Abstract
MicroRNAs are 19-24 nucleotides noncoding RNAs which silence modulate the expression of target genes by binding to the messenger RNAs. Myeloid malignancies include a broad spectrum of acute and chronic disorders originating from from the clonal transformation of a hematopoietic stem cell. Specific genetic abnormalities may define myeloid malignancies, such as translocation t(9;22) that represent the hallmark of chronic myeloid leukemia. Although next-generation sequencing pro-vided new insights in the genetic characterization and pathogenesis of myeloid neoplasms, the molecular mechanisms underlying myeloid neoplasms are lacking in most cases. Recently, several studies have demonstrated that the expression levels of specific miRNAs may vary among patients with myeloid malignancies compared with healthy individuals and partially unveiled how miRNAs participate in the leukemic transformation process. Finally, in vitro experiments and pre-clinical model provided preliminary data of the safety and efficacy of miRNA inhibitory molecules, opening new avenue in the treatment of myeloid hematological malignancies.
Keywords: miRNAs, Acute myeloid leukemia , Myelodysplastic syndrome, Myeloproliferative neoplasms.
1. INTRODUCTION
MicroRNAs (miRNA) are 19-24 nucleotides noncoding RNAs (ncRNA) which regulate the expression of target messenger RNAs (mRNAs) [1, 2]. Mature miRNA are processed from long, capped and polyadenylated precursors which are cleaved in the nucleus, exported to the cytoplasm where duplex miRNA are transformed into single strand miRNA and associated with RISC (RNA-induced silencing complex) [1, 2]. By binding with the seed sequence, mainly at the 3’-untraslated region (3’-UTR) of the target messenger, miRNAs inhibit the translation or favor the destabilization of mRNAs depending on the degree of nucleotide pairing [3, 4].
Recently, several studies have elucidated the physiological roles of miRNAs as key regulators of hematopoiesis [5-7]. The discovery that miRNAs are able to finely tune cell machinery at crucial points suggested the possibility that genetic aberrations at miRNAs gene sequences may contribute to cancer development [8]. Interestingly, miRNAs function, as oncogenes or tumor-suppressors respectively, is strictly dependent on the on the target genes but also may associate with the specific context of normal and tumor cells. The identification of specific miRNA expression profile among normal and tumor tissues has several diagnostic, prognostic and therapeutic implications. Although preliminary, recent findings support the possibility of using miRNAs expression to predict response to specific treatment or outcome among patients with hematological malignancies [9, 10]. Finally, the inhibition of miRNAs by means of antagonist inhibitory molecules in pre-clinical models provided new insights in the understanding of the intricate miRNAs network in the pathogenesis of hematological malignancies and opened a new avenue in their treatment [11].
2. ACUTE MYELOID LEUKEMIA
Acute myeloid leukemia (AML) represents a heterogeneous group of malignancies including subtypes with different biological features and outcomes [12, 13]. The revised World Health Organization (WHO) classification of myeloid malignancies emphasized the crucial role of specific genetic abnormalities in the definition of AML subtypes [13]. However, the availability of more accurate genetic assessment tests allowed us to better understand the molecular mechanisms underlying the most common genetic abnormalities and to provide evidence of the role of new molecular markers that were not unveiled by conventional cytogenetic analysis [12]. Moreover, gene expression profiling (GEP) of patients with normal cytogenetics AML (NC-AML) have shown the presence of recurrent gene mutations explaining the unexpected clinical heterogeneity among patients with NC-AML [14].
In AML the aberrant expression of miRNAs, the association of miRNA expression profiles with cytogenetic characteristics and their impact on treatment response and outcome has been proven by different groups [9, 15-19]. Nevertheless, the pathogenetic mechanisms, i.e. the pathways that miRNAs interfere with in leukemic cells, are in some cases lacking.
In this review we will explore the association between AML and specific miRNAs or miRNA profiles following the main categorization of AML into favorable (core binding factor, CBF leukemias; acute promyelocitic leukemia, APL; AML with mutations of CCAT/enhancer binding protein-alpha, CEBPA or C/EBPα; AML with mutations of nucleophosmin, NPM) and unfavorable AML (AML with fms-like tyrosine kinase receptor 3 internal tandem duplications, FLT3-ITD; AML with abnormalities of the myeloid and lymphoid lineage, MLL gene; AML with deletion 7q or monosomy 7; AML with KIT or BAALC, brain and acute leukemia cytoplasmic, overexpression) [12, 13]. Then we will analyze the role of miRNAs among patients with myelodysplastic syndrome (MDS) and myeloproliferative neoplasms (MPN).
Importantly, the integration of prognostication system with miRNA profile might contribute to better distinguish between low and high risk subtypes addressing clinicians in the choice of therapy strategy among several treatment options including standard chemotherapy, bone marrow transplantation and new biological agents. Furthermore, preclinical models and in vitro experiments suggest that targeting miRNAs can increase leukemia cells susceptibility to chemotherapy.
2.1. AML with Favorable Risk
CBF-AMLs include cases harboring either translocation (t)(8;21) (q22;q22) or inversion (inv)(16) (p13q22)/t(16;16) which lead to the formation of AML1/ETO or RUNX1/ RUNX1T1 and CBFB-MYH11, fusion gene, respectively [20, 21].
It has been reported the miR-126 is upregulated in both t(8;21) and inv(16) leukemias [18, 19, 22]. Following large-scale miRNA expression profiling, including 47 de novo AML, significantly higher levels of miR-126 were observed in CBF leukemias without evidence of amplification at the miR-126 locus (9q34.3). Likely, a lower degree of DNA methylation at the miR-126 locus may explain its upregulation in leukemic blasts from CBF leukemias [22].
The expression levels of miR-193a were found downregulated in cells isolated from patients with t(8;21) AML and associated with event-free (EFS) and overall survival (OS) [23]. In leukemic blasts, AML1/ETO repressed miR-193a and PTEN by binding at the AML1-binding sites and recruiting chromatin-remodelling enzymes. The silenced miR-193a resulted in the loss of the inhibition over the oncoprotein AML1/ETO that miR-193a usually exerts [23]. Furthermore, miR-193a may directly repress not only AML1/ETO but also DNMT3a (DNA methytransferase), HDAC (histone deacetylase), KIT (CD117), the cyclin CCND1 and MDM2 (murine double model 2) genes whereas the tumor suppressor PTEN, a protein involved in the PTEN/PI3K pathway, is upregulated by miR-193a [23]. The miR-193a circuitry provides explanation for the observation that KIT is usually overexpressed and predicts poor outcome in patients with t(8;21) AML [24] and suggests that the enhancement of miR-193a in leukemic blasts might reverse the overexpression of its target genes and restore PTEN activity, apoptosis and cell differentiation [23].
Likewise, either t(8;21) or inv(16) AML overexpressing KIT antigen display downregulation of miR-221/222 and miR-223 compared with normal bone marrow precursors [25]. miR-221/222 binds at the 3’ UTR of the KIT messenger and, thereby, low miR-221/222 levels result in KIT upregulation. Similarly, AML1/ETO binds at the AML1-binding site of the myelopoiesis regulator pre-miR-223 gene silencing its trascription [7]. Interestingly, the ectopic expression of miR-223 in HL60 and SKNO-1 cell lines along with demethylating treatment or RNA inhibitory molecules against AML1/ ETO, reprogrammed myeloid differentiation overcoming maturation block [7].
In blasts from patients with overt t(8;21) AML following myelodysplastic syndrome, RUNX1 (or AML1) may de-localize subnuclearly [26]. The same subnuclear localization was observed in murine myeloid progenitor 32D cell lines following single amino acid substitution. Subnuclear RUNX1 associated with deregulation of miR-24-23-27 cluster and miR-181, enhancement of cell proliferation and blockade of myeloid differentiation via miR-24/MAP kinase pathway. Furthermore, 32D cells transfected with miR-24 and miR-23 and cultured in growth factors enriched medium (IL-3 and G-CSF) showed morphological hallmarks of myeloid differentiation and, limited to miR-24 transfected cells, increased expression of myeloid lineage markers of differentiation (granzyme B, CD11b and myeloperoxidase) suggesting that miR-24/miR-23 may play a crucial role in leukemogenesis of t(8;21) AML [26].
Recently, a dutch group investigated the miRNA profiles from 90 pediatric patients with de novo AML including cases with well characterized cytogenetic abnormalities, that is t(8;21), inv(16), t(15;17), t(7;12) and MLL-rearrangements AML [27]. Despite the selection criteria could have biased the results, miR-9 has been shown downregulated in t(8;21) AML cases (p=0.001). By gain-of-function experiments in leukemic blasts and in xenograft models the authors proved that miR-9 is a tumor suppressor, which can inhibit tumor growth and colony-forming capability, and induce monocytic differentiation [27].
