Abstract
Segmented filamentous bacteria (SFB) are gram positive, anaerobic, spore-forming commensals that reside in the gut of many animal species. Described more than forty years ago, SFB have recently gained interest due to their unique ability to modulate the host immune system through induction of IgA and Th17 cells. Here, we describe a collection of methods to detect and quantify SFB and SFB adhesion in intestinal mucosa, as well as SFB-specific CD4 T cells in the lamina propria. In addition, we describe methods for purification of SFB from fecal material of SFB-monoassociated gnotobiotic mice. Using these methods we examine the kinetics of SFB colonization and Th17 cell induction. We also show that SFB colonize unevenly the intestinal mucosa and that SFB adherence occurs predominantly in the terminal ileum and correlates with an increased representation of SFB-specific Th17 cells.
Keywords: Segmented filamentous bacteria, SFB, Th17 cells, commensal microbiota, intestinal epithelial cells, mucosa-associated bacteria, CD4 T cells
Introduction
Segmented filamentous bacteria (SFB, Candidatus Arthromitus or Candidatus Savagella), are members of the resident murine flora, most closely related to Clostridia (Prakash et al., 2011; Sczesnak et al., 2011). SFB have been described in many vertebrate and invertebrate animal species, as well as in humans (Klaasen et al., 1993; Yin et al., 2013). In contrast to most other commensals, SFB interact directly with intestinal epithelial cells (IECs) in the terminal ileum (Blumershine and Savage, 1978; Klaasen et al., 1992). In contrast to invasive pathogens, SFB do not invade epithelial cells, penetrate the epithelial barrier, or induce intestinal inflammation (Talham et al., 1999). SFB are currently propagated exclusively in SFB-monoassociated gnotobiotic animals (SFB-mono mice) (Klaasen et al., 1991; Umesaki et al., 1995). However, a recent exciting study has described a method for in vitro propagation of the bacteria that promises to allow genetic manipulation of this microorganism in the near future (Schnupf et al., 2015). The original studies in SFB-monoassociated mice showed that SFB have immunomodulatory effects, such as induction of IgA production, recruitment of IELs, and induction of MHCII expression and glycosylation in IECs (Talham et al., 1999; Umesaki et al., 1995), although it remains unclear how specific these functions are to SFB. More recently, SFB were shown to be a specific inducer of Th17 cell differentiation (Ivanov et al., 2009). Even though SFB colonization leads to a general recruitment of CD4 T cells, Th17 cells are the only CD4 T cell subset that is proportionately increased with SFB colonization (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). Characterization of the specificity of the Th17 cell response in SFB-colonized mice, revealed that most intestinal Th17 cells are SFB-specific and that virtually all SFB-specific T cells are Th17 cells (Goto et al., 2014; Yang et al., 2014). Thus, unlike other commensals, SFB are capable of inducing a localized antigen-specific Th17 response. Because of the unique nature of this response, as well as the unique nature of the interaction between SFB and the host, SFB are an excellent model for elucidating fundamental cellular and molecular mechanisms of Th17 cell induction and the effects of mucosa-associated bacteria on host immunity.
Materials and Methods
1.1 Mice
The mice used in this study are C57BL/6 mice obtained from location MP14 at the Jackson laboratory (JAX) and location IBU7 at Taconic Farms (TAC). We have previously reported that JAX mice lack SFB and TAC mice contain SFB as part of their resident microbiota (Ivanov et al., 2009). However, the presence of SFB depends on the particular barrier location at each vendor. Indeed, mice from certain locations at JAX contain SFB and mice from certain locations at TAC do not. Therefore, SFB presence or absence cannot be assumed based solely on vendor source, and we test every animal for SFB upon arrival using the methods described below. Of note, Taconic Farms has recently implemented quarterly screens for SFB as part of their health monitoring program and the results can be found on the corresponding health reports on the Taconic website.
