck WR et al. demonstrated that a number of circRNA species derived from exons of coding genes were present in human fibroblast cells through RNA-seq of libraries from ribosome-depleted RNA with or without the RNase R digestion [28]. In another 2013 paper, Memczak S et al. reported that circRNAs were present with thousands in number in human, mouse, and C. elegans cells [29]. What was more important, they showed indications and lines of evidence that at least certain circRNAs might possess regulatory functions [29]. These findings have immediately inspired enormous interests to uncover circRNA biology in recent years.
The circular form of RNAs in eukaryotic cells may include plant viroids, genome of some single-stranded RNA viruses, circular intermediate of some tRNAs (permutated tRNAs and some excised tRNA introns) [30], loop part of intronic lariats, circRNAs from noncoding genes or antisense transcripts, as well as exonic circRNAs (Fig.1). Currently exonic circRNAs consist of the majority of circRNAs identified in animal cells, and for this review, the term of circRNA refers to exonic circRNAs from back-splicing exons of coding genes unless specified. We would like to summarize and discuss here knowns and unknowns for circRNAs, and hopefully to promote future explorations in this field.
Fig. (1).
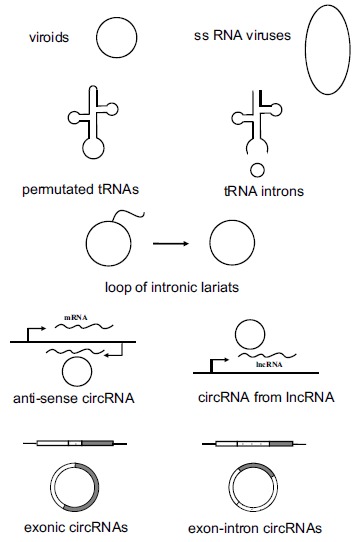
A variety of circular RNAs in eukaryotic cells. The circular RNAs in eukaryotic cells include plant viroids, some single-stranded RNA viruses, permutated tRNAs, some excised tRNA introns, loop of intronic lariats, circRNAs from noncoding genes, circRNAs corresponding to antisense transcripts, exonic circRNAs from coding genes, and exonintron circRNAs from coding genes.
circRNAs IN DIVERSE ORGANISMS
Human and Mammals
circRNAs in human and mice have been extensively profiled [27-29, 31]. In human fibroblasts, it was estimated that 14.4% of actively transcribed genes gave rise to circRNAs, and for some genes the abundance of circRNAs exceeded the corresponding linear mRNA [27]. Analyzing RNA-seq data from the ENCODE project identified ~7,000 human circRNAs with abundance of at least 10% of the transcripts from the corresponding genomic loci [31]. Most circRNAs are expressed with cell specificity [27-29, 31]. Some circRNAs are orthologs between human and mice. For example, Jeck and coauthors identified 69 circRNAs expressed in murine testis that were coded in precisely orthologous genomic sequences to human circRNAs [28]. In another study, it was shown that of about one fifth of the 635 mouse circRNAs annotated were orthologous to human circRNAs, although the sequence conservation of these circRNAs was not higher comparing to that of their neighboring exons [31]. Even most circRNAs are cytoplasmic and some of them possess start codon (e.g. exon 1 is included in the circRNA), the majority of circRNAs should be noncoding as they are not found to associate with polyribosomes [31, 32]. A recent report demonstrated relative higher abundance of circRNAs in murine brain as compared to heart, liver, lung and testis [32].
Drosophila and C. elegans
The first indication about the existence of an exonic circRNA was reported for the Drosophila muscleblind gene in 2006 [24]. A large number of circRNAs have now been identified in Drosophila and also in another important metazoan model organism C. elegans. A survey with over 100 RNA libraries from diverse developmental stages and cell types of Drosophila identified more than 2,500 exonic circRNAs [33]. Across multiple Drosophila species, the generation of circRNAs from hundreds of genes is conserved. RNA-seq with several life stages of C. elegans has recently identified ~1,100 exonic circRNAs [29, 34].
