Abstract
SH2 domain-mediated interactions represent a crucial step in transmembrane signaling by receptor tyrosine kinases. SH2 domains recognize phosphotyrosine (pY) in the context of particular sequence motifs in receptor phosphorylation sites. However, the modest binding affinity of SH2 domains to pY containing peptides may not account for and likely represents an oversimplified mechanism for regulation of selectivity of signaling pathways in living cells. Here we describe the crystal structure of the activated tyrosine kinase domain of FGFR1 in complex with a phospholipase Cγ fragment. The structural and biochemical data and experiments with cultured cells show that the selectivity of phospholipase Cγ binding and signaling via activated FGFR1 are determined by interactions between a secondary binding site on an SH2 domain and a region in FGFR1 kinase domain in a phosphorylation independent manner. These experiments reveal a mechanism for how SH2 domain selectivity is regulated in vivo to mediate a specific cellular process.
INTRODUCTION
In response to ligand stimulation, receptor tyrosine kinases (RTK) recruit and activate a variety of target proteins and substrates by utilizing a unique set of tyrosine autophosphorylation sites in their cytoplasmic regions that serve as binding sites for srchomology-2 (SH2) or phosphotyrosine binding (PTB) domains of target proteins. SH2 domains are small protein modules found in a variety of signaling proteins such as enzymes (e.g. tyrosine kinases, tyrosine phosphatases, phospholipase Cγ), adaptor proteins (e.g. Grb2, Nck, Crk), and transcription factors (i.e. STAT) among many others (Pawson and Nash, 2003; Schlessinger and Lemmon, 2003). Quantitative binding experiments and structural analyses of SH2 domains in complex with phosphotyrosine (pY) containing peptides have shown that the binding specificity of SH2 domains is determined at least in part by interactions with amino acids between at the +1 to +3 positions of phosphotyrosine (pY) containing peptides (Songyang et al., 1993; Waksman et al., 1993; Pawson and Nash, 2003; Schlessinger and Lemmon, 2003). Currently, the prevailing thought is that the cellular specificity and activity of each RTK is determined, for the most part, by the sum of the activities of the complement of signaling proteins that are recruited directly and indirectly to RTKs by interactions mediated by SH2 domains and other protein modules. Although it has been assumed that the recognition of linear peptides by SH2 domains can explain the selectivity of SH2 domain-mediated interactions in living cells, this is likely to be an oversimplification (Piccione et al., 1993; Ladbury and Arnold, 2000). Moreover, the hypothesis that short peptides can recapitulate SH2 domain interactions with their native targets has not been tested and so far no quantitative or structural information is available for the molecular mechanisms that govern selectivity of any SH2 domain with an activated RTK.
Fibroblast growth factors (FGFs) mediate their diverse biological responses by binding to and activating a family of four distinct fibroblast growth factor receptors (FGFRs) (Eswarakumar et al., 2005). Similar to other RTKs, activated FGFRs recruit and phosphorylate a complement of signaling molecules that mediate the cellular activities of FGFs. Numerous biochemical and genetic approaches have demonstrated that FGFR is constitutively bound to the adaptor molecule FRS2 via its juxtamembrane region, whereas the signaling molecule, phospholipase Cγ (PLCγ) is recruited to the C-terminal tail of the receptor upon autophosphorylation of a highly conserved tyrosine (Y766 in case of FGFR1) (Eswarakumar et al., 2005).
Binding of each of the two SH2 domains from PLCγ to pY containing peptides corresponding to PLCγ-binding sites on PDGFR or other RTKs has been analyzed quantitatively and structurally. These studies have indicated that the binding affinities and selectivity of the N-SH2 domain and C-SH2 domain towards pY containing peptides are very similar (Ji et al., 1999; Chattopadhyay et al., 1999). In contrast, in growth factor stimulated cells, the N-SH2 and C-SH2 domains of PLCγ bind to different target proteins. The N-SH2 domain binds preferentially to pY containing sites in the C-terminal tails of PDGFR and other RTKs (Ji et al., 1999; Poulin et al., 2000, 2005). On the other hand, the C-SH2 domain binds selectively to an intramolecular site (pY783) on PLCγ itself (Poulin et al., 2005). This intramolecular interaction triggers a change in the overall structure of PLCγ that together with membrane targeting leads to stimulation of its ability to hydrolyze phosphatidylinositol-4,5-biphosphate (PtdIns(4,5)-P2) (Rhee, 2001), generating the second messengers, diacylglycerol (DAG) and inositol triphosphate (IP3). These molecules, in turn, activate protein kinase C (PKC) and release calcium from intracellular stores, respectively (Rhee, 2001). The molecular mechanism underlying the selectivity of the two SH2 domains of PLCγ towards activated RTKs remains unknown.
In this report, we show that the selectivity of the N-SH2 domain of PLCγ for tyrosine phosphorylated, activated FGFR1 cannot be explained by its recognition of a simple short linear stretch of amino acids. A substantial fraction of the interaction is mediated by phosphorylation-independent interactions between a secondary binding site found exclusively on the N-SH2 domain and a region of the FGFR1 tyrosine kinase domain. These experiments show that pY binding site as the sole determinant cannot explain SH2 specificity in a biological context. The selectivity of an SH2 domain towards a target protein is determined by a secondary site on the SH2 domain that cooperates with the canonical, pY site to mediate a specific and physiologically productive cellular process.
RESULTS
The cytoplasmic region composed of the tyrosine kinase domain and the C-terminal tail containing the PLCγ binding site, pY766, of FGFR1 was expressed and purified (Figure 1A). To obtain a more stable, homogeneously phosphorylated preparation for crystallization, three of the six previously mapped tyrosine phosphorylation sites, Y463, Y583 and Y585, which are dispensable for tyrosine kinase activity of FGFR1 as well as for PLCγ recruitment and activation, (Mohammadi et al., 1996) were mutated to phenylalanine. Following expression and purification, a trisphosphorylated species of FGFR1 in which tyrosines Y653 and Y654 in the activation segment and Y766 in the C-terminal tail were homogenously phosphorylated (FGFR1-3P) was isolated (Figure 1B). We also expressed and purified a PLCγ fragment (PLC-Δ) composed of the two SH2 domains of PLCγ followed by a segment containing Y783 (Figure 1A), an amino acid shown to be required for growth factor stimulation of PLCγ enzymatic activity (Kim et al., 1991; Poulin et al., 2005). A ternary complex composed of FGFR1-3P and PLC-Δ together with the non-hydrolysable ATP analog, AMP-PCP, was prepared and subjected to extensive screening for crystal growth. We obtained crystals belonging to the space group C2, with the asymmetric unit containing a single 1:1 complex of FGFR1 and PLC-Δ and were able to solve the structure at 2.5 Å resolution using molecular replacement (Figure S1; Table S1).
