Summary
It has not been possible to view the transcriptional activity of a single gene within a living eukaryotic cell. It is therefore unclear how long and how frequently a gene is actively transcribed, how this is modulated during differentiation, and how transcriptional events are dynamically coordinated in cell populations. By means of an in vivo RNA detection technique [1–3], we have directly visualized transcription of an endogenous developmental gene. We found discrete “pulses” of gene activity that turn on and off at irregular intervals. Surprisingly, the length and height of these pulses were consistent throughout development. However, there was strong developmental variation in the proportion of cells recruited to the expressing pool. Cells were more likely to reexpress than to initiate new expression, indicating that we directly observe a transcriptional memory. In addition, we used a clustering algorithm to reveal synchronous transcription initiation in neighboring cells. This study represents the first direct visualization of transcriptional pulsing in eukaryotes. Discontinuity of transcription may allow greater flexibility in the gene-expression decisions of a cell.
Results and Discussion
Conventional methods for monitoring transcription utilize microarrays or Northern analysis of RNA on large populations of disrupted cells. These methods, although useful, provide a population average and yield little insight into the specific transcriptional responses of individual cells in their tissue context. Important information about gene activity is lost in this averaged sample. Quantitative studies measuring fluctuations in fluorescent protein expression suggest that gene expression might involve intermittent pulsing [4, 5, 6, 7, 8]. A rigorous single-cell RNA counting study revealed that discontinuous transcription can occur in prokaryotes [9]. However, these transcriptional events have not been directly visualized in eukaryotes. Therefore, it is unclear how long and how frequent transcriptional events are and how they vary during development. In addition, how are transcriptional decisions coordinated in cell populations? Is transcription initiation synchronous, sequential, or asynchronous? What is the balance of reinitiation and de novo expression? We would gain considerable insight into the dynamic nature of transcription if we could view a native gene turning on and turning off in its natural cellular context, subject to all its native cues.
To gain insight into these events in single cells, we used fluorescence microscopy to describe transcriptional activity at a single genomic locus. The system utilizes the high-affinity interaction between a genomic stem loop and the coat protein of the MS2 RNA bacteriophage [1, 2, 3]. Integration of a cassette of MS2 stem loops is targeted into a single endogenous gene. Upon transcription, the MS2 stem loops are read into nascent RNA and detected at the site of transcription with GFP fused to the phage MS2 coat protein (Figure 1A). We used the social amoeba Dictyostelium because of the ease of targeted recombination and the well-described morphological changes as it differentiates. We studied the expression of the discoidin Ia gene, dscA, which is abundantly expressed [10], facilitating detection of transcriptional events, and is reliably induced as cells begin differentiation [11, 12, 13].
Figure 1. Visualizing Transcription of a Developmental Gene in Living Cells.
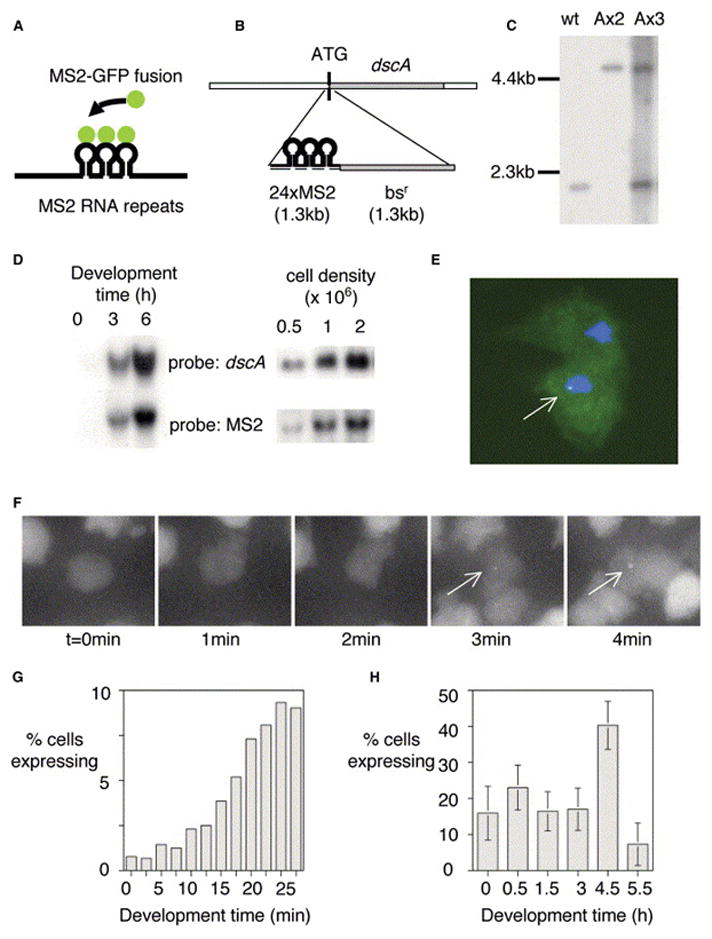
(A) Schematic for RNA detection system.
