Abstract
Cytoplasmic mRNA movements ultimately determine the spatial distribution of protein synthesis. Although some mRNAs are compartmentalized in cytoplasmic regions, most mRNAs, such as housekeeping mRNAs or the poly-adenylated mRNA population, are believed to be distributed throughout the cytoplasm [1–4]. The general mechanism by which all mRNAs may move, and how this may be related to localization, is unknown. Here, we report a method to visualize single mRNA molecules in living mammalian cells, and we report that, regardless of any specific cytoplasmic distribution, individual mRNA molecules exhibit rapid and directional movements on microtubules. Importantly, the ß-actin mRNA zipcode increased both the frequency and length of these movements, providing a common mechanistic basis for both localized and nonlocalized mRNAs. Disruption of the cytoskeleton with drugs showed that microtubules and microfilaments are involved in the types of mRNA movements we have observed, which included complete immobility and corralled and nonrestricted diffusion. Individual mRNA molecules switched frequently among these movements, suggesting that mRNAs undergo continuous cycles of anchoring, diffusion, and active transport.
Results and Discussion
The ability of mRNAs to move in the cytoplasm of eukaryotic cells is essential for sorting sites of synthesis for specific proteins. Studies analyzing the mechanisms of mRNA localization have demonstrated the existence of complex mRNA transport systems. The cytoskeleton is usually required for mRNA localization, and, in a few instances, molecular motors have been shown to be associated with localized mRNA and are required for their movements [5–8]. In yeast, ASH1 mRNPs are associated with a specific myosin motor, and their movements require both this motor and actin filaments [5]. In the Drosophila blastocyst, apically localized mRNAs are associated with dynein, and their movements require both dynein and microtubules [6–8]. These and other studies have provided strong support for the idea that localized mRNAs are actively transported on cytoskeletal filaments.
In contrast to localized mRNAs, nonlocalized ones are thought to be distributed homogeneously throughout the cytoplasm. Nevertheless, they should still be capable of movement. For instance, following nuclear export, they must leave the perinuclear area to be translated throughout the cytoplasm. Studies with inert tracers, such as fluorescent dextrans or ficolls, suggested that particles with sizes equivalent to some mRNPs have limited diffusion in the cytoplasm [9]. Therefore, even nonlocalized mRNA molecules may require an active transport process. Transport of localized and nonlocalized mRNA should differ, to eventually generate their particular distributions.
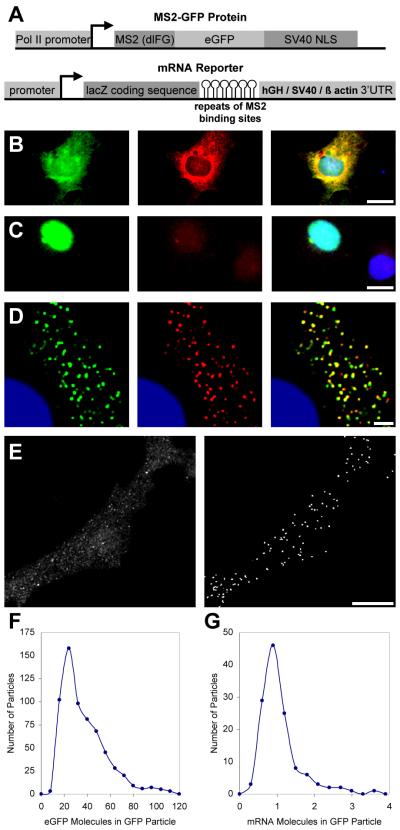
To better understand the mechanisms controlling mRNA movements, we developed a method for visualizing single RNA molecules in living mammalian cells in real time. This method uses a MS2-GFP fusion and a reporter mRNA containing tandemly repeated MS2 binding sites, inserted between LacZ and SV40 sequences [5] (Figure 1A). When the fusion protein was coexpressed with the reporter mRNA, the MS2-GFP bound to the mRNA was exported to the cytoplasm (Figure 1B). When the MS2-GFP fusion protein was expressed alone, a nuclear localization signal confined the protein to the nucleus (Figure 1C). We used COS cells in this study because of their optical properties.
Figure1. Visualization of Single mRNA Molecules in Living Mammalian Cells.
(A) A schematic of the constructs used in this study. The cassettes expressing the MS2-GFP fusion protein and the reporter mRNAs are shown.