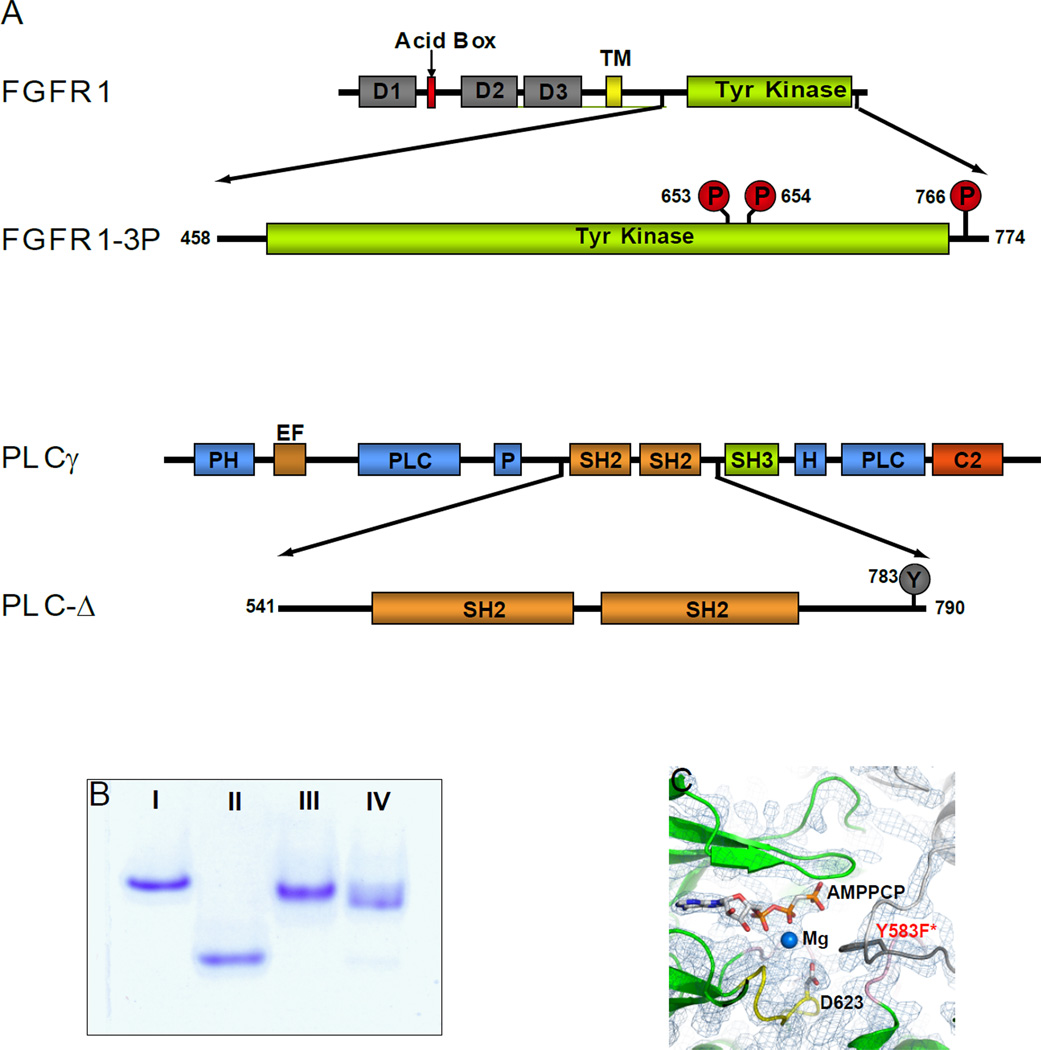
Figure 1. Schematic representation of FGFR1-3P and PLC-Δ.
(A) Schematic representation of wild type FGFR1 and PLCγ as well as their fragments FGFR1-3P and PLC-Δ that were studied in this manuscript. Phosphorylated tyrosine residues are marked by red circles and an unphosphorylated tyrosine by a grey circle. D1, 2 and 3 are Ig-like domains 1, 2 and 3 respectively. TM is transmembrane, PH pleckstrin homology, EF elongation factor, and C2 calcium/lipid-binding domains. (B) Coomassie blue stained native gels (7 %) of purified preparations of unphosphorylated FGFR1 (I), FGFR1-3P (II), PLC-Δ (III) and FGFR1-3P/PLC-Δ complex (IV). (C) The 2Fo-Fc map of AMP-PCP and surrounding kinase region. Ribbon diagrams of FGFR1-3P and the kinase insert region from a symmetry related molecule are shown in green and gray, respectively. AMP-PCP, catalytic base (D623) and the putative substrate (Y583F*; labeled in red) are depicted in a stick representation. Mg ion is shown as a blue sphere.
Overall structure of activated FGFR1 kinase in complex with a PLCγ fragment and with an ATP analog
The overall structure of the ternary complex is shown in Figure 2. The structure consists of a single activated FGFR1 kinase molecule in complex with PLC-Δ and AMP-PCP. The structure of FGFR1-3P presented in Figure 2 shows (more details are presented in Supporting material Figure S2) that pY653 and pY654 maintain the activation segment of the kinase domain in an open configuration virtually identical to the conformations observed in other activated tyrosine kinases including those of insulin receptor (IRK), and insulin-like growth factor receptor-1 (IGF1R) (Hubbard 1997; Favelyukis et al., 2001). Autophosphorylation leads to a large conformational change in the activation segment; T658, one of the amino acids of the activation segment that competes for substrate binding in the autoinhibited conformation (Mohammadi et al., 1996), is moved by approximately 24Å in the active kinase configuration (Figure S2B). Similar to the configurations of other activated tyrosine kinases, pY654 of activated FGFR1 is in contact with an RD pocket that is formed upon autophosphorylation (Johnson et al., 1996). The structure also reveals a well defined nucleotide-binding loop of FGFR1-3P in complex with AMP-PCP together with Mg2+ in a conformation similar to other ATP analog-bound, active kinase structures (Favelyukis et al., 2001; Hubbard, 1997). In contrast to most kinases where the αC moves towards the C-lobe during kinase activation, movement of the entire N-lobe of FGFR1 towards the C-lobe was detected in the structures of the activated conformations (Supporting material: Figure S2A). The structure also shows that the C-terminal segment of PLC-Δ containing the *Y783’ phosphorylation site does not occupy the substrate binding pocket. Instead, this site is occupied by an autophosphorylation site of the kinase insert region (Y583) of a neighboring FGFR-3P molecule that was substituted by a phenylalanine residue (Figure 1C, S2D; S4). The mutated autophosphorylation site Y583F together with additional C-terminal residues interact in an anti-parallel manner with amino acids of the P+1 region of FGFR1-3P activation segment (Figure S2D) as seen in other substrate bound, active kinase structures.
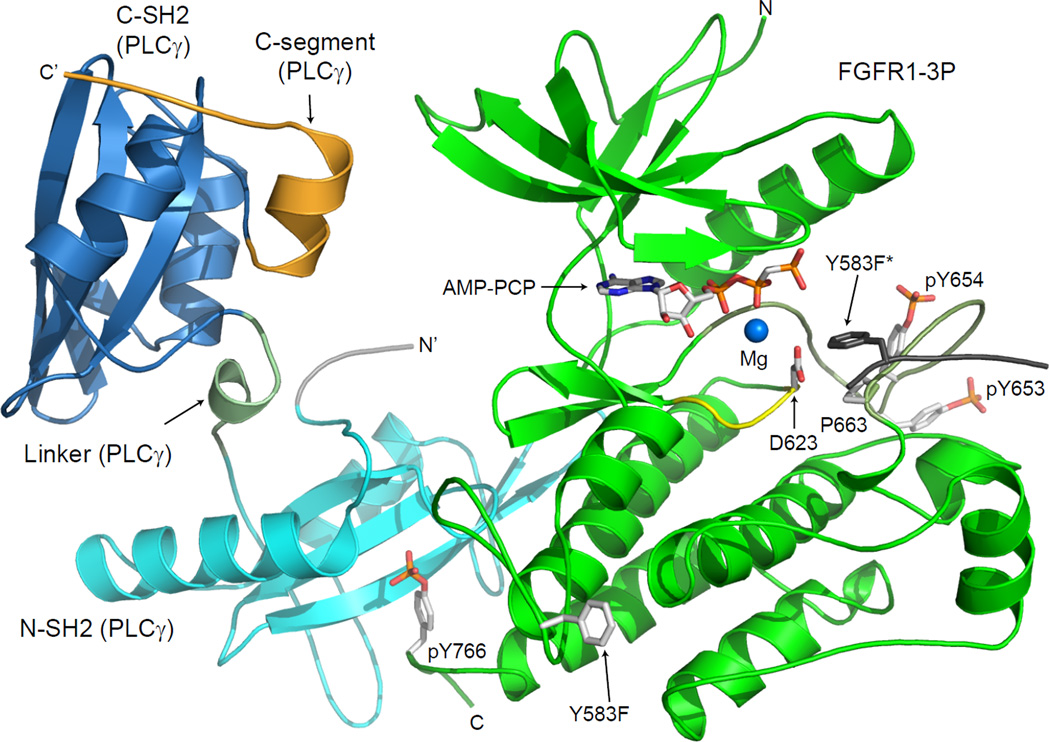
Figure 2. Crystal structure of activated FGFR1 in complex with a PLCγ fragment.
Ribbon diagram of the complex structure FGFR1-3P in green, N-SH2 and C-SH2 domains are in cyan and blue, respectively, the linker connecting the two SH2 domains is in pale green and the C-terminal extension is in orange. AMP-PCP, phosphotyrosines (pY653, pY654 and pY766), Y583F and catalytic base (D623) are shown in a stick representation. Activation segment of FGFR1-3P is colored in pale green and catalytic loop is in yellow. Mg ion is presented as a blue sphere. The kinase insert region from a symmetry related FGFR1 molecule is shown in gray with Y583F* in a stick representation. The color scheme applied in this figure is generally used throughout the figures. Otherwise, differences are indicated.