(B) Schematic for detection of expression of dscA gene.
(C) Southern analysis of dscA-MS2 knockin cell lines. Extra wild-type band for AX3 corresponds to wild-type gene on duplicated portion of chromosome 2.
(D) Northern analysis of dscA-MS2 cells demonstrating appropriate induction by starvation (left) and high culture density during growth (right). The MS2 RNA runs as a single major band at 1.5 kb.
(E) Transformation of MS2-GFP into dscA-MS2 cells reveals a single nuclear spot in expressing cells (arrow).
(F) Visualizing the induction of dscA transcription (arrows, maximal projection of 3D stacks).
(G) Increase in the proportion of expressing cells during the first 30 min of differentiation.
(H) Developmental variation in the proportion of expressing cells. Data are from 30 min movies commencing at the indicated times. 30–40 movies (n = 1500–2000 cells) were collected over 3–4 experimental days for each time point (bars indicate standard deviation).
Twenty-four MS2 stem loops were integrated at the 5′ end of the dscA gene, 6 base pairs downstream of the ATG start codon (Figures 1B and 1C). We used the AX3 strain, as the chromosomal region containing the discoidin I genes is duplicated [14]. Although dscA is dispensible for growth and development in Dictyostelium [15] and there are three almost identical discoidin I genes clustered in the genome, the extra copies mitigate issues of a mutant background. In agreement with this, the growth and development of the MS2-tagged cells are indistinguishable from AX3. Critically, expression of the 1.5 kb MS2 RNA shows the same induction during early development and at high culture densities as discoidin I in wild-type cells (Figure 1D).
Sites of transcription are visualized as a fluorescent nuclear spot in dscA-MS2 cells stably expressing MS2-GFP (Figure 1E), due to the high intensity of the multiple nascent chains at the gene locus. Spots are not observed if MS2-GFP is expressed in wild-type cells lacking the MS2 repeats. Background fluorescence inhibited effective visualization of low or single copy RNA molecules in the cytoplasm. We estimated the number of transcripts at the gene by using FISH with single fluorescent 50-mer probes [16] (see Figure S1 in the Supplemental Data available with this article online). By comparing the intensities of these spots with a dilution series of the probe, we estimate a maximum transcript load of 11 nascent RNA molecules, corresponding to 1 polymerase every 120 bp, in agreement with other analyses [16].
When living dscA-MS2 cells were imaged by fluorescence microscopy, we were able to visualize the time course of gene activation (Figure 1F; see also Movie S1). In the first three frames, the central cell has no detectable transcriptional event. In the fourth frame, a spot becomes visible, which grows in intensity over the next minute. As the observed transcriptional events occur over periods of minutes rather than seconds, we devised an imaging protocol to capture the full range of events. Fields of cells were blind-captured in 3D stacks (0.25 μm z-step), every 2.5 min, for 30 min. At this level of sampling, the nascent RNA could be visualized in multiple z planes. To facilitate acquisition of many data sets, we used a programmable xy-stage for parallel capture of multiple fields. The effects of phototoxicity are cell rounding, impaired motility, and loss of transcription. We eliminated these responses by means of a neutral density filter and a highly sensitive camera to minimize exposure. Cell polarity, motility, and de novo transcription were consistent and appropriate to developmental stage [11, 17] throughout imaging, and the cells formed chemotactic streams, indicative of their well-being.