(B and C) Visualization of the reporter mRNA with the MS2-GFP fusion protein. Cos cells transiently cotransfected with the pMS2GFP and pRSV-Z-6-SV plasmids and hybridized in situ with a probe against the MS2 binding sites are shown. GFP, green; in situ, red; nuclei (DAPI), blue. (B) A cell expressing both the MS2-GFP fusion and the reporter showing a GFP signal in the cytoplasm colocalizing with the probe (yellow). The scale bar represents 20 μm. (C) A cell expressing the MS2-GFP fusion alone, showing only a nuclear signal. The scale bar represents 20 μm.
(D) Improvement of the sensitivity of RNA detection. Cos cells transiently cotransfected with pMS2-GFP and pRSV-Z-24-SV and treated as in (B), except that images were deconvolved. GFP (green) and the in situ hybridization signal using probes to MS2 (red) colocalize in particles (yellow). The scale bar represents 2 μm.
(E–G) Quantification of the number of RNA molecules per particle. Cos cells transiently cotransfected with pMS2-GFP and pGRE-Z-24-hGH.
(E) Automated selection and analysis of the cytoplasmic GFP particles. The scale bar represents 10 μm. Left: acquired image. Right: deconvolved image with automatically selected objects that correspond to the GFP particles.
(F) The histogram depicts the number of GFP molecules per particle. The results are from 8 cells ( >600 particles).
(G) The histogram depicts the number of mRNA molecules per GFP particle detected by in situ hybridization, using a single probe to LacZ. The results are from 5 cells (125 particles).
Discrete particles of GFP-labeled mRNA were resolvable only when 24 MS2 sites were in the reporter mRNA (Figures 1D and 1E), not when either 6 or 12 MS2 sites were present (Figure 1B and data not shown). To show that the GFP particles indeed contained the mRNA reporter, in situ hybridization was performed with a probe recognizing the region between the MS2 binding sites (Figure 1D). More than 90% of cytoplasmic GFP particles colocalized with the signal obtained by in situ hybridization [10] (Figure 1B).
To quantify the number of mRNA molecules in each GFP-labeled particle, we imaged concentrations of purified GFP to determine the amount of light emitted by a given number of GFP molecules, and we calculated the number of GFP molecules in each GFP-labeled mRNA particle [11]. Quantification of the fluorescence from more than 600 GFP-labeled mRNA particles in eight cells showed that most GFP-labeled mRNA particles contained 20–50 molecules of GFP, with an average of 33 (Figures 1E and 1F). Since the MS2 protein binds as a dimer [12], this measurement coincided with the expected number of GFP molecules labeling a single molecule of reporter RNA, assuming that most of the 24 MS2 sites were bound. This quantification procedure was independently validated by using a laco/GFPlaci system for calibration of the GFP signal [13]. To exclude the possibility that clusters of mRNA molecules sparsely labeled with GFP were misidentified as single mRNA molecules, we performed in situ hybridization to a single target on the mRNA reporter [11]. Cells were simultaneously hybridized with a Cy5-labeled probe to a region between MS2 binding sites and a single Cy3-labeled probe to the LacZ portion of the mRNA. GFP, Cy5, and Cy3 were detected concurrently in single cells, allowing quantification of the number of probes colocalized with individual GFP particles [11]. Analysis of 125 multicolor particles in 5 cells showed that the majority of multicolored particles were single molecules, containing 1 Cy3 probe and 8 MS2 probes (Figure 1G). This validates the independent conclusion, from the analysis described above, that the majority of the GFP-labeled mRNA particles detected in the cytoplasm contained a single RNA molecule. There were a small number of GFP particles containing more than a single RNA molecule present in every cell. The single-molecule GFP particles were distinct from the RNA “granules” observed in a variety of cell types, which are much brighter and larger and, although not quantitated, apparently contain multiple RNA molecules [5, 8, 14–18].
The movements of the single mRNA particles were recorded in living cells at a rate of nine images per second. This high frequency was required because some mRNA molecules exceeded 1 µm/s (see below). The mRNAs contained the coding sequence of LacZ, the MS2 sites, and the 3 UTR of either the human growth hormone gene or SV40 (reporters LacZ-24-hGH and LacZ-24-SV, respectively). These 3 UTRs are commonly used for RNA stability and do not contain any localization sequences [2]. They are therefore representative of mRNAs that should be homogeneously dispersed throughout the cell.
