Abstract
cFOS is a pleiotropic transcription factor, which binds to the AP1 site in the promoter of target genes. In the pituitary gonadotropes, cFOS mediates induction of FSHβ and GnRH receptor genes. Herein, we analyzed reproductive function in the cFOS-deficient mice to determine its role in vivo. In the pituitary cFOS is necessary for gonadotropin subunit expression, while TSHβ is unaffected. Additionally, cFOS null animals have the same sex-steroid levels, although gametogenesis is impeded. In the brain, cFOS is not necessary for GnRH neuronal migration, axon targeting, cell number, or mRNA levels. Conversely, cFOS nulls, particularly females, have decreased Kiss1 neuron numbers and lower Kiss1 mRNA levels. Collectively, our novel findings suggest that cFOS plays a cell-specific role at multiple levels of the hypothalamic–pituitary–gonadal axis, affecting gonadotropes but not thyrotropes in the pituitary, and kisspeptin neurons but not GnRH neurons in the hypothalamus, thereby contributing to the overall control of reproduction.
Keywords: cFOS, Kisspeptin, c-Fos/AP-1, GnRH, Gonadotropin
1. Introduction
cFOS is a basic leucine-zipper protein which forms a heterodimer with the cJUN isoform, thus forming an AP1 transcription factor that binds the TPA-response element in the promoter of target genes. cFOS is an immediate-early gene that is activated rapidly and transiently in most cell types. It is induced by a variety of growth factors, cytokines, neurotransmitters, and hormonal signals, as well as environmental stimuli. In turn, cFOS controls a diverse array of cellular processes, including cell proliferation, differentiation, survival, and death. Of all of the eclectic variations of cellular functions in which it is involved, the primary role(s) of cFOS, in any given tissue, is dependent on the cell type and stimuli (Shaulian and Karin, 2002; Wagner and Eferl, 2005). We have shown that although pituitary gonadotropes express receptors for EGF or insulin, which in other cells induce cFOS, in the gonadotrope only GnRH induces cFOS and through it GnRH-target genes (Ely et al., 2011). Thus, cFOS although versatile, has cell-specific and stimulus-specific function in each cell type.
To study the roles of cFOS in vivo, two different cFOS deficient mice were created and their phenotypes were analyzed in mouse strains with mixed backgrounds (129/SvJ × C57Black6J). Similar abnormalities were reported for both mouse strains lacking cFOS (Johnson et al., 1992; Wang et al., 1992). cFOS nulls are born at the proper Mendelian ratio, demonstrating that cFOS is not necessary for embryonic development. However, after 4 weeks of age, cFOS null mice exhibit growth retardation, osteopetrosis, and ultimately, hematopoiesis deficiency. Although cFOS null mice exhibit impairments in peripheral organs such as bone and hematopoietic system, the alteration in the central nervous system (CNS) are cell-specific (Benes et al., 2013; Fleischmann et al., 2003; Yasoshima et al., 2006). For example, adult mice lacking cFOS in the CNS exhibited normal general and emotional behavior but were specifically impaired in hippocampus-dependent spatial and associative learning tasks (Fleischmann et al., 2003). Johnson et al. reported infertility in both sexes, though the cause was not examined (Johnson et al., 1992). Since cFOS null mice are viable, redundancy poses a question and the potential complementary role of a closely related protein, FOSB, was also examined in vivo. Unlike cFOS null mice, FOSB deficient mice were reported to be healthy, viable, fertile, and had a normal life expectancy (Brown et al., 1996). Thus, cFOS is crucial for fertility, whereas FOSB is not. Moreover, intact, functional FOSB is unable to substitute for the loss of cFOS with regards to the reproductive system. This necessitates further examination of the roles of cFOS in modulating the hypothalamic–pituitary–gonadal axis.
cFOS is rapidly induced in gonadotrope cells following GnRH treatment, both in vivo (Padmanabhan et al., 1995) and in model cell lines (Cesnjaj et al., 1994; Wurmbach et al., 2001). cFOS mediates GnRH induction of the FSHβ gonadotropin subunits by binding to the AP1 site in the proximal mouse FSHβ promoter (Coss et al., 2004). Furthermore, cFOS is involved in synergistic induction of FSHβ by GnRH and activin, which is specific for FSHβ and may play a role in differential expression of gonadotropin subunits (Coss et al., 2007). Specific decrease of cFOS protein turnover may contribute to the rise in FSHβ transcription during the time of low GnRH pulse frequency (Reddy et al., 2013). Induction of the GnRH receptor by GnRH is also dependent on cFOS binding to both the AP1 site, to mediate GnRH responsiveness (White et al., 1999), and to the GRAS element where, through interaction with SMAD proteins and FOXL2, mediates the synergy between GnRH and activin (Ellsworth et al., 2003; Norwitz et al., 2002).
In the brain, cFOS serves as a marker of neuronal activation and its expression is increased in GnRH neurons during the preovulatory LH surge and after kisspeptin treatment (Kauffman et al., 2007; Lee et al., 1992). cFOS expression in kisspeptin neurons also coincides with a preovulatory LH surge (Clarkson et al., 2008; Robertson et al., 2009). However, a role of cFOS in GnRH and kisspeptin neurons is still poorly understood. In the gonads, as well, cFOS is expressed in germ cells and granulosa and theca cells in females (Rusovici and LaVoie, 2003), and Sertoli cells in males (Araujo et al., 2009), but its target genes are not known. Thus, a role for cFOS in the testes and ovaries is not elucidated.
The involvement and necessity of cFOS at different levels of the reproductive axis is not well-addressed, and therefore, the underlying cause(s) of infertility in mice lacking cFOS remains unknown. Here, we examined several levels of the hypothalamic–pituitary–gonadal axis in cFOS null mice of both sexes, to ascertain if gene expression impairments exist within the reproductive axis at either the brain, pituitary, and/or gonadal levels.
2. Materials and methods
2.1. cFOS-null mice
The cFOS-null mice were obtained from Jackson Laboratories, where Papaioannou laboratory deposited them, and back crossed to C57Bl6J for six generations. Animals were maintained under a 12-hour light, 12-hour dark cycle and received food and water ad libitum. All experiments were performed with approval from the University of California Animal Care and Use Committee and in accordance with the National Institutes of Health Animal Care and Use Guidelines using 5 and 6 weeks old animals. Genomic DNA was extracted from toe biopsies and analyzed with PCR according to JAX protocol. Animals of both sexes were studied to determine potential sex differences. At least 5 animals per sex per group (WT and null) were analyzed. Males and females were analyzed separately to determine the effect of genotype alone.
2.2. Immunohistochemistry
Tissues from 6-week-old animals were fixed in 4% paraformaldehyde overnight at 4 °C and dehydrated in ethanol/water washes before embedding in paraffin. Embedded tissues were cut into 14-μm coronal sections with a microtome and floated onto SuperFrost Plus slides (Fisher Scientific, Auburn, Alabama). Slides were incubated at 60 °C for 30 minutes, deparaffinized in xylene washes, and rehydrated in ethanol/water washes. Antigen unmasking was performed by heating for 10 minutes in a Tris–EDTA–Tween20 mixture and endogenous peroxidase was quenched by incubating for 10 minutes in 0.3% hydrogen peroxide. After washing in phosphate-buffered saline (PBS), slides were blocked (PBS, 5% goat serum, 0.3% Triton X-100) for 45 minutes and incubated with primary antibodies against GnRH (1:1000, PA1-121, Pierce, Thermo Rockford, IL) overnight at 4 °C. After washing, slides were incubated with biotinylated goat anti-rabbit IgG (1:300, Vector Laboratories) for 30 minutes. The Vectastain ABC elite kit (Vector Laboratories) was used per manufacturer's instructions and incubated for 30 minutes. After washing, the VIP peroxidase kit was used for colorimetric staining for 3 minutes. Slides were dehydrated in an ethyl alcohol series and xylene, and cover-slipped using Vectamount (Vector Laboratories).