The structure of the two SH2 domains of PLCγ in complex with activated FGFR1 shows that each of the two SH2 domains exhibits a typical SH2 domain fold, consisting of two α-helices surrounding an anti-parallel β-sheet composed of three β-strands (Figure 2, 3B). The two SH2 domains are connected to one another via a linker of ten amino acids containing a short α-helical structure at the center of the linker (Figure 2). The linker makes no contacts with the bound FGFR1-3P. The pY binding sites of the two tandem SH2 domains lie on opposite face of the molecule. The N-SH2 domain is bound to the pYLDL sequence within the C-terminal tail of FGFR1-3P (containing pY766) (Figure 2,3 and S5), whereas the C-SH2 binding site is unoccupied and exposed to the solvent. The segment that follows the C-SH2 domain forms a short α-helix followed by an extended stretch of mostly disordered amino acids, including Y783’ phosphorylation site of PLCγ (Figure 2; S6).
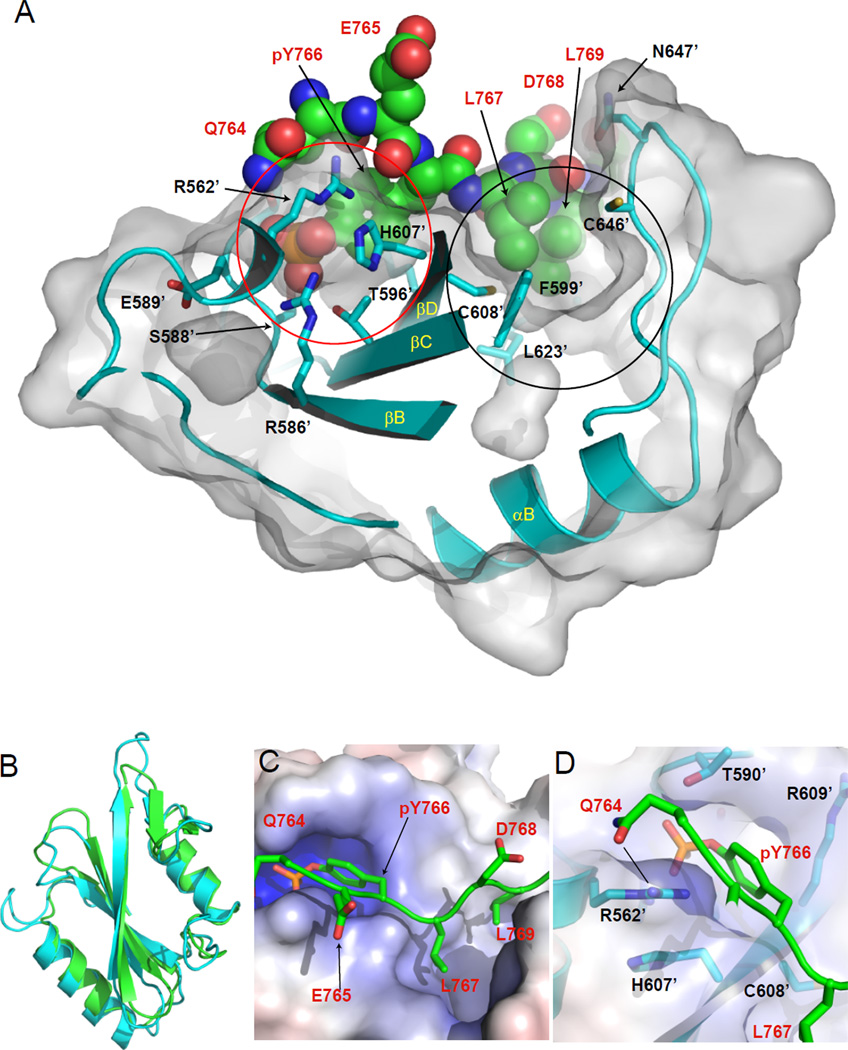
Figure 3. Canonical primary binding site of N-SH2 domain towards pY766 on activated FGFR1.
(A) Surface representation of N-SH2 domain of PLCγ with C-terminal residues of FGFR1-3P containing pY766 shown in spheres. Colors for FGFR in CPK spheres except green for carbon. Ribbon diagram of N-SH2 of PLCγ is shown in cyan. Interacting residues of N-SH2 with the peptide are shown in a stick representation. The pY binding pocket is depicted with red solid circle, and the hydrophobic pocket with black solid circle. T624’ of N-SH2 cannot be seen in this image. Labeled amino acids from FGFR1-3P and N-SH2 domain are shown in red and black, respectively. (B) Ribbon diagrams of superimposed N-SH2 and C-SH2 domains. N-SH2 domain is shown in cyan, and C-SH2 in green. (C) The pockets on the surface of N-SH2 with residues around pY766 of FGFR1-3P. The electrostatic potential surface of N-SH2 is colored in blue (+10 kT) for positive charge and red (-10 kT) for negative charge. The residues around pY766 of FGFR1-3P are depicted with stick representation in red for oxygen, blue for nitrogen, orange for phosphorus and green for carbon. (D) The additional interaction between Q764 (P-2) prior to pY766 of FGFR1-3P and R562’ of N-SH2. The peptide of FGFR1-3P is shown in green with side chains of residues in a stick representation. The interaction between Q764 and R562’ is marked with a black solid line.
Two different sites on the surface of the PLCγ N-SH2 domain mediate complex formation with activated FGFR1. The canonical phosphorylation-dependent two-pronged binding site (primary site) consists of a positively charged patch on the surface of the N-SH2 domain that binds to pY766 and an hydrophobic pocket that accommodates L769 in the pYLDL sequence in the FGFR1 C-terminal tail (Figure 3A). Unexpectedly, the crystal structure also revealed an additional binding site (secondary site) that is partially located on a different surface of the N-SH2 and forms multiple contacts with α-helices on the backside of the C-lobe of the tyrosine kinase domain away from the activation segment of FGFR1 (Figures 2, 4, 5).
Figure 4. Complex formation between activated FGFR1 tyrosine kinase and PLCγ N-SH2 domain.
(A) A view of the molecular surface of FGFR1-3P in complex with PLCγ N-SH2 domain. The same orientation is used in Figure 1C. (B, C) The regions of interactions between N-SH2 domain and FGFR1-3P in the buried surface areas are visualized by pulling apart the two bound molecules. Interactions mediated by the primary binding site of the N-SH2 domain (and the primary binding site itself) are shown in red, and interactions mediated by the secondary binding site of the N-SH2 domain are in blue. Bipartite nature of secondary site is clear in (B).
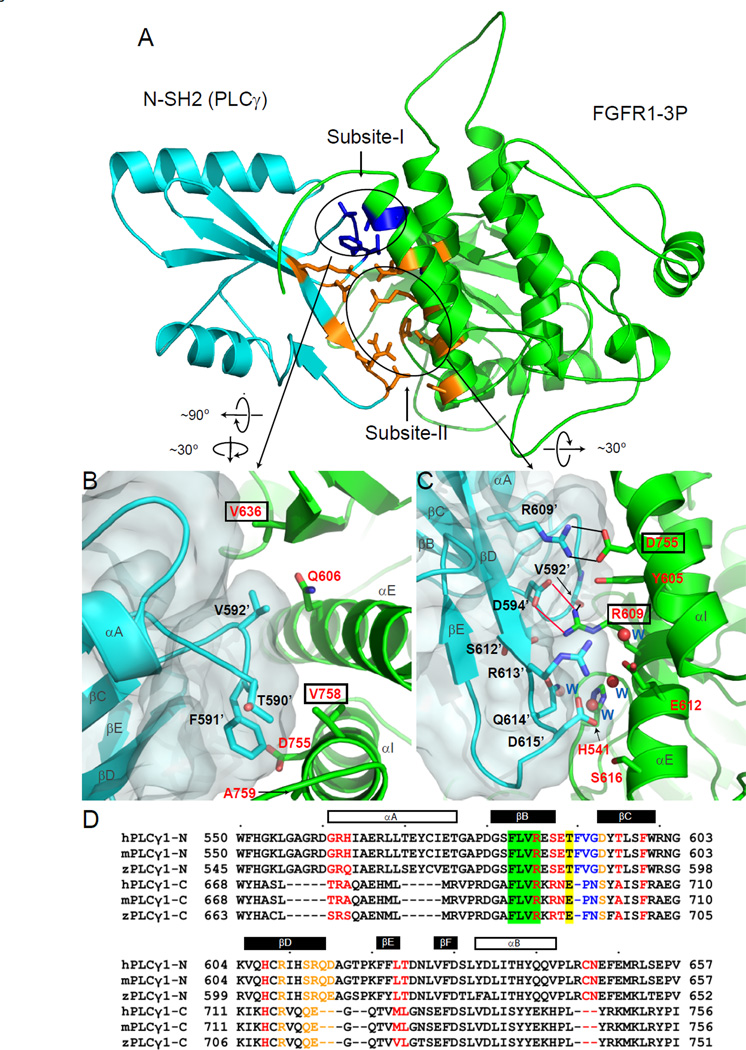
Figure 5. Detailed views of the interface formed by the secondary binding site of the N-SH2 domain with amino acids from the C-lobe of the kinase domain.