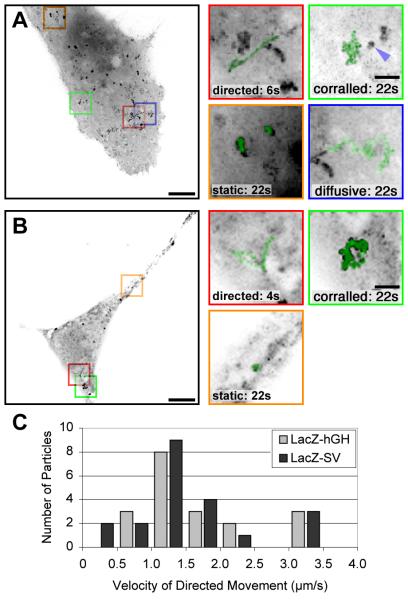
These reporters exhibited four types of motility within a single living cell (Figures 2A and 2B; see the Movies in the Supplementary Material available with this article online). Each category was calculated by examination of a large number of independent series in cells that were stationary for the observed time period (Table S1 in the Supplementary Material, LacZ-24-hGH and LacZ24-SV; both constructs gave equivalent results). One category of particles examined appeared to remain stationary (“static”; 33%–40%). A minority of particles traveled linearly over long distances (> 1.5 µm) and traveled at a constant speed (“directed”; 2%–5%). Occasionally, these particles changed direction abruptly and then followed a second rectilinear path (Figure 2). Since directed movements suggested involvement of a motor, each directed trajectory was analyzed in detail. The net distance traveled by each directed particle ranged from 1.5 μm to 6.5 μm, with a mean of 2.6 μm. The average velocity of directed particles was nonrandom, with a peak between 1 and 1.5 μm/s (Figure 2C). For the longest distances, instantaneous velocities at various intervals were relatively constant during movement (data not shown). The remaining particles moved significantly but never accomplished more than 1.5 μm continuously in a single direction. Among these, some particles moved about a central position, while others displayed a diffusive pattern of movement
Figure 2. Dynamics of mRNA Molecules in the Cytoplasm of Mammalian Cells.
All images were obtained at a rate of nine images per second, for periods of 22 s, and were deconvolved.
(A) Movements of LacZ-24-hGH mRNAs. Cos cells transiently expressing LacZ-24-hGH mRNAs and the MS2-GFP protein were imaged live. Left: a maximum intensity image projection of 200 time frames on 1 image (“maximal projection”). The scale bar represents 10 μm. Right: panel magnifications: the scale bar represents 2 μm. mRNA track superimposed (green) from each of the indicated boxed regions. See Movies 1 and 2 in the Supplementary Material. The blue arrow points to a “static” particle in the vicinity of a “corralled” mRNA.
(B) Movements of LacZ-24-SV mRNAs. Cos cells were transiently cotransfected with pRSV-Z-24-SV and pMS2-GFP and were imaged as in (A). The scale bar represents 10 μm. Right: panel magnifications: track of mRNA movement superimposed (green) on an enlargement from each of the indicated boxed regions. The scale bar represents 2 μm. (See Movies 1 and 2 in the Supplementary Material).
(C) Velocities of directed motion. For each directed movement of either the LacZ-24-hGH or the LacZ-24-SV mRNA particles, the mean velocity was calculated. The corresponding histograms show a peak at 1.0–1.5 μm/s.
The diffusion coefficients of all nonstatic, nondirected mRNAs were extrapolated to determine if the movement of the particles followed a simple diffusion model(x2 4Dt). The mean displacement (x) was calculated by using both a short and a long time scale (0.111 and 10 s, respectively). The diffusion coefficients (D) for short times were similar among all particles examined (about 1 10 9 cm2 /s, Table S2 in the Supplementary Material). In contrast, long-range diffusion coefficients fell into two distinct groups, fast and slow. The diffusion coefficient of the slow group was one-third that of the fast group, indicating restricted diffusion. Displacements of particles with slow long-range diffusion coefficients were essentially unchanged at 25 C, confirming that these movements were due to diffusion. Such restricted diffusion resembled the corralled behavior described in studies of A2RE mRNA granules and neural cell adhesion molecules [19, 20], where particles are able to diffuse until they reach a limiting distance and then change direction. Thus, nondirected, nonstatic particles that showed a slow diffusion coefficient over long time periods were called “corralled” (40%–41%), and the remainder of the nonstatic, nondirected particles were called “diffusional” (15%–25%).
