SUMMARY
In bacteria, the actin-like FtsA protein interacts with the tubulin-like FtsZ protein, helping to assemble the cytokinetic Z ring, anchor it to the cytoplasmic membrane, and recruit other essential divisome proteins. FtsA also interacts with itself, but it is not clear whether this self-interaction is required for its full functionality. Here we describe new dominant negative missense mutations in Escherichia coli ftsA that specifically inhibit FtsA homodimerization and simultaneously cause disruption of Z rings. The negative effects of one mutation, M71A, were suppressed by altering levels of certain division proteins or by additional mutations in ftsA that promote increased integrity of the Z ring. Remarkably, when FtsA, FtsA-M71A, and other mutants of FtsA that compromise self-interaction were connected in a tandem repeat, they were at least partially functional and suppressed defects of an ftsZ84(ts) mutation. This gain of function by FtsA tandems further suggested that FtsA monomers cause deleterious interactions with FtsZ and that increased dimerization or oligomerization of FtsA enhances its ability to promote Z-ring integrity. Therefore, we propose that FtsZ assembly is regulated by the extent of FtsA oligomerization.
Keywords: FtsZ, FtsA, cell division, dimer
INTRODUCTION
In Escherichia coli, the tubulin-like FtsZ protein localizes to the midcell division site and assembles into the Z ring (Bi and Lutkenhaus, 1991; Ma et al., 1996), which then recruits at least ten other proteins that comprise the transmembrane protein complex often called the divisome (Margolin, 2005). These cell division proteins arrive at the Z ring in two stages, with FtsZ, FtsA, and ZipA arriving early, and the remainder (FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, and FtsN) localizing later (Aarsman et al., 2005; Goehring and Beckwith, 2005). The Z ring is probably a network of bundled FtsZ protofilaments, which, like microtubules, assemble in the presence of GTP.
Two essential cell division proteins, FtsA and ZipA, anchor the Z ring to the cytoplasmic membrane. ZipA is a transmembrane protein (Hale and de Boer, 1997), whereas FtsA is a cytoplasmic protein with an amphipathic helix that acts as a membrane targeting sequence (MTS) at its C-terminus (Pichoff and Lutkenhaus, 2005). Both FtsA and ZipA contact FtsZ via a conserved sequence at the C terminus of FtsZ (Hale et al., 2000; Ma and Margolin, 1999; Yan et al., 2000). In the absence of FtsA or ZipA, Z rings can assemble at mid-cell. However, in the absence of both FtsA and ZipA, new Z rings are not formed and pre-formed Z rings are destabilized (Pichoff and Lutkenhaus, 2002), indicating that both FtsA and ZipA enhance the integrity and stability of the Z ring. ZipA can suppress the thermosensitivity of ftsZ84(ts), a mutation in ftsZ that severely destabilizes Z rings at the nonpermissive temperature. ZipA probably accomplishes this by promoting bundling of FtsZ protofilaments (Hale et al., 2000; Raychaudhuri, 1999). FtsA displays some functional overlap with ZipA, but is likely the more important cell division protein because it is more highly conserved and because an ftsA allele (R286W), called FtsA*, can bypass a deletion of zipA (Geissler et al., 2003).
The three-dimensional structure of Thermotoga maritima FtsA (van Den Ent and Lowe, 2000) shows that FtsA is a distant homolog of actin and consists of four domains: 1A, 1C, 2A, and 2B. Domain 1C is required for recruitment of late stage divisome proteins (Corbin et al., 2004; Rico et al., 2004). Domain 2B may be required for interaction with FtsZ (Geissler et al., 2007; Pichoff and Lutkenhaus, 2007). FtsA lacks domain 1B, which is conserved in actin and another bacterial actin homolog MreB (van den Ent et al., 2001), and is required for polymerization into protofilaments.
It is not yet known whether FtsA oligomerizes in vivo like other actin homologs. Nevertheless, E. coli FtsA self-interaction has been demonstrated in yeast two-hybrid (Pichoff and Lutkenhaus, 2007; Rico et al., 2004; Yim et al., 2000) and bacterial two-hybrid (Di Lallo et al., 2003; Karimova et al., 2005) assays. In addition, FtsA formed dimers in Bacillus subtilis (Feucht et al., 2001) and purified Streptococcus pneumoniae FtsA polymerized in vitro in the presence of ATP and Mg2+ (Lara et al., 2005). A recent model for FtsA dimers (Carettoni et al., 2003) predicted that specific portions of domains 1A, 1C, and 2B form a dimer interface. Domain 1C was subsequently shown to be required for recruitment of later division proteins as well as FtsA-FtsA interactions (Rico et al., 2004). To understand more about FtsA-FtsA interactions, without the complication of downstream protein recruitment, we focused on domain 1A, which has not yet been characterized. We first introduced a mutation (M71A) in domain 1A and characterized the mutant FtsA. Here, we provide evidence that domain 1A is specifically involved in FtsA-FtsA interactions, and show that these interactions are crucial for the integrity of the Z ring.
RESULTS
The M71A mutation prevents FtsA function
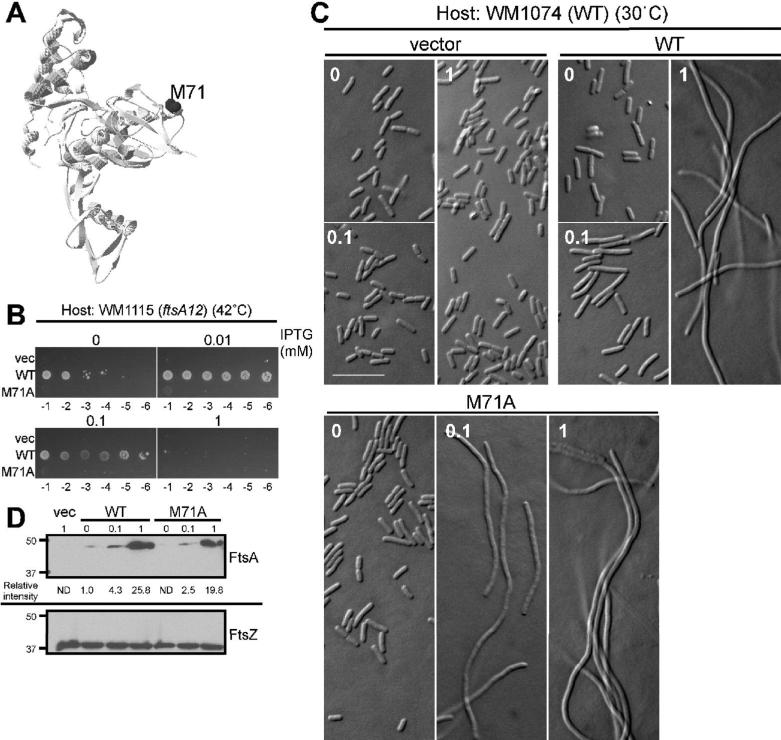
As an first step to test whether dimerization of FtsA is directly required for cell division and whether domain 1A is involved, we replaced Met71 and Val79 in domain 1A, which were postulated to be key residues for dimerization (Carettoni et al., 2003), with Ala (Fig. 1A). The V79A mutation did not affect the function of FtsA (data not shown), whereas FtsA-M71A failed to complement the ftsA12(ts) mutant strain WM1115 (Fig. 1B). Therefore, we characterized FtsA-M71A further.
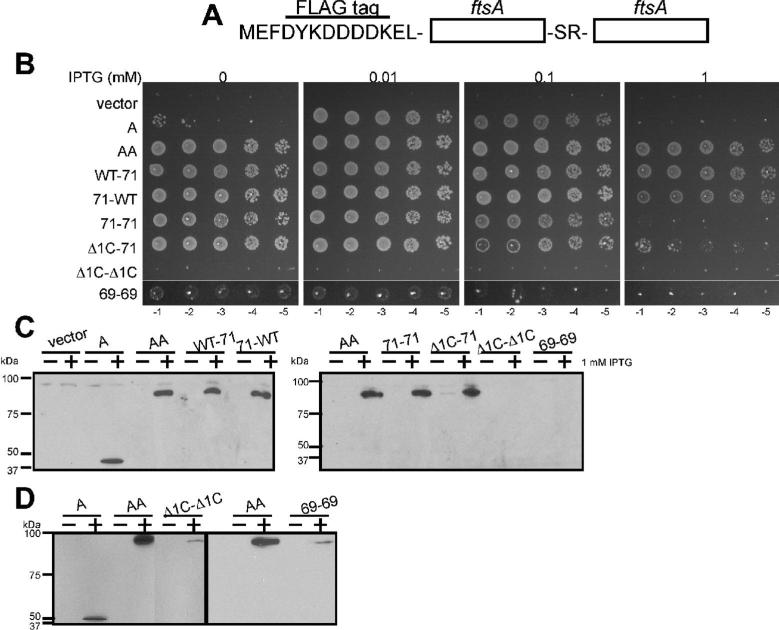
Fig. 1. FtsA-M71A has a dominant negative phenotype.