(A) A bottom view of ribbon diagram of interactions between N-SH2 with C-lobe of the kinase domain. The amino acids mediating the interactions between FGFR1-3P and N-SH2 are depicted in a stick representation. Amino acids of the interface formed by the secondary binding site are shown in blue (subsite-I) and orange (subsite-II). Subsite-I and subsite-II are marked with black circles. (B) A detailed view of the interface formed by subsite-I. Labeled amino acids of FGFR1-3P and N-SH2 domain are in red and black, respectively. Amino acids in FGFR1-3P that were mutated for biochemical experiments are marked by black boxes. Surface of N-SH2 is shown in pale cyan. Secondary structures of the complex are labeled in dark gray. (C) A detailed view of the interface formed by Subsite-II. Water molecules are shown as red spheres. Labeled water molecules are in blue. Interactions of two pairs of core arginine-aspartic acid residues with surrounding residues are shown in black and red solid lines for hydrogen bonds and Van der Waals contacts, respectively (calculated by PDBsum; Laskowski et al., 1997). (D) Structure-based sequence alignment of human, murine and zebrafish PLCγ SH2 domains performed with 3D Coffee (Poirot et al., 2004). Amino acids of the pY binding and hydrophobic pockets of the primary interface are colored in red. Amino acids of subsite-I and –II of the secondary binding site are shown in blue and orange, respectively. Conserved FLVR motif of SH2 domains is in a green box and T590 is colored in yellow. The secondary structures of the N-SH2 domain are indicated.
Canonical phosphorylation dependent binding of the N-SH2 domain to pY766 in activated FGFR1 (primary site)
The pYLDL sequence in the FGFR1 C-terminal tail adopts an extended conformation when bound to the N-SH2 domain of PLCγ (Figure 3A, marked with a red circle; 2C, S7A). The phosphate moiety of pY766 forms hydrogen bonds with the invariant R586’ residue (FLVR motif; βB6) as well as with R562’ (αA2), T590’ (BC2), S588’ (βB7), and T596’ (βC3) of the N-SH2 domain. An additional hydrogen bond is formed between Q764 (P-2 from pY766) and R562’ (αA2), a highly conserved arginine residue among SH2 domains (Liu et al., 2006). The interaction is further stabilized by projection of L767 (at P+1) and L769 (at P+3) of the FGFR1 pYLDL sequence into a hydrophobic pocket on the N-SH2 surface (BG5; Figure 3A, marked with a black circle; 3C, 3D;). Finally, an additional water mediated interaction occurs between D768 (P+2) of FGFR1-3P and N647’ of the βG loop of the N-SH2 domain. The total buried surface area of the interface formed between the phosphorylated C-terminal tail of activated FGFR1 and the canonical pY binding and hydrophobic pockets on the N-SH2 domain is 653 Å2 (PDBsum; Laskowski et al., 1997).
A secondary binding site for N-SH2 domain of PLCγ on the C-lobe of the tyrosine kinase domain of activated FGFR1
In addition to this canonical set of interactions, the N-SH2 domain is bound to activated FGFR1 via a secondary site for FGFR1 that comprises residues from β-strand βD, loops BC and DE of the N-SH2 domain interacting with the αE and αI helices, β8 and the loop between β7 and β8 (β7/β8) of FGFR1-3P. The total buried surface area in the interaction mediated by the secondary site is 533 Å2 (Figures 4, 5 and S7B). On the basis of the hydrophobic and hydrophilic characteristics of regions of the secondary site, we have divided the secondary site into two subsites (Figure 4, 5A).
Subsite–I is composed primarily of amino acids with hydrophobic side chains. Figure 5B shows Van der Waals interactions between F591’ (BC3) of the N-SH2 domain with D755 and A759 in αI of FGFR1-3P and between T590’ of N-SH2 and V758 in αI of FGFR1-3P. In addition, V592’ of the N-SH2 domain, which faces the core of the secondary interface, is in the vicinity of Q606 in αE of FGFR1-3P interact via Van der Waals contacts. V636 in β-strand β8 of FGFR1-3P is also found in the interface bound by V592’, a hydrophobic residue of the N-SH2 domain (Figure 5B).
Subsite-II on the N-SH2 domain is composed primarily of charged residues that form electrostatic interactions with the C-lobe of activated FGFR1 (Figure 4, 5C). Figure 4C illustrates salt bridges between D755 (αI) of FGFR1-3P and R609’ (βD6) of N-SH2 domain as well as interactions between FGFR1 R609 (αE) and D594’ (βC1) of PLCγ. The acid-base pairs (RD pairs) that are located at the center of the interface formed by the secondary site are nearly parallel to one other. R609’ of the N-SH2 makes an additional polar contact with Y605 of FGFR1-3P. In addition, R609 of FGFR1-3P interacts with the backbone carbonyl oxygens of V592’ (BC4) and S612’ of the N-SH2 domain. Additional contacts exist between a hydrophilic region on loop DE on the N-SH2 domain (Figure 5C) in which water-mediated interactions take place between N-SH2 and FGFR1-3P.
Structure based alignment of the amino acid sequences of the N-SH2 and C-SH2 domains (Figure 5D and S8) of PLCγ revealed a high degree of conservation of the amino acids encoding for the pY binding site and for the hydrophobic pocket. The sequence similarity of the residues comprising the pY binding sites (red) is consistent with in vitro binding experiments of isolated N-SH2 or C-SH2 domains to pY containing synthetic peptides corresponding to PLCγ-binding site of PDGFR-β (pY1021) that have demonstrated similar dissociation constants for the two SH2 domains (Larose et al., 1995). The amino acid sequences responsible for mediating the binding of the secondary site of N-SH2 domain to its binding site on the C-lobe of activated FGFR1 (blue and orange) highly diverge from the corresponding regions in the C-SH2 domain. For example, the sequence TFVG (590’–593’) in the N-SH2 domain (in subsite-I) is replaced in the C-SH2 domain by the sequence EPN (698’–700’). Moreover, among the amino acids of subsite-II of N-SH2 domain only R609’ is conserved in the C-SH2 domain (R716’) and none of the four key amino acids that are involved in water-mediated interactions with the N-SH2 are conserved. In addition, D594’ (βC1) of the N-SH2 domain, an amino acid that forms one of the acid-base pairs (RD pairs) at the center of the interface of the secondary site is substituted by S701’ in C-SH2 (Figure 5D).