A rapid elevation in the proportion of cells with detectable transcription occurs during the first 30 min of differentiation (Figure 1G). This increase is progressive, likely reflecting variation in both intrinsic (cell cycle, nutritional status) and extrinsic (proximity to signals) factors. We extended this analysis to look at the entire early differentiation program, with 1500–2000 cells studied per time point (Figure 1H). The proportion of expressing cells initially reaches more than 20%, then falls during the next few hours. Just prior to the onset of aggregation (4.5 hr), a strong increase in the number of cells expressing occurs. The dsc genes are repressed by cAMP signaling [18, 19], and we see marked repression of transcription in chemotactic streams (5.5 hr), although some expression can be detected. However, the level of both MS2 and discoidin RNAs is not diminished at 6 hr (Figure 1D), indicating that the RNA can persist after the repression of transcription. At no point in the differentiation program does the proportion of expressing cells approach 100%. This is consistent with studies that use discoidin I antibodies indicating nonuniform expression during differentiation [20].
Transcriptional pulses have a variable duration (Figures 2A and 2B). The two central cells in Figure 2A both express during the imaging period. The top cell has a detectable site for 4 frames/10 min, while the lower cell expresses for only 2 frames.
Figure 2. Variation in the Length of Transcriptional Pulses.
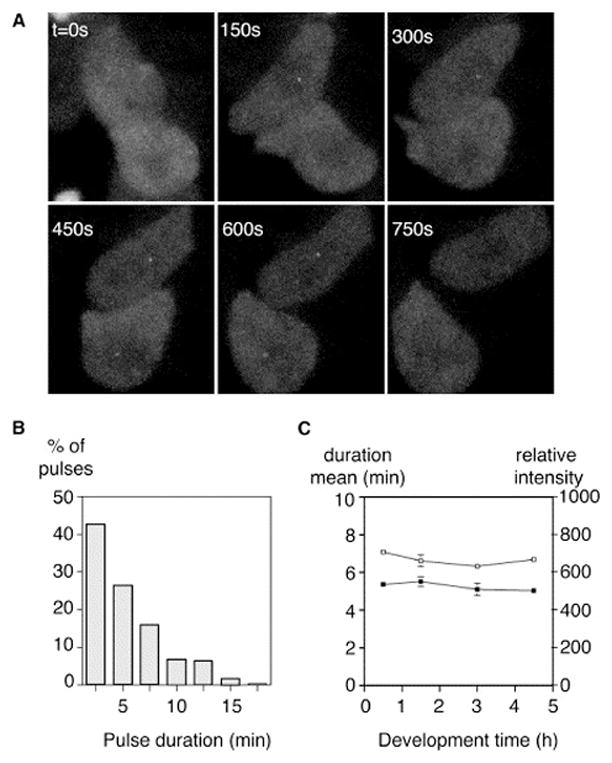
(A) Variation in pulse duration. The top cell has a visible transcriptional event for 4 frames (10 min), whereas the neighboring lower cell expresses for only 2 frames (maximal projection).
(B) Distribution of pulse durations for the 0.5 hr time point. Shorter pulses are more common.
(C) Developmental profile of mean pulse duration (filled symbols) and intensity (open symbols). Pulse duration and intensity are not subject to major developmental variation (bars reflect standard error).
Shorter events (<10 min) are more common, but spots persist in some cells for nearly 20 min (Figure 2B). The mean pulse duration was 5.2 min. When we collate data from a large number of transcriptional pulses, we obtain decay curves that give robust exponential fits (r2 > 0.99, Figure S3). We do not detect significant variation in mean pulse length at different developmental stages (Figure 2C). The mean pulse intensity is also not subject to significant variation during development (Figure 2C).