Acute promyelocitic leukemia (APL) is defined by the t(15;17) translocation resulting in the juxtaposition of the PML and RARA genes, [28]. The backbone of treatment for patients with APL includes all-trans retinoic acid (ATRA) in combination with trioxide arsenic (ATO) or antracycline-based chemotherapy. The ATRA based regimens have significant prolongated the OS with manageable toxicity [29].
Numerous miRNAs are predicted target of PML-RARA response elements by seed sequence pairment [30]. By using a quantitative real time PCR (qRT-PCR) approach, a restricted signature of miRNAs distinguished promyelocitic blasts from normal promyelocytes. Out of 12 granulocyte differentiation signature miRNAs, nine miRNAs resulted overexpressed whereas miR-107, miR-342 and let-7c were downregulated [31]. Additionally, by SNP array study in blasts from 93 patients with APL, the most common copy number alteration (CNAs) was the deletion of 1q31.3 encompassing the coding regions for miR-181a1/b1. Although, mir181a1 and mir181b1 precursors were not differentially expressed in samples with or without del1q31.3, the 1q31.3 deletion was associated with higher number of copy number alterations (CNAs) and increased risk of relapse (HR=28.9, p=0.0031) [32].
Interestingly, downregulation of miR-181b and upregulation of miR-15a, miR-15b, miR-16, miR-107, miR-223, miR-342 and let-7 family were noted following successful treatment with ATRA and chemotherapy [30, 31, 33]. Several mechanisms have been proposed to explain how ATRA acts on miRNAs network in APL blasts [30, 31, 34]. Precisely, ATRA treatment is associated with reduction and/or disappearance of PML/RARA gene-product that, in turn directly regulates the expression of miR-342 and let-7c [31]. Alternatively, following ATRA treatment PU.1 and IRF9 binding at the promoter region restored miR-342 levels and myeloid differentiation [34]. Finally, homeobox-family genes seemed to be modulated by PML/RARA through miRNAs [11, 30].
Few studies have been conducted to validate miRNAs as prognostic markers within large randomized multi-institutional clinical studies (Table 1). The absence of NPM1 mutations and the occurrence of FLT3-ITD are clearly have been clearly associated with worse prognosis among patients with CN-AML [35]. NPM1 mutations that result in cytoplasmic localization of nucleophosmin (NPMc+ AML) have been observed in one third of AML among adults and, in absence of FLT3-ITD, are associated with favorable outcome [14, 35]. Among 85 patients with de novo AML, up-regulation of miR-10a, miR10b, let-7 and miR-29 family was found to clearly distinguish patients with NPMc+ mutated from wild type (wt) NPM1 cases [16]. Notably, among patients with NPMc+ AML miR-10a positively correlated with homeobox-family gene HOXB4 that is located at the same locus of miR-10a in chromosome 17q21 suggesting that both genes may be regulated by the same cis elements [16].
Table 1.
Results of miRNA profiles within clinical trials including patients with AML.
| References | Cell source | Training cohort (n°) | Validation cohort (n°) | Dysregulated miRNAs | Prognostic or Risk-group correlation | Multivariable analysis |
Parameters included at the multivariable analysis |
|---|---|---|---|---|---|---|---|
| Havelange et al, 2014 | Pre-treatment BM | 54 adult patients with de novo AML | 183 elderly patients (≥60 yrs) | miR-10 family | Higher miR-10a and miR-10b levels associated with the odds of achieving CR | Yes | miR-10a and -10b levels, NPM1 mutations, age, unfavorable cytogenetic, CN-AML (training cohort). miR-10a and -10b, NPM1 mutations, BAALC expression (validation cohort) |
| Li et al, 2013 | pre-treatment BM and PB | 81 patients AML-M2 (AML/ETO+ and AML/ETO-) | 89 patients with(8;21) AML | miR-193a | overexpression of miR-193a: better EFS and OS | No | NA |
| Marcucci et al, 2013 | pre-treatment BM and PB | 363 patients with de novo NC-AML | Not included | miR-155 | High miR-155levels were associated with WBC counts, percentage of blasts, FLT3-ITD, RUNX1 mutated, WT1 mutated, high ERG and BAALC High miR-155 had lower odds of achieving CR, shorter DFS and OS |
Yes | Age, race, WBC count, miR-155, NPM1status, FLT3-ITD, BAALC and ERG expression |
| Blum et al, 2010 | pre-treatment BM | 53 previously untreated AML patients ≥60 yrs | Not included | miR-29b | High pre-treatment miR-29b had better response to decitabine | No | NA |
| Schwind et al, 2010 | pre-treatment BM | 187 adult patients with CN-AML < 60 yrs | 122 patients with CN-AML, ≥60 yrs | miR-181a | Higher miR-181a associated with better CR, longer OS and a trend in longer DFS | Yes | Age, WBC count, miR-181a, ERG and BAALCexpressione, CEBPA and NPM1mutation, WT1, FLT3-ITD. |
| Whitman et al, 2010 | pre-treatment BM or PB | 243 adult patients with CN-AML | not included | miR-155, miR-144 miR-451 | Overexpression of miR-155 and underexpression of miR-144 and miR-451 associated with FLT3-ITD | ||
| Garzon et al, 2008 | pre-treatment BM | 85 adult patients with AML | not included | miR-10a, miR-10b, miR-204, miR-155 |
miR-10a, miR-10b, let-7, miR-29 family were upregulated in NPMc+ AML miR-155 was upregulated in FLT3-ITD AML |
No | NA |
| Marcucci et al, 2008 | pre-treatment BM | 64 patients with high molecular risk CN-AML < 60 yrs | 55 patients with high molecular risk CN-AML | miR-181 family, miR-124, miR-128, miR-194, miR-219, miR-220a, miR-320 |
miR-181 family was inversely associated with EFS (death, lack of CR, or relapse) miR-124, miR-128, miR 194, miR-219, miR-220a, miR-320 were positively with EFS |
Yes | MicroRNA summary value, ratio of FLT3-ITD to FLT3 (high vs low) and WBC count (log), age, sex, race, Hb level, platelet count, percentage of blasts, the presence or absence of extramedullary involvement, NPM1 mutation, and BAALC and ERG expression |
Annotations: CN-AML, cytogenetics normal, acute myeloid leukemia; yrs, years; EFS, event free survival; OS, overall survival, WBC, white blood cell; FLT3-ITD, fms-like tyrosine receptor internal tandem duplications; WT1, Wilm’s tumor gene; BAALC, brain and acute leukemia cytoplasmic gene; CR, complete response; DFS, disease free survival; NPM1, nuclephosmin; CEBPA, CCAAT/enhancer binding protein-alpha gene; NPMc+, nucleophosmin cytoplasmic positive.
Recently, it has been reported that high baseline miR-10 family members expression levels predict the probability of complete remission among 54 previously untreated patients with AML at a single center (p=0.002) [36]. At the multivariable analysis including miR-10-5p, NPM1 mutational status, age, unfavorable prognostic group, normal cytogenetics) miR-10-5p expression levels and NPM1 mutational status were significantly associated with survival (p=0.019 and p=0.005, respectively). Similar results were noted among a validation cohort of 183 patients of 60 yrs or older with de novo CN-AML treated on Cancer and Leukemia Group B (CALGB)/ALLIANCE protocol [36]. Despite the previous demonstration of a strong positive correlation between HOX-related genes and miR-10 and miR-20a [37], the authors failed to prove miR-10-5p affecting apoptosis and cell proliferation at the baseline and after chemotherapy in cell lines and primary blasts by functional experiments [36].
On the contrary, miR-204 is downregulated in patients with NPMc + AML. By performing western blotting in cell lines with high expression of HOXA9, HOXA10 and MEIS1 (NPM1+ OCI-AML3 cell line and MEG-01 cell line), transfection with miR-204 lentivirus has been shown to robustly downregulate HOXA10 and MEIS1 but not HOXA9 protein. Accordingly, HOXA10 and MEIS1 protein levels were reversed after an antisense oligonucleotide against miR-204 was introduced [14].
MiR-181 family members have been found overexpressed among patients with CN-AML and CEBPA mutations and associated with event-free (p<0.001), disease-free (p=0.004) and overall survival (p=0.009) independently from other molecular and clinical features [38]. Indeed, the truncated C/EBPα-p30 isoform, that derives from CEBPA mutated gene, has been demonstrated to bind miR-181a-1 promoter and upregulate its transcription [39].