1.2 Colonization of mice with SFB by oral gavage
Fecal pellets for oral gavage are collected from live SFB-positive mice and frozen at −80° until use. Alternatively, pellets may be collected and homogenized immediately for gavage as described below. Frozen feces are transported to the vivarium on dry ice and homogenized in water using a 20G needle. Solids are separated by gravity before the cleared supernatant is transferred to a 50 ml conical tube. Cleared supernatants are introduced into mice (200–500 μl/animal) by oral gavage. We generally use the equivalent of the contents of 1–2 fecal pellets per recipient mouse. In our hands this procedure results in colonization of most animals when using feces from SFB-positive specific pathogen free (SPF) mice (90–100%). For better results, especially when using feces from SFB-monocolonized mice, the gavage can be repeated after 4–8 hours (colonization with SFB-mono feces after a single gavage can be variable and seems to depend on host genetics and endogenous microbiota). In all cases, SFB colonization must be confirmed by Q-PCR. In successfully colonized animals, SFB can be detected in feces on day 4 post colonization. We usually confirm SFB levels by Q-PCR on day 4 and day 8 after the last gavage.
1.3 Genomic DNA extraction from feces and quantitative real-time PCR (Q-PCR)
Fresh or frozen fecal pellets are weighed prior to DNA extraction. Individual fecal pellets are weighed and placed in 2 ml polypropylene tubes (Sarstedt) together with 0.5 ml of 0.1 mm zirconia beads (BioSpec), 500 μl of DNA extraction buffer (200mM Tris, 200mM NaCl and 20mM EDTA), 210 μl of 20% SDS and 500 μl of Phenol-Chloroform-Isoamyl alcohol (P:C:I) (25:24:1). Bacterial cells in the sample are lysed using a FastPrep 24 bead-beater (MP Biomedical) for 1 minute at maximum speed. Note that addition of more than 100 mg of feces results in incomplete lysis and extraction of bacterial DNA and therefore we do not recommend using more than a single fecal pellet per tube. After lysis, the samples are centrifuged at 16,000 g for 5 minutes at 4°C and the DNA-containing aqueous phase is collected and subjected to two additional P:C:I extractions to further remove organic contaminants. DNA is precipitated with 0.1 volume 3M sodium acetate, pH 5.2 and two volumes of 100% ethanol at −80°C for 10–20 minutes followed by centrifugation for 20 minutes at 16,000 g and 4°C. After washing in 70% ethanol, the DNA pellet is resuspended in low strength Tris-EDTA buffer. Contaminating RNA is removed by treatment with 0.1mg/mL of RNAse A (Thermo Fisher) for 5 minutes at room temperature (RT).
Q-PCR is conducted on the fecal DNA to determine the amount of SFB and total Eubacteria present in each sample by amplifying 16S rRNA genes using the following bacteria-specific primers (Barman et al., 2008): 5′-GACGCTGAGGCATGAGAGCAT-3′ and 5′-GACGGCACGGATTGTTATTCA-3′ for SFB, and 5′-ACTCCTACGGGAGGCAGCAGT-3′ and 5′-ATTACCGCGGCTGCTGCG-3′ for Eubacteria (UNI). Q-PCR is performed on a Roche LightCycler 480 with SYBR Green reagent. The PCR conditions include a pre-incubation step at 95° for 5 minutes, 40 amplification cycles of 95°C for 10 s and annealing at 62°C for 45 s, and a melting curve.
1.4 Isolation of mucosa-associated bacteria
A 0.5 cm piece of the intestine is removed after sacrifice, opened longitudinally and flushed vigorously with sterile PBS to remove mucus, intestinal contents and any loosely associated bacteria before being placed in cold, sterile PBS. The epithelial layer is loosened by incubation in cold 30 mM EDTA in PBS or HBSS for 15 minutes. The tissue is then placed in cold sterile PBS and the epithelial layer is separated from the underlying basal membrane and muscularis mucosae by gently scraping with a bent sterile 28G needle under a dissecting microscope. The epithelial sheets containing mucosa-associated bacteria are collected and subjected to genomic DNA extraction as described in section 1.3.
1.5 Assessing SFB-specific T cell responses using an ex vivo proliferation assay
Lamina propria lymphocytes (LPL) are isolated as previously described (Goto et al., 2014). CD4 T cells are purified from this preparation by fluorescence-activated cell sorting (FACS) or magnetic beads (MACS) and labeled with CFSE or CellTrace Violet proliferation dye (Life Technologies). For assessment of SFB-specific responses, 5 × 104 purified CD4 T cells are co-cultured in T cell medium (RPMI containing 10% FCS, 1% non-essential amino acids, 1% sodium pyruvate, 1% L-glutamine, 1% Pen/Strep, 20 mM HEPES and 0.001% β-mercaptoethanol) in 96 well U-bottom plates with either 5 × 104 MACS purified splenic CD11c+ dendritic cells or 2 × 105 total TCRα-KO splenocytes as antigen presenting cells in the presence or absence of autoclaved bacterial lysates (prepared as described in section 1.6). Specific proliferation in response to presented antigens is determined by flow cytometry at 72h by calculating the number of live CD4 T cells with dye dilution, or of live blasting (FSChigh) CD4 T cells.