2.3. qPCR analysis
Tissues from 5-week-old animals were dissected, total RNA extracted and reverse transcribed using Superscript III (Invitrogen, CA). qPCR was performed using an iQ SYBR Green Supermix and an IQ5 real-time PCR machine (Bio-Rad Laboratories, Hercules, CA), with primers listed in Table 1, under the following conditions: 95 °C for 5 min, followed by 40 cycles at 95 °C for 20 s, 56 °C for 30 s, and 72 °C for 30 s. A standard curve with dilutions of 10 pg/well, 1 pg/well, 100 fg/well, and 10 fg/well of a plasmid containing LHβ, FSHβ, or GAPDH cDNA was generated in each run with the samples. The amount of the gene of interest was calculated by comparing threshold cycle obtained for each sample with the standard curve generated in the same run. Replicates were averaged and divided by the mean value of GAPDH in the same sample. To quantify expression of genes for which cDNA-containing plasmid was unavailable to generate a standard curve, relative gene expression was calculated using 2−ΔΔCt. After each run, a melting curve analysis was performed to confirm that a single amplicon was generated. Five animals per group were used and males and females were analyzed separately. Statistical differences in expression between genotypes were determined by Student's T-test, with Tukey–Kramer post hoc HSD for multiple comparisons using JMP software (SAS Institute; Cary, North Carolina).
Table 1.
| Antibody | Cat # | Dilution | Provider |
|---|---|---|---|
| GnRH | PA1-121 | 1:1000 | Pierce, Thermo Scientific |
| Primers | Forward | Reverse |
|---|---|---|
| LH-R | AATTCACCAGCCTACTGGTTG | CCACTGAGTTCATTCTCCTCA |
| FSH-R | TTTGGAAGAATTGCCTGATGAT | CATGACAAACTTGTCTAGACTA |
| LHb | CTGTCAACGCAACTCTGG | ACAGGAGGCAAAGCAGC |
| FSHb | GCCGTTTCTGCATAAGC | CAATCTTACGGTCTCGTATACC |
| aGSU | ATTCTGGTCATGCTGTCCATGT | CAGCCCATACACTGGTAGATGG |
| GnRH-R | GCCCCTTGCTGTACAAAGC | CCGTCTGCTAGGTAGATCATCC |
| TSHb | AAGAGCTGGGGTTGTTCAAA | ACAAGCAAGAGCAAAAAGCAC |
| GAPDH | TGCACCACCAACTGCTTAG | GGATGCAGGGATGATGTTC |
2.4. Serum collection
For serum collection, 5-week-old mice were sacrificed by isoflurane inhalation and blood was obtained from the inferior vena cava. The blood was left to coagulate for 15 minutes at room temperature, and then centrifuged at 2000 RCF for 15 minutes for serum separation. Hormone assays were performed by University of Virginia, Ligand Core. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core is a fee-for-service core facility and is in part supported by the Eunice Kennedy Shriver NICHD/NIH (SCCPIR) Grant U54-HD28934. LH was analyzed using a sensitive two-site sandwich immunoassay (Haavisto et al., 1993), and mouse LH reference prep (AFP5306A; provided by Dr. A.F. Parlow and the National Hormone and Peptide program) was used as standard. FSH was assayed by RIA using reagents provided by Dr. A.F. Parlow and the National Hormone and Peptide Program, as previously described (Gay et al., 1970). Mouse FSH reference prep AFP5308D was used for assay standards. Steroid hormone levels were analyzed using validated commercially available assays, information for which can be found on the core's website: http://www.medicine.virginia.edu/research/institutes-and-programs/crr/lab-facilities/assay-methods-page and reported in Haisenleder et al. (2011). Limits of detection were 0.24 ng/ml for LH, 2.4 ng/ml for FSH, 3 pg/ml for estradiol, and 10 ng/dl for testosterone. Intraand inter-assay coefficients of variation were 6.4%/8.0%, 6.9%/7.5%, 6.0%/11.4% and 4.4%/6.4% for the LH, FSH, estrogen (E2) and testosterone (T), respectively. For the assays used for this paper, inter-assay coefficients of variation data are the result of 30 assays for LH and FSH, and 60 assays for E2 and T. Five animals per group were used for each hormone analysis and, males and females were analyzed separately to determine differences due to genotype. Statistical differences in hormone levels between wild-type and null groups were determined by Student's T-test, and Tukey–Kramer post hoc HSD for multiple comparisons using JMP software (SAS Institute; Cary, North Carolina).
2.5. Ovarian stimulation and histology
For harvest of primary mouse granulosa cells, ovaries were dissected from 6-week-old female mice in diestrus. Ovarian follicles were punctured with needles to release granulosa cells and oocytes. Cells were separated from debris by filtering through a 100-μm filter and subsequently, oocytes were removed from the granulosa cells by passing the cell suspension through 40-μm nylon mesh cell strainer. Granulosa cells were seeded at 0.2 × 106 cells per well (24-well plate) and cultured in serum-free McCoys 5A culture media containing antibiotics and incubated at 37 °C, 5% CO2 for at least 2 hours, prior to treatment with 50 ng/ml ovine FSH for 1 hour. Ovine FSH was obtained from Dr. A. F. Parlow at the National Hormone and Peptide Program of the National Institute of Diabetes and Digestive and Kidney Diseases. After removal of media, cells were lysed, cAMP measured according to the manufacturer's protocol using cAMP-Glo Assay kit (Promega Cat # V1501, Madison, Wisconsin, USA) and concentration was calculated using a standard curve. To stimulate ovulation, 6-week-old female mice were injected i.p. with 5 IU PMSG, followed by 5 IU hCG i.p. injection 48 hours later (both from Sigma-Aldrich, St. Louis, Missouri). The following day, ovaries were collected for histological analysis and hematoxylin and eosin staining of paraffin sections. The Institutional Animal Care and Use Committee at the University of California, Riverside, approved all animal protocols.
2.6. In situ hybridization
For Gnrh and Kiss1 gene expression analysis, brains from 6-week-old mice were collected at sacrifice, frozen immediately on dry ice, and stored at −80 °C. Brains were sectioned on a cryostat into five coronal series of 20 μm sections which were thaw-mounted onto Superfrost-plus slides and stored at −80 °C. Single-label in situ hybridization was performed as previously described (Poling and Kauffman, 2012; Semaan et al., 2010). Briefly, slide-mounted brain sections encompassing the entire preoptic area and hypothalamus from one of the 5 sets of serial brain sections were fixed in 4% paraformaldehyde, pretreated with acetic anhydride, rinsed in 2× SSC (sodium citrate, sodium chloride), delipidated in chloroform, dehydrated in ethanol, and air-dried. Radiolabeled (33P) Kiss1 or Gnrh antisense riboprobe (0.04 pmol/ml) was combined with tRNA, heat-denatured, added to hybridization buffer, and applied to each slide (100 μl/slide). Slides were cover-slipped and placed in a 55 °C humidity chamber overnight. The slides were then washed in 4× SSC and placed into RNAse A treatment for 30 min at 37 °C, then in RNAse buffer without RNase at 37 °C for 30 min. After washing in 2× SSC at room temperature, slides were washed in 0.1× SSC at 62 °C for 1 hour, dehydrated in ethanol, and air-dried. Slides were then dipped in Kodak NTB emulsion, air-dried, and stored at 4 °C for 4–5 days (depending on the assay) before being developed and cover-slipped.