2.2. AML with Unfavorable Risk
Several studies observed that miR-155 expression levels are independently associated with high risk AML and poor outcome. (Table 2) [9, 15] The role that miR-155 plays in hematopoiesis and hematological malignancies was described by Mallardo et al. [85]
Table 2.
miRNA profiles associated with specific molecular or cytogenetic abnormalities in acute myeloid leukemia and their targets.
| Disease and Specific Molecular Abnormalities | Dysregulated miRNAs | Predicted Target | Reference |
|---|---|---|---|
| CBF [t(8;21) and inv(16)] | miR-126/miR-126* (upregulated) | PLK2, SPRED1 | Li et al, 2008 |
| miR-193 (downregulated) | AMl1/ETO, DNMT3a, HDAC, KIT, CCND1, MDM2 | Li et al, 2013 | |
| miR221/222 (downregulated) | KIT | Brioschi et al, 2010 | |
| miR-9 (downregulated) | LIN28B/HMGA2 | Emmrich et al, 2014 | |
| miR-223 (downregulated) | Fazi et al, 2007 | ||
| t(15;17) | miR-224, miR-368, miR-382 (upregulated) | Notevaluated | Li et al, 2008 |
| miR-181 (downregulated) | Notevaluated | Nowak et al, 2012 | |
| Disease and Specific Molecular Abnormalities | Dysregulated miRNAs | Predicted Target | Reference |
|
miR-15a, miR-15b, miR-16, miR-142, miR-181b, miR-223, let-7a, let-7d (upregulated); miR-107, miR-342, let-7c (downregulated) |
Not evaluated | Careccia et al, 2009 | |
| MLL-rearrangements | miR-17-5p, miR17-3p, miR-18a, miR-19a, miR-20a, miR-19b, miR-92 (polycistronic) (upregulated) | not evaluated | Li et al, 2008 |
|
miR-191 (upregulated) miR-29 (downregulated) |
not evaluated | Garzon et al, 2008 | |
| NPMc+ | miR-10a, miR-10b, let-7, miR-29 family, miR-15a-16-1, miR-17-18a-19a-20a cluster(upregulated) | HOXAB4 (miR-10); HAXA10 MEIS1 (miR-204) | Garzon et al, 2008 |
| FLT3-ITD | miR-155 (upregulated) | Garzon et al, 2008 | |
| miR-181 family, miR-124, miR-128, miR 194, miR-219, miR-220a, miR-320(upregulated) | Toll-like receptors, IL1-beta, CARD (caspase recruitment domain) | Marcucci et al, 2008 | |
| RUNX1mutated |
let-7, miR-223, miR-99a, miR-100 (downregulated) miR-211, miR-220, miR-595(upregulated) |
Not evaluated | Mendler et al, 2012 |
| BAALCoverexpressed | miR-3151(upregulated) | TP53 pathway | Eisfeld et al, 2014 |
Annotations: CBF, core binding factor; MLL, myeloid and lymphoid gene; NPMc+, nucleophosmin cytoplasmic positive; FLT3-ITD, fms-like tyrosine receptor internal tandem duplications; BAALC, brain and acute leukemia cytoplasmic.
By using miRNA expression profiling, 12 (miR-181 family, miR-124, miR-128-1, miR-194, miR-219-5p, miR-220a, miR-320) out of 305 miRNAs were differently associated with event-free survival (EFS) among patients younger than 60 yrs and with normal karyotype, wild type NPM1 and FLT3-ITD [17]. The miRNA signature that was derived from 64 patients treated in the CALGB 19808 multicenter study, was confirmed by the analysis of a validation cohort including 55 patients from the CALGB 9621 study who received similar treatment but significantly differed from training group in the white-cell count, the grade of blast bone marrow infiltration, the percentage of circulating blasts, and finally the proportion of patients with high levels of ERG gene expression in leukemic cells. In the confirmatory group, the miRNA summary value was confirmed to correlate with EFS (p=0.03) and the percentage of circulating blasts (p=0.004) [33]. The CALGB group confirmed miR-181 as prognostic markers in a larger population of patients with CN-AML, especially within poor molecular risk patients with FLT3-ITD and/or wtNPM1 [40]. Mir-181 was measured at the baseline in bone marrow cells from 187 younger adults (< 60 yrs) treated with intensive induction therapy and consolidation autologous bone-marrow transplantation according with CALGB9621 and 19808 and in 122 older patients (≥ 60 yrs) (validation cohort). Higher miR-181 expression levels at the baseline were significantly associated with better CR rate (p=0.04) and longer OS (p=0.01). In both reports, the multivariable analysis including the most reliable molecular markers, miR-181 retained its prognostic power [17, 40]. Interestingly, the expression levels of miR-181 family were inversely associated with the risk of event [17] and were associated with the expression levels of predicted target genes encoding proteins that mediate the intracellular transduction following the activation by toll-like receptors and interleukin-1β [17, 18, 40].
The CALGB group analyzed the gene expression signature of 72 patients aged ≥ 60 yrs with primary CN-AML harboring FLT3-ITD and treated frontline with intensive chemotherapy [41]. Although the treatment included in some cases investigational drugs, patients with FLT3-ITD had significantly shorter DFS and OS compared with FLT3-WT (p 0.007 and < 0.001, respectively). However, the difference disappeared among patients aged ≥ 70 yrs. As expected, FLT3-ITD samples correlated with increased expression of miR-155 and miR-125b-2. Additionally, miR-144, miR-451, miR-488, miR-486-5p were encountered among the most downregulated miRNAs in the same group of patients [41].
RUNX-1 mutations in elderly with wtNPM1 are associated with poor outcome [42, 43]. Recently, a study has shown that samples with RUNX1 mutations yielded lower levels of the let-7 family, miR-223 (a positive regulator of granulopoiesis) miR-99a and miR-100 whereas miR-211, miR-220 and miR-595 were upregulated [44].
By gene-expression profiling, 91 pediatric patients with AML exhibited a reciprocal pattern of HOXA9 (belonging to the homeobox-family) and growth factor independent 1 factor (GFI1) profiles with HOXA9-like signature clustering 11q23 (MLL1) cases and GFI1-like signature defining t(15;17), t(8;21) and inv(16) cases [11]. Surprisingly, among the validation cohort including 460 adults with AML and MDS, either MLL translocations leukemias or NPM1 mutant cases showed HOXA-9 like signature [11]. HOXA9 interacts with MEIS1 and PBX1 resulting in the formation of a transcriptional factor complex that regulates multiple genes. In mice GFI1 directly interfere with the expression of HOXA9, PBX1 and MEIS1 during maturation of myeloid precursor [45]. HOXA9 was shown to target miR-21 and miR-196b that were up-regulated in patients with 11q23 abnormalities compared with t(8,21) cases [11].
AML with MLL translocations were associated with upregulation of miR-191 and down regulation of miR-29 family members [16]. In AML cell lines and primary samples the restoration of nomal levels of miR-29b induces apoptosis and shrinkage of tumors. Furthermore, disappearance of tumors were observed in xenograft mouse model that were treated by injection of syntethic miR-29b after inoculation of K562 cells in mice flanks [46]. By GEP analysis in K562 transfected with miR-29b or scrambled oligonucleotides, several genes related to apoptosis (MCL-1, TRA4, and MYBL2), cell cycle regulation (CDK4, CDK6 and CCND2) and cell proliferation (JAK2 and IGF1) were found deregulated according to miR-29 expression levels. Furthermore, in primary cell lines from patients with AML, miR-29a and miR-29b correlated with specific signatures and approximately 42% of the miR-29a-correlated genes were also associated with the expression levels of miR-29b. The genes that were found to be positively or negatively associated with miR-29 belonged to different pathways including apoptosis, protein metabolism and modification processes [46].
Aberrant hypermethylation and histone modifications may epigenetically alter the expression of genes that participate to normal hematopoiesis [47]. In AML cell lines (Kasumi-1, MV4-11 and K562) miR-29b lentivirus infection resulted in a decrease of global DNA methylation [46]. Although a significant reduction of the expression of all three DNA methytransferases DNMT1, DNMT3A and DNMT3B at both RNA and protein levels was observed, miR-29b has been shown to directly interact with DNMT3A and DNMT3B 3’-UTR regions whereas the inhibition of DNMT1 was mediated by its transactivator Sp1 [46]. In KIT-driven AML, the down regulation of miR-29b and therefore the increased levels of Sp1, enhance the activity of the Sp1/NFκB/HDAC complex. The Sp1/NFκB complex has been shown to favor the expression of BAALC gene and miR-3151 that are located on chromosome 8q22 [48]. Increased expression of BAALC and miR-3151 is associated with inferior outcome in patients with AML [49, 50]. The miR-3151 binds at the 3’-UTR of TP53, thereby affecting the TP53-mediated apoptosis pathway in leukemic cells. Furthermore, the expression of BAALC is under the control of RUNX1, that is frequently translocated or mutated in AML [48]. The intrigued network involving miR-29b and miR-3151 and their interactions with Sp1/NFκB, KIT, TP53, DNMT and BAALC provide us new elements to better understand the mechanisms of leukemic transformation and suggest that miRNAs are key-regulators of various cellular pathways (Fig. 1).