(A) Mapping of Met71 of E. coli FtsA on the crystal structure of T. maritima FtsA. (B) Complementation of WM1115 (ftsA12ts) by FtsA (pWM2669) or FtsA-M71A (pWM2671) at 42°C. Each strain was grown at 30°C for 4 h in the absence of IPTG prior to spotting on plates, which contained indicated amounts of IPTG and were incubated at 42°C overnight. (C) DIC micrographs of WM1074 cells carrying pWM2780 (vector), pWM2669 (FtsA), or pWM2671 (FtsA-M71A) in the presence of the indicated concentrations of IPTG (mM). Scale bar is 10 μm. (D) Immunoblot of WM1074 cells pWM2780 (vector), pWM2669 (FtsA), or pWM2671 (FtsA-M71A) grown with the indicated concentrations of IPTG (mM) was probed with anti-FLAG (M2) (top) and anti-FtsZ (bottom).
WT ftsA and ftsA-M71A genes with N-terminal FLAG tags were expressed from an IPTG-inducible promoter in plasmids pWM2669 and pWM2671, respectively. The tags, which were used for immunodetection, did not detectably affect function, and for simplicity they will not be included in the descriptions of FtsA derivatives. As shown in Fig. 1B, WT FtsA complemented WM1115 at 42°C in the presence of 0.01 and 0.1 mM IPTG, but not at higher levels of FtsA (1 mM IPTG). This was expected, as the toxicity of FtsA overproduction is well known (Dai and Lutkenhaus, 1992; Dewar et al., 1992). Unlike WT FtsA, however, FtsA-M71A failed to complement ftsA12(ts) at any concentration of IPTG (Fig. 1B).
To address whether FtsA-M71A is more toxic than equivalent levels of WT FtsA, we observed the cell division phenotype of WT cells (WM1074) producing extra WT FtsA or FtsA-M71A (Fig. 1C and Table 1). Even in the absence of IPTG, WM1074 cells carrying pWM2671 (FtsA-M71A) were slightly but significantly longer than cells carrying pWM2780 (vector) or pWM2669 (WT FtsA). With 0.1 mM IPTG, cells producing FtsA-M71A became highly filamentous, whereas cells producing FtsA were only slightly longer than those in the absence of IPTG. With 1 mM IPTG, however, even cells producing FtsA became highly filamentous. Immunoblot analysis confirmed that protein levels of FtsA-M71A were equivalent to or slightly less than those of FtsA at each IPTG concentration (Fig. 1D, top), indicating that FtsA-M71A inhibits cell division at significantly lower protein levels than WT FtsA. These results suggest that FtsA-M71A failed to complement ftsA12(ts) because of a partial loss of function.
Table 1.
Average cell length(a) (μm) of WM1074 cells (WT) producing extra FtsA or mutant FtsA proteins.
| IPTG (mM) | 0 | 0.01 | 0.1 | 1 |
|---|---|---|---|---|
| Vector (pWM2780) | 3.23±0.68 (n=109) | 2.78±0.52 (n=113) | 2.86±0.52 (n=105) | 2.76±0.57 (n=110) |
| FtsA (WT) (pWM2669) | 3.24±0.59 (n=107) | 3.33±0.72 (n=113) | 5.29±1.62 (n=120) | ND |
| M71A (pWM2671) | 4.00±1.09 (n=125) | 5.22±1.53 (n=151) | 22.99±12.70 (n=108) | ND |
Cells that had a visible septum were not counted.
ND: not determined
To determine whether the failure of FtsA-M71A to complement was a result of an inability to localize to the Z ring, we examined localization of GFP-FtsA-M71A. This fusion localized proficiently to the Z ring in the absence of functional FtsA (Fig. S1), indicating that the M71A mutation did not significantly affect the normal targeting of FtsA to the Z ring. Next, to determine whether the M71A mutation affected recruitment of later divisome proteins, we examined whether FtsA-M71A is able to recruit the late-stage protein FtsN by using a DivIVA-FtsA polar recruitment assay (Corbin et al., 2004). When DivIVA-FtsA was overproduced and localized to cell poles, GFP-FtsN also was forced to localize to poles as expected (Fig. S2A). Also as expected, DivIVA-FtsA-Δ1C did not recruit GFP-FtsN to cell poles, because domain 1C is required for recruitment of FtsN (Corbin et al., 2004; Rico et al., 2004). Immunoblotting indicated that DivIVA-FtsA-Δ1C was produced at levels similar to DivIVA-FtsA (Fig. S2B). Like DivIVA-FtsA, DiviVA-FtsA-M71A was able to re-localize GFP-FtsN to cell poles and was produced at levels similar to DivIVA-FtsA (Fig. S2A-B), indicating that the M71A mutation did not significantly inhibit the recruitment of FtsN.
We also used a bacterial two-hybrid assay (see below) to examine the interaction between FtsA and FtsN. We detected an interaction between FtsN and either FtsA or FtsA-M71A, although the FtsN interaction with FtsA-M71A was slightly weaker than its interaction with FtsA (Table 2). We cannot rule out the possibility that one or more other later divisome proteins are not recruited by FtsA-M71A. Nevertheless, the results suggest that the failure of FtsA-M71A to complement is not simply because it cannot recruit later proteins.
Table 2.
Summary of the bacterial two-hybrid assays
| T18 | T25 | β-galactosidase activity (unit)a Mean±SE | P valueb |
|---|---|---|---|
| none (pUT18C) | none (pKT25) | 199.2±12.20 (n=20) | NA |
| FtsA self-interactions | |||
| WT | WT | 1777±132.9 (n=15) | <0.0001 (vs. none-none) |
| M71A | M71A | 1133±88.52 (n=18) | <0.0001 (vs. none-none) |
| 0.0002 (vs. WT-WT) | |||
| FtsA* | FtsA* | 5150±1417 (n=9) | <0.0001 (vs. none-none) |
| 0.0054 (vs. WT-WT) | |||
| M71A+A* | M71A+A* | 2704±255.9 (n=9) | <0.0001 (vs. none-none) |
| 0.0018 (vs. WT-WT) | |||
| Δ1C | Δ1C | 232.7±10.52 (n=9) | 0.0988 (vs. none-none) |
| E69P | E69P | 569.5±54.20 (n=20) | <0.0001 (vs. none-none) |
| <0.0001 (vs. WT-WT) | |||
| <0.0001 (vs. M71A-M71A) | |||
| FtsA-FtsZ interactions | |||
| none (pUT18C) | FtsZ | 225.2±12.03 (n=10) | NA |
| WT FtsA | FtsZ | 1478±76.73 (n=10) | NA |
| FtsA-M71A | FtsZ | 1368±55.68 (n=10) | 0.2639 (vs. FtsA-FtsZ) |
| FtsA* | FtsZ | 2267±115.1 (n=10) | <0.0001 (vs. FtsA-FtsZ) |
| FtsA-M71A+A* | FtsZ | 1821±68.68 (n=10) | 0.0037 (vs. FtsA-FtsZ) |
| FtsA-Δ1C | FtsZ | 707.5±41.61 (n=10) | <0.0001 (vs. FtsA-FtsZ) |
| FtsA-E69P | FtsZ | 1081±140.5 (n=10) | 0.0233 (vs. FtsA-FtsZ) |
| FtsA-FtsN interactions | |||
| none (pUT18C) | FtsN | 249.6±22.20 (n=18) | NA |
| WT FtsA | FtsN | 945.1±106.3 (n=18) | <0.0001 (vs. none-FtsN) |
| FtsA-M71A | FtsN | 672.9±74.68 (n=18) | <0.0001 (vs. none-FtsN) |
| 0.0437 (vs. FtsA-FtsN) | |||
| FtsA-Δ1C | FtsN | 226.8±20.53 (n=8) | 0.4026 (vs. none-FtsN) |
| FtsA-E69P | FtsN | 271.0±35.21 (n=18) | 0.6109 (vs. none-FtsN) |
Interactions between the indicated hybrid proteins were quantified by measuring β-galactosidase activities (Miller Units) in suspensions of DHM1 cells carrying the corresponding plasmids.
P values were determined by unpaired T test. P<0.05 was considered significantly different from the indicated interaction.