ISH slides were analyzed with an automated image processing system (Dr. Don Clifton, University of Washington) by a person blinded to the treatment group (Chowen et al., 1990). The software counts the number of silver grain clusters representing Kiss1 or Gnrh cells, as well as the number of silver grains over each individual cell, which provides a semi-quantitative count of Kiss1 or Gnrh mRNA expressed per cell. Cells were considered Kiss1 or Gnrh positive when the number of silver grains in a cluster exceeded that of background by 3-fold. Neuron numbers and mRNA levels per cell from males and females were analyzed with two-factor ANOVA and Tukey–Kramer HSD post hoc test, with a significance p < 0.05, using JMP software (SAS Institute; Cary, North Carolina).
3. Results
3.1. Gonadotrope gene expression is lower in cFOS null mice
We determined previously that cFOS is a critical transcription factor through which GnRH induces FSHβ gene in the LβT2 gonadotrope model cell line (Coss et al., 2004). It is also involved in differential expression of gonadotropin subunits that are necessary for reproductive fitness (Coss et al., 2007; Reddy et al., 2013). In this study, we analyzed a role of cFOS in reproduction in vivo, using cFOS null animals. As reported before (Johnson et al., 1992), we observed that heterozygous crosses result in the Mendelian ratio of offspring at birth. There was no gene dosage effect, since heterozygous animals were not different from the wild types in any paradigm we examined; thus, we present only wild-type and null results.
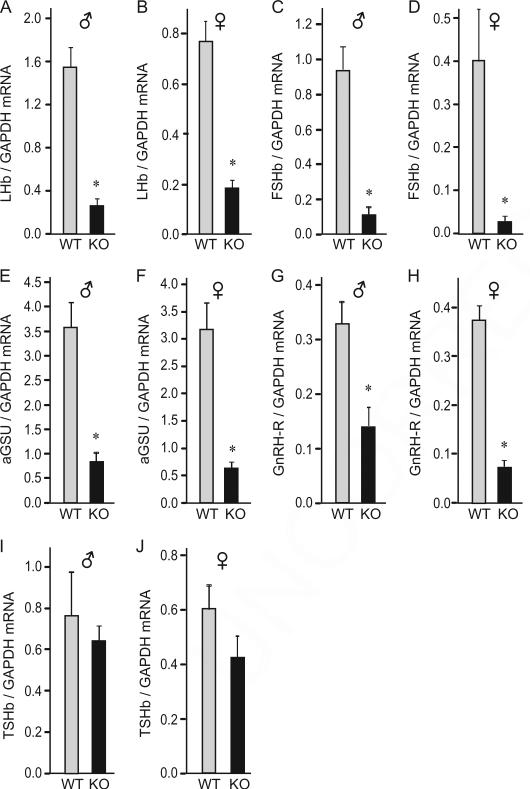
Due to our interest in gonadotrope gene expression and a known role for cFOS in FSHβ and GnRH receptor induction by GnRH, we started our analysis of cFOS null animals by assessing gonadotrope gene expression (Fig. 1A–H). In both sexes at postnatal day 35 (p35), cFOS deficient mice had lower Lhb, Fshb, Cga (α-GSU) subunit, and Gnrhr mRNA expression than wild-types. Lhb was 83% lower in males and 76% lower in females; Fshb was 88% lower in males and 93% lower in females; Cga was 77% and 80% lower in males and females respectively, and Gnrhr was 57% and 80% lower, respectively. Since cFOS deficiency affected gonadotrope specific gene expression, and both gonadotropes and thyrotropes express a common α-GSU which heterodimerizes with the hormone-specific β subunits, we analyzed the expression of the third glycoprotein hormone β subunit. Tshb mRNA levels were the same in wild-type and cFOS deficient animals in both males (Fig. 1I) and females (Fig. 1J). Thus, cFOS is necessary for gonadotrope gene expression, but not for thyrotrope gene expression.
Fig. 1.
Gonadotrope and thyrotrope gene expression in cFOS null mice. Quantitative PCR of p35 pituitaries shows that gonadotrope gene expression is significantly reduced in cFOS-null mice. Total RNA was purified from 5 male and 5 female mice per WT or KO group, reverse transcribed, and the level of hormone expression assayed by real-time PCR. In each sample, the amount of hormone mRNA, calculated from the standard curve, was compared to the amount of Gapdh, and presented as a ratio. Light bars, wild type (WT); black bars, cFOS nulls (KO). Data are presented as the group mean ± SEM; asterisks (*) indicate significant difference (p < 0.05) in the expression in the cFOS-null animals from the wild type animals.
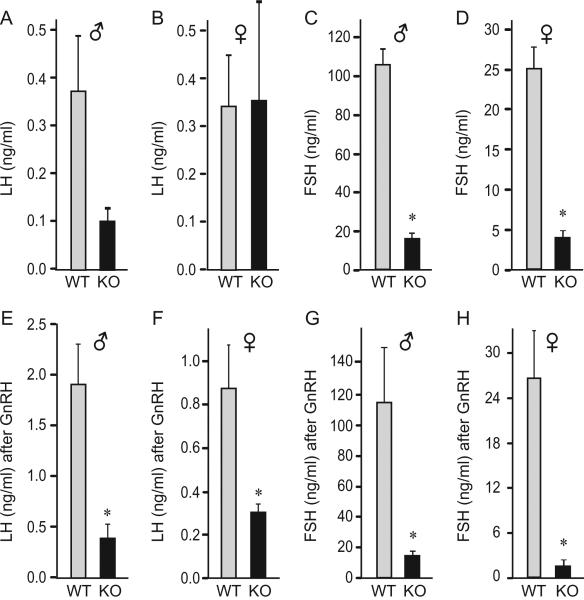
Serum concentrations of circulating gonadotropin hormones were assayed to determine whether reduction in gonadotropin gene expression resulted in lower circulating hormone levels. Difference in serum baseline LH concentration in male animals did not reach statistical significance due to variability, although there was a trend for lower level in the nulls (Fig. 2A). There was no difference in baseline LH levels between wild-type and null females (Fig. 2B). FSH was 85% lower in mutants of both sexes (Fig. 2C and D). To determine pituitary responsiveness to GnRH, we analyzed serum gonadotropin levels 10 minutes following subcutaneous injection of GnRH. Absolute values of GnRH-induced LH were significantly lower in nulls compared to wild-type males (Fig. 2E), due to a different baseline levels. Interestingly, although null male mice started with lower baseline LH levels, both wild-type and null males exhibited a comparable 5-fold increase in LH serum concentration following GnRH injection (compare Fig. 2A and E). Thus, in null males LH concentration increased in response to GnRH treatment. The response to GnRH in nulls implies sufficient expression of GnRH receptors in null males. In females, wild-types responded to GnRH with increased LH levels (Fig. 2F), while nulls retained the same level as prior to the GnRH treatment (compare Fig. 2B and F), revealing a significant difference in GnRH-responsiveness between wild-type and null females. This sex difference in GnRH responsiveness may stem from the alterations in GnRH receptor expression that is more severe in null females (Fig. 1H). Although circulating FSH levels were not dramatically increased following GnRH injection, perhaps due to timing of blood collection, null mice of both sexes had 96% lower FSH in the circulation than the wild-types after the GnRH treatment (Fig. 4G and H). Collectively, both pituitary mRNA levels and serum hormone analyses indicate that cFOS is necessary for normal levels of gonadotropins, particularly FSH.