Fig. (1).
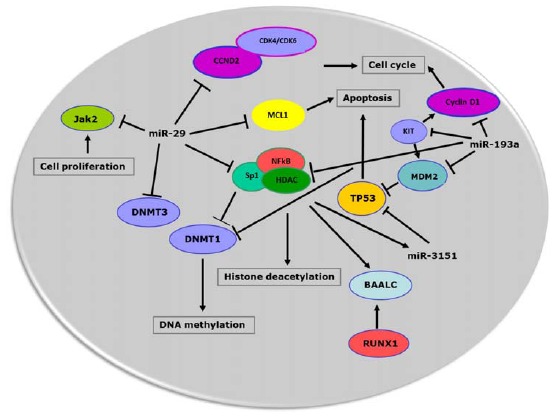
MiRNAs network in acute myeloid cell. miRNAs interfere with multiple pathways in malignant cell resulting in dysregulation of cell cycle, apoptosis, cell proliferation and epigenetics (DNA methylation and histone deacetylation). Furthermore, different miRNAs may converge on the same cellular pathways suggesting a possible cooperation effect among miRNAs that are aberrantly expressed in leukemic cells.
High risk AML group includes cases with del(7q) or monosomy 7 [13]. Mir-29b-1/miR29a clusters at chromosome 7q32 and in primary AML samples with monosomy 7 miR-29a and miR-29b, but not miR-29c, that resides on chromosome 1q32.2, expression levels were lower than cases with other genetic abnormalities [46]. The observation that the rate of clincial response among patients with del(7q)/monosomy 7 who received decitabine were higher compared with historical population, suggested that hypomethylating agents might counteract the inhibition on DNMT enzymes secondary to low miR-29 expression levels in this subgroup of patients [51, 52].
3. MYELODYSPLASTIC SYNDROMES
Myelodsyplastic syndromes (MDS) are usually believed pre-leukemic conditions and may precede overt AML. Recently, the availability of next generation sequencing (NGS) allowed to prove the molecular heterogeneity of MDS providing a deeper insight in the pathogenesis of this disease and explanation of different clinical outcomes among affected patients [53]. Studies of miRNA expression in MDS failed in some circumstances to reveal specific miRNA signatures as it has been clearly observed in AML. However, these results may be related to the use of unpurified stem cells as source of tumor cells in studies aiming to analyze miRNA profiles in MDS patients.
Similarly with AML cases, miR-10, miR-181 and miR-155 were found to be prognosticaly relevant among patients with MDS where they are more likely associated with specific risk categories [54, 55]. However, in contrast with AML, miR-181 has been correlated to shorter survival in patients with either high risk (International prognostic score system, IPSS, Int-2 or high risk) or low risk MDS [55]. Furthermore, MDS carrying trisomy 8 and del(5q) were more likely to express specific miRNA profiles compared with MDS with normal cytogenetics or with different abnormalities [39, 56].
In mice model with additional sex combs-like 1 (ASXL1) mutations displaying myelodysplastic-like features, it has been reported that miR-125a and HOXA9 are derepressed by mutant ASXL1 through inhibition of polycomb repressive complex 2-mediated (PRC2-mediated) methylation and histone H3K27 [57]. miR-125 inhibited the expression of C-type lectin domain family 5 (Clec5a), which is involved in myeloid differentiation [57].
Recently, it has been observed that miRNAs expression levels can be modulated in cells as well as in body fluid [58]. Using a high-throughput digital quantification technology to study the miRNA profiles in plasma samples from 77 patients with MDS, a “7 miRNA” signature emerged as an independent prognostic factor of survival within patients with normal cytogenetics (p=0.008) [59].
4. CHRONIC MYELOID LEUKEMIA (CML) AND MYELOPROLIFERATIVE NEOPLASMA (MPD)
Chronic myeloid leukemia (CML) is a myeloproliferative disorder originating from the hematopoietic stem cell (HSC) and characterized by exuberant cell proliferation and abnormal differentiation of the myeloid precursors. The hallmark of CML is the Philadelphia chromosome (Ph) that results from the translocation of the long arms of cromosomes 9 and 22. The t(9;22) causes the formation of the oncoprotein BCR-ABL1, a constitutively active tyrosine kinase that confers growth factor-independent proliferation and to the hematopoietic progenitors., [60].
Although the vast majority of patients achive hematological, cytogenetic and molecular response by TKIs treatment, a small proportion of patients with CML will eventually fail into because of the occurrence of resistance to TKIs or progression to blast crisis (BC) [61]. The mutations at the drug binding site are responsible of the occurrence of drug resistance in most cases [62]. However, the mechanism(s) of resistance to TKIs are not completely known. In patients with CML harboring the deletion in 9q34, miR-199b and miR-219-2 were downregulated compared with patients without the deletion. However, only miR-199b out of the two miRNAs associated with imatinib drug resistance [63]. Hence, the deletion of miR-199b located at 9q34.11 may provide explanation of the poor prognosis and suggest the possible mechanism of drug resistance in patients with del(9q) CML [64].
In a model of CML, miR-328 upregulated the translation of the C/EBPα mRNA through a regulatory RNA-binding protein (hnRNP E2) that represses CEBPA identifying a novel decoy regulatory mechanism [65]. MiR-328 is markedly reduced in BC-CML CD34+ cells when compared with CP-CML CD34+ cells suggesting that the downregulation of miR-328 might contribute to disease progression. Accordingly, the re-expression of miR-328 levels in BC-CML CD34+ cells restored myeloid differentiation in response to stimulation with G-CSF through the upregulation of C/EBPα protein and of PIM1 serine-threonine kinase [65].
In patients with newly diagnosed CML and following two weeks of treatment, miR-150 and miR146 were found increased whereas miR-142-3p and miR-199b-5p were decreased at the same time point compared with pre-treatment levels. Furthermore, miR-150 resulted downregulated in blasts from patients with overt CB-CML [66]. Despite the paucity of samples, the authors documented positive correlation between miRNAs levels at diagnosis and specific clinical features [66]. Importantly, miR-142-3p correlated with Sokal score (p< 0.05) and miR-18 fold change between day 0 and day 14, inversely associated with the time to initial complete hematological response (r=-0.7198, p=0.0125). The results were confirmed in a validation cohort of 12 patients [66]. Interestingly, imatinib may induce demethylation of miR-203 which targets BCR/ABL1, suggesting that the switch-off exerted by imatinib on constitutively activated ABL kinase might rely on additional mechanisms other than the inhibition at the ATP-binding pocket [67].
In imatinib resistant CML cell lines miR-181 was significantly repressed compared with drug-sensitive cell lines [68]. Lyn kinase (Lyn) turns off miR-181 that targets myeloid cell-leukemia 1 (MCL-1), an antiapoptotic protein whose expression may be upregulated in TKIs resistant cells. According to this model, the mechanism of resistance in cells overexpressing Lyn and Mcl-1 is mediated by miR-181 [68].
Myeloproliferative neoplasms (MPD) include primary myelofibrosis, essential trombocytemia and polycytemia vera [69]. Using NGS, in a training cohort of 22 patients with MPN comparing the genomic DNA of granulocytes with in vitro expanded CD3+ T-lymphocytes, as germline control, 141 somatic mutations were identified [70]. Recurrent mutations occurred at 8 gene loci including miR-662 that was mutated in 7.4% of cases. Similarly, miR-17, miR-19a, miR-542 and miR-663a were characterized by missense mutations in their stem-loop coding region (miR-662,-663,-542) or downstream the seed sequence (miR-17-5p) [70].
The thrombopoietin receptor (MPL) mutations have been found in primary myelofibrosis and essential thrombocytemia resulting in constitutive activation of the janus kinase 2 (JAK2) signaling [71-73]. In platelets from patient with MPN, miR-28 is overexpressed compared with stable levels in platelets from healthy individuals [74]. It has been proposed that miR-28 exerts its action targeting MPL as well as other proteins essential for megakaryocyte differentiation. Furthermore, the ectopic expression of miR-28 transfected CD34-derived megakaryocytes inhibited terminal differentiation.