NA: not applicable
Negative effects of overproduced FtsA and FtsA-M71A on the Z ring
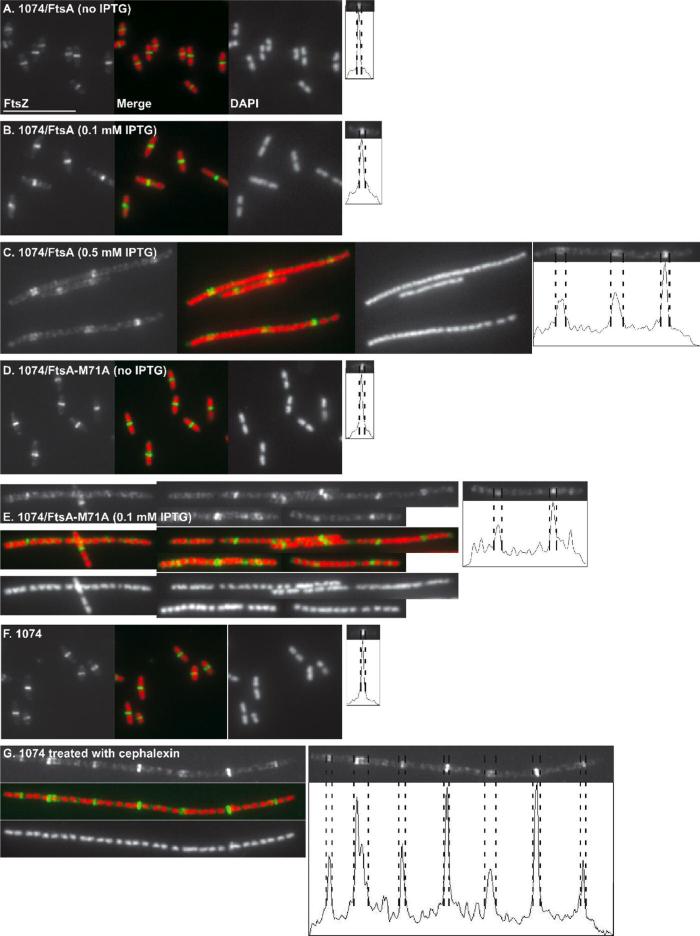
Because FtsA-M71A localized efficiently to Z rings, we reasoned that its toxicity might be caused by an altered interaction with FtsZ. Moreover, filamentous cells produced upon production of FtsA-M71A exhibited no signs of constrictions, supporting the idea that the inhibition of cell division was at an early stage. To investigate the mechanism of this inhibition, we examined Z rings by immunofluorescence microscopy under conditions of FtsA or FtsA-M71A overproduction. With either 0 or 0.1 mM IPTG driving the production of WT FtsA, Z rings assembled proficiently at potential division sites between nucleoids, and cells were of normal length (Fig. 2A-B). However, when FtsA was overproduced with 0.5 mM IPTG, many Z rings failed to localize between nucleoids in the filamentous cells, and FtsZ staining that was localized between nucleoids tended to be more diffuse compared with cells producing FtsA with 0.1 mM IPTG (Fig. 2C, compare peak widths in the intensity scans at the right). To rule out the possibility that the diffuse FtsZ banding pattern was from being in filamentous cells and not a direct result of FtsA overproduction, we induced filamentation of WT WM1074 cells with cephalexin, which inhibits activity of PBP3 (FtsI) and hence inhibits a later stage of cell division. Many Z rings failed to localize at potential division sites (i.e. between nucleoids) upon treatment with cephalexin (Fig. 2F and G), in agreement with previous results (Pogliano et al., 1997). However, a significantly higher proportion of Z rings present between nucleoids were sharper and well defined in these filamentous cells than those in filamentous cells overproducing FtsA (compare Fig. 2C and G for typical examples), suggesting that overproduction of FtsA destabilizes the Z ring.
Fig. 2. Inhibition of Z-ring assembly by overproduction of FtsA or FtsA-M71A.
FtsZ (green) and nucleoids (red) were visualized by immunofluorescence and DAPI staining of WM1074 cells carrying pWM2669 (FtsA) (A-C) or pWM2771 (FtsA-M71A) (D, E) in the presence of the indicated concentrations of IPTG. Localization of the Z rings in WM1074 cells treated without (F) or with (G) cephalexin. Representative cells and widths of Z rings are shown at the right. Scale bar = 10 μm.
Next, we examined the effect of FtsA-M71A on Z rings. In the absence of IPTG, Z rings were localized as sharp bands between two nucleoids in short cells (Fig. 2D). Unlike WT FtsA, FtsA-M71A caused cell filamentation at 0.1 mM IPTG, and most of the Z rings became diffuse (Fig. 2E), resembling those from cells producing WT FtsA at higher levels (Fig. 2C). FtsZ levels in various induction conditions were unchanged (Fig. 1D, bottom). We can conclude that FtsA-M71A destabilizes the Z-ring when overproduced, but at a lower concentration than WT FtsA.
Intergenic suppression of M71A
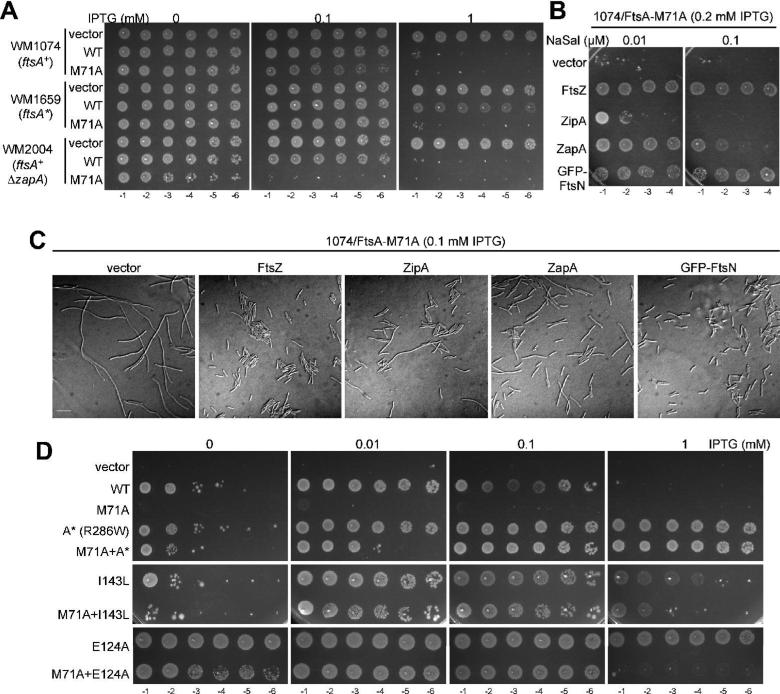
The integrity of the Z ring is balanced by positive and negative factors affecting FtsZ assembly (Romberg and Levin, 2003). If FtsA-M71A destabilizes the Z-ring as suggested by the above results, we reasoned that decreasing overall Z-ring integrity might exacerbate the negative effects of FtsA-M71A, whereas increasing Z-ring integrity might suppress its negative effects. ZapA (YgfE) is a nonessential protein that increases the integrity of the Z ring (Gueiros-Filho and Losick, 2002). We found that equivalent overproduction of FtsA-M71A (0.1 mM IPTG) was more toxic in WM2004 (ΔzapA) than in WM1074 (zapA+) cells (Fig. 3A), consistent with the idea that FtsA-M71A decreases Z-ring integrity. Likewise, a strain with a chromosomal ftsA* (R286W) allele, which maps to the top of FtsA structure in domain 2B (see also Fig. 4) and increases Z-ring integrity (Geissler et al., 2003; Geissler et al., 2007), partially suppressed the toxicity of FtsA-M71A (Fig. 3A).
Fig. 3. Suppression of toxicity caused by overproduction of FtsA-M71A.
(A) Colony viabilities of WM1074 (ftsA+), WM1659 (ftsA*) or WM2004 (ftsA+ ΔzapA) carrying either pWM2780 (vector), pWM2669 (FtsA), or pWM2671 (FtsA-M71A) in the presence of the indicated concentrations of IPTG. Plates were incubated at 30°C overnight. (B) Colony viabilities of WM1074 carrying pWM2671 (FtsA-M71A) plus either the compatible plasmids pKG110, pWM2765 (FtsZ), pWM3073 (ZipA), pWM3074 (ZapA) or pWM3075 (GFP-FtsN) in the presence of 0.2 mM IPTG (to induce production of FtsA-M71A) and the indicated concentrations of sodium salicylate (NaSal) to induce production of the other cell division proteins. (C) DIC micrographs of the same strains described in (B) in the presence of 0.1 mM IPTG and the indicated concentrations of sodium salicylate (NaSal). Scale bar = 10 μm. (D) Colony viabilities of WM1115 (ftsA12ts) carrying either pWM2780 (vector), pWM2669 (FtsA), pWM2671 (FtsA-M71A), pWM2781 (FtsA*), pWM2782 (FtsA-M71A+A*), pWM3077 (FtsA-I143L), pWM3078 (FtsA-M71A+I143L), pWM2702 (FtsA-E124A), or pWM3076 (FtsA-M71A+E124A) as described for Fig. 1B.