Fig. 2.
Circulatory levels of gonadotropin hormones. A–D, Gonadotropin hormone levels in serum from the p35 day old animals, 5 per group, presented as the mean ± SEM. E–H, Animals were injected subcutaneously with 200 ng/kg GnRH and blood was collected 10 minutes post injection. Asterisks (*) indicate significant difference, p < 0.05, in the levels between cFOS-null animals and the wild-type animals.
Fig. 4.
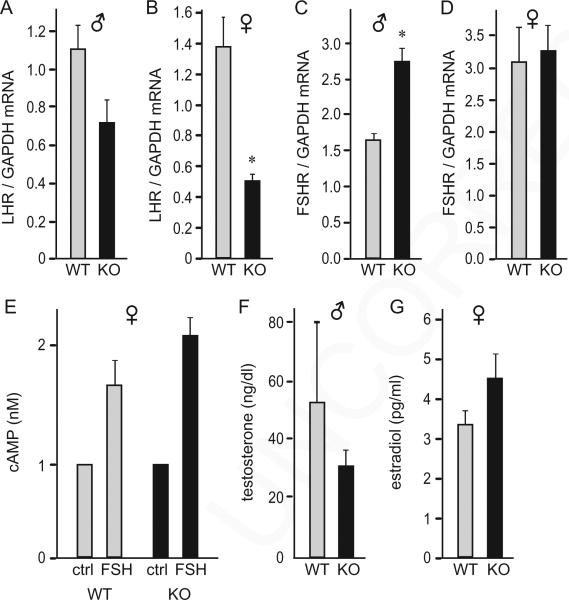
Normal sex steroid hormone levels in cFOS null mice. A–D, Total RNA was extracted from 6-week-old mice, 5 male and 5 female per WT or KO group, reverse transcribed, and the level of gene expression was assayed by real-time PCR. In each sample, the amount of mRNA of interest was compared to the amount of Gapdh, and presented as a ratio. Light bars, wild type (WT); black bars, cFOS nulls (KO). Results are presented as the group mean ± SEM and asterisks (*) indicate significant difference in the expression in the cFOS-null animals from the wild type animals. E, Primary granulosa cell cultures from 6-week-old wild-type (WT) and null female (KO) were treated with 50 ng/ml ovine FSH for 1 hour, to elicit an increase in intracellular cAMP levels. The experiment was repeated 3 times and results, presented as the mean ± SEM, indicate no difference between phenotypes. F–G, Serum estradiol and testosterone levels from 6-week-old mice (5 per group) exhibit no difference between phenotypes.
3.2. cFOS is not required for steroidogenesis
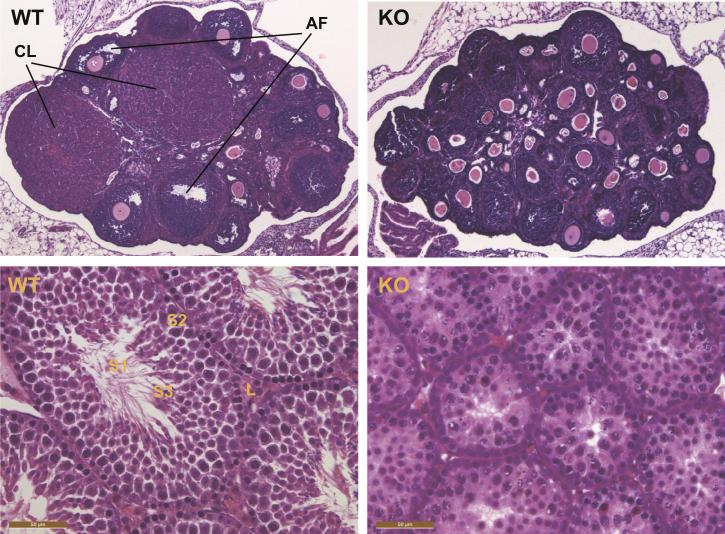
Given that gonadotropin gene expression was diminished in null animals, we analyzed the effects of lower FSH levels on the gonads. Histological analysis of gonads at 6 weeks of age revealed that cFOS null females exhibited a block in folliculogenesis, reminiscent of a phenotype observed in FSHβ deficient mice (Kumar et al., 1997). Specifically, female nulls lacked antral follicles and corpora lutea, indicating a lack of ovulation, while both antral follicles (AF) and corpora lutea (CL) were present in wild-type littermates (Fig. 3 top panels). Homozygous null ovaries contained normal primordial follicles, a greater number of secondary follicles with normal oocytes, and a thick granulosa cell layer, indicating normal early follicular development and typical granulosa cell proliferation, but no antrum development (Fig. 3 top right panel). Testes analysis revealed that most seminiferous tubules in cFOS null males lacked spermatozoa (S1, Fig. 3 bottom panel). Spermatogonia were present (S2), but there was a smaller number of elongated spermatids (S3), indicating potential meiosis problems. Testes also contained a lower number of interstitial Leydig cells (L, Fig. 3 bottom panel). Further examination revealed that cFOS null females do not exhibit vaginal opening, an external measure of puberty, by 42 days of age (6 weeks), while wild-type littermates had a mean onset of vaginal opening at p29.2 (data not shown). In the cFOS null males, there was no mature sperm in the epididymis by 6 weeks of age, while wild-type males presented with 4.7 × 106/ml sperm count (data not shown). Therefore, cFOS is necessary for late folliculogenesis and spermatogenesis, but not for early germ cell development.
Fig. 3.
Histology of cFOS-deficient gonads. Top panels, Hematoxylin and eosin staining of sectioned ovaries collected from wild-type (WT) and cFOS-null mice (KO) at 6 weeks of age demonstrated a lack of corpora lutea (CL) and antral follicles (AF), which were present in wild-type littermates. Bottom panels, Testes from null males at 6 weeks of age revealed reduced Leydig cells (L) and diminished spermatogenesis, indicated with a lack of spermatozoa (S1) and lower number of spermatids (S3), but a presence of spermatogonia (S2).
Since gonadal histology differed between the genotypes, we examined sex steroid hormone levels and expression of gonadotropin hormone receptors in the gonads. Lhr expression was 62% lower, specifically in female null mice (Fig. 4B), while in the null males there was a trend for a decrease that did not reach statistical significance (Fig. 4A). On the other hand, Fshr was significantly higher in null males than wild-types (1.66-fold, Fig. 4C), while null and wild-type females had the same expression of FSH receptor (Fig. 4D). To determine if ovaries can respond to FSH, since FSH treatment induces cFOS in granulosa cells (Ness and Kasson, 1992; Rusovici and LaVoie, 2003), we stimulated primary cultures of granulosa cells with FSH to elicit an increase in intracellular cAMP levels. Granulosa cells from both wild-type and null mice exhibited similar increase in cAMP following 1-hour treatment with 50 ng/ml FSH, indicating that the FSH receptors are expressed and functional in the granulosa cells from null animals (Fig. 4E). However, stimulation with pregnant mare serum gonadotropins (PMSG) followed by human chorionic gonadotropin (hCG) 48 hours later did not elicit ovulation in null females, determined by a lack of corpora lutea in the ovarian histological sections (data not shown). We then analyzed concentration of sex-steroid hormones in the circulation. In males, serum testosterone was similar between genotypes (Fig. 4F). Although null males exhibited a trend for lower testosterone levels, the difference did not reach statistical significance, as with the level of LHR expression in males. In females, serum estrogen was the same between genotypes (Fig. 4G). Collectively, these results indicate that cFOS is necessary for gametogenesis, but not for steroidogenesis or gonadotropin hormone receptor expression in the gonads.