1.6 Gradient purification of SFB from feces
Fresh or frozen fecal pellets from SFB-monocolonized mice are homogenized in sterile PBS. After extensive vortexing, the supernatant is cleared of debris by a short low-speed centrifugation (if necessary the extraction can be repeated). Next, bacteria are pelleted by centrifugation at 4,000 g and washed twice with PBS. The SFB filaments are then resuspended in 6 ml of PBS and transferred to a 15 ml conical tube. 3 ml of 60% w/v NycoPrep Universal (Accurate Chemical) is carefully underlayed beneath the bacterial fraction and the gradient is centrifuged at 1800 g for 20 min at RT without brake. The interphase containing bacteria should be visible as a white ring between the NycoPrep and PBS layers. This fraction is collected and washed twice with PBS by centrifugation at 4000 g. The pellet containing SFB is resuspended in PBS, aliquoted, and autoclaved for generation of SFB lysates used for stimulation of LP CD4 T cells (described in section 1.5). This protocol, and the one in section 1.7, works well on frozen feces from SFB-mono mice.
1.7 FACS sorting of gradient-purified SFB
NycoPrep-enriched SFB filaments are re-suspended in sterile PBS and stained with the bacterial viability dye SYTO®9 (Invitrogen) at a final concentration of 5 μM for 15 min at RT. After incubation, the suspension is diluted with 50 volumes of PBS, filtered through a 40uM mesh, and sorted on a FACS Aria II (BD) using a 70 μm-nozzle. SFB filaments are sorted based on SYTO9 positive cells with high SSC-W, and the filaments are collected directly into 15 ml tubes with PBS. Sorted SFB are pelleted by centrifugation at 3,000 rpm for 15 min at 4 °C, re-suspended in appropriate amount of buffer and used for downstream applications.
1.8 Flow Cytometry
The following antibodies were used for flow cytometry: CD4-PE-Cy7 (RM4-5), TCRβ-PerCP-Cy5.5 (H57-597), IL-17A-PE (eBio17B7), IFNγ-APC (XMG1.2), all from eBioscience, and Vβ14-biotin (14-2) from BD. Fixable viability dye (Invitrogen) was used to gate on live cells, and intracellular cytokine staining was performed using Cytofix/Cytoperm reagents (BD). Samples were collected on an LSR II or Fortessa (BD) and analyzed using FlowJo software (TreeStar).
Results
2.1 Geographical localization of SFB in SPF mice
Pioneering studies in the early 1970’s identified SFB in the feces by Gram stain, however SFB may preferentially localize to, and mediate unique functions in, different parts of the gastrointestinal tract (Davis and Savage, 1974). To investigate the distribution of SFB and their interaction with IECs we purified bacterial genomic DNA from descending segments of the small intestines of SFB-positive C57BL/6 mice. To determine if SFB was present, genomic DNA was prepared from the luminal contents of each section. To compare the interaction between SFB and epithelial cells, genomic DNA was prepared from intestinal epithelial sheets from each segment (see Methods). SFB were present in luminal contents throughout the gastrointestinal tract, but progressively increased in the proximal to distal direction. Although, we detected SFB in luminal contents, SFB levels in the duodenum were extremely low and in some animals were below the threshold of detection of our Q-PCR assay. SFB levels increased by almost two orders of magnitude in the jejunum and another 20 fold in the terminal ileum (Figure 1A). SFB levels per mg of luminal contents were further increased in colonic contents and feces (data not shown). However, the percentage of SFB amongst total bacteria was less than 0.5% in jejunum and feces. In contrast, in the terminal ileum, SFB represented 10–15% of the bacterial DNA in the lumen (Figure 1C and data not shown).
Figure 1. Geographical localization of SFB in SPF mice.