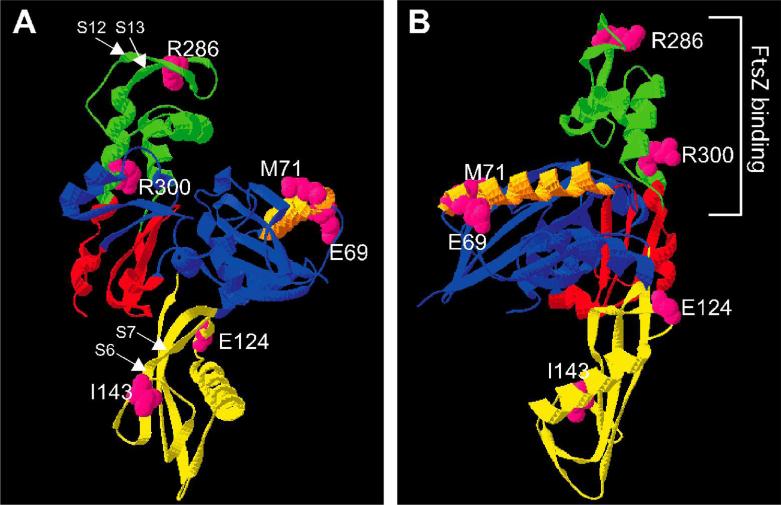
Fig. 4. Mapping of E. coli FtsA-M71, E69 and intragenic missense suppressors of M71A on the three-dimensional structure of T. maritima FtsA.
Front view (A) and rotated view (B). Each domain is shown in a different color: 1A (blue), 1C (yellow), 2A (red), and 2B (green). Alpha helix H1 is shown in orange. Strands S6, S7, S12, and S13 are highlighted by arrows.
We then investigated whether overproduction of proteins that increase Z-ring integrity could suppress the toxicity from overproduction of FtsA-M71A. When produced from a salicylate-inducible promoter in a compatible plasmid, FtsZ, ZipA, ZapA, or a functional GFP-FtsN fusion suppressed, to different degrees, the toxicity and cell filamentation caused by overproduction of FtsA-M71A (Fig. 3B-C). Whereas growth of cells producing FtsA-M71A was rescued by GFP-FtsN at two different salicylate concentrations, rescue by ZipA or ZapA was apparent only at the lower salicylate concentration (Fig. 3B), probably because higher levels of ZipA or ZapA themselves are toxic (Geissler et al., 2003) (and data not shown). The suppression by extra FtsZ was expected, as it is known to suppress the toxicity of excess WT FtsA (Dai and Lutkenhaus, 1992; Dewar et al., 1992). These results are all consistent with the idea that FtsA-M71A decreases the integrity of the Z-ring.
Intragenic suppression of M71A
To further test this hypothesis, we determined whether FtsA* or two other ftsA gain-of-function mutant alleles could restore function to FtsA-M71A and suppress overproduction toxicity when present on the same molecule. FtsA-I143L suppresses defects in FtsQ, although it cannot completely bypass FtsQ (Goehring et al., 2007); R286W can also suppress similar defects in FtsQ. FtsA-E124A bypasses the lack of FtsN or FtsK, although it does not efficiently compensate for the lack of ZipA (Bernard et al., 2007).
FtsA-M71A+R286W complemented the ftsA12(ts) strain (Fig. 3D, top panels), and did not have a dominant-negative phenotype in WT cells (data not shown), indicating that FtsA* (R286W) can rescue the defects of the M71A mutation. Interestingly, the I143L and E124A mutations also suppressed the M71A mutation (Fig. 3D, middle and bottom panels), although not as efficiently. The degree of suppression by the various alleles is remarkable given that the M71A mutation by itself permitted no growth under all the conditions tested (Fig. 3D, top panels).
The M71A mutation reduces FtsA interactions with itself but not with FtsZ
To investigate the effects of the M71A mutation on FtsA dimerization, we used a bacterial two-hybrid system based on an adenylate cyclase interaction-mediated reconstitution of a cyclic AMP (cAMP) signaling cascade (Karimova et al., 1998). FtsA and its mutant derivatives were fused to the C-termini of the T25 or T18 domains of adenylate cyclase. The self-interaction for FtsA-Δ1C exhibited the same low basal β-galactosidase activity as the vector control (Table 2), consistent with the requirement for domain 1C in FtsA self-interaction observed in a yeast two-hybrid system (Rico et al., 2004). In contrast, WT FtsA derivatives exhibited a strong positive activity, confirming previous results (Karimova et al., 2005).
Interestingly, the measured interaction activity of FtsA-M71A averaged 36% lower than that for WT FtsA. This difference was statistically significant, although the mutant proteins still had ~5 fold more activity than the vector control (Table 2). In contrast, the activity of FtsA* increased approximately 3-fold over that of WT FtsA, suggesting that this mutation increases the ability of FtsA to form dimers. Moreover, FtsA* rescued the ability of FtsA-M71A to self-interact in this assay (Table 2), consistent with the intragenic suppression results.
We also asked whether the M71A mutation affected the interaction between FtsA and FtsZ in the same bacterial two-hybrid system. There was no significant difference between WT FtsA-FtsZ and FtsA-M71A-FtsZ interactions (Table 2). On the other hand, interactions between FtsZ and either FtsA* or FtsA-M71A-A* were significantly stronger than with WT FtsA, which is consistent with previous results with the yeast two-hybrid system (Geissler et al., 2007). These results suggest that whereas the M71A mutation inhibits FtsA self-interactions, it does not significantly affect the interaction between FtsA and FtsZ, which is consistent with the ability of FtsA-M71A to localize to the Z ring.
Other residues in alpha-helix H1 are important for FtsA self-interaction
The negative effects conferred by the M71A mutation prompted us to explore whether other residue changes in alpha-helix H1 that contains Met71 might also have similar effects. We randomly mutagenized Met71 and nearby surface residues and screened for toxicity or complementation of ftsA12(ts).
We isolated three different classes of mutants: WT FtsA-like mutants (D66R, D66A, Q67W, Q67G, E69V and M71L), which complemented the ftsA12 strain at 42°C and were toxic when overproduced; M71A-like mutants (D66I, D66L, Q67D and E69P), which did not complement but were toxic; and FtsA*-like mutants (D66G and E69R), which complemented and were not toxic (Table 3). All of these mutant proteins were produced at the same levels as WT FtsA, except for FtsA-E69P, which was at slightly lower levels (data not shown).
Table 3.
Classes of growth phenotypesa of mutations in alpha-helix H1 of FtsA.
| 30°C | 42°C | |||
|---|---|---|---|---|
| ftsA allele | no IPTG | 1 mM IPTG | no IPTG | 1 mM IPTG |
| Vector (pWM2784) | +++ | +++ | - | - |
| ftsA (WT) | +++ | - | +++ | + |
| ftsA-D66R | +++ | - | +++ | + |
| ftsA-D66A | +++ | - | +++ | ++ |
| ftsA-Q67W | +++ | - | +++ | - |
| ftsA-Q67G | +++ | - | +++ | - |
| ftsA-E69V | +++ | - | +++ | + |
| ftsA-M71L | +++ | - | +++ | - |
| ftsA-M71A | +++ | - | - | - |
| ftsA-D66I | +++ | - | - | - |
| ftsA-D66L | +++ | - | - | - |
| ftsA-Q67D | +++ | - | - | - |
| ftsA-E69P | + | - | - | - |
| ftsA-Δ1C | +++ | - | - | - |
| ftsA* | +++ | +++ | +++ | +++ |
| ftsA-D66G | +++ | +++ | +++ | +++ |
| ftsA-E69R | +++ | + | +++ | ++ |
WM1115 (ftsA12) was used as host strain for this test.
+++, growth indistinguishable from WT at 30°C in the absence of IPTG; ++, poorer growth than WT at 30°C in the absence of IPTG; +, poor but still detectable growth; -, no growth.
Asp66 was tolerant to charge-reversing mutations, because FtsA-D66R, D66A, and D66G were functional. However, FtsA-D66I and D66L, like FtsA-M71A, were nonfunctional and more toxic than WT FtsA, indicating that replacing Asp66 with hydrophobic residues disturbed FtsA function. Introducing a negatively charged residue (Asp) at Gln67 (Q67D) prevented complementation, although replacement of Gln67 with Gly had no obvious effect. As with Asp66, alterations of Glu69 had disparate effects. FtsA-E69V and FtsA-E69R were functional, whereas FtsA-E69P was unable to complement ftsA12(ts) and was more toxic than FtsA-M71A (Table 3 and Fig. S3).