Transcription is discontinuous (Figure 3A). We observe multiple pulses of transcription in a large proportion of cells. The cell in Figure 3A expresses for the first four frames, and then the gene is inactive for the next three frames before reinitiating again for two frames at the end of the movie. Intensity time series plots for typical examples of pulsing cells are displayed in Figure S2. The interval size between transcriptional pulses is subject to large variation (Figure 3B): shorter intervals (<10 min) are more common, but gaps as long as 25 min were recorded. Again, the decay curves give strong exponential fits (r2 > 0.99). The mean pulse interval is 5.8 min and is not subject to significant developmental variation (Figure 3C). It is unlikely that the discontinuity reflects pulsing in the detection system. The MS2 RNA-MS2 protein interaction has a nanomolar affinity and is highly stable, as cytoplasmic RNA particles retain bound MS2-GFP even if the unbound fraction is restricted to the nucleus [21]. The pulsing effect is also not a consequence of bleaching during imaging. The illumination strength was diminished by a strong neutral density filter, and the illumination time (7 s total) was minimized to maintain cell health. While optimizing the imaging protocol, we found that the illumination dose for detectable bleaching results in high phototoxicity. In addition, the intensity of individual pulses can increase over time (Figure S2). The discontinuity is also unlikely to result from RNA destruction at the transcription site. The stability of the MS2 RNA is apparent in previous work on both prokaryotes and eukaryotes [2, 9]. Stability is also evident in Dictyostelium, as both discoidin and MS2 RNAs increase strongly during development, remaining at high levels during aggregation, when fresh transcription is repressed (Figures 1D and 1H).
Figure 3. Transcription Is Discontinuous.
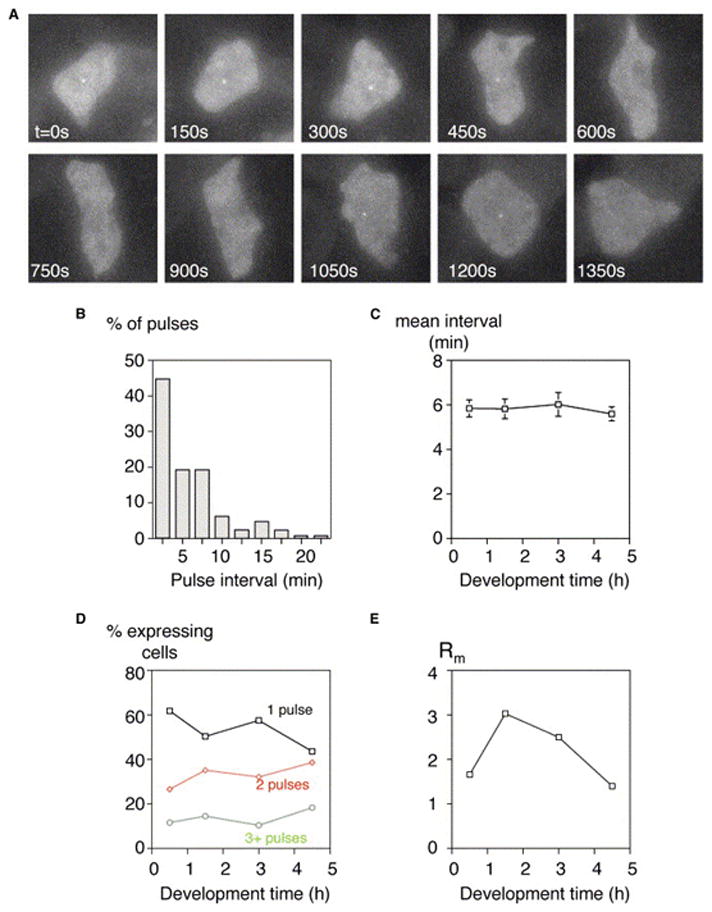
(A) Multiple pulses of dscA transcription. The central cell initially transcribes for 5 frames, then is inactive for 3 frames before reinitiating and expressing for a further 2 frames (maximal projection).
(B) Variation in pulse spacing. The plot displays the spacing (interval) between pulses for all cells in the 0.5 hr time point.
(C) The mean interval between pulses is approximately 6 min and is not subject to large developmental variation (bars reflect standard error).