The secondary binding site on the N-SH2 domain is responsible for mediating enhanced binding affinity for activated FGFR1
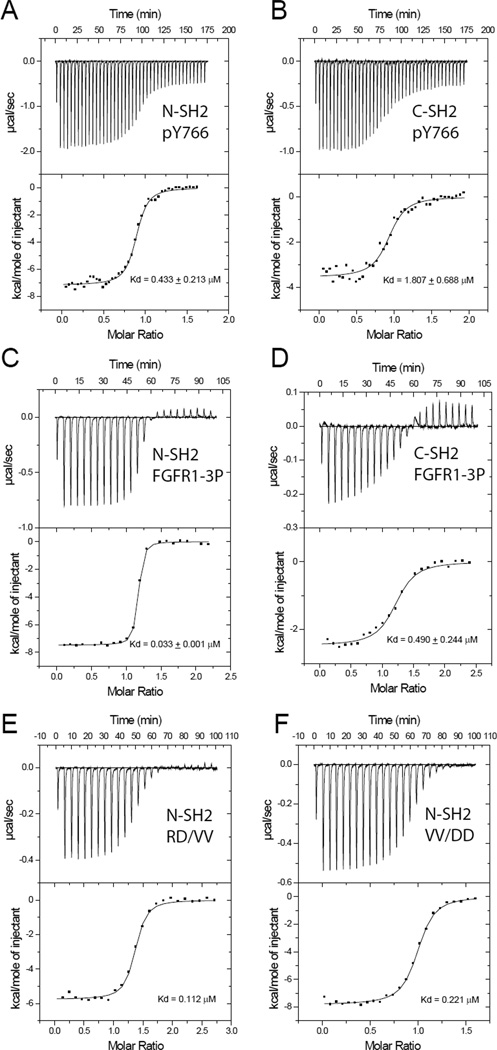
We applied two methods to compare the dissociation constants of each of the SH2 domains both to a synthetic phosphopeptide containing pY766 corresponding to the C-terminal region of FGFR1 (TSNQEpYLDLSM, pY766 peptide) and to the intact kinase domain of activated FGFR1 (FGFR1-3P). We first used isothermal titration calorimetry (ITC) to measure the dissociation constants for pY766 peptide binding to isolated N-SH2 and C-SH2 domains (Figure 6 and Table S2). The N-SH2 and C-SH2 domains bind to the pY766 peptide with dissociation constants of approximately 0.4 µM and 1.8 µM respectively. The C-SH2 domain binds to the intact FGFR1-3P kinase domain with a similar dissociation constant of 0.5 µM. In stark contrast, the N-SH2 domain binds 10 – 40 times more tightly to FGFR1-3P than the binding of the N-SH2 or C-SH2 domain to the pY766 peptide, with a dissociation constant of 33 ± 1 nM.
Figure 6. Isothermal titration calorimetry experiments demonstrate that PLCγ N-SH2 domain secondary binding site is responsible for mediating high affinity interactions.
ITC curves for the interaction of the SH2 domains of PLCγ with either an FGFR1-derived phosphopeptide (pY766) (A, B) or intact phosphorylated FGFR1 kinase domain (FGFR1-3P) (C, D). For the interaction with the pY766 peptide, 42 7-µL injections of biotin-XXTSNQEpYLDLSM-NH2 were titrated into (A) the N-SH2 domain or (B) the C-SH2 domain of PLCγ in the isothermal cell (1.4 mL). For the interaction with the intact kinase domain, 24 12.5-µL injections of (C) the N-SH2 domain or (D) the C-SH2 domain of PLCγ were titrated into FGFR1-3P in the isothermal cell (1.4 mL). The top panels show the baseline corrected data collected at 25 °C in PBS plus 2.5 mM DTT, while the bottom panels show the integrated heats released as a function of the molar ratio of titrant to macromolecule in the cell. The data was corrected for the heat of dilution of the titrant and subsequently, fit to a one-site model by a nonlinear least squares algorithm. (E, F). ITC curves for the interaction of the N-SH2 domain of PLCγ with FGFR1-3P harboring double point mutations FGFR1-3P (RD/VV) (E) or FGFR1-3P (VV/DD) (F). For both runs, 24 12.5-µL injections of the N-SH2 domain of PLCγ were titrated into mutated FGFR1-3P in the isothermal cell (1.4 mL). The extensive thermodynamic parameters obtained are given in Table S2, but briefly, the affinities obtained are as follows: (A) Binding of N-SH2 domain to pY766 peptide: KD = 0.433 ± 0.213 µM, (B) C-SH2 domain to pY766 peptide: KD= 1.807 ± 0.688 µM (C) N-SH2 domain to FGFR1-3P: KD= 0.033 ± 0.001 µM (D) C-SH2 domain to FGFR1-3P: KD = 0.490 ± 0.244 µM. (E) N-SH2 domain to FGFR1-3P (RD/VV): KD = 0.112 µM, and (F) 21 N-SH2 domain to FGFR1-3P (VV/DD): KD = 0.221 µM.
We next asked whether the secondary binding site plays a key role in defining the high affinity binding of the N-SH2 domain for FGFR1-3P. ITC measurements were used to compare N-SH2 domain binding to wild-type FGFR1-3P and to a mutated FGFR1-3P in which critical amino acids in the C-lobe that contact the N-SH2 secondary binding site were altered (Figure 6E, 6F). In one FGFR1-3P variant, V636 and V758, (in the binding site for subsite-I), were replaced by aspartic acid (FGFR1-3P(VV/DD)). In a second mutant, R609 and D755 of FGFR1-3P, (in the binding site for subsite-II), were replaced by valines (FGFR1-3P(RD/VV)). The amino acids are highlighted in Figure 5 with black boxes. Neither alteration altered the FGFR1-3P tyrosine kinase activity: both variants are phosphorylated completely at Y653, Y654, and Y766 (data not shown). The ITC measurements showed that the N-SH2 domain binds to the FGFR1-3P(VV/DD) mutant with a dissociation constant of 0.22 µM and to the FGFR1-3P(RD/VV) mutant with a dissociation constant of 0.11 µM. Thus, altering the target of subsite-I or -II alone in this way reduces N-SH2 binding affinity by 3–6 fold, indicating that both subsites of the secondary interface contribute to the high affinity binding of N-SH2 to activated FGFR1-3P.
In similar experiments using surface plasmon resonance (SPR) spectroscopy, we also found that N-SH2 from PLCγ binds significantly more tightly (~ 40 fold) to FGFR1-3P (0.142 ± 0.02 µM by Biacore 1000; 0.044 µM by Biacore T100) than to an immobilized biotin-conjugated pY766 peptide (5.67 ± 0.60 µM by Biacore 1000). SPR studies also indicated that the C-SH2 domain binds with a similar affinity to the pY766 peptide and FGFR1-3P (Figure S9 and Table S3). This represents an approximately 40 – 70 fold increase in the binding affinity of FGFR1-3P to N-SH2 domain, as compared to binding of the N-SH2 or C-SH2 domains to the pY766 peptide.
While the absolute values of dissociation constants determined by ITC or SPR measurements vary, both methods consistently demonstrated that the binding affinity of the N-SH2 domain to FGFR1-3P is 10 – 70 fold higher than the binding affinities of N-SH2 or C-SH2 domains to the pY766 peptide (Figure S9). Moreover, the binding affinity of FGFR1-3P mutant (in the region responsible for mediating secondary binding site on N-SH2 domain) to isolated C-SH2 or N-SH2 domains is similar to the binding affinities of the two individual SH2 domains to the pY766 peptide. These experiments demonstrate that the high affinity binding of N-SH2 domain to the FGFR1-3P is mediated by the secondary binding site of the PLC-Δ N-SH2 domain.