Because the increased toxicity of FtsA-M71A correlated with decreased self-interaction, we used the bacterial two-hybrid assay to test whether FtsA-D66I, D66L, Q67D, and E69P were also defective in self-interaction. Importantly, the β-galactosidase activity for FtsA-E69P was significantly lower than that for WT FtsA, and about half the levels of FtsA-M71A, but higher than the vector control and FtsA-Δ1C (Table 2). The β-galactosidase activities of other FtsA mutants (D66I, D66L, and Q67D) were similar to that of FtsA-M71A (data not shown). GFP-FtsA-E69P localized to the Z ring in the absence of functional FtsA (Fig. S1) and FtsA-E69P retained the ability to interact with FtsZ in the bacterial two-hybrid assay (Table 2). Although we could not detect protein levels of any T18- or T25-FtsA derivatives because of high backgrounds on immunoblots using anti-FtsA or anti-adenylate cyclase, it is reasonable to assume that the E69P fusion was produced at levels similar to the others because it could interact with FtsZ. Moreover, fusing DivIVA and GFP to its N terminus seemed to stabilize FtsA-E69P (Fig. S1 and S2). These results indicate that the interaction of FtsA-E69P with FtsZ was not significantly disrupted, and that the E69P mutation probably alters the structure of alpha-helix H1 but not the FtsZ binding domain. The data are consistent with the idea that FtsA-E69P interacts with FtsZ mainly as a monomer, resulting in decreased integrity of the Z ring.
To address whether the toxicity of FtsA-E69P was also a result of failure to recruit later divisome proteins, we tested whether FtsA-E69P interacts with FtsN. The DiviVA assay (Fig. S2A) and the bacterial two-hybrid assay (Table 2) showed that FtsA-E69P failed to interact with FtsN even though DivIVA-FtsA-E69P was produced at equivalent levels (Fig. S2B). These results suggest that dimerization of FtsA may be required for recruitment of FtsN, although we cannot rule out the possibility that Glu69 has a specific role in recruitment. In either case, we conclude that potential sequestration of FtsN and possibly other late divisomal proteins is probably not the cause of the severe dominant negative effects of FtsA-E69P. Interestingly, Glu69 is highly conserved among different species and Met71 is partially conserved (Fig. S4). Moreover, altering Glu69 to three different residues (E69V, E69P, and E69R) yielded three different phenotypic classes, suggesting that subtle changes at this residue can either increase or decrease FtsA-FtsA interactions (Table 3). Overall, we conclude that alpha-helix H1 is crucial for FtsA function, probably because this helix is important for proper homodimerization.
Our data suggest that the amino acids in alpha-helix H1 are involved in FtsA self-interactions. We then asked whether the residues known to suppress defects in self-interaction were located near other portions of the proposed dimer interface. We mapped E124, I143, and R286 (FtsA*) in addition to the alpha-helix H1 residues E69 and M71 on the three-dimensional structure of T. maritima FtsA (Fig. 4A and B). All residues were at or near the proposed dimer interface, which is consistent with their ability to suppress defects of FtsA-M71A.
A tandem FtsA repeat is functional
So far, our results with FtsA mutants suggest that inhibiting FtsA homodimerization results in less Z-ring integrity and stimulating homodimerization of FtsA (e.g., FtsA*) increases Z-ring integrity. However, we could not rule out the possibility that the various mutations could also cause other changes in FtsA separate from alterations in dimerization. We reasoned that forcing FtsA to dimerize by fusing two FtsA monomers in a head to tail configuration might address this issue, and provide additional support for the model.
Therefore, we constructed a tandem repeat of WT FtsA (referred to as AA), which has a single FLAG tag at the N-terminus (Fig. 5A). Remarkably, AA produced from pWM3079 complemented the ftsA12(ts) strain (Fig. 5B) as well as an ftsA depletion strain at the nonpermissive temperature of 42°C (Fig. S5), and cells were normal in length (data not shown). This indicates that AA is fully functional in the absence of WT FtsA. Protein levels of AA were comparable with those of FtsA, and no degradation products were detected (Fig. 5C-D), suggesting that the AAtandem (~88 kDa) is the functional unit. The lack of detectable FtsA monomer under these conditions also suggests that a monomer-dimer transition is not necessary for FtsA function.
Fig. 5. FtsA tandems are functional in vivo.
(A) Schematic illustration of an FtsA tandem repeat (AA), which has an FLAG tag at the N-terminus of AA and a Ser-Arg dipeptide between FtsA subunits. (B) Complementation of WM1115 (ftsA12ts) by FtsA (pWM2669), AA (pWM3079), WT-71 (pWM3080), 71-WT (pWM3081), 71-71 (pWM3082), Δ1C-71 (pWM3083), Δ1C-Δ1C (pWM3084), or 69-69 (pWM3085), as described for Fig. 1B. (C) Immunoblots of representative cultures used for the spot assay (B) were probed with anti-FLAG (M2). Both blots show cultures producing AA (~88 kDa) as controls. (D) Immunoblots of WM1115 carrying pWM2785 (FtsA), pWM2777 (AA), pWM3036 (Δ1C-Δ1C), or pWM3086 (69-69) in the presence and absence of 1 mM IPTG were probed with anti-FLAG (M2).
Fusing M71A mutant FtsA proteins in tandem restores function
We then asked whether forcing the defective FtsA-M71A to exist as a tandem (71-71) would rescue its function and suppress its overproduction toxicity. Remarkably, 71-71 complemented ftsA12(ts) and the FtsA depletion strain at 42°C (Fig. 5B and S5), indicating that fusing FtsA-M71A in tandem rescued its function. However, 71-71 was still approximately as toxic as WT FtsA and more toxic than AA when overproduced (Fig. 5B and S3), suggesting that 71-71 retains some dominant negative activity of the original FtsA-M71A (see Discussion).
To test whether halving the number of M71A subunits per tandem might suppress this toxicity, we constructed WT-71 and 71-WT tandems. These mixed tandems were both functional and permitted growth at 1 mM IPTG, indicating that they were less toxic than 71-71 at equivalent levels (Fig. 5B-C). No difference in function could be detected between fusions carrying the M71A subunit at the N-terminal or C-terminal end. We confirmed that there were no detectable degradation products of WT-71, 71-WT, and 71-71 (Fig. 5C). Even if very small amounts of degraded monomeric FtsA-M71A were present, 71-71 must be functional as a tandem because non-tandem FtsA-M71A does not complement ftsA mutants, as shown earlier.
A tandem of the more severely defective E69P mutant (69-69) did not complement the ftsA12(ts) strain WM1115 nearly as effectively as AA and 71-71 (Fig. 5B). However, when grown in broth, the WM1115 strain producing 69-69 was a mixture of short cells and filaments, compared to all filaments with the vector control after 2 h at 42°C (data not shown), indicating that 69-69 had some complementing activity. Unlike with AA, we could not detect 69-69 by immunoblotting when it was produced from pWM3085, but could detect low levels of 69-69 when it was produced from pWM3086, which has a stronger promoter (Fig. 5C-D). These low levels of 69-69 remained inefficient for complementation. As even lower (undetectable) levels of AA could still complement ftsA12(ts) (Fig. 5B-C), the results suggest that the defects of FtsA-E69P are sufficiently severe that connecting them in tandem cannot efficiently rescue them.
FtsA-Δ1C can function only if tethered to a WT subunit
Because FtsA-Δ1C is defective in dimerization and recruitment of later division proteins, we asked whether a tandem might rescue its function. However, Δ1C-Δ1C did not complement either, and unlike 69-69, could not form even sick colonies at several IPTG concentrations (Fig. 5B). Although cellular levels of Δ1C-Δ1C protein were equivalent to those of 69-69 and therefore significantly lower than those of AA (Fig. 5C-D), the residual complementation activity of 69-69 compared to Δ1C-Δ1C suggests that the failure of Δ1C-Δ1C to function probably is not solely caused by its low protein levels.
To circumvent the problem of low protein levels and to test whether having one subunit with an intact 1C domain could rescue the defects of Δ1C-Δ1C, we constructed a Δ1C-71 tandem. This fusion was produced at higher levels equivalent to those of AA, and was capable of rescuing complementation of ftsA12(ts) at all IPTG levels except the highest (Fig. 5B-C). Therefore, only one intact 1C domain per FtsA tandem is needed for full function.
FtsA tandems enhance Z-ring integrity
FtsA* (R286W) increases the integrity of the Z-ring and accelerates ring reassembly (Geissler et al., 2003; Geissler et al., 2007). Because FtsA* also seems to stimulate dimerization, we asked whether tandem FtsA repeats might also increase Z ring integrity. We initially tested this idea by determining whether AA or 71-71 could suppress the thermosensitivity of an ftsZ84 (ts) strain. The FtsZ84 protein has a much lower GTP binding and hydrolysis activity than WT FtsZ (de Boer et al., 1992; Raychaudhuri and Park, 1992), and Z rings disappear rapidly at 42°C in the mutant strain (Addinall et al., 1997). However, increased gene dosage of ZipA can efficiently suppress ftsZ84 thermosensitivity (Raychaudhuri, 1999).