These results shed light on the mechanisms by which miRNAs contribute to the onset in the pathogenesis of myeloproliferative neoplasms [74] and on the mechanism(s) of resistance of TKIs other than mutations of the active binding site.
5. miRNAs AND LEUKEMIC STEM CELLS (LSCs) IN MYELOID HAEMATOLOGICAL MALIGNACIES
Leukemic stem-cells (LSCs) represent the natural reservoir in patients with AML, CML and MDS and are responsible of clinical minimal residual disease (MRD) positivity, relapse and/or chemo-resistance [75, 76].
An interesting finding was that following transfection into CD34+ AML cell line KG1a of miR-9, miR-24 and miR-125a oligonucleotides, CD34 protein expression levels measured by Western blotting were significantly reduced suggesting that low expression of CD34 on the surface of NPMc+ blasts [77] may be due to the up-regulation of specific miRNAs [15]. Recently, distinct miRNA expression profiles were identified in LSCs from adults with de novo CN-AML [78]. Among 364 patients with CN-AML, the expression of 44 activated genes in LSCs was associated with high risk molecular features (FLT3-ITD, WT1 and RUNX1 mutations, wtCEBPA and TET2, high ERG, miR-155 and BAALC gene expression levels) and shorter disease-free and overall survival (p<0.001 for both), even at the multivariable analysis including the most common prognostic markers. The observation that miRNAs which are relevant for stem cell function might be upregulated in leukemic blasts suggests the implementation and the inclusion of miRNAs inhibitory molecules in order to eradicate the LSC reservoir, therefore improving the disease-free and overall survival of patients with AML [78].
In CML as in other myeloid malignancies, the persistence of quiescent CML HSCs might contribute to disease progression because leukemic stem cell proliferation and survival are apparently resistant to TKIs inhibition [76]. In patients with CML, the expression of miR-17~92 polycistronic pri-mRNA transcripts and mature miRNAs (miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b, miR-92-1) were found upregulated in chronic phase (CP) but not in BC-CML CD34+ [79, 80]. Accordingly, in 3 BCR/ABL1 positive cell lines, a downregulation of miRNA encoded within the polycistrionic miR-17-92 cluster was observed following imatinib treatment or the addition of anti BCR/ABL1 RNA interference (RNAi). The polycistrionic miR-17-92 cluster is transcriptionally regulated by c-MYC [81] and c-MYC participates with BCR-ABL in the leukemic transformation process [82].
In primary myelofibrosis CD34+ cells, several miRNAs involved in trascriptional control and chromatin remodeling were found deregulated [83]. In particular, overexpression of miR-155-5p impaired the activity of JARID2 resulting in increased formation of CD41+ megakariocyte precursors [83].
6. MicroRNAs AS PREDICTIVE MARKERS OF RESPONSE AND THERAPY TARGETS
So far, few studies within clinical trials have identified miRNAs or miRNA signature to predict chemo-sensitivity in AML [9, 38, 44, 51].
In a phase 2 study with frontline single agent decitibine in a single cohort of older patients (≥ 60 yrs) who were not fit or refused other treatments, higher levels of miR-29 were predictive of clinical response (p=0.02) [51]. The observation than high miR-29 expression levels favor clinical response to standard treatment, suggested to investigate whether the delivery of miR-29 molecules into leukemic cells has antileukemic effects. One concern when small molecules are introduced into body fluids is that they can be degraded. Transferrin-conjugated nanoparticles containing miR-29b have shown significant results in terms of apoptosis induction and cell growth inhibition [84]. More impressive is the fact that mice who were treated with the combination of miR-29b nanoparticles followed by decitabine have significantly better outcome compared with mice treated with decitabine as single agent [84]. A similar “priming” effect was noted with the novel HDAC inhibitor AR-42 which sensitized leukemic cells to decitabine by removing the inhibition exerted by HDAC on miR-29b expression [52].
MiRNA inhibitory molecules represent a challenging tool for the treatment of patients with hematological malignancies. Experimental data in vitro and in pre-clinical models suggest that the interference of miRNAs in combination with standard treatment might contribute to chemo-sensitivity and leukemia control [11]. In a xenograft model of MLL leukemia, the injection of miR-21 and miR-196b antagomirs in combination with daunorubicin and aracytin (that resembles the D3+A7 scheme used as standard induction treatment of human AML) significantly improved the life span of the mice compared with recipients treated with control antagomirs (p < 0.01) or AML1/ETO leukemia models [11]. Furthermore, by using lentivirus-delivered CXCR4 shRNA the expression levels of let-7a were found significantly increased in OCI-AML3 contributing to sensitize cells to aracytin, a backbone drug in the treatment of AML. The reduction of let-7a target genes, such as the antiapoptotic protein BCL-XL and MYC, would be responsible for inhibition of cell proliferation and chemo-sensitivity [85]. Lenalidomide, an anti-angiogenic drug that has been approved for treatment of myelodysplastic syndrome with del(5q) and of multiple myeloma, may enhance the translation of the C/EBPα-p30 isoform in myeloid blasts from patients with CN-AML and CEBPA mutations. The induction of the C/EBPα-p30 isoform results in higher expression of miR-181a and inhibition of leukemic growth [39]. Therefore, specific miRNA signature may explain the better chemo sensitivity of certain subtype of AML, such as those with CEBPA mutations [9].
Although further studies are warranted to confirm that the RNA inhibitor molecules are effective and not harmful for patient safety, these results suggest that the repression of miRNAs may serve for treatment of certain patients with AML.
7. CONCLUSIONS AND FUTURE DIRECTIONS
Over the last decade, several studies underscored the pivotal role that miRNAs have in myeloid malignancies, including acute myeloid leukemia, myelodysplastic syndrome and myeloproliferative neoplasms. MiRNAs may act as tumor suppressor or oncogene interfering with multiple pathways involved in cell cycle, tumor growth and apoptosis. Interestingly, different leukemic subtypes have distinct miRNAs expression profiles. Furthermore, the identification that miRNA expression profiles may predict outcome in myeloid malignancies or chemo-sensitivity to specific drug suggest to include miRNAs in the prognostication system of these broad spectrum of diseases. Finally, in vitro experiments and pre-clinical models provided promising results of the safety and the efficacy of miRNAs inhibitory molecules. Further studies and clinical trials are warranted to confirm the prognostic and predict power of miRNAs and to test the miRNAs inhibitory molecules.