Individual particles were sometimes observed to change their behavior during the time period examined (Figure 2). Movies lasting 15 s and taken at 3-min intervals were used to analyze static particles. After 3 and 6 min, 68% and 41%, respectively, of the static particles could still be identified (n 22). Thus, the behavior of many of the molecules could be sustained over this time period, but most of them eventually moved, demonstrating that particles are able to switch among movement categories. In addition, the behavior of mRNAs did not appear to depend on a particular area of the cell, because different movements could be found next to each other at the same time (see Figure 2A, the “directed” particle becomes “diffusive”).
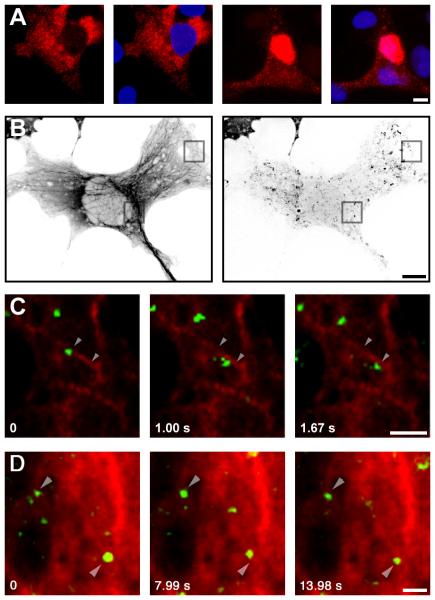
The cytoskeleton would be expected to play an important role in mRNA movements: anchoring static particles, providing tracks for directional motion, or limiting diffusion of corralled particles. Association of mRNA with the cytoskeleton has been demonstrated for localized mRNA, and for the bulk of poly-adenylated mRNA [4], but was never analyzed for a specific, nonlocalized mRNA species. To examine this point, cells were transfected with the LacZ-SV40 reporter, were triton extracted before fixation, and the presence of the reporter mRNA in the cytoskeletal-associated fraction was then assessed by quantitative in situ hybridization. Fifteen percent of the reporter mRNA was retained in the cell after triton extraction (Figure 3A). This value is lower than the 40% of static particles we observed, but some mRNA anchored on the cytoskeleton may be lost during permeabilization. To further examine the association of the reporter mRNA with the cytoskeleton, and to determine which type of filament could be involved, microtubules and microfilaments were disrupted. Destruction of microtubules by colcemid (Table S1) or nocodazole (not shown) had a strong effect: the number of static particles decreased by more than half, the number of corralled particles decreased by 30%, while the number of diffusional particles increased over 3-fold (Table S1, GH/Colc). Directed particles could not be observed following microtubule disruption, and hence this movement appeared to require microtubules. To confirm this, the reporter was detected with a MS2-YFP fusion protein in a cell expressing CFP-tubulin, and both were observed in living cells (Figure 3B). In this sequence, a number of static particles can be observed superimposed on microtubules (12 out of 21), suggesting that they were indeed anchored there (Figures 3B and 3D). Importantly, we could occasionally observe an mRNA molecule moving along the projection of a single microtubule (Figure 3C). Particle movements were not always directly centered on the microtubule, which could be due to a significant distance between the GFP and the motor binding sites on the mRNA reporter or the presence of an adaptor protein between the mRNA and a molecular motor.
Figure 3. Role of the Cytoskeleton in Movement of Nonlocalized mRNA.
(A) Retention of the LacZ-hGH mRNA after triton extraction. Cos cells transfected with pGRE-Z-24-GH were extracted with 0.1% triton for 1 min at room temperature (right) or were fixed directly (left), and they were hybridized in situ with an MS2 probe. Unextracted cells show a stronger cytoplasmic staining (6.5x ). (68 68 μm).
(B–D) Simultaneous localization of microtubules and mRNA movements in live cells. Cos cells were transiently cotransfected with pMS2-YFP, pCFP-tubulin, and pGRE-Z-24-hGH and were imaged live in the CFP and YFP wavelengths. Images were captured in the CFP channel, immediately followed by a movie in the YFP channel (3 images per second for 15 s) and, again, a CFP image. This reduced the artifacts due to cytoskeletal remodeling during recording of mRNA movements.