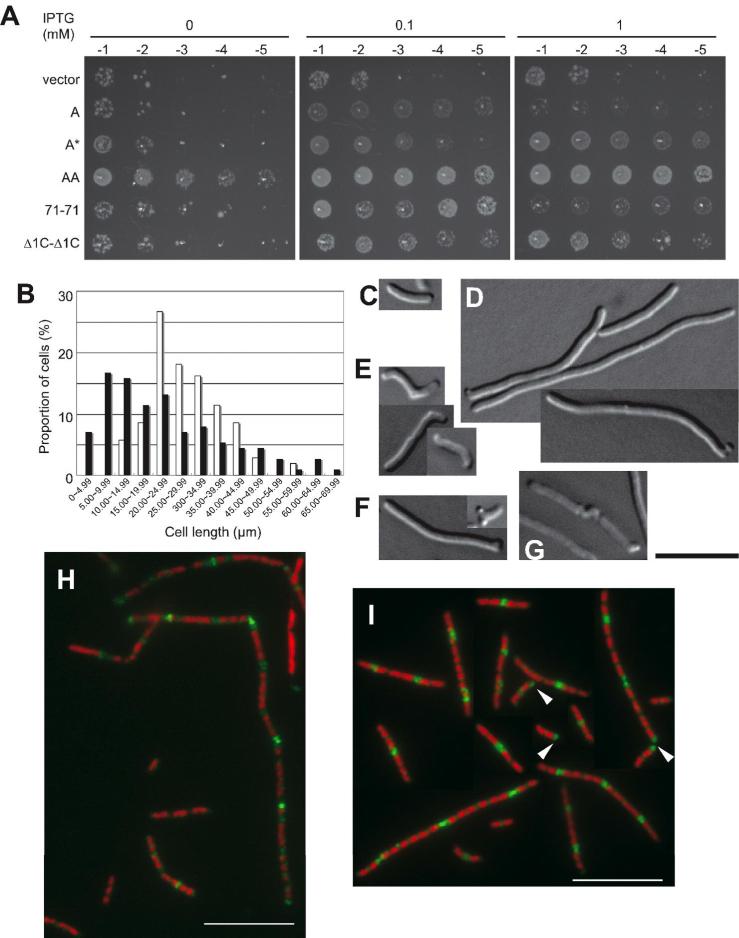
As shown in Fig. 6A, AA efficiently suppressed the ftsZ84(ts) strain WM1125. Weak suppression was exhibited by 71-71 (0.1 mM IPTG), FtsA* (1 mM IPTG), and even WT FtsA (0.1 mM IPTG) compared to the vector control. The Δ1C-Δ1C fusion behaved like FtsA*, with partial suppression at higher IPTG concentrations (Fig. 6A). One possible way that Δ1C-Δ1C might suppress is by forming a trimer with the endogenous FtsA. It is also possible that Δ1C-Δ1C stabilizes FtsZ84 independently of the endogenous FtsA and then the endogenous FtsA interacts with the FtsZ84 ring stabilized by Δ1C-Δ1C. In any case, these results suggest that AA was the most effective suppressor of ftsZ84(ts), and that 71-71 and Δ1C-Δ1C also have some suppression activities.
Fig. 6. FtsA tandems support growth of WM1125 (ftsZ84) at 42°C.
(A) Colony viabilities of WM1125 (ftsZ84ts) carrying either pWM2780 (vector), pWM2669 (FtsA), pWM2781 (FtsA*), pWM3079 (AA), pWM3082 (71-71), or pWM3084 (Δ1C-Δ1C) as described for Fig. 1B. (B) Distribution of lengths of WM1125 cells producing AA without IPTG (open bars) or with 0.1 mM IPTG (filled bars). (C-G) DIC micrographs of representative cells of WM1125 producing AA in the presence of 0.1 mM IPTG: normal (C), filaments and minicells (D), bent (E), branched (F), and twisted (G). (H-I) Localization of Z rings in WM1125 producing AA in the absence (H) and presence of 0.1 mM IPTG (I). FtsZ (green) was visualized by immunofluorescence microscopy, and nucleoids (pseudocolored red) by DAPI staining. Arrowheads highlight FtsZ polar localization. Scale bar = 10 μm.
We then investigated the extent of the suppression. WM1125 cells producing AA with 0 or 0.1 mM IPTG at 42°C in broth did not divide normally, despite the viability of colonies on the plates. Although there was no major difference in average cell length at different levels of IPTG (29.03 ± 9.77 μm with no IPTG vs. 22.76 ± 15.70 with 0.1 mM IPTG), the proportion of shorter (<10 μm) cells in 0.1 mM IPTG was higher (~25%) than in its absence (0%) (Fig. 6B). Therefore, higher levels of AA more efficiently suppressed the ftsZ84 cell division defect at 42°C, consistent with the increased viability at higher IPTG levels (Fig. 6A). Cellular levels of FtsZ84 were unchanged (data not shown).
We examined the morphology and Z rings of WM1125 cells producing AA at 42°C. In the absence of IPTG, cells were filamentous and FtsZ staining was mostly diffuse, with only occasional FtsZ staining at potential division sites between nucleoids (Fig. 6H). With 0.1 mM IPTG, the culture was a mixture of normal cells (Fig. 6C), minicells and filaments (Fig. 6D), and cells with unusual bends, branches, or twisted division septa (Fig. 6E-G). FtsZ staining at mid-cell was observed in shorter cells (Fig. 6I), but FtsZ bands in filamentous cells were generally not as sharp as those in the cephalexin-treated ftsZ+ strain (compare with Fig. 2F). FtsZ also often localized at cell poles (Fig. 6I, arrowheads), possibly because AA prevented disassembly of used FtsZ84 rings (Yu and Margolin, 2000). Therefore, although AA could not fully suppress the defects of ftsZ84, it clearly stimulated function of Z rings under conditions normally completely nonpermissive for Z rings.
This gain of function behavior prompted us to ask whether AA could compensate for the loss of other essential cell division proteins. Indeed, AA partially compensated for the loss of zipA, ftsK, or ftsE, but did not significantly compensate for the loss of ftsN or ftsQ (D. Shiomi and W. Margolin, unpublished results). Overall, we propose that increased dimerization of FtsA, either via tandem fusions or by a mutation such as FtsA*, not only suppresses the negative effects of FtsA mutants that are defective in self-interaction, but also enhances the functionality of WT FtsA.
Only one available FtsZ-binding site per FtsA dimer is sufficient for cell division
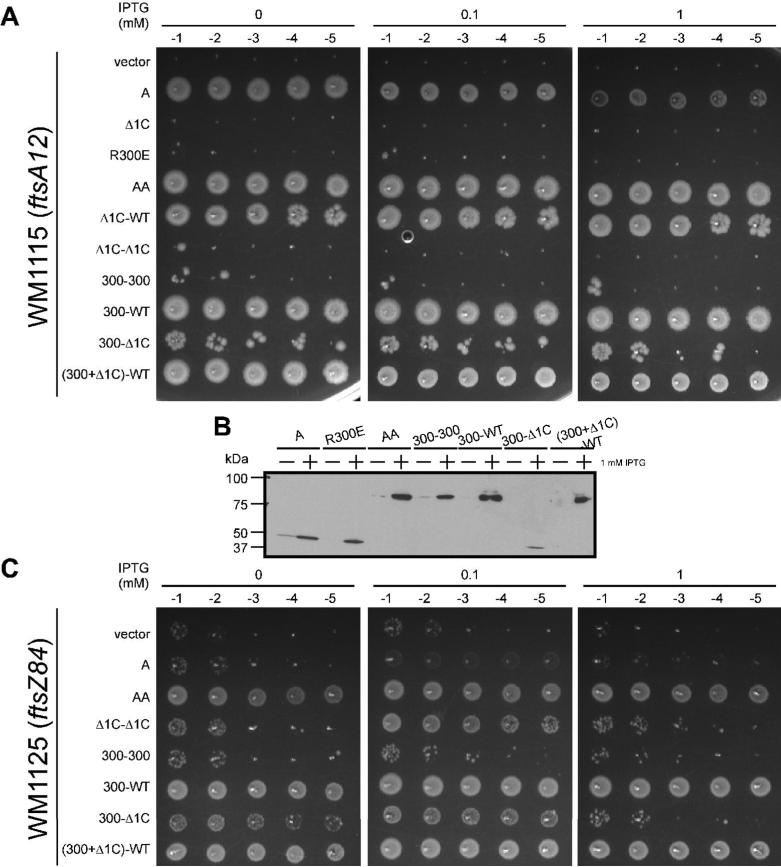
FtsA point mutants were recently found that are specifically defective in binding to FtsZ (Pichoff and Lutkenhaus, 2007). These mutants, including R300E, cluster at the back of the crystal structure shown in Fig. 4A or at the right of the rotated structure in Fig. 4B, suggesting that they represent a binding interface. Interestingly, Carettoni et al. (2003) predicted that R300 also is involved in dimerization of FtsA. To gain additional insight into whether the two subunits of FtsA dimers might have distinct activities, we explored whether a tandem FtsA repeat with one monomer unable to bind FtsZ could remain functional.