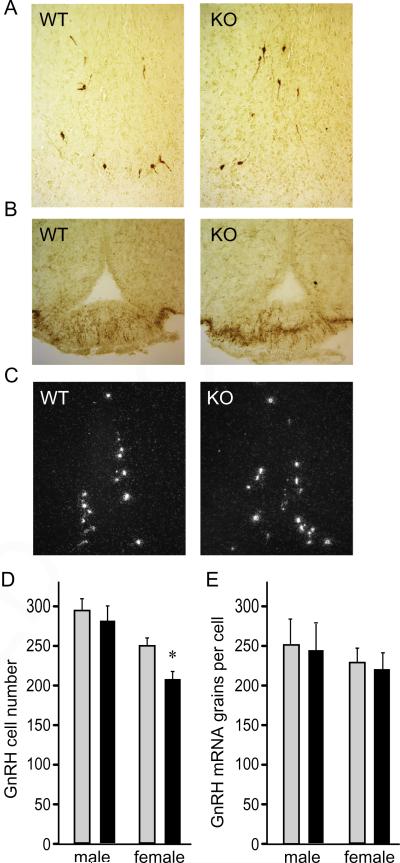
3.3. GnRH neuron number and migration are not altered due to a lack of cFOS
Given that both gonadotropin hormones, which are synthesized and secreted in response to GnRH, were reduced in null animals, we next examined GnRH expression and the number of GnRH neurons in the hypothalamus in 6 week old animals. Using immunohistochemistry, we elucidated that GnRH neurons are present in both genotypes, in both sexes, in a normal pattern in the preoptic area of the hypothalamus (Fig. 5A), indicating that cFOS is not necessary for developmental migration of GnRH neurons from the olfactory placode to their final adult location. Likewise, the median eminence in both genotypes were stained with GnRH, demonstrating that axon targeting of GnRH neurons is not affected by deletion of cFOS (Fig. 5B). In situ hybridization was used to determine GnRH neuron number and mRNA expression levels (Fig. 5C). There was no difference in GnRH neuron number in males, while null females exhibited a 17% decrease compared to wild-types (Fig. 4D). Although statistically significant, this decrease is unlikely to affect fertility, since only 34% of the entire GnRH neuron population is sufficient for full reproductive function (Herbison et al., 2008). Expression levels of GnRH mRNA per cell were also the same in both genotypes in both sexes (Fig. 5E), showing that cFOS is not necessary for normal GnRH gene expression. Therefore, although cFOS is activated by a variety of stimuli in GnRH neurons, GnRH neuron migration, axon targeting, cell number, and gene expression are not altered by the deletion of cFOS.
Fig. 5.
GnRH is not affected with cFOS deficiency. A, Immunohistochemistry of the preoptic area with GnRH antibody was used to identify GnRH neurons in the hypothalami of 6-week-old wild-type (WT) and null (KO) mice. B, Median eminence was stained for GnRH. C, In situ hybridization was performed to analyze GnRH neurons and quantification of neuron number presented as the group mean ± SEM of 5 animals per group in D, while quantification of the grains per cells to analyze mRNA expression is presented in E. Light bars, wild type; black bars, cFOS nulls in D, E. Results of quantifications were presented as the mean ± SEM and asterisks (*) indicate significant difference (p < 0.05) in null females in GnRH neuron number, as determined by two-factor ANOVA and Tukey–Kramer HSD post hoc test.
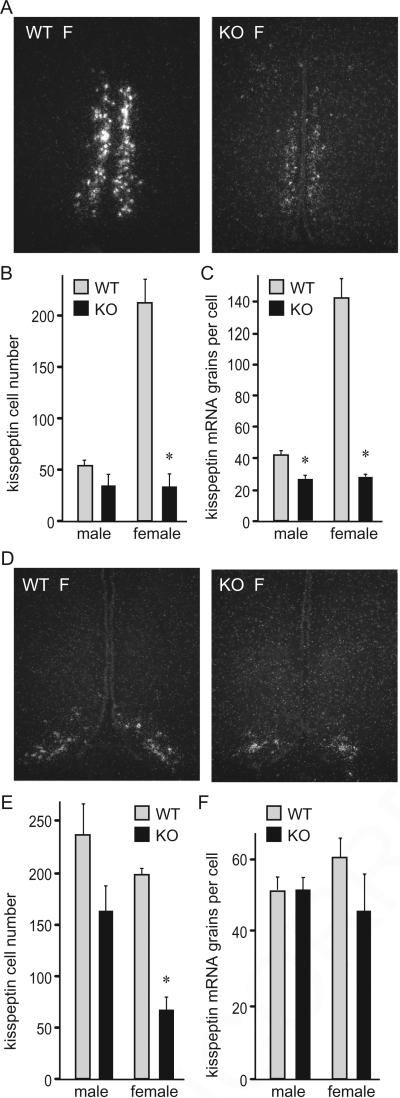
3.4. cFOS is necessary for kisspeptin neurons in female mice
cFOS deficiency resulted in diminished levels of both gonadotropin hormones, but no major alteration was found in GnRH number and mRNA expression levels. Therefore, we examined kisspeptin neurons because kisspeptin is necessary for GnRH secretion. Kisspeptin neuron number and mRNA expression levels in the anteroventral periventricular nucleus (AVPV) were sexually dimorphic, as previously shown, with females having a larger number of Kiss1 neurons (in situ hybridization of female (F) AVPV shown in Fig. 6A). cFOS deficiency resulted in a dramatic 84% decrease in the number of Kiss1 neurons in females (Fig. 6A and B), despite no genotype difference in circulating estrogen. In males, the decrease did not reach significance. Expression of Kiss1 mRNA levels per cell in AVPV neurons was 38% lower in males and 80% lower in females (Fig. 6C). In the arcuate nucleus (ARC), again only female null mice demonstrated a significant decrease, with 66% fewer Kiss1 neurons detected versus wild-types (Fig. 6D and E). Contrary to AVPV neurons, kisspeptin neurons in the ARC of both sexes in both genotypes had the same levels of Kiss1 mRNA per cell (Fig. 6F). Together, our results indicate that cFOS is necessary for kisspeptin gene expression in both hypothalamic nuclei, with dramatic reductions observed primarily in females.
Fig. 6.
Lower number of Kiss1 neurons in female cFOS null mice determined by in situ hybridization. A, Representative image of the AVPV area in p42 day old female (F) mice. WT, wild type; KO, cFOS-deficient mice. B, Quantification of Kiss1 neuron numbers from 5 animals per group shows a decrease specifically in cFOS null female mice compared to wild-type females (light bars, wild type (WT); black bars, cFOS nulls (KO)). C, Expression levels of Kiss1 mRNA per cell, as determined by grains per cell, is decreased in both cFOS null males and females. Results of quantifications were presented as the mean ± SEM. *, A significant decrease in cFOS nulls compared to the wild type within each sex. D, Representative image of the arcuate nucleus (ARC) in females. E, Kiss1 neuron number in the ARC is lower in female cFOS null mice. F, No genotype difference in Kiss1 mRNA levels per cell in the ARC in either male or female mice.