Complex formation between activated FGFR1 and PLCγ in stimulated cells and FGF stimulation of PtdIns(4,5)P2 hydrolysis depend upon binding of FGFR1 to the secondary binding site on N-SH2 domain
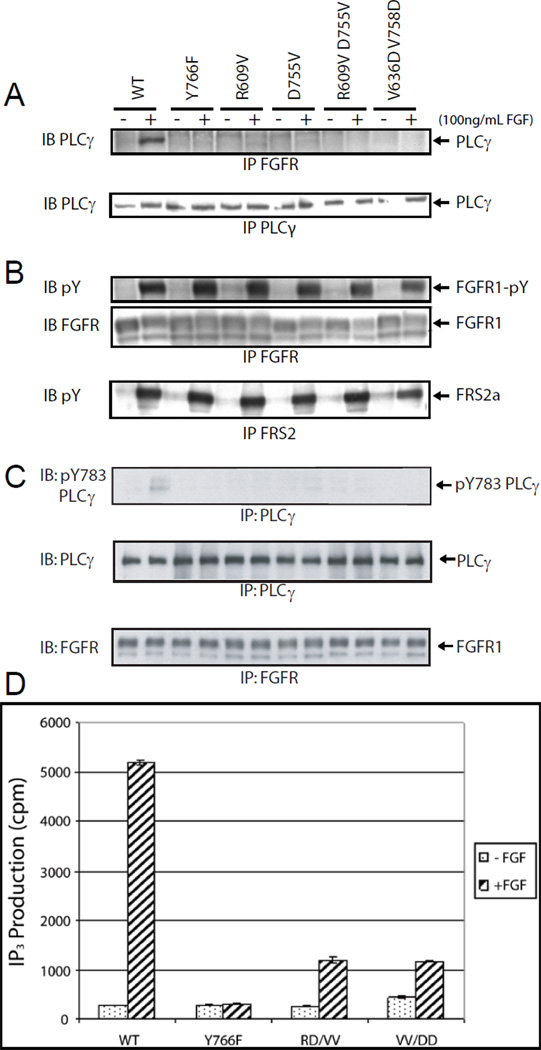
We next tested the physiological significance of the interaction between the C-lobe of FGFR1 and the secondary site on N-SH2 domain by analyzing FGF1-induced biological responses in L6 cells (devoid of endogenous FGFRs) expressing either wild type FGFR1 or FGFR1 mutants with decreased affinity for the N-SH2 domain (Figure 7). We generated L6 myoblasts matched for stable expression levels of wild type or the following FGFR1 mutants; Y766F in the primary binding site for the N-SH2 domain, and single or double mutations in the FGFR1 C-lobe including FGFR1(R609V), FGFR1(D755V), FGFR1(RD/VV) and FGFR1(VV/DD). Lysates from FGF1-stimulated or unstimulated cells were subjected to immunoprecipitation with anti-FGFR1 antibodies or anti-PLCγ antibodies followed by immunoblotting with anti-PLCγ antibodies. The lysates were also subjected to immunoprecipitation with anti-FGFR1 antibodies followed by immunoblotting with either anti-FGFR1 or anti-pY antibodies as well as to immunoprecipitation with anti-FRS2 antibodies followed by immunoblotting with antipY antibodies. Although FGF-stimulation promotes robust complex formation between activated WT FGFR1 and PLCγ, (Figure 7A, top left two lanes), complex formation with PLCγ could not be detected in FGF1 stimulated cells expressing the FGFR1(Y766F) mutant or FGFR1 with mutations in amino acids of the C-lobe (secondary interface) that function as binding sites for the secondary binding sites on the N-SH2 domain (Figure 7A). Tyrosine autophosphorylation of each FGFR1 mutant was similar to that seen for WT FGFR1 (Figure 7B), and similar levels of FGF1-induced stimulation of FRS2 tyrosine phosphorylation were seen in cells expressing WT or FGFR1 mutants. Thus, neither the single nor double mutations affected the integrity and tyrosine kinase activity of FGFR1.
Figure 7. PLCγ N-SH2 domain secondary site is required for complex formation with FGFR1, tyrosine phosphorylation of PLCγ and for FGF induced PtdIns(4,5)P2 hydrolysis.
L6 myoblasts stably expressing WT FGFR1 or FGFR1 harboring single point mutations FGFR1(Y766F), FGFR1(R609V), FGFR1(D755V) or double point mutations FGFR1(RD/VV) or FGFR1(VV/DD) were stimulated with 100 ng/mL FGF for 5 min at 37 °C. (A). Lysates from unstimulated or FGF1-stimulated cells were subject to immunoprecipitation with anti-FGFR1 or anti-PLCγ antibodies followed by SDS-PAGE and immunoblotting with anti-PLCγ antibodies. (B) Lysates from unstimulated or FGF1-stimulated cells were subjected to immunoprecipitation with anti-FGFR1 or anti-FRS2 antibodies followed by SDS-PAGE and immunoblotting with anti-FGFR1 or anti-phosphotyrosine antibodies as indicated. (C) Lysates from unstimulated or FGF1-stimulated cells were subject to immunoprecipitation with anti-FGFR1 or anti-PLCγ antibodies followed by SDS-PAGE and immunoblotting with anti-pY783 (PLCγ), anti-PLCγ, or anti-FGFR1 antibodies as indicated. (D) Mutations of amino acids in the C-lobe of FGFR1 that interact with PLCγ N-SH2 domain result in decreased FGF1-mediated inositol-(1,4,5)-trisphosphate (IP3) generation. IP3 accumulation (Kim et al. 1991) in L6 myoblast cells stably transfected with either WT FGFR1, FGFR1 harboring a single point mutation, FGFR1(Y766F), or double point mutations, FGFR1(VV/DD) or FGFR1 (RD/VV). Cells were left unstimulated (dotted bar) or stimulated (striped bar) with 100 ng/mL acidic FGF1 for 30 min at 37 °C. Data represent the average of triplicate experiments and shown is the average IP3 generation ± standard error.
We also examined the tyrosine phosphorylation of PLCγ in complex with activated FGFR1 by subjecting lysates from unstimulated or FGF1 stimulated cells to immunoprecipitation with anti-FGFR1 antibodies followed by immunoblotting with either anti-PLCγ or antibodies directed against a synthetic peptide corresponding to pY783 of PLCγ. The experiment presented in Figure 7C shows tyrosine phosphorylation of PLCγ in FGF1 stimulated cells expressing wild type FGFR1 while negligible tyrosine phosphorylation of PLCγ was detected in FGF1 stimulated cells expressing the FGFR1(Y766F) mutant or the FGFR1 mutants in which critical amino acids in the C-lobe of the tyrosine kinase domain that serve as binding sites for the N-SH2 domain of PLCγ were mutated. We therefore conclude that both the primary and secondary binding sites of N-SH2 domain with FGFR1 are required for FGF1 induced recruitment and tyrosine phosphorylation of PLCγ by activated FGFR1 in living cells. Moreover, binding of activated FGFR1 to each of the primary or secondary binding sites on N-SH2 is required for complex formation with and tyrosine phosphorylation of PLCγ.
We next asked whether interactions between the C-lobe of FGFR1 and the secondary site on N-SH2 domain are required for PLCγ activation, by comparing FGF1-induced stimulation of PtdIns(4,5)P2 hydrolysis in L6 cells expressing WT FGFR1, FGFR1(Y766F), FGFR1(RD/VV) or the FGFR1(VV/DD) mutant. L6 cells were labeled with [3H]inositol in DMEM (Dulbecco’s Modified Eagle’s Medium) containing 0.5 % FBS for 24 hours at 37 °C and incubated with LiCl for 20 minutes prior to addition of FGF1 or buffer alone for additional 30 minutes at 37 °C. The experiment presented in Figure 7D shows robust FGF1-induced inositol-1,4,5-trisphosphate (IP3) accumulation in L6 expressing WT FGFR1. In contrast, FGF1 induced IP3 accumulation was strongly compromised in cells expressing either the FGFR1(Y766F) mutant, in which the target for the primary binding site is mutated, or in cells expressing the FGFR1(RD/VV) or FGFR1(VV/DD) mutants, where targets for each of the subsites of the secondary binding site on N-SH2 are mutated.
These experiments show that FGF1 stimulation of PLCγ activation and IP3 accumulation depend upon PLCγ recruitment by activated FGFR1 through binding and interactions mediated by the primary and secondary binding sites of the N-SH2 domain; binding to each of the sites is required for complex formation with and tyrosine phosphorylation and activation of PLCγ. However, while the initial recognition of activated FGFR is likely mediated by weak interactions mediated by the primary binding site of the SH2 domains, the selectivity of the N-SH2 towards complex formation with activated FGFR1 is determined by interactions between the secondary binding site of NSH2 domain with amino acids on the C-lobe of the activated FGFR1.
DISCUSSION
An important step in cell signaling by RTKs is recruitment and activation of cellular signaling proteins by means of SH2 domain binding to tyrosine phosphorylation sites in the cytoplasmic regions of RTKs or to tyrosine phosphorylation sites on closely associated docking proteins. It is widely accepted that a conserved binding site on all SH2 domains is responsible for binding to phosphorylated tyrosine residues (pY) and that the specificity of complex formation with pY containing peptides is mediated – at least in part - by binding of a variable region in the SH2 domain to side-chains of amino acids C-terminal to the pY moiety primarily at the P+1 and P+3 positions. It has previously been proposed that the modest selectivity of SH2 domain towards pY containing peptides (5 – 20 fold) may not account for the selectivity of SH2 domain containing cell signaling proteins towards binding to activated RTKs in vivo in their natural environment. Indeed, the dissociation constants of the N-SH2 or the C-SH2 domains of PLCγ to a variety of pY containing peptides (with a limited degree of sequence similarity) corresponding to tyrosine phosphorylation sites of PDGFR, FGFR and EGFR are all within the micromolar (µM) range. In contrast, in vivo, the N-SH2 and C-SH2 domain of PLCγ exhibit a very strict selectivity towards tyrosine phosphorylation sites on two different proteins. While the N-SH2 domain of PLCγ binds selectively to a pY containing sites in the C-terminal tails of PDGFR, FGFR, and other activated RTKs, the C-SH2 domain of PLCγ binds intramolecularly to pY783 on PLCγ; a step shown to be required for a conformational change essential for stimulation of PLCγ enzymatic activity.