FtsA-R300E or a tandem repeat of FtsA-R300E (300-300) did not complement ftsA12(ts) (Fig. 7A), which is not surprising as neither should bind to FtsZ. However, a mixed tandem of WT FtsA and R300E (300-WT) was functional (Fig. 7A), suggesting that only one available FtsZ-binding site per FtsA dimer is sufficient for function. We then constructed mixed tandems of R300E and Δ1C [300-Δ1C and (300+Δ1C)-WT] to address whether the same subunit is responsible for binding FtsZ and recruitment of later divisome proteins. Whereas (300+Δ1C)-WT complemented ftsA12(ts) efficiently, 300-Δ1C complemented poorly (Fig. 7A), again suggesting that one intact FtsA subunit per dimer is sufficient for function. The few colonies present might be suppressors, although we did not observe any colonies for Δ1C-Δ1C under all conditions tested (Fig. 6). Cellular levels of 300-300, 300-WT, and (300+Δ1C)-WT were similar to those of AA (Fig. 7B). The 300-Δ1C protein band was undetectable, suggesting that low levels caused the weak complementation. However, when cellular levels of 300-Δ1C were increased to those of AA with a stronger promoter, the weak complementation was still observed (D. Shiomi and W. Margolin, unpublished data).
Fig. 7. Functionality of FtsA mutant tandems with defects in binding to FtsZ and/or recruitment of later divisome proteins.
(A) Complementation of WM1115 (ftsA12ts) by FtsA (pWM2785), Δ1C (pWM2789), R300E (pWM3121), AA (pWM2777), Δ1C-WT (pWM3034), Δ1C-Δ1C (pWM3036), 300-300 (pWM3122), 300-WT (pWM3123), 300-Δ1C (pWM3124), or (300+Δ1C)-WT (pWM3125, as described for Fig. 1B. (B) An immunoblot of representative cultures used for the spot assay (A) probed with anti-FLAG (M2). (C) Suppression of WM1125 (ftsZ84ts) carrying either pWM2784 (vector), pWM2785 (FtsA), pWM2777 (AA), pWM3036 (Δ1C-Δ1C), pWM3122 (300-300), pWM3123 (300-WT), pWM3124 (300-Δ1C), or pWM3125 [(300+Δ1C)-WT] as described for Fig. 1B.
We then asked whether tandem repeats containing R300E had gain-of-function properties. As expected, 300-300 did not suppress ftsZ84(ts), presumably because FtsA-FtsZ interaction is required for the suppression (Fig. 7C). However, other tandem repeats containing only one R300E mutant subunit suppressed ftsZ84(ts), including 300-Δ1C (Fig. 7C). These results suggest that an FtsA dimer needs only one FtsZ binding site per dimer to increase integrity of normally thermosensitive Z rings. The evidence also suggests that if one FtsA subunit in a dimer is defective in binding to FtsZ or recruiting downstream proteins, the other subunit must be functional for the cognate activity to permit full function.
DISCUSSION
Physiological significance of FtsA dimerization/oligomerization
Because of its structural similarity to actin, it is not surprising that FtsA interacts with itself, based on yeast two-hybrid experiments (Pichoff and Lutkenhaus, 2007; Yan et al., 2000; Yim et al., 2000) bacterial two-hybrid experiments (Di Lallo et al., 2003; Karimova et al., 2005), self-assembly of FtsA from S. pneumoniae into helical filaments in vitro (Lara et al., 2005), and detection of dimers of B. subtilis FtsA in vivo (Feucht et al., 2001). The crystal structure of FtsA from T. maritima is of the monomer form (van Den Ent and Lowe, 2000), and therefore provides little structural insight into oligomers. However, Carettoni et al. subsequently used phage display and modeling methods to propose a possible structure of an FtsA dimer with a head to tail arrangement of FtsA monomers rotated 180° with respect to each other (Carettoni et al., 2003). One of the predicted dimer interfaces, domain 1C, has been shown by several studies to be required for self-interaction of FtsA and recruitment of later divisome proteins but not for FtsA interactions with FtsZ (Carettoni et al., 2003; Corbin et al., 2004; Rico et al., 2004). The C-terminal membrane targeting sequence of FtsA is required for function and is also involved in FtsA-FtsA interactions (Yim et al., 2000), but its effects on dimerization are not yet understood.
In this study, we used point mutants in alpha helix H1 in domain 1A of E. coli FtsA, part of the putative dimer interface, to test directly whether FtsA dimerization or oligomerization is required for its function in cell division. Interestingly, of the 13 point mutations constructed, 6 behaved like WT FtsA, 5 were dominant negative and were not able to complement ftsA12(ts) under any conditions tested, and 2 were significantly less toxic than WT FtsA when overproduced, suggesting that they were gain of function mutants like FtsA*. We focused on two of the dominant negative mutants for further study, because bacterial two-hybrid analysis indicated that these mutations, M71A and E69P, inhibited FtsA-FtsA interactions, whereas experiments with GFP fusions revealed that their interactions with FtsZ were not significantly changed. Our results support the idea that inefficient dimerization of FtsA-M71A results in dominant negative effects directly on FtsZ assembly.
The most compelling support for the idea that the non-complementing and dominant negative phenotypes of these mutants result from their inability to dimerize efficiently comes from the suppression of these phenotypes by tandem FtsA repeats. For example, tethering FtsA-M71A monomers in tandem restored complementation of ftsA12(ts) or an ftsA null mutation, and mitigated the dominant negative effects. Mutants that were more toxic than M71A. such as E69P, functioned much less well in tandem, although the E69P tandem was still more functional and less toxic than the untethered form.
We conclude that dimerization or oligomerization is critical for FtsA function, and that alpha helix H1 is required for this self-interaction. It is likely that Met71 is an important contact at the dimer interface, and changing it to Ala significantly weakens the contact whereas the more conservative change to Leu does not. The strong effect of E69P probably occurs because the change to Pro breaks the alpha helix, disrupting the dimer interface. Although future studies will be needed, we propose that the other potential gain of function mutations that we found in this helix act oppositely from E69P or M71A and strengthen the dimer interface.
Potential mechanism of inhibition of Z rings by FtsA monomers
Because decreased FtsA dimerization seems to correlate with increased toxicity, we propose that when FtsA is overproduced, a significant fraction is in the monomer form, which causes the toxicity. Because of the high rate of turnover between the cytoplasmic pool of FtsZ and the FtsZ in the ring (Anderson et al., 2004; Stricker et al., 2002) and because FtsZ and FtsA interact with each other independently of the ring (Jensen et al., 2005), excess FtsA might sequester FtsZ subunits, destabilizing the ring. Excess FtsA might also compete with other FtsZ-stabilizing factors such as ZipA and ZapA for the same binding site at the C terminus of FtsZ. The fact that excess FtsZ restores normal cell division in the presence of excess WT FtsA is consistent with such a mechanism.
Nevertheless, the results with the M71A mutant suggest that competition or sequestration may not be sole explanation for the observed effects. For example, if FtsA competes for binding sites on FtsZ, then how can FtsA-M71A, which seems to have similar affinity for FtsZ as WT FtsA, exert the same negative effect on Z ring assembly at significantly lower FtsA levels? The same may be true for E69P. Therefore, we favor the idea that an FtsA monomer has an innate inhibitory activity on FtsZ assembly that is masked in the FtsA dimer.
This inhibitory activity might originate in domain 1A of FtsA, because it has some structural similarity to the N-terminus of MinC (Cordell et al., 2001), the major domain of MinC involved in its inhibition of FtsZ assembly (Hu and Lutkenhaus, 2000). If domain 1A of the FtsA monomer functions as an inhibitor of FtsZ assembly, then increased dimerization of FtsA may sterically block the negative activity, consistent with our results. Interestingly, helix H1 of FtsA is located within this structural similarity with MinC (Cordell et al., 2001), suggesting that the M71A or E69P mutations may strengthen the potential FtsZ-inhibitory activity of FtsA in addition to inhibiting FtsA dimerization.