(D) Developmental profile of pulse frequency (pulses per 30 min movie). Cells were scored for 1, 2, or 3+ pulses, at the indicated developmental times. There is significant variation between the proportions of the different pulse classes between 0.5 hr and 4.5 hr time points (χ2: p < 0.001).
(E) High tendency for reinitiation is observed at time points where low numbers of cells express. Ratio of the probability of a second pulse to the probability of the first (Rm) plotted against developmental time. The expected Rm value is 1 in a situation where reinitiation is not favored over de novo transcription. Cells at the 1.5 hr and 3 hr time points have a significant elevation of Rm over expected levels (χ2: p < 0.001) and are significantly different from the 4.5 hr time point (χ2: p < 0.001).
The pulsing is irregular, contrasting the periodic associations of transcriptional components with ER-responsive genes [22] and oscillations in RNA levels observed in E. coli due to the cell’s pumping out of the gene inducer [23]. The pulses are considerably slower than the oscillations in p53 protein levels (mean duration 5 hr), which occur after DNA damage [6]. Therefore, our observations may reflect a distinct underlying process. Our data are more reminiscent of a recent study in E. coli [9]. By means of a lac-derived promotor inducible with IPTG, Golding et al. [9] observed a staggered increase in the number of RNA molecules that could be counted in the cell. These studies estimated a gene-on time of approximately 6 min, similar to the present study, but a considerably longer off time (37 min). Differences likely reflect the manner of observation (visualizing nascent RNA rather than RNA counting), eukaryote versus prokaryote, and the nature of the gene studied.
During the capture periods, we observed cells with up to five pulses of transcription, although lower pulse frequencies were most common (Figure 3D). The pulse frequency (defined as the number of pulses occurring in the 30 min capture period) shows significant variation between different developmental stages (Figure 3D). Although there is no clear-cut developmental trend, at 4.5 hr there are a higher proportion of multiple pulsers than at 0.5 hr (χ2: p < 0.001). However, the pulse frequencies at 1.5 hr are only slightly different from 4.5 hr, in spite of the large difference in the recruitment of expressing cells (Figure 1H).
The basic properties of pulses (length, height) are consistent throughout development, and the changes in frequency are modest. This is striking given the dramatic changes in dscA transcriptional cues, from cell-autonomous signals (starvation) to strong extracellular signaling (cAMP and CMF [20]). Variation occurs not in these properties of the pulses, but in the number of cells recruited into the expressing population. In this context, these pulses have some parallels with the p53 protein oscillations observed by Lahav et al. [6]. These have a duration and height that does not vary with strength of stimulus (irradiation), but the proportion of cells that respond increases greatly with dosage. Together, these observations imply a simple threshold model. Below stimulus threshold, no expression occurs, yet above this threshold, stimulated cells display a standard range of transcriptional response, regardless of developmental time, nature of signal, or signal strength. This model implies an absence of continuous refinement in transcriptional regulation during differentiation, for example by chromatin modification or by altering the wiring of intracellular signaling.
Reinitiation is more common than de novo initiation at low population expression levels (1.5 hr and 3 hr). Where few cells initiate expression, cells that are expressing are more likely to have a second pulse than a nonexpressing cell is to begin expression. We quantified this via a simple ratio of the probability of a second pulse to the probability of the first. A value greater than 1 would exist if reinitiation were favored over de novo transcription. We calculated this ratio (Rm) for our data, at different developmental stages. The value is higher than 1 for all stages (Figure 3E; p < 0.001 for 1.5 hr). This indicates, particularly during the midstarvation phase of differentiation, that reinitiation is more common than de novo transcription. This effect could imply some form of “transcriptional memory” where cells use the memory of past transcriptional events to enhance new transcription. The frequency plot for pulse intervals associated with the strongest memory effect (1.5 hr) can fit a simple exponential decay function (Figure S3). In other words, the likelihood of reinitiation diminishes as time elapses after an event. Although more complex models are still possible, the memory effect could involve reinitiation from a preexisting transcription complex and/or cotranscriptional chromatin modification temporarily maintaining template accessibility [24]. Alternatively, it may reflect preexisting differences between cells such as cell-type differentiation or passive fluctuations in upstream factors. Discoidin I protein is expressed in only a proportion of cells during development. It is therefore likely that this is a direct view of the sustained transcriptional decision underlying cell-type differentiation.