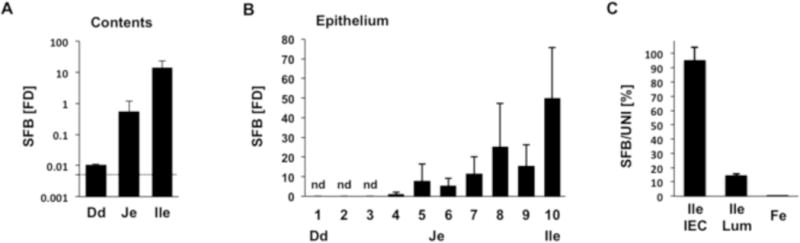
(A) Luminal contents from Dd, duodenum (most proximal 1 cm); Je, jejunum (mid-point); and Ile, ileum (most distal 1 cm) of the small intestine (SI) of SFB-positive C57BL/6 mice were collected and the relative amount of SFB determined by qPCR. (B) The small intestine of SFB-positive C57BL/6 mice was divided into ten equal segments (numbered from 1 to 10) and intestinal epithelial cells (IECs) were isolated from a 0.5 cm piece of each segment. Relative amount of mucosa-associated SFB was determined for each segment by Q-PCR. Dd, duodenum, segment 1; Je, jejunum, segment 5–6; Ile, terminal ileum, segment 10. (C) Luminal contents and IECs were collected from terminal ileum (segment 10) and feces (Fe) of SFB-positive C57BL/6 mice and SFB and total Eubacteria quantified by qPCR. SFB is expressed as a percentage of total Eubacteria. Error bars, standard deviation of the mean. Q-PCR data in A and B are represented as relative fold difference (FD) to one of the jejunum samples.
In the mucosa-associated fraction, SFB attachment was first detected around the mid-point of the SI, but increased significantly in the ileum, reaching highest levels in the last 1/10 of the SI (Figure 1B). Moreover, in the terminal ileum, SFB represented more than 95% of the bacteria in the mucosa-associated fraction (Figure 1C). We did not detect any mucosa-associated SFB in either the cecum or colon, although both locations contained an abundance of SFB in the lumen (data not shown). Thus, in healthy laboratory mice, SFB are virtually the only ileal mucosa-associated resident bacteria, and therefore, represent a unique model to study the mechanisms of interaction of such bacteria with the host.
2.2. Kinetics of SFB colonization and induction of Th17 cells
We recently demonstrated that long-term colonization with SFB specifically induces Th17 cells in the SI LP (Ivanov et al., 2009). SFB are currently the only known gut-resident, non-pathogenic bacteria capable of inducing Th17 cells. We examined the kinetics of SFB colonization and Th17 cell induction following introduction into conventionally-raised SPF hosts. SFB-negative SPF mice from our colony received a fecal transplant from SFB-positive Taconic B6 mice as described in the Methods. SFB levels were followed in feces by Q-PCR and Th17 levels were followed in the small intestinal lamina propria (SI LP) by flow cytometry. As shown in Figure 2A, SFB DNA was detected in feces as early as day 2 post gavage, but remains low between day 1 and day 3. A significant spike in SFB levels occurs at day 4 and we use this time point to predict long-term SFB colonization. SFB levels may slightly increase, but reach equilibrium by day 5–7 post-colonization (Figure 2A). Th17 cell induction followed delayed kinetics compared to SFB colonization. A significant increase in Th17 cells was not detected in the LP on day 4 after introduction of SFB. A slight increase may be present on day 6, however, robust Th17 cells induction is not detected until d7 after SFB gavage and Th17 cells quickly reach steady state levels (Figure 2B). To allow for this equilibrium to be established, we usually analyze Th17 levels at 2 weeks post gavage, though in our experience, if high SFB colonization is confirmed on day 4, similar frequencies of Th17 cells are observed if the mice are examined at anytime after day 7.
Figure 2. Kinetics of SFB colonization and Th17 cell induction.