The Other Eukaryotic Organisms
A survey for circRNAs in yeasts (Schizosaccharomyces pombe and Saccharomyces cerevisiae), protists (Plasmodium falciparum and Dictyostelium discoideum) and a plant (Arabidopsis thaliana) has been conducted [35]. A substantial number of exonic circRNAs are identified in all these species. Two points of particular interest are the relative small introns in S. cerevisiae, S. pombe, Dictyostelium, P. falciparum and Arabidopsis and the very rare alternative splicing in S. pombe and P. falciparum. These two features are related to mechanisms of circRNA biogenesis in the following discussion.
The Prokaryotes
circRNAs may have a deep root in the evolution as they are also found in Archaea. It was reported that the archaeon Sulfolobus solfataricus P2 has circRNAs as excised tRNA introns, rRNA processing intermediates, and a large portion of circular molecules corresponding to ncRNAs (e.g. C/D box RNAs and RNase P RNA), although it seems that no circRNA is generated from coding genes in this archaeon [36, 37]. Many of these circRNAs were conserved in another archaeon Sulfolobus acidocaldarius [36]. There is essentially no circRNA in eubacteria with only several of interesting candidates corresponding to rRNAs and tRNAs [37].
circRNA BIOINFORMATICS
Genome-wide identification of circRNAs relies on an efficient bioinformatic analysis. There are multiple pipelines developed to focus on reads from the back-spliced junctions for the identification of circRNAs from RNA-seq data. These include find_circ [29], Segemehl [38], CIRCexplorer [39], CIRC [40] and PFOR [41, 42]. All of them except PFOR depend on the reference genome. In brief, reads that aligned contiguously to the reference genome are filtered out, then the unmapped reads are analyzed further for possible dual alignments as two segments mapped to two genomic sites in the reverse (head-to-tail) order. The resulting reads are considered as circRNA junctions. PFOR is independent on the genome, suitable for de novo assembly of circRNAs based on the overlapped reads to pinpoint the full sequences of circRNAs from small RNA-seq data [41, 42].
circRNA BIOGENESIS AND STABILITY
Biogenesis of circRNAs has been recent research hotspot. It had been noticed that relatively long introns and the complementary repeat sequences flanking the region of circularization were associated respectively with the circularization of circEts-1 and circSry [13, 43]. Several genome-wide bioinformatic analyses along with some experimental data now confirmed the association of flanking long introns and the presence of bracketing intronic repeats such as Alu elements to the biogenesis of mammalian circRNA [28, 39, 44, 45]. The association of flanking long introns and complementary sequences with most backsplicing exons has two important indications for mammalian circRNA biogenesis. First, the regions of circularization are marked with hard-to-splice long introns. Secondly, the two splicing sites of backsplicing are brought together by the flanking complementary sequences or some other interactions (Fig. 2). There are also many exceptions in which circRNAs are formed without association with long boarding introns and intronic repeats [44-46]. Besides flanking intronic sequences, the exonic sequences involved in backsplicing may be crucial also for some circRNAs [44-46]. It seems that the exonic sequences and intronic repeats must collaborate with one another for circRNA biogenesis [44].
Fig. (2).
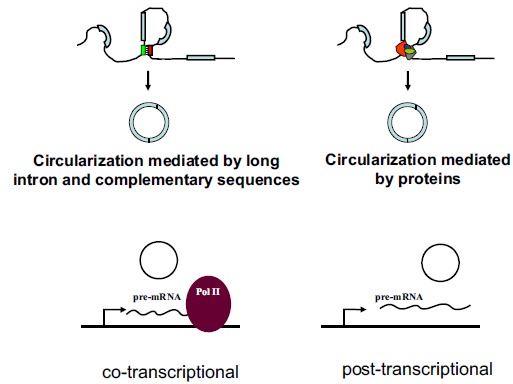
circRNA Biogenesis. Biogenesis of circRNAs may be mediated by flanking long introns and intronic complementary sequences. RNA binding proteins such as QKI, MBL and ADAR are shown to associate with the biogenesis of certain circRNAs. circRNAs may be generated co-transcriptionally or post-transcriptionally.