ACKNOWLEDGEMENTS
Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is also supported as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar, and by the CLL Global Research Foundation. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, the Department of Defense, a Developmental Research Award in Leukemia SPORE, a SINF MDACC-DKFZ grant in CLL, the Duncan Family Institutional Seed Funds, The Blanton-Davis Ovarian Cancer - 2013 Sprint for Life Research Award, the Laura and John Arnold Foundation, the RGK Foundation, and the Estate of C. G. Johnson, Jr. Dr. Ciccone is supported by AIL-Associazione Italiana contro le Leucemie, Sezione di Ferrara.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen M.T., Häger M., Glenthøj A., Asmar F., Clemmensen S.N., Mora-Jensen H., Borregaard N., Cowland J.B. miRNA-130a regulates C/EBP-ε expression during granulopoiesis. Blood. 2014;123(7):1079–1089. doi: 10.1182/blood-2013-08-523233. [DOI] [PubMed] [Google Scholar]
- 7.Fazi F., Racanicchi S., Zardo G., Starnes L.M., Mancini M., Travaglini L., Diverio D., Ammatuna E., Cimino G., Lo-Coco F., Grignani F., Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12(5):457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Berindan-Neagoe I., Monroig Pdel.C., Pasculli B., Calin G.A. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014;64(5):311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcucci G., Maharry K.S., Metzeler K.H., Volinia S., Wu Y.Z., Mrózek K., Nicolet D., Kohlschmidt J., Whitman S.P., Mendler J.H., Schwind S., Becker H., Eisfeld A.K., Carroll A.J., Powell B.L., Kolitz J.E., Garzon R., Caligiuri M.A., Stone R.M., Bloomfield C.D. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J. Clin. Oncol. 2013;31(17):2086–2093. doi: 10.1200/JCO.2012.45.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrajoli A., Shanafelt T.D., Ivan C., Shimizu M., Rabe K.G., Nouraee N., Ikuo M., Ghosh A.K., Lerner S., Rassenti L.Z., Xiao L., Hu J., Reuben J.M., Calin S., You M.J., Manning J.T., Wierda W.G., Estrov Z., O’Brien S., Kipps T.J., Keating M.J., Kay N.E., Calin G.A. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122(11):1891–1899. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velu C.S., Chaubey A., Phelan J.D., Horman S.R., Wunderlich M., Guzman M.L., Jegga A.G., Zeleznik-Le N.J., Chen J., Mulloy J.C., Cancelas J.A., Jordan C.T., Aronow B.J., Marcucci G., Bhat B., Gebelein B., Grimes H.L. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Invest. 2014;124(1):222–236. doi: 10.1172/JCI66005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel J.P., Gönen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., Huberman K., Cheng J., Viale A., Socci N.D., Heguy A., Cherry A., Vance G., Higgins R.R., Ketterling R.P., Gallagher R.E., Litzow M., van den Brink M.R., Lazarus H.M., Rowe J.M., Luger S., Ferrando A., Paietta E., Tallman M.S., Melnick A., Abdel-Wahab O., Levine R.L. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vardiman J.W., Thiele J., Arber D.A., Brunning R.D., Borowitz M.J., Porwit A., Harris N.L., Le Beau M.M., Hellström-Lindberg E., Tefferi A., Bloomfield C.D. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 14.Schlenk R.F., Döhner K., Krauter J., Fröhling S., Corbacioglu A., Bullinger L., Habdank M., Späth D., Morgan M., Benner A., Schlegelberger B., Heil G., Ganser A., Döhner H., German-Austrian Acute Myeloid Leukemia Study Group Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 15.Garzon R., Garofalo M., Martelli M.P., Briesewitz R., Wang L., Fernandez-Cymering C., Volinia S., Liu C.G., Schnittger S., Haferlach T., Liso A., Diverio D., Mancini M., Meloni G., Foa R., Martelli M.F., Mecucci C., Croce C.M., Falini B. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA. 2008;105(10):3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon R., Volinia S., Liu C.G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., Flomenberg N., Marcucci G., Calin G.A., Kornblau S.M., Kantarjian H., Bloomfield C.D., Andreeff M., Croce C.M. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcucci G., Radmacher M.D., Maharry K., Mrózek K., Ruppert A.S., Paschka P., Vukosavljevic T., Whitman S.P., Baldus C.D., Langer C., Liu C.G., Carroll A.J., Powell B.L., Garzon R., Croce C.M., Kolitz J.E., Caligiuri M.A., Larson R.A., Bloomfield C.D. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 18.Jongen-Lavrencic M., Sun S.M., Dijkstra M.K., Valk P.J., Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 19.Cammarata G., Augugliaro L., Salemi D., Agueli C., La Rosa M., Dagnino L., Civiletto G., Messana F., Marfia A., Bica M.G., Cascio L., Floridia P.M., Mineo A.M., Russo M., Fabbiano F., Santoro A. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am. J. Hematol. 2010;85(5):331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 20.Kwong Y.L., Chan V., Wong K.F., Chan T.K. Use of the polymerase chain reaction in the detection of AML1/ETO fusion transcript in t(8;21). Cancer. 1995;75(3):821–825. doi: 10.1002/1097-0142(19950201)75:3<821::AID-CNCR2820750312>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 21.Tobal K., Johnson P.R., Saunders M.J., Harrison C.J., Liu Yin J.A. Detection of CBFB/MYH11 transcripts in patients with inversion and other abnormalities of chromosome 16 at presentation and remission. Br. J. Haematol. 1995;91(1):104–108. doi: 10.1111/j.1365-2141.1995.tb05253.x. [DOI] [PubMed] [Google Scholar]
- 22.Li Z., Lu J., Sun M., Mi S., Zhang H., Luo R.T., Chen P., Wang Y., Yan M., Qian Z., Neilly M.B., Jin J., Zhang Y., Bohlander S.K., Zhang D.E., Larson R.A., Le Beau M.M., Thirman M.J., Golub T.R., Rowley J.D., Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl. Acad. Sci. USA. 2008;105(40):15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Gao L., Luo X., Wang L., Gao X., Wang W., Sun J., Dou L., Li J., Xu C., Wang L., Zhou M., Jiang M., Zhou J., Caligiuri M.A., Nervi C., Bloomfield C.D., Marcucci G., Yu L. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y.Y., Zhao L.J., Wu C.F., Liu P., Shi L., Liang Y., Xiong S.M., Mi J.Q., Chen Z., Ren R., Chen S.J. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc. Natl. Acad. Sci. USA. 2011;108(6):2450–2455. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brioschi M., Fischer J., Cairoli R., Rossetti S., Pezzetti L., Nichelatti M., Turrini M., Corlazzoli F., Scarpati B., Morra E., Sacchi N., Beghini A. Down-regulation of microRNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010;12(11):866–876. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidi S.K., Dowdy C.R., van Wijnen A.J., Lian J.B., Raza A., Stein J.L., Croce C.M., Stein G.S. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69(21):8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emmrich S., Katsman-Kuipers J.E., Henke K., Khatib M.E., Jammal R., Engeland F., Dasci F., Zwaan C.M., den Boer M.L., Verboon L., Stary J., Baruchel A., de Haas V., Danen-van Oorschot A.A., Fornerod M., Pieters R., Reinhardt D., Klusmann J.H., van den Heuvel-Eibrink M.M. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia. 2014;28(5):1022–1032. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 28.de Thé H., Le Bras M., Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 2012;198(1):11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo-Coco F., Avvisati G., Vignetti M., Thiede C., Orlando S.M., Iacobelli S., Ferrara F., Fazi P., Cicconi L., Di Bona E., Specchia G., Sica S., Divona M., Levis A., Fiedler W., Cerqui E., Breccia M., Fioritoni G., Salih H.R., Cazzola M., Melillo L., Carella A.M., Brandts C.H., Morra E., von Lilienfeld-Toal M., Hertenstein B., Wattad M., Lübbert M., Hänel M., Schmitz N., Link H., Kropp M.G., Rambaldi A., La Nasa G., Luppi M., Ciceri F., Finizio O., Venditti A., Fabbiano F., Döhner K., Sauer M., Ganser A., Amadori S., Mandelli F., Döhner H., Ehninger G., Schlenk R.F., Platzbecker U., Gruppo Italiano Malattie Ematologiche dell’Adulto. German-Austrian Acute Myeloid Leukemia Study Group. Study Alliance Leukemia Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013;369(2):111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- 30.Saumet A., Vetter G., Bouttier M., Portales-Casamar E., Wasserman W.W., Maurin T., Mari B., Barbry P., Vallar L., Friederich E., Arar K., Cassinat B., Chomienne C., Lecellier C.H. Transcriptional repression of microRNA genes by PML-RARA increases expression of key cancer proteins in acute promyelocytic leukemia. Blood. 2009;113(2):412–421. doi: 10.1182/blood-2008-05-158139. [DOI] [PubMed] [Google Scholar]
- 31.Careccia S., Mainardi S., Pelosi A., Gurtner A., Diverio D., Riccioni R., Testa U., Pelosi E., Piaggio G., Sacchi A., Lavorgna S., Lo-Coco F., Blandino G., Levrero M., Rizzo M.G. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene. 2009;28(45):4034–4040. doi: 10.1038/onc.2009.255. [DOI] [PubMed] [Google Scholar]