4. Discussion
Despite extensive studies on regulation of the gonadotropin subunit genes using cell models and dispersed pituitary cultures, little is known about in vivo roles of intermediary immediate early genes, such as cFOS, that are proposed to be involved in GnRH induction of gonadotrope specific genes. Studies using cell lines indicate that cFOS is a direct target of GnRH signaling that, upon induction, activates FSHβ and GnRH receptor gene transcription (Coss et al., 2004; White et al., 1999). In the brain, cFOS is also often used as a marker of neuronal activation, in excitation of GnRH neurons and kisspeptin neurons (Clarkson et al., 2008; Lee et al., 1992). The pleio-tropic transcription factor, cFOS, is induced by a variety of stimuli in various other cell types, but exhibits cell-specific and stimuli-specific function in each tissue examined. Herein, we examined the role of cFOS in hypothalamic–pituitary–gonadal (HPG) axis gene expression in vivo, using cFOS-deficient mice, and demonstrate that there is a specificity of cFOS necessity for pituitary gene expression and neuronal gene induction.
In the original report with cFOS null animals, Johnson et al. reported infertility due to impaired gametogenesis (Johnson et al., 1992). Although lethality at 2 months of age is due to impaired hematopoiesis in adults, at birth, cFOS null mice are viable without gross developmental deficits, indicating that cFOS is not required for embryonic development (Johnson et al., 1992). Several reports analyzing cFOS nulls in the central nervous system identified cell-specific effects, without severe global defects (Benes et al., 2013; Fleischmann et al., 2003; Yasoshima et al., 2006). Thus, although we cannot discount possible compensatory mechanisms or nonspecific defects that impinge on the reproductive axis, our results indicate cell-specific and/or sex-dependent functions of cFOS in the HPG.
As predicted from studies using gonadotrope-derived cell lines (Coss et al., 2004; White et al., 1999), we show here in vivo that cFOS is necessary for expression of the FSHβ subunit and the GnRH receptor. Additionally, we determined that cFOS is necessary in vivo for LHβ and αGSU gene expression. Since LHβ and αGSU gene promoters have been analyzed in detail and cFOS/AP1 response elements have not been identified (Jorgensen et al., 2004); nor does cFOS overexpression induce their promoters in the cell line (data not shown), the lower levels of these genes in null mice may stem from a decreased expression of the GnRH receptor. Lower FSHβ expression may either stem from a direct effect of cFOS on the FSHβ promoter, as has been shown in cell lines, or due to a decrease in GnRH receptor expression. A decrease in the GnRH receptor was a likely cause of the lack of LH secretion in null females following GnRH stimulus. Although LHβ gene expression and LH levels in response to GnRH are lower in nulls, basal circulatory levels of LH are the same between wild-type and null females. LHβ mRNA has a very long half-life (Bouamoud et al., 1992), allowing for translational regulation (Nguyen et al., 2004) that may be compensatory in knockouts, resulting in the similar levels of LH in circulation. On the other hand, cFOS is not necessary for expression of a third pituitary glycoprotein hormone, TSHβ. Thus, in the pituitary, cFOS is required for gene expression in the gonadotrope, but not in thyrotrope.
Corresponding to relatively minor effects on gonadotropin hormone receptor gene expression in the gonads, sex steroid levels are the same between wild type and null mice, in males and females. LH receptor was affected in a sex-dependent manner, exhibiting a decrease only in females. Effect only in females would suggest a role for estrogen, however estrogen levels were the same in wild-type and null mice. Furthermore, estrogen receptor α knockout animals and aromatase null mice both exhibit an increase in LH receptor expression, pointing to a negative regulation of the LH receptor by estrogen (Fisher et al., 1998; Schomberg et al., 1999). Thus, cFOS may regulate LH receptor expression directly in a sex-specific manner. In contrast to LH receptor, cFOS deficiency did not have an effect on the FSH receptor in females, but FSH receptor expression was increased in null males. This result is surprising, since a cFOS binding site was identified in the FSH receptor promoter and considered to play a role in FSH induction of its own receptor (Griswold et al., 2001). Increased expression in null males points to a repressive role of cFOS for FSH receptor gene. Unexpectedly, sex steroid hormone levels were unchanged, implying that cFOS does not play a critical role in expression of steroidogenic enzymes. Contrary to steroid hormone levels, epididymides of male nulls exhibit absence of mature sperm, and ovaries of female nulls lack antral follicles and do not ovulate, even in response to exogenous gonadotropin treatment. In the testes, cFOS is expressed in spermatogonia, spermatocytes, and Sertoli cells (Araujo et al., 2009). A role for cFOS in germ cells is not known, while in Sertoli cells multiple roles have been postulated, including proliferation and tight junction formation (Lui et al., 2006; Musnier et al., 2012). Due to a lack of known cFOS target genes in germ cells, we were not able to delineate the cause of the block in spermatogenesis. In the ovaries, cFOS is present in granulosa cells, theca cells and oocytes (Dias et al., 2013; Rusovici and LaVoie, 2003). In granulosa cells, cFOS is induced by FSH treatment (Ness and Kasson, 1992). The amount of cFOS decreases with the growth of the follicle and diminishes after luteinization (Rusovici and LaVoie, 2003). It was therefore postulated that cFOS plays a role in granulosa cell proliferation. Our results, however, do not agree with a role for cFOS in granulosa cell proliferation, since follicles in null animals had multiple layers of granulosa cells, but indicate a role for cFOS in antrum formation. Due to necessary intrafollicle signaling between theca, granulosa and oocyte, throughout maturation (Richards and Pangas, 2010), we were not able to explain the cause of anovulation. Estrogen levels in females, and testosterone in males, were the same between genotypes, indicating that although cFOS is necessary for gametogenesis, it is superfluous for steroidogenesis.
There was no effect of cFOS deficiency on GnRH neuron migration, number of GnRH neurons, or GnRH transcription. The minor decrease observed in null females is unlikely to be physiologically significant since the number of GnRH neurons can be substantially reduced without impairing fertility (Herbison et al., 2008). The observed minor decrease in cFOS null females may be caused by a reduction in upstream kisspeptin signaling (Choe et al., 2013; Kauffman et al., 2007), since we detected significant decreases in Kiss1 neuron number and mRNA levels specifically in females. Although cFOS is activated by numerous neurotransmitters and neuropeptides, and is used as a marker for neuronal activation including in GnRH neurons, GnRH gene expression is not dramatically affected in cFOS nulls. This implies that the GnRH gene is not transcriptionally regulated by any of the signals that activate the neuron through cFOS induction and that cFOS is not necessary for proper GnRH transcription. Lack of major effect on GnRH expression points that the alteration in GnRH secretion rather than transcription is a cause of downstream reproductive consequences. Furthermore, despite a decrease in kisspeptin, the absence of an effect on GnRH expression indicates that kisspeptin does not regulate GnRH gene at the transcriptional level, since lower kisspeptin, particularly in females, is not reflected in dramatically lower GnRH expression at the level of GnRH neuron. Thus, these findings indicate that kisspeptin plays a major role on GnRH secretion or neuronal activation, but not transcription. Thus, this is a likely cause for the observed deficits in pituitary and gametogenesis.