Is there any protein specific for circRNA biogenesis? Recently, the RNA-binding protein quaking I 5 (QKI5) was shown to participate in the circularization of some exons [47]. Another interesting example is the “self-regulation” of circRNA biogenesis by muscleblind (MBL) protein on circMbl. circMbl and its flanking introns contain MBL binding sites, and thus MBL can modulate the generation of circMbl [48]. As both QKI5 and MBL are known factors of alternative splicing for selective mRNAs, there is a concern about involuntary involvement of these proteins in backsplicing [49].
circRNAs are ubiquitously present in essentially all mammalian cell types, and for this reason, any essential factor for circRNA biogenesis should be present in all cells. On the other hand, there is no doubt that circRNAs are expressed with cell-type and gene specificity and are not piggybacking byproducts of alternative splicing [27-29, 50]. The molecular ratio of circRNAs to mRNAs for individual genes has also to be managed for different cells under different conditions [45, 50]. Molecular mechanisms of these regulations are waiting for further investigations.
As splicing is generally co-transcriptional, the backsplicing for the generation of circRNAs may also be co-transcriptional. Ashwal-Fluss R et al. provided data to show the co-transcriptional produce of circRNAs; backsplicing of circRNA and linear splicing of mRNA may compete against each other [48]. On the other hand, Liang and Wilusz found with overexpressing plasmids that the pre-mRNA requires a stable 3′ end for the favor of circRNA biogenesis, arguing strongly that circRNA biogenesis may be post-transcriptional at least in their experimental system [44] (Fig. 3).
Fig. (3).
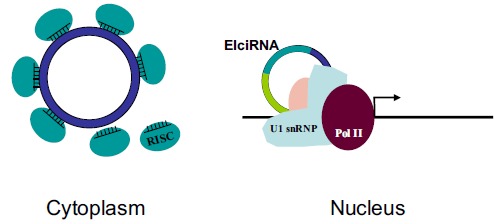
circRNA Function & Functional mechanism. Two circRNAs (CDR1as and circSry) play roles as microRNA sponge in the cytoplasm, and EIciRNAs regulate transcription in the nucleus.
Researches on circRNAs in organisms other than mammals provide additional insights to circRNA biogenesis. It was found that C. elegans circRNAs were significantly enriched with flanking reverse complementary sequences in introns, and the double-strand RNA-editing enzyme ADAR1 mitigated circRNA expression presumably via melting double-stranded structures formed by these complementary sequences [34]. The facts that yeasts, protists, and Arabidopsis having exonic circRNAs but without long intron and the lack of pervasive alternative splicing in S. pombe and P. falciparum indicate strongly that circRNAs may have a splicing independent biogenesis in these species.
The amount of each circRNA and the expression profile of circRNAs are decided presumably together by circRNA biogenesis and degradation. How circRNAs are degraded is largely an unexplored subject. circRNAs are more stable due to their inaccessibility to exonucleases such as RNase R [9, 10]. Questions such as how circRNAs are degraded intracellularly (and/or intercellularly), whether and how this process is regulated, and could circRNA degradation be regulated for individual circRNAs are waiting for future investigations.
circRNA FUNCTIONS AND FUNCTIONAL MECHANISMS
Multiple lines of evidence suggest strongly that circRNAs may play crucial regulatory roles in eukaryotic cells. From yeast to human cells, circRNAs are shown to demonstrate specific expression profiles corresponding to specific cellular conditions, developmental stages, and cell types [27-29, 33, 34, 45, 50].
The first serious circRNA functional studies demonstrated that two circRNAs function as microRNA sponge [29, 51]. The mammalian circRNA CDR1as harbors over 70 conserved binding sites for miR-7, and indeed overexpression CDR1as suppressed miR-7 function [29, 51]. The murine circRNA circSry might be a microRNA sponge with more than ten miR-138 binding sites, although these sites are missing in human circSry [31, 51]. Both CDR1as and circSry are cytoplasmic, which is in consistence with their functional role as microRNA sponge. Most circRNAs are found in the cytoplasm, and findings about these two circRNAs indicated immediately a regulatory mechanism as microRNA sponge as a common way for circRNA function. Although later analysis showed that only several out of all the thousands human circRNAs harbor multiple computational binding sites for individual microRNAs, it is unlikely for human circRNAs to play a general role as microRNA sponge [31]. Furthermore, P. falciparum and S. cerevisiae lack microRNA pathway but both have circRNAs [35]. Interestingly, Drosophila circRNAs do harbor over a thousand predicted miRNA sites conserved across the genus [33].