- 32.Nowak D., Klaumuenzer M., Hanfstein B., Mossner M., Nolte F., Nowak V., Oblaender J., Hecht A., Hütter G., Ogawa S., Kohlmann A., Haferlach C., Schlegelberger B., Braess J., Seifarth W., Fabarius A., Erben P., Saussele S., Müller M.C., Reiter A., Buechner T., Weiss C., Hofmann W.K., Lengfelder E. SNP array analysis of acute promyelocytic leukemia may be of prognostic relevance and identifies a potential high risk group with recurrent deletions on chromosomal subband 1q31.3. Genes Chromosomes Cancer. 2012;51(8):756–767. doi: 10.1002/gcc.21961. [DOI] [PubMed] [Google Scholar]
- 33.Garzon R., Pichiorri F., Palumbo T., Visentini M., Aqeilan R., Cimmino A., Wang H., Sun H., Volinia S., Alder H., Calin G.A., Liu C.G., Andreeff M., Croce C.M. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26(28):4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 34.De Marchis M.L., Ballarino M., Salvatori B., Puzzolo M.C., Bozzoni I., Fatica A. A new molecular network comprising PU.1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009;23(5):856–862. doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- 35.Falini B., Martelli M.P., Bolli N., Bonasso R., Ghia E., Pallotta M.T., Diverio D., Nicoletti I., Pacini R., Tabarrini A., Galletti B.V., Mannucci R., Roti G., Rosati R., Specchia G., Liso A., Tiacci E., Alcalay M., Luzi L., Volorio S., Bernard L., Guarini A., Amadori S., Mandelli F., Pane F., Lo-Coco F., Saglio G., Pelicci P.G., Martelli M.F., Mecucci C. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108(6):1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 36.Havelange V., Ranganathan P., Geyer S., Nicolet D., Huang X., Yu X., Volinia S., Kornblau S.M., Andreeff M., Croce C.M., Marcucci G., Bloomfield C.D., Garzon R. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood. 2014;123(15):2412–2415. doi: 10.1182/blood-2013-10-532374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Havelange V., Stauffer N., Heaphy C.C., Volinia S., Andreeff M., Marcucci G., Croce C.M., Garzon R. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger RNA expression profiling. Cancer. 2011;117(20):4696–4706. doi: 10.1002/cncr.26096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcucci G., Maharry K., Radmacher M.D., Mrózek K., Vukosavljevic T., Paschka P., Whitman S.P., Langer C., Baldus C.D., Liu C.G., Ruppert A.S., Powell B.L., Carroll A.J., Caligiuri M.A., Kolitz J.E., Larson R.A., Bloomfield C.D. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008;26(31):5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickey C.J., Schwind S., Radomska H.S., Dorrance A.M., Santhanam R., Mishra A., Wu Y.Z., Alachkar H., Maharry K., Nicolet D., Mrózek K., Walker A., Eiring A.M., Whitman S.P., Becker H., Perrotti D., Wu L.C., Zhao X., Fehniger T.A., Vij R., Byrd J.C., Blum W., Lee L.J., Caligiuri M.A., Bloomfield C.D., Garzon R., Marcucci G. Lenalidomide-mediated enhanced translation of C/EBPα-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood. 2013;121(1):159–169. doi: 10.1182/blood-2012-05-428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwind S., Maharry K., Radmacher M.D., Mrózek K., Holland K.B., Margeson D., Whitman S.P., Hickey C., Becker H., Metzeler K.H., Paschka P., Baldus C.D., Liu S., Garzon R., Powell B.L., Kolitz J.E., Carroll A.J., Caligiuri M.A., Larson R.A., Marcucci G., Bloomfield C.D. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2010;28(36):5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitman S.P., Maharry K., Radmacher M.D., Becker H., Mrózek K., Margeson D., Holland K.B., Wu Y.Z., Schwind S., Metzeler K.H., Wen J., Baer M.R., Powell B.L., Carter T.H., Kolitz J.E., Wetzler M., Moore J.O., Stone R.M., Carroll A.J., Larson R.A., Caligiuri M.A., Marcucci G., Bloomfield C.D. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(18):3622–3626. doi: 10.1182/blood-2010-05-283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnittger S., Dicker F., Kern W., Wendland N., Sundermann J., Alpermann T., Haferlach C., Haferlach T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117(8):2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 43.Gaidzik V.I., Bullinger L., Schlenk R.F., Zimmermann A.S., Röck J., Paschka P., Corbacioglu A., Krauter J., Schlegelberger B., Ganser A., Späth D., Kündgen A., Schmidt-Wolf I.G., Götze K., Nachbaur D., Pfreundschuh M., Horst H.A., Döhner H., Döhner K. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J. Clin. Oncol. 2011;29(10):1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 44.Mendler J.H., Maharry K., Radmacher M.D., Mrózek K., Becker H., Metzeler K.H., Schwind S., Whitman S.P., Khalife J., Kohlschmidt J., Nicolet D., Powell B.L., Carter T.H., Wetzler M., Moore J.O., Kolitz J.E., Baer M.R., Carroll A.J., Larson R.A., Caligiuri M.A., Marcucci G., Bloomfield C.D. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012;30(25):3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horman S.R., Velu C.S., Chaubey A., Bourdeau T., Zhu J., Paul W.E., Gebelein B., Grimes H.L. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113(22):5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., Havelange V., Volinia S., Blum W., Rush L.J., Perrotti D., Andreeff M., Bloomfield C.D., Byrd J.C., Chan K., Wu L.C., Croce C.M., Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcucci G., Yan P., Maharry K., Frankhouser D., Nicolet D., Metzeler K.H., Kohlschmidt J., Mrózek K., Wu Y.Z., Bucci D., Curfman J.P., Whitman S.P., Eisfeld A.K., Mendler J.H., Schwind S., Becker H., Bär C., Carroll A.J., Baer M.R., Wetzler M., Carter T.H., Powell B.L., Kolitz J.E., Byrd J.C., Plass C., Garzon R., Caligiuri M.A., Stone R.M., Volinia S., Bundschuh R., Bloomfield C.D. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J. Clin. Oncol. 2014;32(6):548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisfeld A.K., Schwind S., Patel R., Huang X., Santhanam R., Walker C.J., Markowitz J., Hoag K.W., Jarvinen T.M., Leffel B., Perrotti D., Carson W.E., III, Marcucci G., Bloomfield C.D., de la Chapelle A. Intronic miR-3151 within BAALC drives leukemogenesis by deregulating the TP53 pathway. Sci. Signal. 2014;7(321):ra36. doi: 10.1126/scisignal.2004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwind S., Marcucci G., Maharry K., Radmacher M.D., Mrózek K., Holland K.B., Margeson D., Becker H., Whitman S.P., Wu Y.Z., Metzeler K.H., Powell B.L., Kolitz J.E., Carter T.H., Moore J.O., Baer M.R., Carroll A.J., Caligiuri M.A., Larson R.A., Bloomfield C.D. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(25):5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baldus C.D., Tanner S.M., Ruppert A.S., Whitman S.P., Archer K.J., Marcucci G., Caligiuri M.A., Carroll A.J., Vardiman J.W., Powell B.L., Allen S.L., Moore J.O., Larson R.A., Kolitz J.E., de la Chapelle A., Bloomfield C.D. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood. 2003;102(5):1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 51.Blum W., Schwind S., Tarighat S.S., Geyer S., Eisfeld A.K., Whitman S., Walker A., Klisovic R., Byrd J.C., Santhanam R., Wang H., Curfman J.P., Devine S.M., Jacob S., Garr C., Kefauver C., Perrotti D., Chan K.K., Bloomfield C.D., Caligiuri M.A., Grever M.R., Garzon R., Marcucci G. Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood. 2012;119(25):6025–6031. doi: 10.1182/blood-2012-03-413898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mims A., Walker A.R., Huang X., Sun J., Wang H., Santhanam R., Dorrance A.M., Walker C., Hoellerbauer P., Tarighat S.S., Chan K.K., Klisovic R.B., Perrotti D., Caligiuri M.A., Byrd J.C., Chen C.S., James Lee L., Jacob S., Mrózek K., Bloomfield C.D., Blum W., Garzon R., Schwind S., Marcucci G. Increased anti-leukemic activity of decitabine via AR-42-induced upregulation of miR-29b: a novel epigenetic-targeting approach in acute myeloid leukemia. Leukemia. 2013;27(4):871–878. doi: 10.1038/leu.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T., Yoshida K., Roller A., Nadarajah N., Shiraishi Y., Shiozawa Y., Chiba K., Tanaka H., Koeffler H.P., Klein H.U., Dugas M., Aburatani H., Kohlmann A., Miyano S., Haferlach C., Kern W., Ogawa S. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pons A., Nomdedeu B., Navarro A., Gaya A., Gel B., Diaz T., Valera S., Rozman M., Belkaid M., Montserrat E., Monzo M. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk. Lymphoma. 2009;50(11):1854–1859. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- 55.Sokol L., Caceres G., Volinia S., Alder H., Nuovo G.J., Liu C.G., McGraw K., Clark J.A., Sigua C.A., Chen D.T., Moscinski L., Croce C.M., List A.F. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br. J. Haematol. 2011;153(1):24–32. doi: 10.1111/j.1365-2141.2011.08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussein K., Theophile K., Büsche G., Schlegelberger B., Göhring G., Kreipe H., Bock O. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leuk. Res. 2010;34(9):1169–1174. doi: 10.1016/j.leukres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 57.Inoue D., Kitaura J., Togami K., Nishimura K., Enomoto Y., Uchida T., Kagiyama Y., Kawabata K.C., Nakahara F., Izawa K., Oki T., Maehara A., Isobe M., Tsuchiya A., Harada Y., Harada H., Ochiya T., Aburatani H., Kimura H., Thol F., Heuser M., Levine R.L., Abdel-Wahab O., Kitamura T. Myelodysplastic syndromes are induced by histone methylation–altering ASXL1 mutations. J. Clin. Invest. 2013;123(11):4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014;11(3):145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 59.Zuo Z., Maiti S., Hu S., Loghavi S., Calin G.A., Garcia-Manero G., Kantarjian H.M., Medeiros L.J., Cooper L.J., Bueso-Ramos C.E. Plasma circulating-microRNA profiles are useful for assessing prognosis in patients with cytogenetically normal myelodysplastic syndromes. Mod. Pathol. 2015;28(3):373–382. doi: 10.1038/modpathol.2014.108. [DOI] [PubMed] [Google Scholar]