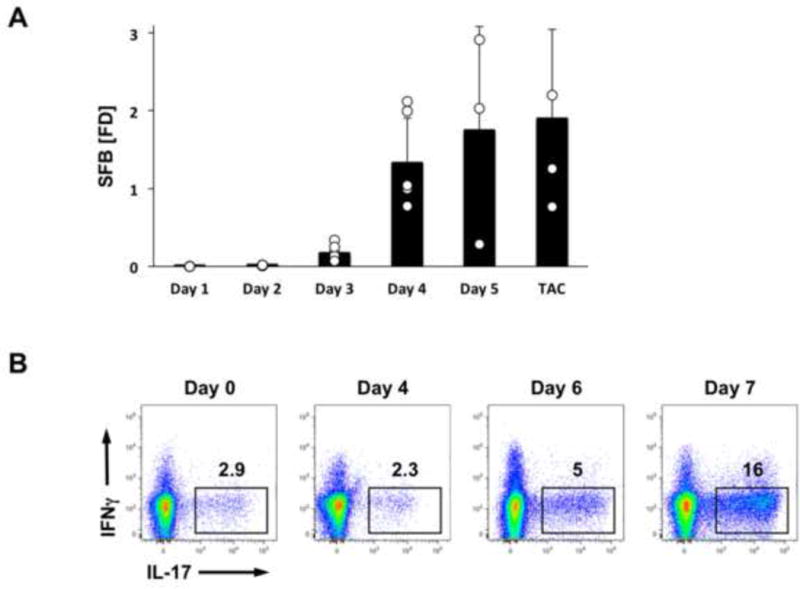
SFB-negative C57BL/6 mice were colonized on Day 0 with SFB by oral gavage. (A) SFB colonization in feces at the indicated time points quantified by Q-PCR. (B) Th17 cell proportions in SI LP at the indicated time points. FACS plots are gated on TCRβ+CD4+ cells. TAC, endogenously colonized mice from Taconic Farms. SFB levels are normalized to fecal weight and are represented as relative fold difference (FD) to one of the samples.
2.3. Specificity of SFB-induced Th17 cells
We also examined the specificity of SFB-induced Th17 cells. As we reported recently, most induced Th17 cells recognize SFB antigens (Goto et al., 2014). SFB-specific CD4 T cells can be detected in mice by examining their proliferative response to SFB antigens as described in the Methods. The SFB-specific T cell response can be detected when CD4 T cells purified from small intestinal lamina propria are incubated in vitro with antigen-presenting cells and SFB antigens. As shown in Figures 3A and 3B, proliferation of CD4 T cells in response to SFB antigens is detected after 72 hours only in CD4 T cells isolated from SFB-positive mice, but not in CD4 T cells from mice that lack SFB (Figure 3A,B). When LP CD4 T cells were separated into Th17 and non-Th17 cells based on IL-17 expression in IL-17-GFP-reporter mice, only Th17 cells responded to SFB antigens, showing that almost all SFB-specific CD4 T cells are Th17 cells (Figure 3C,D and (Goto et al., 2014)). An analysis of the SFB induced Th17 cell repertoire showed a specific enrichment for Vβ14 TCRs (Goto et al., 2014; Yang et al., 2014) and an immunodominant SFB antigen is specifically recognized by Vβ14+ Th17 TCRs (Yang et al., 2014). Very few Vβ14+ Th17 cells are present among the small fraction of microbiota induced Th17 cells in SFB-negative conventionally raised mice, (e.g. Jackson C57BL/6J mice) and are induced upon SFB colonization (Figure 3E). Therefore, expansion or enrichment of Vβ14+IL-17+ cells can be used as an initial readout for the presence of an SFB-specific response following SFB colonization (Figure 3), although a definitive demonstration of the presence of SFB-specific cells requires a functional assay, such as ELISPOT, SFB-tetramer staining, or the in vitro proliferation method described here (Goto et al., 2014; Yang et al., 2014).
Figure 3. SFB-induced Th17 cells are SFB-specific.

(A,B) Response of MACS purified SI CD4 T cells to antigens prepared from SFB-monoassociated mice (SFB), SFB-negative Jackson C57BL/6 mice (Jax) and germ-free C57BL/6 mice (GF). CD4 T cells were incubated with antigen-presenting cells (APCs) and bacterial antigens for 72 hours as described in Methods. (A) Quantification of proliferating blasts in the CD4 T cell gate (FSChi). (B) Total numbers of live proliferated CD4 T cells. (C,D) Response of FACS-purified CD4 T cells isolated from the SI LP of SFB-positive IL-17A-GFP reporter mice to different bacterial antigens. GFP+ (Th17) and GFP− (non-Th17) CD4 T cells were cultured with bacterial lysates from E. coli, C. perfingens (Clost.), and mouse intestinal bacteria (MIB) as described in Methods. The total number of proliferated cells per well was determined by counting the cells within the live CD4+TCRβ+FSC-AhiCellTracelow gate in (C). (E) TCRVβ14+IL-17+ cells in the SI LP of SFB-negative and SFB-positive mice. Plots are gated on TCRβ+CD4+ cells. **p ≤0.005, unpaired t-test. Error bars on all figures represent the standard deviation of the mean.