A specific subtype of circRNAs called exon-intron circRNA (EIciRNA) is found to localize predominantly in the nucleus [45]. EIcircRNAs are also distinct in their sequences; unlike the other exonic circRNAs with all introns spliced out, EIciRNAs are intron-retained or composed with both exonic and intronic sequences. EIciRNA promotes the transcription of RNA polymerase II (pol II) in cis via RNA-RNA interaction between EIciRNA and U1 snRNA. The interacting site of EIciRNA and U1 snRNA is the 5’ splicing site of the EIciRNA retained intron [45].
Cellular localization would be linked directly with physiological function and functional mechanism of circRNAs. The pervasive distribution in the cytoplasm for circRNAs may be passive due to the leaky and breakdown of nuclear membrane in mitosis. This simple assumption can not explain the nuclear localization of EIciRNA and the cytoplasmic localization of circRNAs in non-dividing cells (e.g. neurons) [32]. Interestingly, punctae enriched with circRNAs are noticed recently in the neuronal dendrites [32]. All these suggest strongly that active transporting or localization mechanisms for circRNAs must exist, correlated with physiological functions of specific circRNAs and maybe with some cell specificity.
DIVERSITY OF circRNAs
The way to view exonic circRNAs as a family with common features in their sequences, biogenesis, cellular localization, functions and functional mechanisms may need to be abandoned. The discovery of EIciRNA with its function and functional mechanism provides a paradigm of circRNA diversity [45]. The composition of both exonic and intronic sequences makes EIciRNA distinct from the other circRNAs. The inclusion of intron against intron exclusion may be subjected to regulation in EIciRNA biogenesis. The nuclear localization of EIciRNAs may be due to their association with nuclear proteins (e.g. proteins of U1 snRNP), or some other mechanism may be involved. Finally the function and functional mechanism as transcriptional regulator may be unique to EIciRNAs but not to the other circRNAs. We see several discrepancies or even disputes in circRNA biology such as roles of flanking long introns and reverse complementary sequences in circRNA biogenesis, cotranscriptional or posttranscriptional generation of circRNAs, and being microRNA sponge or not. It is possible that these are not really discrepancies but rather indications of great diversity within circRNAs. The only common feature of all circRNAs may just be that they are circular.
circRNAs AND HUMAN DISEASES
There are indications about the association between some human diseases and circRNAs. Upon the finding of circRNA corresponding to the INK4/ARF-associated ncRNA, it was noticed that this circRNA might have some correlation with risk of atherosclerosis [25]. The discovery of CDR1as an miR-7 sponge promoted thoughts about the link between circRNAs and neurodegenerative diseases (NDs) such as Alzheimer's disease (AD), as miR-7 and some other microRNAs are directly related to NDs [52-55]. Several studies showed association of circRNAs with cancers [56-59]. The overall circRNA abundance is lower in tumor cells and correlates negatively with cell proliferation [59]. Interestingly, levels of circRNAs increase relative to linear mRNAs during aging [33, 34]. There is a database for SNP (single nucleotide polymorphism) associated with diseases for genomic sites with corresponding circRNAs [60]. Exactly to how much degree that circRNAs are involved in human diseases and the actual roles of circRNAs underneath the etiology of particular disease remain for further studies.
CONCLUSION
A large number of circRNAs have been identified in eukaryotes. With multiple lines of evidence, these molecules are believed to be crucial regulatory elements and are not splicing noise as previously thought. The field of circRNA has bloomed in recent years with researches related to their expressing profiles, biogenesis, physiological relevance and functional mechanisms. However, we are still at the beginning of this field with a lot of unknowns waiting for further investigations.