- 60.Rowley J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 61.Hochhaus A., O’Brien S.G., Guilhot F., Druker B.J., Branford S., Foroni L., Goldman J.M., Müller M.C., Radich J.P., Rudoltz M., Mone M., Gathmann I., Hughes T.P., Larson R.A., IRIS Investigators Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23(6):1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- 62.Hughes T., Saglio G., Branford S., Soverini S., Kim D.W., Müller M.C., Martinelli G., Cortes J., Beppu L., Gottardi E., Kim D., Erben P., Shou Y., Haque A., Gallagher N., Radich J., Hochhaus A. Impact of baseline BCR-ABL mutations on response to nilotinib in patients with chronic myeloid leukemia in chronic phase. J. Clin. Oncol. 2009;27(25):4204–4210. doi: 10.1200/JCO.2009.21.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joshi D., Chandrakala S., Korgaonkar S., Ghosh K., Vundinti B.R. Down-regulation of miR-199b associated with imatinib drug resistance in 9q34.1 deleted BCR/ABL positive CML patients. Gene. 2014;542(2):109–112. doi: 10.1016/j.gene.2014.03.049. [DOI] [PubMed] [Google Scholar]
- 64.Calabrese G., Fantasia D., Di Gianfilippo R., Stuppia L., Di Lorenzo R., Palka G. Fluorescence in situ hybridization analysis of minimal residual disease and the relevance of the der(9) deletion in imatinib-treated patients with chronic myeloid leukemia. Haematologica. 2006;91(7):994–995. [PubMed] [Google Scholar]
- 65.Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C.J., Becker H., Chandler J.C., Andino R., Cortes J., Hokland P., Huettner C.S., Bhatia R., Roy D.C., Liebhaber S.A., Caligiuri M.A., Marcucci G., Garzon R., Croce C.M., Calin G.A., Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140(5):652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flamant S., Ritchie W., Guilhot J., Holst J., Bonnet M.L., Chomel J.C., Guilhot F., Turhan A.G., Rasko J.E. Micro-RNA response to imatinib mesylate in patients with chronic myeloid leukemia. Haematologica. 2010;95(8):1325–1333. doi: 10.3324/haematol.2009.020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shibuta T., Honda E., Shiotsu H., Tanaka Y., Vellasamy S., Shiratsuchi M., Umemura T. Imatinib induces demethylation of miR-203 gene: an epigenetic mechanism of anti-tumor effect of imatinib. Leuk. Res. 2013;37(10):1278–1286. doi: 10.1016/j.leukres.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 68.Zimmerman E.I., Dollins C.M., Crawford M., Grant S., Nana-Sinkam S.P., Richards K.L., Hammond S.M., Graves L.M. Lyn kinase-dependent regulation of miR181 and myeloid cell leukemia-1 expression: implications for drug resistance in myelogenous leukemia. Mol. Pharmacol. 2010;78(5):811–817. doi: 10.1124/mol.110.066258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tefferi A. Classification, diagnosis and management of myeloproliferative disorders in the JAK2V617F era. Hematology (Am Soc Hematol Educ Program) 2006;•••:240–245. doi: 10.1182/asheducation-2006.1.240. [DOI] [PubMed] [Google Scholar]
- 70.Tenedini E., Bernardis I., Artusi V., Artuso L., Roncaglia E., Guglielmelli P., Pieri L., Bogani C., Biamonte F., Rotunno G., Mannarelli C., Bianchi E., Pancrazzi A., Fanelli T., Malagoli Tagliazucchi G., Ferrari S., Manfredini R., Vannucchi A.M., Tagliafico E., AGIMM investigators Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia. 2014;28(5):1052–1059. doi: 10.1038/leu.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pardanani A.D., Levine R.L., Lasho T., Pikman Y., Mesa R.A., Wadleigh M., Steensma D.P., Elliott M.A., Wolanskyj A.P., Hogan W.J., McClure R.F., Litzow M.R., Gilliland D.G., Tefferi A. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108(10):3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- 72.Pikman Y., Lee B.H., Mercher T., McDowell E., Ebert B.L., Gozo M., Cuker A., Wernig G., Moore S., Galinsky I., DeAngelo D.J., Clark J.J., Lee S.J., Golub T.R., Wadleigh M., Gilliland D.G., Levine R.L. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staerk J., Lacout C., Sato T., Smith S.O., Vainchenker W., Constantinescu S.N. An amphipathic motif at the transmembrane-cytoplasmic junction prevents autonomous activation of the thrombopoietin receptor. Blood. 2006;107(5):1864–1871. doi: 10.1182/blood-2005-06-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Girardot M., Pecquet C., Boukour S., Knoops L., Ferrant A., Vainchenker W., Giraudier S., Constantinescu S.N. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood. 2010;116(3):437–445. doi: 10.1182/blood-2008-06-165985. [DOI] [PubMed] [Google Scholar]
- 75.Wouters R., Cucchi D., Kaspers G.J., Schuurhuis G.J., Cloos J. Relevance of leukemic stem cells in acute myeloid leukemia: heterogeneity and influence on disease monitoring, prognosis and treatment design. Expert Rev. Hematol. 2014;7(6):791–805. doi: 10.1586/17474086.2014.959921. [DOI] [PubMed] [Google Scholar]
- 76.Pellicano F., Sinclair A., Holyoake T.L. In search of CML stem cells’ deadly weakness. Curr. Hematol. Malig. Rep. 2011;6(2):82–87. doi: 10.1007/s11899-011-0085-y. [DOI] [PubMed] [Google Scholar]
- 77.Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., Bigerna B., Pacini R., Pucciarini A., Liso A., Vignetti M., Fazi P., Meani N., Pettirossi V., Saglio G., Mandelli F., Lo-Coco F., Pelicci P.G., Martelli M.F., GIMEMA Acute Leukemia Working Party Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 78.Metzeler K.H., Maharry K., Kohlschmidt J., Volinia S., Mrózek K., Becker H., Nicolet D., Whitman S.P., Mendler J.H., Schwind S., Eisfeld A.K., Wu Y.Z., Powell B.L., Carter T.H., Wetzler M., Kolitz J.E., Baer M.R., Carroll A.J., Stone R.M., Caligiuri M.A., Marcucci G., Bloomfield C.D. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 2013;27(10):2023–2031. doi: 10.1038/leu.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Venturini L., Battmer K., Castoldi M., Schultheis B., Hochhaus A., Muckenthaler M.U., Ganser A., Eder M., Scherr M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 80.Ciccone M., Calin G.A., Perrotti D. From the PP2A to the PADs in Hematological Malignancies. Front. Oncol. 2015;16(5):1–10. doi: 10.3389/fonc.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 82.Sawyers C.L., Callahan W., Witte O.N. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70(6):901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 83.Norfo R., Zini R., Pennucci V., Bianchi E., Salati S., Guglielmelli P., Bogani C., Fanelli T., Mannarelli C., Rosti V., Pietra D., Salmoiraghi S., Bisognin A., Ruberti S., Rontauroli S., Sacchi G., Prudente Z., Barosi G., Cazzola M., Rambaldi A., Bortoluzzi S., Ferrari S., Tagliafico E., Vannucchi A.M., Manfredini R., Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014;124(13):e21–e32. doi: 10.1182/blood-2013-12-544197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang X., Schwind S., Yu B., Santhanam R., Wang H., Hoellerbauer P., Mims A., Klisovic R., Walker A.R., Chan K.K., Blum W., Perrotti D., Byrd J.C., Bloomfield C.D., Caligiuri M.A., Lee R.J., Garzon R., Muthusamy N., Lee L.J., Marcucci G. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013;19(9):2355–2367. doi: 10.1158/1078-0432.CCR-12-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y., Jacamo R., Konopleva M., Garzon R., Croce C., Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J. Clin. Invest. 2013;123(6):2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mallardo M., Poltronieri P., D’Urso O.F. Non-protein coding RNA biomarkers and differential expression in cancers: a review. J. Exp. Clin. Cancer Res. 2008;27:19. doi: 10.1186/1756-9966-27-19. [DOI] [PMC free article] [PubMed] [Google Scholar]