Here we describe the molecular basis underlying the selectivity of the N-SH2 of PLCγ for activated FGFR1 by determining the crystal structure of tyrosine phosphorylated, activated FGFR1 in complex with a fragment of PLCγ composed of the two tandem SH2 domains with a segment containing the phosphorylation site Y783. The structure shows that, in addition to the canonical pY binding site found in all SH2 domains, the N-SH2 domain of PLCγ utilizes an additional region separate from its canonical pY binding site for binding to a region in the C-lobe of the tyrosine kinase domain of FGFR1 in a phosphorylation-independent manner. The buried surface area in this secondary site is similar to that occluded in the canonical primary binding site that binds pY. We also employed isothermal titration calorimetry and surface plasmon resonance measurements to demonstrate that the dissociation constant of the N-SH2 domain to activated FGFR1 domain is as low as 33 nM, which is 10 – 70 fold tighter than the N-SH2 or C-SH2 domain binding to a pY766-containing peptide. Activated FGFR1 variants harboring mutations in C-lobe amino acids that bindthe N-SH2 domain secondary binding site show significantly reduced N-SH2 binding affinity. Moreover, when these interactions are similarly disrupted in cells, tyrosine phosphorylation of PLCγ and complex formation between PLCγ and activated FGFR1 in FGF1 stimulated cells were strongly compromised. The loss of these interactions was accompanied by inhibition of FGF-stimulated PtdIns(4,5)P2 hydrolysis, which we show requires the integrity of the secondary binding site on the PLCγ N-SH2 domain.
We propose that the mechanism described in this report that controls the binding affinity and selectivity of the N-SH2 and C-SH2 domains towards their specific pY binding sites on activated FGFR1 and tyrosine phosphorylated PLCγ may also control the binding affinity and selectivity of other SH2 domains towards activated RTKs, tyrosine phosphorylated docking proteins and other targets of SH2 domains. Indeed, structure based alignment of amino acid sequences of SH2 domains has shown that the amino acids that function as the secondary binding site on the N-SH2 domain of PLCγ are among the most variable regions in SH2 domain sequences (Figure 5D and S8). The variability in the amino acid residues that constitute the secondary binding sites on SH2 domain may provide sufficient diversity to enable binding to different sites in proteins in a phosphorylation independent manner to cooperate with interactions mediated by the canonical SH2 domain binding site to pY sites resulting in establishment of selective, stable and physiologically relevant interactions. In this model, the primary role of tyrosine phosphorylaton (and the primary binding site on the SH2 domain) might be to define temporal specificity, with the secondary site playing a crucial role in target specificity.
On the basis of the structural analysis, quantitative binding experiments, and comparison of the biological activities of wild type and FGFR1 mutants it is possible to draw several molecular steps that may take place during PLCγ activation in response to FGF1 stimulation. Accordingly, following FGF stimulation and pY766 autophosphorylation, PLCγ forms a complex with the C-terminal tail of FGFR1 through binding of either the N-SH2 or the C-SH2 domains to pY766. Since the C-SH2 domain of PLCγ is devoid of a secondary binding site, all PLCγ molecules bound to pY766 via their C-SH2 domain will eventually dissociate. Complex formation between FGFR1 and PLCγ at steady state is therefore entirely mediated by binding of the N-SH2 domain to the activated FGFR1 through cooperation between the primary and secondary binding sites to pY766 and to the C-lobe of the tyrosine kinase domain of FGFR1, respectively. The enhanced catalytic activity of FGFR1 caused by autophosphorylation of Y654 in the activation segment (Furdui et al., 2006) leads to tyrosine phosphorylation of Y783 on PLCγ; a pY site which binds to C-SH2 domain intramolecularly resulting in a conformational change essential for stimulation of PLCγ enzymatic activity.
It is possible that the selectivity of PLCγ C-SH2 domain to pY783 is aided by additional interactions between a secondary binding site on C-SH2 domain with an as yet to be identified region on PLCγ However, this may not be necessary as a dissociation constant in a µM range may be sufficient to drive interactions of pY783 and its binding partner, the C-SH2 domain, that is located on the same PLCγ molecule. The high local concentration caused by reduced dimensionality of C-SH2 domain binding to pY783 on the same molecule may substitute for the high affinity interactions generated by the cooperative interactions mediated by binding of the primary and secondary sites of PLCγ N-SH2 domain to activated FGFR1. While some SH2 domains may require cooperation between their primary and secondary binding sites in order to be able to mediate a specific biological response, other SH2 domains may cooperate with other targeting signals on the same or another signaling protein in order to achieve high affinity binding (Donaldson et al, 2002; Groesch et al, 2006; Chan et al, 2003; Filippakopoulos et al, 2008). Such additional interactions mediated by SH3, PTB and PH domains on the same signaling molecule may decrease the overall apparent dissociation constant of the signaling molecules from µM (SH2 domain mediated canonical interactions) to low nM or even pM range. It is worth drawing analogy with the high affinity binding of the SH3 domain of Fyn to a PxxP motif on HIV-1 Nef and by interactions between the RT loop of the SH3 domain with an additional site on Nef (Lee et al, 1996). Finally, the experiments presented in this report demonstrating that short pY peptides do not recapitulate SH2 domain mediated interactions with native targets and an SH2 mediated cell signaling event in living cells raise doubts about the biological significance of the establishment of RTK dependent signaling networks determined by proteomic micro array analysis of SH2 domain interactions with short pY peptides (Jones et al, 2006).
MATERIALS AND METHODS
Protein expression and Purification
Expression and purification of unphosphorylated FGFR1 (aa458-774), FGFR1-3P, and FGFR1 mutants were performed as previously described (Furdui et al., 2006). For the PLCγ construct, the cDNA encoding residues 541’ – 790’ of the rat PLCγ was sub-cloned into the expression vector pET41b+ (Novagen) with the restriction sites, NdeI and HindIII. The pET41b+(2SH2-PLCγ) was transformed into E. coli strain BL21, and cultures were grown in terrific broth (TB) media at 37 °C to an OD600 of 0.8 and induced with 1 mM isopropyl-thiogalactopyranoside (IPTG) at 18 °C for 10 hours. Cells were harvested and resuspended in lysis buffer (20 mM Tris-HCl pH 8.0, 20 mM NaCl, and 2mM phenylmethyl-sulphonyl fluoride (PMSF)) then lysed by French press followed by centrifugation to remove cellular debris. The protein was first isolated by ion-exchange chromatography on Source Q beads (GE Healthcare) and eluted with a NaCl gradient up to 1M. The eluted sample was subsequently subjected to size-exclusion chromatography using Superdex-200 (S200, GE Healthcare), and further purified by Mono-Q (GE Healthcare) ion-exchange chromatography. The purity and mass of the purified protein was verified by electrospray mass spectroscopy.