We may need to learn from the development in the research field of linear lncRNA to answer the fundamental questions such as how circRNAs are generated, degraded, localized, and what are their functions and functional mechanisms. The first examples of lncRNAs such as XIST had been identified as potent epigenetic regulator [61]. Researches within the past two decades have now demonstrated that lncRNAs are a diverse family with thousands of members functioning with a plethora of molecular mechanisms in an array of physiological relevancies [62-66]. The circRNA field may need to realize the potentially great diversity among circRNAs, and have to start from detailed studies of circRNA subtypes (e.g. EIciRNAs). The full picture of circRNA biology may not be able to assemble until specific subtypes of circRNAs are explored.
circRNAs may be directly linked to future biomedical research for disease etiology, diagnosis and treatment, and more understanding in circRNAs would be essential for the development of RNA biology and biomedicine.
BIOINFORMATIC RESOURCE
find_circ, http://circbase.org/cgi-bin/downloads.cgi
Segemehl, http://www.bioinf.uni-leipzig.de/Software/segemehl/
CIRCexplorer, https://github.com/YangLab/CIRCexplorer
CIRC, https://sourceforge.net/projects/ciri/
PFOR2, http://journals.plos.org/plospathogens/article?id= 10.1371/journal.ppat.1004553
Circ2Traits, http://gyanxet-beta.com/circdb/
ACKNOWLEDGEMENTS
The authors thank Dr. Xueru Zhang, Mr. Wanchen Hu, and other members of the Ge Shan lab for discussions and supports. This work was supported by the National Basic Research Program of China (2011CBA01103 and 2015CB943000), National Natural Science Foundation of China (31471225, 81171074, and 91232702), CAS (KJZD-EW-L01-2), and the Fundamental Research Funds for the Central Universities (WK2070000034).
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Crick F.H. On Protein Synthesis. Symp. Soc. Exp. Biol. 1956;XII:139–163. [PubMed] [Google Scholar]
- 2.Geiduschek E.P., Haselkorn R. Messenger RNA. Annu. Rev. Biochem. 1969;38:647–676. doi: 10.1146/annurev.bi.38.070169.003243. [DOI] [PubMed] [Google Scholar]
- 3.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gould A.R. Studies on encapsidated viroid-like RNA. II. Purification and characterization of a viroid-like RNA associated with velvet tobacco mottle virus (VTMoV). Virology. 1981;108(1):123–133. doi: 10.1016/0042-6822(81)90532-8. [DOI] [PubMed] [Google Scholar]
- 5.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 6.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 7.Arnberg A.C., Van Ommen G.J., Grivell L.A., Van Bruggen E.F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19(2):313–319. doi: 10.1016/0092-8674(80)90505-X. [DOI] [PubMed] [Google Scholar]
- 8.Cech T.R. RNA splicing: three themes with variations. Cell. 1983;34(3):713–716. doi: 10.1016/0092-8674(83)90527-5. [DOI] [PubMed] [Google Scholar]
- 9.Vincent H.A., Deutscher M.P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281(40):29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 13.Cocquerelle C., Daubersies P., Majérus M.A., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11(3):1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 15.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 16.Zaphiropoulos P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. USA. 1996;93(13):6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaphiropoulos P.G. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol. Cell. Biol. 1997;17(6):2985–2993. doi: 10.1128/MCB.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailleul B. During in vivo maturation of eukaryotic nuclear mRNA, splicing yields excised exon circles. Nucleic Acids Res. 1996;24(6):1015–1019. doi: 10.1093/nar/24.6.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldas C., So C.W., MacGregor A., Ford A.M., McDonald B., Chan L.C., Wiedemann L.M. Exon scrambling of MLL transcripts occur commonly and mimic partial genomic duplication of the gene. Gene. 1998;208(2):167–176. doi: 10.1016/S0378-1119(97)00640-9. [DOI] [PubMed] [Google Scholar]
- 20.Chao C.W., Chan D.C., Kuo A., Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 1998;4(9):614–628. [PMC free article] [PubMed] [Google Scholar]
- 21.Surono A., Takeshima Y., Wibawa T., Ikezawa M., Nonaka I., Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum. Mol. Genet. 1999;8(3):493–500. doi: 10.1093/hmg/8.3.493. [DOI] [PubMed] [Google Scholar]
- 22.Li X.F., Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 1999;274(12):8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- 23.Gualandi F., Trabanelli C., Rimessi P., Calzolari E., Toffolatti L., Patarnello T., Kunz G., Muntoni F., Ferlini A. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J. Med. Genet. 2003;40(8):e100. doi: 10.1136/jmg.40.8.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houseley J.M., Garcia-Casado Z., Pascual M., Paricio N., O’Dell K.M., Monckton D.G., Artero R.D. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J. Hered. 2006;97(3):253–260. doi: 10.1093/jhered/esj037. [DOI] [PubMed] [Google Scholar]