2.4. Purification of SFB from SFB-monocolonized mice
To purify SFB for the generation of SFB antigens and other applications, we developed a two-step protocol using a Nycoprep density gradient to enrich SFB filaments from fecal suspensions, followed by obtaining highly pure SFB by cell sorting. SFB were purified from feces or cecal contents of SFB-monocolonized (SFB-mono) mice as described in the Methods. In our experience fecal pellets yield a higher quality preparations compared to cecal contents. This method also allows for isolation of a considerable proportion of live cells as demonstrated by Live/Dead staining (Figure 4A). Therefore, this method is also useful for downstream applications requiring live bacteria and, indeed, a similar protocol was recently utilized to expand SFB in vitro (Schnupf et al., 2015).
Figure 4. Bacterial cell sorting of SFB from feces.
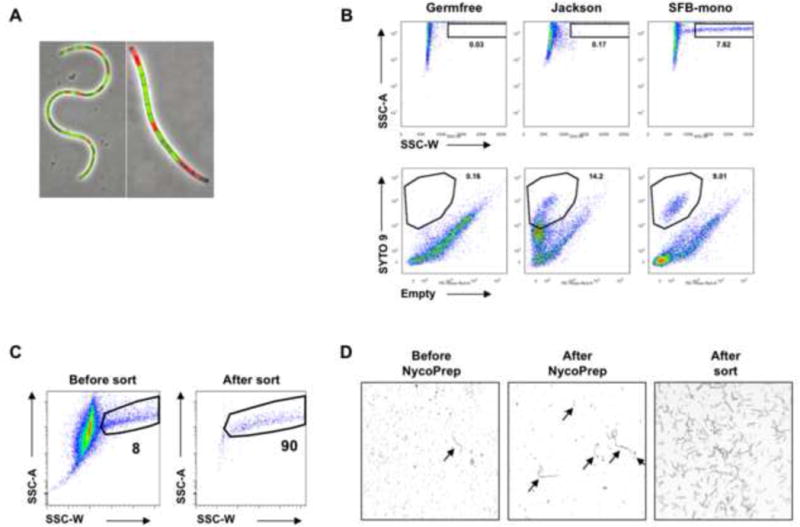
(A) SFB filaments after NycoPrep density gradient purification were stained with LIVE/DEAD BacLight Bacterial Viability kit (Invitrogen) and examined under a fluorescent microscope. (B,C) Bacteria were isolated from the feces of SFB-mono, SFB-negative Jackson C57BL/6, and germfree mice as control and pre-cleared using a NycoPrep gradient as described in Methods. Bacteria were stained with SYTO-9 viability dye and SYTO-9+SSC-Whi SFB filaments were purified by cell sorting (C). (D) Phase-contrast images of SFB filaments from SFB-Mono feces at different stages of the sorting protocol. Arrows indicate SFB filaments. Data are representative of 4–5 independent experiments.
Following The Nycoprep gradient, highly pure SFB filaments can be obtained by cell sorting (FACS). Because SFB form filaments that often reach 100 um in length, SFB are clearly distinguishable by flow cytometry in feces of SFB-monocolonized mice and form a distinct population, not present in similarly prepared fecal contents from germfree and SFB-negative Jackson B6 mice (Figure 4B). FACS results in a highly purified fraction, containing SFB filaments and free of additional debris that can be used for downstream applications (Figure 4C and D). For example, bacterial lysates prepared from FACS purified SFB were highly efficient in inducing proliferation of SFB-specific T cells (Goto et al., 2014).