Crystallization and structure determination
For crystallization, FGFR1-3P and PLC-Δ were mixed in a 1:1.2 molar ratio and concentrated to 10 mg/ml. Protein sample was transferred to the buffer containing 1mM AMP-PCP, 6mM MgCl2, 2mM tris[2-carboxyethyl]phosphine hydrochloride (TCEP-HCl), 20 mM Tris pH 8.0, 100 mM NaCl, and 2 mM sodium orthovanadate (Na3VO4) and subjected to screening and optimization. Crystals of the ternary complex were obtained at 4 °C in 7 days using the hanging drop technique containing equal volumes of protein solution and reservoir buffer (6 – 8 % [w/v] polyethylene glycol 8000, 100 mM Tris pH 8.0 – 8.8, 10 mM taurine). The crystals belonged to the centered monoclinic space group C2 with unit cell dimensions of a = 133.9 Å, b = 64.6 Å, c = 94.2 Å, and β = 96.5°. There was one 1:1 FGFR1-3P/PLC-Δ complex bound with an AMP-PCP and one Mg2+ in the asymmetric unit. The solvent content of the complex was around 45%. Crystals were transferred into the cryoprotectant containing reservoir buffer with 25% glycerol then flash frozen in liquid nitrogen. Data were collected on the beamline X25 at the National Synchrotron Light Source and were processed by using HKL2000 (Minor et al., 2000). A molecular replacement solution for FGFR1 was found with Phaser (McCoy et al., 2007) using the structures of inactive FGFR1 (Mohammadi et al., 1996; PDB code: 1FRK) and the unpublished PLC-Δ structure as the search models (the search model of PLC-Δ is unfinished due to the data quality). The coordinates of AMP-PCP and decavanadate (DVT) were obtained from HIC-Up server to be docked into the structure (Kleywegt, 2007). Strong difference density near the N-lobe of FGFR1-3P was observed. We assigned it as a DVT cluster. Multiple rounds of the model building and the refinement of FGFR1-3P/PLC-Δ were carried out with Coot (Emsley and Cowtan, 2004) and CNS (Brünger et al., 1998) to a crystallographic R and Rfree of 24.9 % and 28.9 %, respectively. Four amino acids (541’–544’) on the N-terminus and twenty amino acids at the C terminus (771’–790’) of PLC-Δ are disordered. Figures were prepared using PYMOL (www.pymol.org). Interacting interface areas for the primary and secondary interfaces between FGFR1-3P/N-SH2 as well as for pY766 (residues from 762 to 770):N-SH2 were calculated with PDBsum (Laskowski et al., 1997). Detailed structure determination and refinement procedure are described in supplemental material. Data and refinement statistics are summarized in Table S1.
Isothermal Titration Calorimetry
ITC studies were performed on a MicroCal VP-ITC (Northampton, MA) at 25 °C. An initial delay of 60s was set with an additional 250s delay between injections to allow for signal to return to baseline. In general, proteins were dialyzed in PBS plus 2.5 mM DTT overnight, and peptide was subsequently dissolved in same buffer. For peptide experiments, an initial 3.5 µL titration of phoshopeptide was performed, which was followed by 42.7 µL titrations of phosphopeptide (800-µM) into the macromolecule [N-SH2 (110.6 µM) or C-SH2 domain (92.7 µM)] in a 1.4 mL isothermal cell. For binding to the intact FGFR1-3P-WT or mutant kinase domain, an initial 5 µL injection of either the N-SH2 domain (311 µM) or the C-SH2 domain (340 µM) was titrated into FGFR1-3P (34 µM), followed by 24 – 12.5 µL injections. The data was fit by a non-linear least squares method to a single independent site binding model using ORIGIN 5.0. Background reaction enthalpy due to titrant dilution or buffer dilution of macromolecule was determined from the last 5 – 10 injections, and data were corrected for background heat release prior to curve fitting. Concentrations of both protein and phosphopeptide were determined by amino acid analysis.
Cell culture, Immunoprecipitation and Immunoblotting experiments
A retroviral vector, pBABE, containing a puromycin resistance gene was utilized to make stable cell lines expressing FGFR1-WT and various FGFR1 mutants in L6 Myoblasts. Full details are described in Figure legends and in previous publications (Lax et al., 2002).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01-AR051448, R01-AR051886, and P50-AR054086 to J.S. We thank Titus Boggon, Mark Lemmon, Kate Ferguson, Benjamin Turk, David Calderwood, Valsan Mandiyan, Ewa Folta-Stogniew, Cristina Furdui and Steve Hubbard for their support. and members of Brookhaven National Laboratory Beamlines X6, X25, and X29 for their support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The prime mark is used throughout the text to distinguish PLC-Δ residues.
REFERENCES
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS, et al. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A, Vecchi M, Ji Q, Mernaugh R, Carpenter G. The role of individual SH2 domains in mediating association of phospholipase C-γ1 with the activated EGF receptor. J. Biol. Chem. 1999;274:26091–26097. doi: 10.1074/jbc.274.37.26091. [DOI] [PubMed] [Google Scholar]
- Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat. Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and the Crk-derived phophopeptide. Proc. Natl. Acad. Sci. USA. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;D(60):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat. Struct. Biol. 2001;8:1038–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Kofler M, Hantschel O, Gish GD, Grebien F, Salah E, Neudecker P, Kay LE, Turk BE, Superti-Furga, et al. Structural coupling of SH2-kinase domains links FES and Abl substrate recoginition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 Kinase Is Mediated by a Sequential and Precisely Ordered Reaction. Mol. Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Groesch TD, Zhou F, Mattila S, Geahlen RL, Post CB. Structural Basis for the requirement of two phosphotyrosine residues in signaling mediated by Sky tyrosine kinase. J. Mol. Bio. 2006;356:1222–1236. doi: 10.1016/j.jmb.2005.11.095. [DOI] [PubMed] [Google Scholar]
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Chattopadhyay A, Vecchi M, Carpenter G. Physiological requirement for both SH2 domains for phospholipase C-γ1 function and interaction with platelet-derived growth factor receptors. Mol. Cell. Biol. 1999;19:4961–4970. doi: 10.1128/mcb.19.7.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN, Noble MEM, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85(2):149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB3 receptors using protein micro arrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- Kim HK, Kim JW, Zilberstein A, Margolis B, Kim JG, Schlessinger J, Rhee SG. PDGF stimulation of inositol phospholipid hydrolysis requires PLC-gamma 1 phosphorylation on tyrosine residues 783 and 1254. Cell. 1991;65(3):435–441. doi: 10.1016/0092-8674(91)90461-7. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ. Crystallographic refinement of ligand complexes. Acta Cryst. 2007;D(63):94–100. doi: 10.1107/S0907444906022657. (CCP4 Proceedings) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury JE, Arold S. Searching for specificity in SH domains. Chem. Biol. 2000;7(1):R3–R8. doi: 10.1016/s1074-5521(00)00067-3. [DOI] [PubMed] [Google Scholar]
- Larose L, Gish G, Pawson T. Construction of an SH2 domain-binding site with mixed specificity. J. Biol. Chem. 1995;270(8):3858–3862. doi: 10.1074/jbc.270.8.3858. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: A Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- Lax I, Wong A, Lamothe B, Lee A, Frost A, Hawes J, Schlessinger J. The docking protein FRS2alpha controls a MAP kinase-meidated negative feedback mechnism for signaling by FGF receptors. Mol. Cell. 2002;10(4):709–719. doi: 10.1016/s1097-2765(02)00689-5. [DOI] [PubMed] [Google Scholar]
- Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins--establishing the boundaries of pY signaling. Mol. Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Gross-Kunstleve RW, Adams PD, Winn MD, Stroni LC, Read RJ. Phaser Crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Tomchick D, Otwinowski Z. Strategies for macromolecular synchrotron crystallography. Structure. 2000;8:R105–R110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Piccione E, Case R, Domchek S, Hu P, Chaudhuri M, Backer J, Schlessinger J, Shoelsen S. Phosphatidylinositol 3-kinase p85 SH2 domain specificity defined by direct phosphopeptide/SH2 domain binding. Biochemistry. 1993;32:3197–3202. doi: 10.1021/bi00064a001. [DOI] [PubMed] [Google Scholar]
- Poirot O, O’Toole E, Notredame C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31(13):3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin B, Sekiya F, Rhee SG. Differential roles of the Src homology 2 domains of phospholipase C-γ 1 (PLC-γ 1) in platelet-derived growth factor-induced activation of PLC-γ 1 in intact cells. J. Biol. Chem. 2000;275:6411–6416. doi: 10.1074/jbc.275.9.6411. [DOI] [PubMed] [Google Scholar]
- Poulin B, Sekiya F, Rhee SG. Intramolecular interaction between phosphorylated tyrosine-783 and the C-terminal Src homology 2 domain activates phospholipase C-γ 1. Proc. Natl. Acad. Sci. USA. 2005;102:4276–4281. doi: 10.1073/pnas.0409590102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG. Regulation of phsophoinositide-specific phopholipase C. Annu. Rev. Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE. re12. 2003 doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.