To assay the contribution of cell signaling to dscA induction during differentiation, we studied the relative distribution of dscA-expressing cells by means of an algorithm that can detect nonrandom clustering of cells based on expression. Community effect signaling would be reflected by close proximity of expressers, and sparseness of expressing cells would indicate lateral inhibition. An intermediate distribution would reflect the dominance of the intrinsic stimulus (e.g., starvation) or uniform concentrations of a global signal. We noticed that expressing cells are frequently close to other expressing cells (Figure 4A). To investigate the significance of this, we compared the pair-wise distances between transcription sites and the pair-wise distances between all cell centroids (Figure 4B). Expressing cells at the onset of differentiation (0 hr) show weakly significant clustering. Expressing preaggregative cells (4.5 hr) show strongly significant clustering. These data likely reflect a community effect or similarity in access to global signaling cues. We extended the analysis further, as our data allow the assessment of the dynamics of transcription in cell groups. The observed clustering is yet more significant if the analysis is restricted to coordinates obtained at pulse initiation (Figure 4B), implying concerted transcription induction. This approach therefore allows detection of the dynamic organization of transcription in cell populations and distinction between synchronous and asynchronous models of cell signaling. Our observations imply transient yet sharp variations in extracellular signaling and a sharp transition in the cellular response. A preaggregative cell can experience the peak and trough of a wave of extracellular cAMP within 2–3 min [25]. The simultaneous exposure of neighboring cells to fluctuations in this repressive signal, or a positive signal, such as the secreted glycoprotein CMF [20], may be responsible for simultaneous transcription initiation.
Figure 4. Expression of dscA in Cell Clusters.
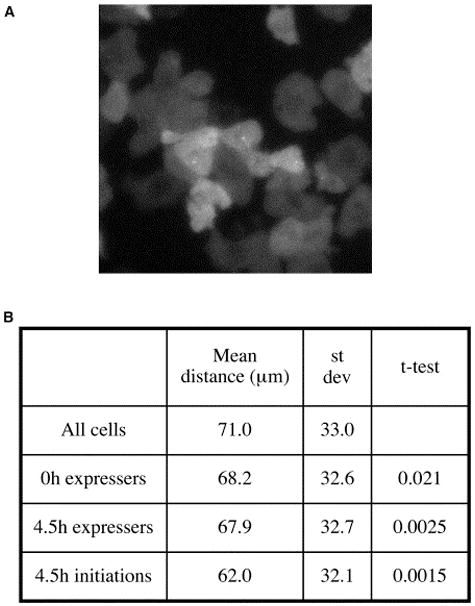
(A) Example image of the expression of dscA restricted to neighboring cells (maximal projection).
(B) Significant clustering of dscA expressers. Pairwise distances between all transcription sites in a field were compared with pairwise distances between the centroids of all the cells (>105total cell:cell comparisons) in a field. These data were analyzed from cells at the onset of differentiation (0 hr) and preaggregation (4.5 hr). Most significant clustering was detected when only the initiating frame of the pulse was used, suggestive of coordinated transcription induction.
Conclusions
We have directly viewed the intrinsic transcriptional behavior of a developmentally regulated genetic locus. We have provided direct visual demonstration of discontinuous eukaryotic transcription. The pulses of transcription are irregular in length and spacing, although mean gene-on and gene-off times were estimated to be 5–6 min, in transcribing cells. The basic properties of pulses (length, intensity) are consistent throughout development, and changes in frequency are modest. This is surprising in the light of the strong changes in transcriptional stimuli occurring through differentiation and implies rigidity in signaling circuits regulating a gene. Variation occurs not in these properties of the pulses but in the number of cells recruited into the expressing population. The advantage to the cell of pulsatile rather than continuous transcription may be sensitivity in the control of gene expression. Pulsing permits greater flexibility in transcriptional decisions—the cell is less committed to a particular program if it does not make all the required RNA in one burst.