2.5. SFB adhesion correlates with SFB-specific Th17 cell induction
As shown in Figure 1B, SFB unevenly colonize the gastrointestinal tract. In the small intestine, SFB preferentially attach to epithelial cells in the terminal ileum. In order to examine whether the geography of SFB localization correlates with Th17 cell induction, we examined the frequency of Th17 cells in different parts of the small intestine of SFB-negative and SFB-positive mice housed in our colony (Figure 5). Th17 cell induction was evident in all parts of the intestine in SFB-positive compared to SFB-negative mice, however Th17 cells represented a larger proportion of CD4 T cells in the ileum, and especially in the terminal ileum (Figure 5A,B). More importantly, a significant induction of Vβ14+ Th17 cells was only observed in the ileum and was most robust in the terminal ileum, suggesting that SFB-specific Th17 cells are preferentially induced in this location (Figure 5A,C). Thus, induction of SFB-specific Th17 cells occurs preferentially in the terminal ileum and correlates with SFB attachment.
Figure 5. Localization of Th17 cells in the small intestine.
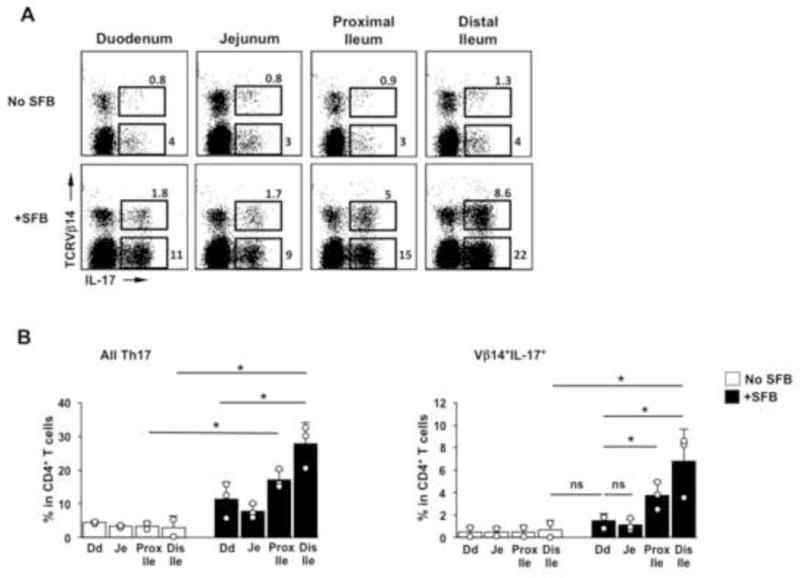
SI of SFB-negative C57BL/6 mice (No SFB) and SFB-positive C57BL/6 mice (+SFB) were divided into four equal segments and LPL isolated from each segment. Representative FACS plots (A) and statistics (B) of total Th17 cells, and SFB-specific (Vβ14+) Th17 cells in each segment. FACS plots gated on TCRβ+CD4+ cells. *p ≤.05, Student t-test. Error bars represent standard deviation of the mean.
Discussion
SFB-like filamentous bacteria were first described in the late 19th century in the intestines of arthropods by Joseph Leidy, who also gave them the collective name Arthromitus (Leidy, 1849). In vertebrates, SFB were extensively studied in the 1970’s and 1990’s due to their unique morphology and interaction with the host epithelium. More recently, we and others showed that SFB play a unique role in induction of Th17 cells in the small intestinal lamina propria (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). In follow-up studies we showed that SFB induce exclusively SFB-specific Th17 cells (Goto et al., 2014). In contrast, neither Th17 nor non-Th17 CD4 T cells respond to antigens derived from other commensal bacteria (Figure 3). This is in agreement with other studies demonstrating that antigen-specific responses to luminal bacteria are not normally present in the intestinal mucosa, a phenomenon sometimes refer to as immunological ignorance (Hooper and Macpherson, 2010). SFB are, therefore, unique among gut-resident commensals not only because they induce a Th17 cell response, but also because they induce a localized mucosal antigen-specific response. Indeed, studies by Littman et al. have identified SFB immunodominant antigens and corresponding T cell receptors (Yang et al., 2014). Therefore, SFB represent unique model for studying the effects of mucosa-associated bacteria on the host, induction of local mucosal antigen specific responses, and induction of Th17 cell responses.