The observed decrease in the Kiss1 gene expression in both the AVPV and ARC, primarily in females, is intriguing. Molecular mechanisms of Kiss1 gene expression in the hypothalamus have not been elucidated and our results implicate cFOS as an important regulator of Kiss1 transcription in vivo. Li et al. reported that the human kisspeptin promoter contains two cFOS/AP1 sites at positions −1272 and −418 (Li et al., 2007), but did not examine whether these sites are functional, i.e. whether kisspeptin expression would be diminished if cFOS/AP1 elements were mutated. It is possible that cFOS binds these elements to induce Kiss1 gene expression. By what means these elements respond in a sex-dependent manner or more strikingly in the AVPV is not clear. We postulate that cFOS is also involved in estrogen regulation of Kiss1 expression in AVPV. Explicitly, Kiss1 mRNA levels were decreased only in the AVPV, a brain site where Kiss1 is strongly upregulated by estradiol as part of estrogen positive feedback during the preovulatory LH surge (Smith et al., 2005). cFOS involvement in estrogen signaling has been shown in multiple tissues. cFOS is recruited to the estrogen response element in promoters that are induced by estrogen treatment (Gaub et al., 1990). Additionally, estrogen responsive genes that do not contain estrogen responsive elements in their promoters are activated via estrogen receptor recruitment by cFOS to the AP1 site (Jakacka et al., 2001; Kushner et al., 2000). It is entirely possible that similar mechanisms, i.e. estrogen receptor recruitment to the AP1 site, are at play on the kisspeptin promoter in the AVPV. cFOS is very rapidly regulated and involvement of cFOS in the estrogen signaling and upregulation of AVPV Kiss1 expression may be required for a rapid increase of kisspeptin synthesis prior to the surge.
In the ARC, female nulls also had lower kisspeptin cell numbers. The ARC may be involved in estrogen negative feedback, and estrogen treatment lowers Kiss1 expression in this region (Smith et al., 2005). Thus, if cFOS is involved in estrogen signaling in ARC by recruiting estrogen receptor to the AP1 site, it is expected that cFOS nulls would have a higher Kiss1 expression. More recently, Dubois et al., however, determined that negative feedback does not require estrogen receptor α (Dubois et al., 2015), which would agree with our results. Our data indicate that cFOS plays a role in specification of kisspeptin expressing cell in the ARC, likely due to binding to the putative cFOS/AP1 site in the promoter of the kisspeptin gene. However, to further delineate whether the effect on kisspeptin observed herein is due to direct cFOS binding to the Kiss1 promoter, promoter analysis using reporter assays or chromatin immunoprecipitation in isolated kisspeptin neurons should be performed.
In summary, our analysis of the cFOS deficient mice reveals several important gene targets of this transcription factor in the hypothalamic–pituitary–gonadal axis, which contribute to impaired gametogenesis and infertility in these mice. We determined that gonadotropin gene expression and kisspeptin neuron numbers were diminished, while GnRH neuron migration and numbers were unaffected. Interestingly, several targets in gonads, pituitary and hypothalamus, including kisspeptin neurons, exhibited sex-specific effects, which will be a focus of future studies.
5. Conclusions
We determined that cFOS plays a cell-specific role at multiple levels of the hypothalamic–pituitary–gonadal axis in vivo. In the pituitary, cFOS is required for gene expression in the gonadotrope, but not in thyrotrope. Furthermore, cFOS is necessary for spermato-genesis and ovulation, but not for early gametogenesis or sexsteroid hormone synthesis. In the brain, cFOS is essential for kisspeptin expression and kisspeptin neuron number specifically in females, but not for GnRH neuron migration to the hypothalamus, axon targeting to median eminence, or GnRH gene expression. Striking effect on the Kiss1 gene expression in AVPV may implicate cFOS in the estrogen positive feedback, which is necessary for the preovulatory surge.
Acknowledgements
The authors thank Kristen Tolson and Matthew Poling for their help.
This research was supported by NIH grants R01 HD057549 and R21 HD058752 (to DC); R01 HD065856 (to ASK). University of Virginia, Ligand Core is supported by U54-HD28934.
References
- Araujo FC, Oliveira CA, Reis AB, Del Puerto HL, Martins AS, Reis FM. Expression of the proto-oncogene c-fos and the immunolocalization of c-fos, phosphorylated c-fos and estrogen receptor beta in the human testis. Histol. Histopathol. 2009;24:1515–1522. doi: 10.14670/HH-24.1515. [DOI] [PubMed] [Google Scholar]
- Benes J, Mravec B, Kvetnansky R, Myslivecek J. The restructuring of muscarinic receptor subtype gene transcripts in c-fos knock-out mice. Brain Res. Bull. 2013;94:30–39. doi: 10.1016/j.brainresbull.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Bouamoud N, Lerrant Y, Ribot G, Counis R. Differential stability of mRNAs coding for alpha and gonadotropin beta subunits in cultured rat pituitary cells. Mol. Cell. Endocrinol. 1992;88:143–451. doi: 10.1016/0303-7207(92)90019-3. [DOI] [PubMed] [Google Scholar]
- Brown JR, Ye H, Bronson RT, Dikkes P, Greenberg ME. A defect in nurturing in mice lacking the immediate early gene fosB. Cell. 1996;86:297–309. doi: 10.1016/s0092-8674(00)80101-4. [DOI] [PubMed] [Google Scholar]
- Cesnjaj M, Catt KJ, Stojilkovic SS. Coordinate actions of calcium and protein kinase-C in the expression of primary response genes in pituitary gonadotrophs. Endocrinology. 1994;135:692–701. doi: 10.1210/endo.135.2.7518388. [DOI] [PubMed] [Google Scholar]
- Choe HK, Kim HD, Park SH, Lee HW, Park JY, Seong JY, et al. Synchronous activation of gonadotropin-releasing hormone gene transcription and secretion by pulsatile kisspeptin stimulation. PNAS. 2013;110:5677–5682. doi: 10.1073/pnas.1213594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowen JA, Argente J, Vician L, Clifton DK, Steiner RA. Proopiomelanocortin messenger RNA in hypothalamic neurons is increased by testosterone through aromatization to estradiol. Neuroendocrinology. 1990;52:581–588. doi: 10.1159/000125647. [DOI] [PubMed] [Google Scholar]
- Clarkson J, d'Anglemont de Tassigny X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J. Neurosci. 2008;28:8691–8697. doi: 10.1523/JNEUROSCI.1775-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coss D, Jacobs SB, Bender CE, Mellon PL. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J. Biol. Chem. 2004;279:152–162. doi: 10.1074/jbc.M304697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coss D, Hand CM, Yaphockun KK, Ely HA, Mellon PL. p38 mitogen-activated kinase is critical for synergistic induction of the FSH beta gene by gonadotropin-releasing hormone and activin through augmentation of c-Fos induction and Smad phosphorylation. Mol. Endocrinol. 2007;21:3071–3086. doi: 10.1210/me.2007-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias FC, Khan MI, Sirard MA, Adams GP, Singh J. Differential gene expression of granulosa cells after ovarian superstimulation in beef cattle. Reproduction. 2013;146:181–191. doi: 10.1530/REP-13-0114. [DOI] [PubMed] [Google Scholar]