The ability to view transcription of a gene in vivo has also allowed direct observation of important developmental phenomena as they occur. We observe a transcriptional memory—in other words a sustained transcriptional decision. This is the basis of all cell-type differentiation. We can also view the transcriptional effects of signaling events in cell populations as they occur. The ability to view a living cell as it makes a transcriptional decision will illuminate the effect of the microenvironment on gene expression.
Experimental Procedures
Tagging the dscA Gene
A 2 kb genomic fragment spanning the dscA locus was assembled from cloned genomic DNA into pBluescript. A fragment bearing 24 MS2 repeats upstream of the blasticidin resistance cassette [26] was inserted into the AccI site adjacent to the ATG of the coding sequence. Polylinker NotI and ApaI sites were used to yield a targeting fragment from this vector, which was transformed as described [27]. Southern blotting of EcoRV digested genomic DNA gave a band shift of 2.6 kb in appropriately targeted clones. In AX3, correct insertion was confirmed by PCR with primers outside the region spanned by the targeting vector in combination with primers to the resistance cassette. For expression of the MS2-GFP fusion, MS2 protein sequence was cloned upstream of GFP into the pDEXH 82 expression vector [28] previously modified to encode a STOP codon after the GFP sequence. This is an integrating vector expressing MS2-GFP without an NLS, via the Dictyostelium Actin15 promotor. Selection of stable clones expressing MS2-GFP was achieved by a selection of 10 μg/ml G418. We obtained several stable clones where expressing cells had only a 2-to 3-fold variation in absolute fluorescence levels. Nonexpressers represented <1% of total cells. Northern analysis was carried out as described [27].
Imaging of MS2-dscA Expression
We used log-phase growth cells maintained 4–6 days in HL5 media [29] in the absence of selection. Culture, differentiation, and imaging were carried out in temperature-controlled rooms at 22°C. Cells were washed free of media and plated in KK2 buffer (20 mM KPO4 [pH 6.2]) on thin KK2–2% agar at a density of 0.4 × 106 cells/cm2. After 5 min for cells to settle, buffer was removed and cells incubated in a humidified chamber. At selected times during differentiation, 1 cm2 squares of agar were excised and inverted onto a Bioptechs ΔTPG imaging dish and covered with mineral oil to prevent dessication.
Cells were imaged on an IX81 inverted microscope with a PlanApo 60x, 1.4 NA objective (Olympus) coupled with a Lambda DG-4/OF30 light source (Sutter Instrument) with a 1.0 OD neutral density filter (#ND10A, Thor Labs) to attenuate excitation and a GFP filter set (#41017, Chroma Technology). Cells were imaged with a Cascade 512B CCD camera (Photometrics) in 3D by IPLab (Windows v3.7, Scanalytics). 38 z-slices were acquired with 250 nm step size with an exposure time of 30 ms. Stacks were collected every 2.5 min for 30 min and multiple fields were collected in parallel by means of a motorized xy stage (MS2000-XYPZ, Applied Scientific Instrumentation). By spatially oversampling, transcriptional events were visible in several z-planes and occupied multiple pixels in individual z-frames. We therefore set the minimum threshold for an event as two adjacent pixels brighter than any other pixels in the cell. These pixels could be directly over each other in z or adjacent in xy. Pixel intensities were quantified by a program written to calculate the mean intensity of a 5 pixel circle (267 nm/pixel) around a transcriptional event. Background levels of GFP were obtained by calculating the mean intensity of a 1 pixel circle surrounding the initial circle. Values were averaged over 5 z-planes. Measures of pulse length and intervals were taken from complete events only. Pulses/intervals tailing into the beginning or end of movies were excluded because of the lack of complete information. Curve fitting was carried out with Table curve 2D version 5.01 (Systat Software Inc.).
Supplementary Material
The frame interval is 2.5 min (total movie length is 30 min) and images were captured, in 3D, by a 1.35NA 100× objective. Image sequence was acquired and converted to avi format by means of IPlab software.
Acknowledgments
This work was funded in part by a Medical Research Council Career Development Award to J.R.C. We would like to thank Professors C. Weijer and R. Insall for advice on the imaging of developing cells.