(B) Left: CFP image; right: maximum intensity image projection of YFP movie. The scale bar represents 10 μm.
(C) Magnification of merged images of microtubules (red) with YFP (green). The scale bar represents 2 μm. Arrows point to the center of mass of a “directed” particle at t 0 and t 1.67 s. The distance between the center of mass at these time points is 1.3 μm (particle speed 780 nm/s).
(D) Magnification of merged images of microtubules (red) with YFP (green). The scale bar represents 2 μm. Arrows point to the center of mass of two “static” particles that colocalize with microtubules.
Microfilaments were disrupted with swinholide, an actin polymerization inhibitor [21]. Following a 2-hr treatment, altered cell morphology made the mRNA particles difficult to track, though mRNA appeared more mobile. The fraction of static particles decreased to 13%, while that of corralled particles was not affected; the number of diffusing particles increased correspondingly to 42% (Table S1, GH/Swh). Altogether, these data indicate that, in Cos cells, microtubules are actively involved in mRNA movements and that either actin filaments or microtubules can anchor mRNAs.
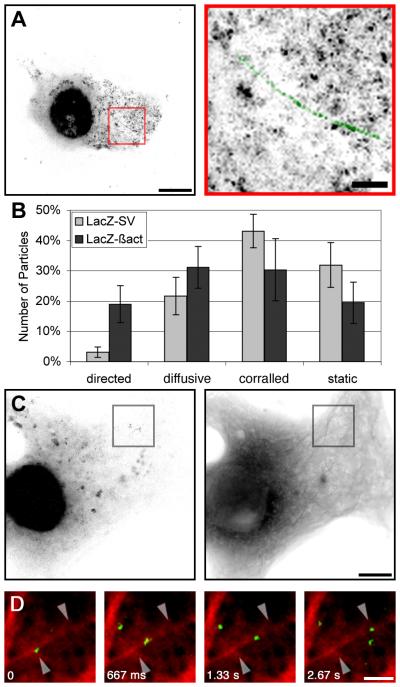
It was then possible to compare these results with the presence of a specific “zipcode” sequence on the dynamics of single molecules. The zipcode sequence is responsible for ß-actin mRNA localization to the peripheral cytoplasm, such as the leading edge of fibroblasts [2] or the growth cone of axons [22]. Following insertion of the zipcode into the reporter mRNA, significantly more cytoplasmic particles could be seen moving, and these showed longer linear movements (as much as 16.6 μm, or three times the maximal distance observed with non-zipcode-containing reporters; Figures 4A and 4B; Table S1, LacZ-24-ßact; and Movies 1–4). To quantitatively assess mRNA motility, a scoring method that used maximal image projections was developed; thus, we did not need to analyze every frame of each movie. Directedness scoring was designed to evaluate the number of particles that moved more than 1.5 μm, and directedness scores for the zipcode reporter were significantly increased compared to those for the non-zipcode reporter (p < 0.001). The average speed of both reporters was the same, approximately 1–1.5 μm/sec. Therefore zipcode-containing reporters appeared to move not only more frequently, but also over longer distances than control mRNA. Although Cos cells are not attenuated in their polarity like fibroblasts or neurons, they are derived from polarized cells, show asymmetry in their morphology, and have some of the machinery of the localization mechanism in the presence of the trans-acting factor required for ß-actin localization, ZBP1 [23].
Figure 4. Cytoplasmic Movements of ß-actin mRNA Reporter.
(A) Movements of LacZ-24-ßact mRNAs. Cos cell transiently cotransfected with pMS2-GFP and pRSV-Z-24-ßact, imaged live. Left: a maximum intensity image projection of 200 frames. The scale bar represents 10 μm. Right: panel magnifications: track of mRNA movement superimposed (green) on an enlargement from each of the indicated boxed regions. The scale bar represents 2 μm. (See Movies 3 and 4 in the Supplementary Material).
(B) Influence of a zipcode sequence on moving particles. Moving particles of GFP-labeled RNA, observed in the image sequences acquired, were classified as either directed, diffusional, corralled, or static. The average distribution from cells transfected with an mRNA reporter either containing (LacZ-24-ßact) or not containing (LacZ-24-hGH, LacZ-24-SV) the ß-actin zipcode sequence are shown. (n 9 image sequences each, 162 particles. The bars report the standard errors).