- Dubois SL, Acosta-Martinez M, DeJoseph MR, Wolfe A, Radovick S, Boehm U, et al. Positive, but not negative feedback actions of estradiol in adult female mice require estrogen receptor alpha in kisspeptin neurons. Endocrinology. 2015;156:1111–1120. doi: 10.1210/en.2014-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, Clay CM. The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol. Cell. Endocrinol. 2003;206:93–111. doi: 10.1016/s0303-7207(03)00235-1. [DOI] [PubMed] [Google Scholar]
- Ely HA, Mellon PL, Coss D. GnRH induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol. Endocrinol. 2011;25:669–680. doi: 10.1210/me.2010-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CR, Graves KH, Parlow AF, Simpson ER. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. PNAS. 1998;95:6965–6970. doi: 10.1073/pnas.95.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J. Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaub MP, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell. 1990;63:1267–1276. doi: 10.1016/0092-8674(90)90422-b. [DOI] [PubMed] [Google Scholar]
- Gay VL, Midgley AR, Jr., Niswender GD. Patterns of gonadotrophin secretion associated with ovulation. Fed. Proc. 1970;29:1880–1887. [PubMed] [Google Scholar]
- Griswold MD, Kim JS, Tribley WA. Mechanisms involved in the homologous down-regulation of transcription of the follicle-stimulating hormone receptor gene in Sertoli cells. Mol. Cell. Endocrinol. 2001;173:95–107. doi: 10.1016/s0303-7207(00)00412-3. [DOI] [PubMed] [Google Scholar]
- Haavisto AM, Pettersson K, Bergendahl M, Perheentupa A, Roser JF, Huhtaniemi I. A supersensitive immunofluorometric assay for rat luteinizing hormone. Endocrinology. 1993;132:1687–1691. doi: 10.1210/endo.132.4.8462469. [DOI] [PubMed] [Google Scholar]
- Haisenleder DJ, Schoenfelder AH, Marcinko ES, Geddis LM, Marshall JC. Estimation of estradiol in mouse serum samples: evaluation of commercial estradiol immunoassays. Endocrinology. 2011;152:4443–4447. doi: 10.1210/en.2011-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison AE, Porteous R, Pape JR, Mora JM, Hurst PR. Gonadotropin-releasing hormone (GnRH) neuron requirements for puberty, ovulation and fertility. Endocrinology. 2008;149:597–604. doi: 10.1210/en.2007-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J. Biol. Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- Jorgensen JS, Quirk CC, Nilson JH. Multiple and overlapping combinatorial codes orchestrate hormonal responsiveness and dictate cell-specific expression of the genes encoding luteinizing hormone. Endocr. Rev. 2004;25:521–542. doi: 10.1210/er.2003-0029. [DOI] [PubMed] [Google Scholar]
- Kauffman AS, Park JH, McPhie-Lalmansingh AA, Gottsch ML, Bodo C, Hohmann JG, et al. The kisspeptin receptor GPR54 is required for sexual differentiation of the brain and behavior. J. Neurosci. 2007;27:8826–8835. doi: 10.1523/JNEUROSCI.2099-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, et al. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Lee WS, Smith MS, Hoffman GE. cFos activity identifies recruitment of luteinizing hormone-releasing hormone neurons during the ascending phase of the proestrous luteinizing hormone surge. J. Neuroendocrinol. 1992;4:161–166. doi: 10.1111/j.1365-2826.1992.tb00154.x. [DOI] [PubMed] [Google Scholar]
- Li D, Mitchell D, Luo J, Yi Z, Cho SG, Guo J, et al. Estrogen regulates KiSS1 gene expression through estrogen receptor alpha and SP protein complexes. Endocrinology. 2007;148:4821–4828. doi: 10.1210/en.2007-0154. [DOI] [PubMed] [Google Scholar]
- Lui WY, Sze KL, Lee WM. Nectin-2 expression in testicular cells is controlled via the functional cooperation between transcription factors of the Sp1, CREB, and AP-1 families. J. Cell. Physiol. 2006;207:144–157. doi: 10.1002/jcp.20545. [DOI] [PubMed] [Google Scholar]
- Musnier A, Leon K, Morales J, Reiter E, Boulo T, Costache V, et al. mRNA-selective translation induced by FSH in primary Sertoli cells. Mol. Endocrinol. 2012;26:669–680. doi: 10.1210/me.2011-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness JM, Kasson BG. Gonadotropin regulation of c-fos and c-jun messenger ribonucleic acids in cultured rat granulosa cells. Mol. Cell. Endocrinol. 1992;90:17–25. doi: 10.1016/0303-7207(92)90096-o. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Santos SJ, Kreidel MK, Diaz AL, Rey R, Lawson MA. Acute regulation of translation initiation by gonadotropin-releasing hormone in the gonadotrope cell line LbetaT2. Mol. Endocrinol. 2004;18:1301–1312. doi: 10.1210/me.2003-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwitz ER, Xu S, Xu J, Spiryda LB, Park JS, Jeong KH, et al. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both GnRH- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J. Biol. Chem. 2002;277:37469–37478. doi: 10.1074/jbc.M206571200. [DOI] [PubMed] [Google Scholar]
- Padmanabhan V, Dalkin A, Yasin M, Haisenleder DJ, Marshall JC, Landefeld TD. Are immediate early genes involved in gonadotropin-releasing hormone receptor gene regulation? Characterization of changes in GnRH receptor (GnRH-R), c-fos, and c-jun messenger ribonucleic acids during the ovine estrous cycle. Biol. Reprod. 1995;53:263–269. doi: 10.1095/biolreprod53.2.263. [DOI] [PubMed] [Google Scholar]
- Poling MC, Kauffman AS. Sexually dimorphic testosterone secretion in prenatal and neonatal mice is independent of kisspeptin-Kiss1r and GnRH signaling. Endocrinology. 2012;153:782–793. doi: 10.1210/en.2011-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GR, Xie C, Lindaman LL, Coss D. GnRH increases c-Fos half-life contributing to higher FSHbeta induction. Mol. Endocrinol. 2013;27:253–265. doi: 10.1210/me.2012-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J. Clin. Invest. 2010;120:963–972. doi: 10.1172/JCI41350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteinizing hormone surge. Endocrinology. 2009;150:3664–3671. doi: 10.1210/en.2009-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusovici R, LaVoie HA. Expression and distribution of AP-1 transcription factors in the porcine ovary. Biol. Reprod. 2003;69:64–74. doi: 10.1095/biolreprod.102.013995. [DOI] [PubMed] [Google Scholar]
- Schomberg DW, Couse JF, Mukherjee A, Lubahn DB, Sar M, Mayo KE, et al. Targeted disruption of the estrogen receptor-alpha gene in female mice: characterization of ovarian responses and phenotype in the adult. Endocrinology. 1999;140:2733–2744. doi: 10.1210/endo.140.6.6823. [DOI] [PubMed] [Google Scholar]
- Semaan SJ, Murray EK, Poling MC, Dhamija S, Forger NG, Kauffman AS. BAX-dependent and BAX-independent regulation of Kiss1 neuron development in mice. Endocrinology. 2010;151:5807–5817. doi: 10.1210/en.2010-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- Wang Z-Q, Ovitt C, Grigoriadis AE, Möhle-Steinlein U, Rüther U, Wagner EF. Bone and haematopoietic defects in mice lacking c-fos. Nature. 1992;360:741–745. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- White BR, Duval DL, Mulvaney JM, Roberson MS, Clay CM. Homologous regulation of the gonadotropin-releasing hormone receptor gene is partially mediated by protein kinase C activation of an Activator Protein-1 element. Mol. Endocrinol. 1999;13:566–577. doi: 10.1210/mend.13.4.0262. [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J. Biol. Chem. 2001;276:47195–47201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- Yasoshima Y, Sako N, Senba E, Yamamoto T. Acute suppression, but not chronic genetic deficiency, of c-fos gene expression impairs long-term memory in aversive taste learning. PNAS. 2006;103:7106–7111. doi: 10.1073/pnas.0600869103. [DOI] [PMC free article] [PubMed] [Google Scholar]