Studying the effects of SFB in vivo is critical to understanding how the bacteria modulate the immune system. Here, we describe a collection of methods to quantify SFB in both luminal contents and intestinal epithelium, to purify SFB and isolate SFB antigens, and to detect SFB-specific Th17 cells in vivo. We also examine the kinetics and distribution of SFB colonization and Th17 cell induction. We find that although SFB are present throughout the gastrointestinal tract, their relative representation increases from proximal to distal direction and is highest in the terminal ileum. Moreover, SFB attachment to the intestinal mucosa occurs only in the second half of the small intestine, with a distinct preference for the terminal ileum, where SFB represent almost 100% of mucosa-associated bacteria in healthy laboratory mice. Interestingly, SFB-specific Th17 cell induction follows delayed kinetics and occurs a few days after expansion of SFB is detected in feces. Although we did not directly examine their response to SFB antigens, the increase in Vβ14+IL-17+ cells in the terminal ileum suggests that SFB-specific Th17 cells are also enriched in the distal parts of the small intestine and correlate with SFB adhesion. Therefore, SFB adhesion to the intestinal epithelium may be a crucial factor for induction of localized, commensal antigen-specific Th17 cell responses. How this is achieved will be important to elucidate in future studies. LP CD4 T cell responses are induced in secondary lymphoid organs, such as the mesenteric lymph nodes (MLNs), where antigen-specific T cells are primed and imprinted with homing receptors to the intestine. However, SFB adhesion may be important for expansion of SFB-specific T cells locally in the terminal ileum or participate in the establishment or maintenance of their Th17 cell differentiation program. In general, signals in the local gut environment seem to be dominant for the presence of SFB-specific Th17 cells. In support of this, we have shown that IL-17+ CD4 T cells are not present in MLNs after SFB colonization and expansion of Th17 cells in response to SFB occurs mainly in the LP (Goto et al., 2014). Where SFB-specific T cells are primed is also currently unclear. Although this is presumed to occur in MLNs, SFB-specific Th17 cell responses were induced normally in MLN and Peyer’s patch-deficient LTα-KO mice (Goto et al., 2014). Furthermore, SFB have been shown to induce the formation of tertiary lymphoid tissue, and subsequent IgA production, in the absence of both lymph nodes and Peyer’s patches, and these lymphoid structures may serve as alternative local sites for Th17 cell priming by SFB (Lécuyer et al., 2014).
SFB are immunomodulatory bacteria with a unique relationship with the host. The presence of SFB has major effects on immune homeostasis and the susceptibility or progression of disease (Heczko et al., 2000; Ivanov et al., 2009; Kriegel et al., 2011; Lee et al., 2011; Wu et al., 2010). The mechanisms of SFB adhesion and induction of localized mucosal Th17 cell antigen-specific responses will be important to elucidate in future studies. This may involve conceptually novel ways of immune activation by mucosa-associated commensals, such as specific induction of signaling pathways and cytokine production by IECs or delivery of commensal antigens to, and activation of, particular host innate immune cell subsets exclusively in the terminal ileum.
Acknowledgments
This work was supported by the National Institutes of Health R01-DK098378 to I.I.I. and by the Crohn’s and Colitis Foundation of America SRA#259540 to I.I.I. I.I.I. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trust.
References
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumershine RV, Savage DC. Filamentous microbes indigenous to the murine small bowel: a scanning electron microscopic study of their morphology and attachment to the epithelium. Microb Ecol. 1978:95–103. doi: 10.1007/BF02014280. [DOI] [PubMed] [Google Scholar]
- Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heczko U, Abe A, Finlay BB. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. The Journal of infectious diseases. 2000;181:1027–1033. doi: 10.1086/315348. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS microbiology reviews. 1992;8:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Laboratory animals. 1993;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Van Wezel HP, Beynen AC. Mono-association of mice with non-cultivable, intestinal, segmented, filamentous bacteria. Archives of microbiology. 1991;156:148–151. doi: 10.1007/BF00290989. [DOI] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E, Rakotobe S, Lengliné-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, Reynaud CA, Cerf-Bensussan N, Gaboriau-Routhiau V. Segmented Filamentous Bacterium Uses Secondary and Tertiary Lymphoid Tissues to Induce Gut IgA and Specific T Helper 17 Cell Responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidy J. On the existence of entophyta in healthy animals, as a natural condition. Proc Acad Nat Sci Phila. 1849;4:225–233. [Google Scholar]
- Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, Taylor TD, Ohno H, Umesaki Y, Hattori M. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell host & microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015 doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host & Microbe. 2011;10:1–13. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infection and immunity. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, Sczesnak A, Liao JJ, Torres VJ, Jenkins MK, Lafaille JJ, Littman DR. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M, Zhu B, Yu HD, Xiang C, Wang X. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. Isme J. 2013;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]