- 25.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 30.Soma A., Onodera A., Sugahara J., Kanai A., Yachie N., Tomita M., Kawamura F., Sekine Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318(5849):450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- 31.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C., Wang X., Hou J., Liu H., Sun W., Sambandan S., Chen T., Schuman E.M., Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18(4):603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Reports. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Reports. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danan M., Schwartz S., Edelheit S., Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40(7):3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doose G., Alexis M., Kirsch R., Findeiß S., Langenberger D., Machné R., Mörl M., Hoffmann S., Stadler P.F. Mapping the RNA-Seq trash bin: unusual transcripts in prokaryotic transcriptome sequencing data. RNA Biol. 2013;10(7):1204–1210. doi: 10.4161/rna.24972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann S., Otto C., Doose G., Tanzer A., Langenberger D., Christ S., Kunz M., Holdt L.M., Teupser D., Hackermüller J., Stadler P.F. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15(2):R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16(1):4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q., Wang Y., Cao M., Pantaleo V., Burgyan J., Li W.X., Ding S.W. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. USA. 2012;109(10):3938–3943. doi: 10.1073/pnas.1117815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Qi S., Tang N., Zhang X., Chen S., Zhu P., Ma L., Cheng J., Xu Y., Lu M., Wang H., Ding S.W., Li S., Wu Q. Discovery of replicating circular RNAs by RNA-seq and computational algorithms. PLoS Pathog. 2014;10(12):e1004553. doi: 10.1371/journal.ppat.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubin R.A., Kazmi M.A., Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167(1-2):245–248. doi: 10.1016/0378-1119(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 44.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., Zhu P., Chang Z., Wu Q., Zhao Y., Jia Y., Xu P., Liu H., Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 46.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H., Bindereif A. Exon circularization requires canonical splice signals. Cell Reports. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Chen L., Shan G. Circular RNAs remain peculiarly unclear in biogenesis and function. Sci. China Life Sci. 2015;58(6):616–618. doi: 10.1007/s11427-015-4855-y. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 50.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 52.Junn E., Lee K.W., Jeong B.S., Chan T.W., Im J.Y., Mouradian M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA. 2009;106(31):13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau P., de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin. Cell Dev. Biol. 2010;21(7):768–773. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill J.M., Lukiw W.J., Bhattacharjee S., Zhao Y.H., Alexandrov P.N., Dua P., Pogue A.I. Deficits in the natural circular RNA (circRNA) ‘sponge’ for miRNA-7 (cirs7) in Alzheimer's Disease (AD): miRNA-7 up-regulation, and down-regulation of the key phagocytosis protein Ubiquitin Ligase A (UBE2A). Alzheimers Dement. 2014;10(4):785–786. doi: 10.1016/j.jalz.2014.05.1520. [DOI] [Google Scholar]
- 56.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 57.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li P., Chen S., Chen H., Mo X., Li T., Shao Y., Xiao B., Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 59.Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosal S., Das S., Sen R., Basak P., Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12(8):542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 62.Fu X.D. Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 2014;1(2):190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu H., Wang X., Wang H.D., Wu J., Ren J., Meng L., Wu Q., Dong H., Wu J., Kao T.Y., Ge Q., Wu Z.X., Yuh C.H., Shan G. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 2012;3:1073. doi: 10.1038/ncomms2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu S., Wu J., Chen L., Shan G. Signals from noncoding RNAs: unconventional roles for conventional pol III transcripts. Int. J. Biochem. Cell Biol. 2012;44(11):1847–1851. doi: 10.1016/j.biocel.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 65.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]