References
- 1.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci USA. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 3.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 5.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 6.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 7.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 9.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 10.Rowekamp W, Poole S, Firtel RA. Analysis of the multigene family coding the developmentally regulated carbohydrate-binding protein discoidin-I in D. discoideum. Cell. 1980;20:495–505. doi: 10.1016/0092-8674(80)90636-4. [DOI] [PubMed] [Google Scholar]
- 11.Devine JM, Tsang AS, Williams JG. Differential expression of the members of the discoidin I multigene family during growth and development of Dictyostelium discoideum. Cell. 1982;28:793–800. doi: 10.1016/0092-8674(82)90058-7. [DOI] [PubMed] [Google Scholar]
- 12.Clarke M, Kayman SC, Riley K. Density-dependent induction of discoidin-I synthesis in exponentially growing cells of Dictyostelium discoideum. Differentiation. 1987;34:79–87. doi: 10.1111/j.1432-0436.1987.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 13.Poole S, Firtel RA, Lamar E, Rowekamp W. Sequence and expression of the discoidin I gene family in Dictyostelium discoideum. J Mol Biol. 1981;153:273–289. doi: 10.1016/0022-2836(81)90278-3. [DOI] [PubMed] [Google Scholar]
- 14.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander S, Shinnick TM, Lerner RA. Mutants of Dictyostelium discoideum blocked in expression of all members of the developmentally regulated discoidin multigene family. Cell. 1983;34:467–475. doi: 10.1016/0092-8674(83)90380-x. [DOI] [PubMed] [Google Scholar]
- 16.Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH. Visualization of single molecules of mRNA in situ. Methods Enzymol. 2003;361:245–304. doi: 10.1016/s0076-6879(03)61015-3. [DOI] [PubMed] [Google Scholar]
- 17.Varnum B, Edwards KB, Soll DR. The developmental regulation of single-cell motility in Dictyostelium discoideum. Dev Biol. 1986;113:218–227. doi: 10.1016/0012-1606(86)90124-7. [DOI] [PubMed] [Google Scholar]
- 18.Williams JG, Tsang AS, Mahbubani H. A change in the rate of transcription of a eukaryotic gene in response to cyclic AMP. Proc Natl Acad Sci USA. 1980;77:7171–7175. doi: 10.1073/pnas.77.12.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blusch J, Alexander S, Nellen W. Multiple signal transduction pathways regulate discoidin I gene expression in Dictyostelium discoideum. Differentiation. 1995;58:253–260. doi: 10.1046/j.1432-0436.1995.5840253.x. [DOI] [PubMed] [Google Scholar]
- 20.Clarke M, Gomer RH. PSF and CMF, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia. 1995;51:1124–1134. doi: 10.1007/BF01944730. [DOI] [PubMed] [Google Scholar]
- 21.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 23.Le TT, Harlepp S, Guet CC, Dittmar K, Emonet T, Pan T, Cluzel P. Real-time RNA profiling within a single bacterium. Proc Natl Acad Sci USA. 2005;102:9160–9164. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 25.Durston AJ. Pacemaker activity during aggregation in Dictyostelium discoideum. Dev Biol. 1974;37:225–235. doi: 10.1016/0012-1606(74)90144-4. [DOI] [PubMed] [Google Scholar]
- 26.Sutoh K. A transformation vector for dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- 27.Chubb JR, Bloomfield G, Xu Q, Kaller M, Ivens A, Skelton J, Turner BM, Nellen W, Shaulsky G, Kay RR, et al. Developmental timing in Dictyostelium is regulated by the Set1 histone methyltransferase. Dev Biol. 2006;292:519–532. doi: 10.1016/j.ydbio.2005.12.054. [DOI] [PubMed] [Google Scholar]
- 28.Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussman R, Sussman M. Cultivation of Dictyostelium discoideum in axenic medium. Biochem Biophys Res Commun. 1967;29:53–55. doi: 10.1016/0006-291x(67)90539-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The frame interval is 2.5 min (total movie length is 30 min) and images were captured, in 3D, by a 1.35NA 100× objective. Image sequence was acquired and converted to avi format by means of IPlab software.