(C and D) Colocalization of directed motion with microtubules.
(C) Cos cells transiently cotransfected with pMS2-YFP, pCFP-tubulin, and pRSV-Z-24act, imaged live. Left: maximum intensity image projection of YFP; right, CFP. The scale bar represents 10 μm.
(D) Merged images of CFP microtubules (red) with sequences of movement of the RNA labeled with MS2-YFP (green). The scale bar represents 2 μm. The arrowheads point to the particle center of mass at t 0 and t 2.67 s. The distance between the arrowheads is 2.7 μm (particle speed 990 nm/s).
We also investigated whether the zipcode-containing reporter also used microtubules for movement. Following transfection of Cos cells with CFP-tubulin, MS2-YFP, and the LacZ-24-ßact reporter, we observed that the trajectory of the zipcode-containing reporter also occurred on the projections of microtubules (Figures 4C and 4D; Movies). The trajectory of one particle appeared to follow the axis of a single tubule and then cross orthogonally onto what may have been a second tubule (Movie 4). The movement of both reporters, with or without the zipcode, suggests a stochastic model: mRNA molecules do not have a constant behavior, but they switch among a variety of possible movements (static, directed, or diffusional) with specific probabilities for each, depending on the sequences involved.
Almost half the molecules never moved at all, suggesting that they were structurally anchored over the time window examined. Indeed, disruption of either microtubules or microfilaments released some of these static molecules. A second fraction of mRNA molecules was corralled and stayed within small cytoplasmic areas ( <5 μm2) over the time frame investigated. Corralled mRNA did not fit simple diffusional models: over a long time scale, diffusion of these molecules was severely impaired. Recent data analyzing movement of lipid droplets in the cytoplasm of Cos cells showed that, for all observed time scales, many of the droplets did not fit a simple diffusion curve because of the viscoelastic behavior of the cytoplasm [24]. Restriction of mRNA diffusion could be due to steric hindrance of the cytoskeletal network and associated organelles on mRNA movements, resulting in a dynamic molecular sieve that would create transient cytoplasmic microdomains [24– 26]. Consistent with this, 30% were released upon disruption of microtubules. Confinement of mRNA within the cytoskeletal network may increase association with translation initiation factors and ribosomes [27, 28]. A population of mRNA molecules moved with the characteristics of diffusion. Either over a long or a short time scale, these molecules had similar values for an extrapolated diffusion coefficient (about 1 10 9 cm2 /s), and these values closely resembled predicted values (see the Supplementary Material). Experiments with inert tracers or lipid droplets showed that the cytoplasm is heterogeneous and contains areas that permit diffusion of large molecules [24–26]. Messenger RNA may diffuse through such channels, and this might explain their apparent “wandering” path.
A smaller fraction of reporter mRNAs (2%–4%, without the zipcode) had directed motion. These mRNA molecules traveled considerable distances in a short time (up to 6.5 μm in 4 s, at 1.5 μm/sec). This mode of transport was considerably more efficient than diffusion: a diffusing molecule would take 235 s to travel 6.5 μm (D 0.45 × 10 9 cm2 /s). The proportion of directional movements is low, but it may be an infrequent event that is efficient, due to the rapidity of the transport process. We believe the directional motion is indicative of an RNA-motor complex for several reasons. First, the trajectory of the particles was linear, and the speed was constant during movement. Second, individual velocities were not highly variable; most of them ranged between 1 and 1.5 μm/s. Third, they required microtubules, and, fourth, mRNA molecules could be visualized moving close to, or perhaps along, microtubules. It is likely that these directional movements are a general property of mRNAs: they were observed both with and without the zipcode. A non-sequence-specific mRNA transport mechanism would counterbalance the poor diffusional mobility of the general population of mRNAs. In addition, it would account for a number of previous observations that showed nonlocalized mRNAs at the same location as localized mRNAs, although at lower levels [2, 29]. Targeting of “housekeeping” mRNAs to sites distant from the nucleus may ensure that these mRNAs distribute throughout the cell and synthesize products necessary for cellular organelles.
RNAs containing the ß-actin mRNA “zipcode” [2] also displayed directional motion on microtubules (22%), similar to nonlocalizing mRNA. Importantly, these were increased in both frequency and length. Thus, the ability to visualize movements of single mRNA molecules demonstrates for the first time that mRNA molecules can move directly on cytoskeletal cables. It also shows that mRNAs are continuously subjected to cycles of anchoring, diffusion, and transport, and it reveals the sequence specificity of mRNA kinetics, by which the probability of directed movement is regulated. Therefore, we propose that a common probablistic mechanism moves all mRNAs (and perhaps all RNAs in the cytoplasm), but that the movement of particular, localized RNAs is biased for longer motor-directed trajectories, as supported by previous observations [16]. This may bring these RNAs into cellular regions where they are more likely to anchor. Possibly, the stochastic movements may explain the long times needed for localization in oocytes [18].
Experimental Procedures
Plasmids
The plasmid pSL-MS2x6, which contains six tandemly repeated MS2 binding sites, has been previously described [5]. Repeats of the six sites were obtained as described [5], and this generated pSL-MS2x12 and pSL-MS2x24.
In Situ Hybridization, Imaging, and Deconvolutions
Fluorescent hybridization was performed as previously described [10], with Cy3-conjugated or Cy5-conjugated oligonucleotide probes against the MS2 binding site or a region between every two MS2 binding sites, respectively [5]. Images were acquired either on a DMRA microscope equipped with epifluorescence and a motorized stage (Leica) or with an Olympus BX51 with a piezo stepper motor and a planapo 100 n.a. 1.35 objective lens. Digital images were recorded with a 12 bit C4795-NR CCD camera (Hamamatsu) or a Photometrics Cool-Snap Camera. Three-dimensional deconvolution was performed with the software Huygens2 (Bitplane) or with EPR (Exhaustive Photon Reassignment, Scanalytics), with stacks of 20–40 images taken with steps of 0.1 μm in the z axis.
To quantitate GFP fluorescent signals, we followed the procedure described [11], with the following modifications. Cos7 cells expressing both the MS2-GFP fusion and the LacZ-24-hGH reporter were fixed, imaged, and deconvolved [11]. Using Metamorph, the three dimensional images were then thresholded to remove the background haze, leaving only the GFP particles containing the mRNA reporter molecules. A typical threshold was set to about 25% of the intensities within the RNA particles. The pixels below the threshold were set to zero, while the others retained their original value. A mask was then used to set to zero all the pixels within the cell nucleus, and all the remaining cytoplasmic objects were sorted and analyzed with Voxelshopro (Bitplane). These objects comprised some remaining background, well-resolved GFP particles, and aggregates of such particles in regions of high RNA concentration. A manual survey of a number of GFP particles showed that their size ranged from 20 to 80 voxels; therefore, only the objects within this range were selected for further analysis. The total fluorescence intensity (TFI) was then automatically calculated for each object, and the data from several images were pooled together.
The validity of our GFP quantification procedure was confirmed with a yeast strain that expresses a GFP-laci fusion and contains an array of 256 laci sites integrated in a single genomic locus. To force binding to saturation, the GFP-laci fusion was overexpressed from a 2 μm vector. Under these conditions, the laci repeat was visible as a fluorescent dot, together with variable amounts of free GFP-laci in the nucleus. Yeast cells grown on selective media were mounted in low-melting agarose and were imaged live. To eliminate errors due to the replication of the laci array, only cells in G1 (without bud), or with two well-resolved fluorescent particles, were analyzed. Quantitation of the signals from three-dimensional, deconvolved images showed that the particles contained an average of 464 195 GFP-laci molecules (n 51). This is the expected value if most of the laci sites were bound (the laci mutant used here binds as a dimer).
For imaging of live cells, coverslips were mounted in an FCS2 chamber (Bioptechs), which allowed both temperature control and renewal of the media within the chamber. The objective was also heated to 37 C. Digital images were recorded with a 12 bit C4795NR CCD camera (Hamamatsu) at a rate of nine images per second. To increase the quality of the movies, we used a novel deconvolution algorithm (Huygens2, Bitplane) that works on single planes within the depth of field of the objective. This resulted in a significant increase in the signal-to-noise ratio, from about 1.2–2 in the original images to about 8–12, and this proved important for tracking mRNA molecules. To easily display the behavior of mRNA particles during a movie, we used maximal image projections. In this case, the threedimensional stack (x, y, t) of two-dimensional images (x, y) is projected along the time axis, but only the brightest light value along t is retained for each (x, y) space coordinate. This particular mode of projection results in excellent visualization of the trajectory of mRNA particles. Movies taken from three experiments were analyzed in detail by visual examination. In each experiment, the behavior of more than 300 RNA particles, distributed in at least 15 cells, was classified in 1 of the 4 categories described in the text.
Supplementary Material
Acknowledgments
We thank A. Femino for her help with the quantification of the number of probes per molecule, and we also thank Pierre Travo and the Imaging Facility of IFR 24. This work was supported by grants from L’Association pour la Recherche contre le Cancer (l’ARC) (9043) and the Ministe` re de la Recherche France aise et de la Technologie (Action Concertee Incitative) to E.B. and by National Institutes of Health grant GM54887 to R.H.S. N.A. was supported by a fellowship from l’ARC, La Ligue, and Sidaction. D.F. is supported by the Albert Einstein Medical Scientist Training Program. The authors declare that they have no competing financial interests.
Footnotes
Supplementary Material
Supplementary Material including additional details of the methodology, quantitation of the categories of movement for mRNAs of different sequences and conditions, calculations to determine diffusion coefficients, and movies of particle movement is available at http://images.cellpress.com/supmat/supmatin.htm.
References
- 1.Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu. Rev. Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- 2.Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3 -untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassell G, Singer RH. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- 4.Taneja KL, Lifshitz LM, Fay FS, Singer RH. Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J. Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 6.Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106:35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- 7.Schnorrer F, Bohmann K, Nusslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- 8.Wilkie GS, Davis I. Drosophila wingless and pairrule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 9.Luby-Phelps K, Castle PE, Taylor DL, Lanni F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc. Natl. Acad. Sci. USA. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartrand P, Bertrand E, Singer RH, Long RM. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. doi: 10.1016/s0076-6879(00)18072-3. [DOI] [PubMed] [Google Scholar]
- 11.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 12.Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 13.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis I, Ish-Horowicz D. Apical localization of pairrule transcripts requires 3 sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 15.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theurkauf WE, Hazelrigg TI. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development. 1998;125:3655–3666. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- 17.Rook MS, Lu M, Kosik KS. CaMKIIalpha 3 untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 19.Carson JH, Cui H, Barbarese E. The balance of power in RNA trafficking. Curr. Opin. Neurobiol. 2001;11:558–563. doi: 10.1016/s0959-4388(00)00249-x. [DOI] [PubMed] [Google Scholar]
- 20.Simson R, Yang B, Moore SE, Doherty P, Walsh FS, Jacobson KA. Structural mosaicism on the submicron scale in the plasma membrane. Biophys. J. 1998;74:297–308. doi: 10.1016/S0006-3495(98)77787-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bubb MR, Spector I, Bershadsky AD, Korn ED. Swinholide A is a microfilament disrupting marine toxin that stabilizes actin dimers and severs actin filaments. J. Biol. Chem. 1995;270:3463–3466. doi: 10.1074/jbc.270.8.3463. [DOI] [PubMed] [Google Scholar]
- 22.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J. Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada S, Wirtz D, Kuo SC. Mechanics of living cells measured by laser tracking microrheology. Biophys. J. 2000;78:1736–1747. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luby-Phelps K. Effect of cytoarchitecture on the transport and localization of protein synthetic machinery. J. Cell. Biochem. 1993;52:140–147. doi: 10.1002/jcb.240520205. [DOI] [PubMed] [Google Scholar]
- 26.Janson LW, Ragsdale K, Luby-Phelps K. Mechanism and size cutoff for steric exclusion from actin-rich cytoplasmic domains. Biophys. J. 1996;71:1228–1234. doi: 10.1016/S0006-3495(96)79367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minton AP. Confinement as a determinant of macromolecular structure and reactivity. II. Effects of weakly attractive interactions between confined macrosolutes and confining structures. Biophys. J. 1995;68:1311–1322. doi: 10.1016/S0006-3495(95)80304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howe JG, Hershey JW. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell. 1984;37:85–93. doi: 10.1016/0092-8674(84)90303-9. [DOI] [PubMed] [Google Scholar]
- 29.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.