One of the more striking results in this study was the suppression of the dominant-negative effects of the M71A mutant by second-site suppressor mutations or by increased expression of other cell division genes. Carettoni et al. (2003) proposed that alpha-helix H1 (shown in orange in Fig. 4) of one FtsA monomer contacts subdomain 1C (shown in yellow in Fig. 4) of the next FtsA monomer. Their model places strand S12-S13, at the top of FtsA molecule containing residue R286, in close proximity to residues 126-133 in subdomain 1C, which in turn is adjacent to residue E124, strand S6-S7 (containing I143), and alpha-helix H1 in subdomain 1A. Therefore, suppression of the M71A dimerization defect by FtsA* (R286W), I143L, and E124A may occur because they are all at or near the dimer interface. The increased dimerization exhibited by R286W in the bacterial two-hybrid assay is consistent with this idea.
Suppression by overproduction of other cell division proteins, particularly ZipA, ZapA or GFP-FtsN, is likely to be indirect. ZapA and ZipA may suppress M71A toxicity by competing with FtsA for binding sites on FtsZ as mentioned above. However, it is equally plausible that they may antagonize the postulated inhibitory activity of monomeric FtsA, perhaps by bundling FtsZ protofilaments (Gueiros-Filho and Losick, 2002; Hale et al., 2000; Raychaudhuri, 1999). Unlike ZipA or ZapA, GFP-FtsN is not likely to bind directly to FtsZ, and probably stabilizes the divisome by an unknown mechanism (Bernard et al., 2007; Dai et al., 1993).
Is this postulated FtsZ-inhibitory activity of FtsA relevant to normal cell division? The total FtsA:FtsZ ratio remains constant during the cell cycle of E. coli (Rueda et al., 2003). Nonetheless, because significant proportions of FtsZ and probably FtsA are present outside the ring (Stricker et al., 2002; Thanedar and Margolin, 2004), it is not known what the FtsA:FtsZ ratio is at the Z ring or how it changes during the cell division cycle. If the contraction of the Z ring is accompanied by the loss of FtsZ subunits (Sun and Margolin, 1998), then the combination of rapid turnover of FtsZ in the ring and attachment of FtsA to the membrane may increase the local FtsA:FtsZ ratio as the Z-ring contracts. This could act as a positive feedback loop to drive ring contraction to completion. The full function of the tandem repeats in the absence of any detectable monomeric FtsA suggests either that (i) some of the tandem repeats act simply as tethered monomers and can function in this negative role, (ii) the negative activity has no role in cell division, or (iii) it has a role but is redundant, possibly because other factors such as MinC also help to contract the Z ring.
FtsA dimerization/oligomerization increases Z-ring integrity
Remarkably, the tandem repeat fusions AA, 71-71 and Δ1C-Δ1C could restore the ability of an ftsZ84 mutant to grow and divide at the non-permissive temperature, similar to the effect of increased ZipA levels (Raychaudhuri, 1999). As was observed in that study, we found that the consequences of this restoration were the production of minicells and cells with altered septal morphology, suggesting that increasing Z-ring integrity causes reduced sensitivity to MinC and perhaps other negative regulators of FtsZ assembly. On the other hand, tandem FtsA constructs defective in binding to FtsZ, such as 300-300, were not able to suppress the thermosensitivity of ftsZ84 significantly, indicating that this suppression is likely mediated via a direct FtsA-FtsZ interaction.
The gain of function displayed by the FtsA tandem repeats and their rescue of the M71A defects suggests that despite being linked by just a dipeptide, the FtsA monomers may contact each other occur at the normal dimer interface. The poor function of 69-69 suggests that either E69P has effects in addition to inhibiting dimerization, or normal dimerization did not occur efficiently in the 69-69 fusions, possibly because of disrupted secondary structure. Both N and C termini of FtsA were not part of the solved crystal structure, leaving about 40 amino acid residues undefined. This length might be sufficiently flexible to allow vertical orientation of the monomers as postulated by Carettoni et al., but it is also possible the length is not sufficient and/or that monomers are also arranged horizontally. If FtsA forms large filaments/oligomers as suggested in vitro (Lara et al., 2005), then FtsA could self-interact vertically and laterally. Lateral contacts may contribute to FtsA-FtsA interactions and help stabilize longitudinal contacts within a putative FtsA lattice.
A new model for FtsA-FtsZ interactions at the Z ring
Our results suggest a new model for how FtsA dimers or oligomers might increase the integrity of the Z ring (Fig. 8A). Because the two FtsA monomers in the predicted homodimer are rotated 180° with respect to each other, and FtsZ may bind to one side of each monomer (Pichoff and Lutkenhaus, 2007), then one FtsZ subunit could bind to the front of the top FtsA subunit, and another FtsZ subunit could bind to the back of the bottom FtsA subunit. Whereas an FtsA monomer would bind only to one FtsZ protofilament, a single FtsA homodimer could crosslink two different FtsZ protofilaments, similar to the proposed function of ZapA (Low et al., 2004). If FtsA assembles into oligomers (Lara et al., 2005), then FtsA filaments might form a membrane-bound scaffold to reinforce FtsZ protofilaments (Fig. 8A), which might explain the stabilization of FtsZ84 rings. Stimulation of ATP hydrolysis might induce FtsA disassembly into monomers, which would in turn reveal the putative MinC-like FtsZ disassembly activity necessary to trigger Z-ring contraction.
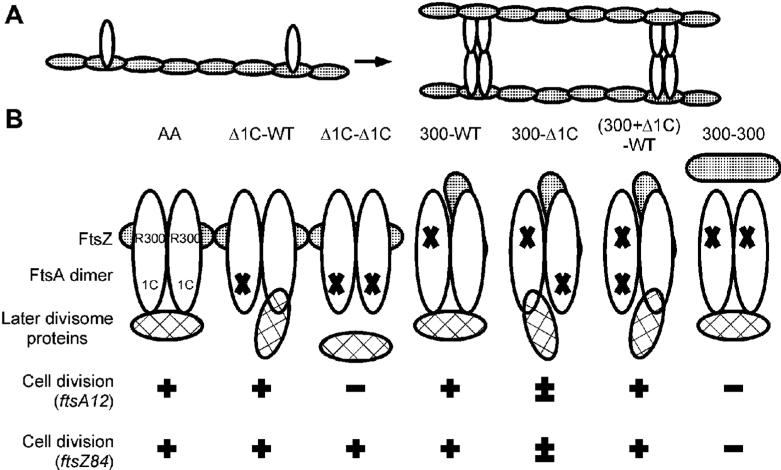
Fig. 8. FtsA dimerization/oligomerization and cell division.
(A) Proposed interaction of FtsA dimers/oligomers with FtsZ and the Z ring. FtsZ protofilaments (speckled ovals) initially form a scaffold for the binding of FtsA monomers (white ovals). Subsequently, FtsA dimerizes and/or oligomerizes, crosslinking FtsZ protofilaments and increasing the integrity of the Z ring. (B) Model of FtsA dimers (white ovals) and their potential interactions with FtsZ and later divisome proteins. Residue R300 is required for binding to FtsZ (speckled ovals), and domain 1C is required for recruitment of late proteins (hatched ovals). The ability of each potential FtsA dimer to promote cell division of an ftsA(ts) or ftsZ(ts) mutant is shown below.
The results with mixed tandems containing one subunit unable to bind to FtsZ and the other unable to recruit downstream proteins suggest that there may be no division of labor between the two subunits of the dimer, and that one fully functional subunit per dimer is sufficient both for FtsA function and for promoting integrity of the Z ring (Fig. 8B). The latter point is underscored by the ability of the (300+Δ1C)-WT tandem, which has only one subunit that can bind to FtsZ and recruit downstream proteins, to complement ftsA12(ts) and to suppress ftsZ84(ts). If this derivative only formed dimers, however, it would probably not be able to link two FtsZ molecules (Fig. 8B). This suggests that these FtsA tandem repeats form oligomers, and that it is the oligomeric state of FtsA, even when partially disabled for other functions, that stimulates the integrity of the Z ring in a way that non-oligomeric FtsA cannot.
EXPERIMENTAL PROCEDURES
Strains, plasmids, and growth conditions
E. coli strains and plasmids used in this study are listed in Table 4. Bacteria were grown in LB (Luria-Bertani) medium containing 0.5% NaCl supplemented with appropriate antibiotics at 30°C, unless otherwise indicated. Ampicillin (Amp), kanamycin (Kan) or chloramphenicol (Cm) was added at 100 μg, 50 μg or 20 μg per ml, respectively, as needed. Strain XL1-Blue (Stratagene) was used for plasmid constructions.
Table 4.
Strains and plasmids used in this study.
| Strains | Description | Source/Reference |
|---|---|---|
| W3110 | wild-type strain | Lab collection |
| WM1074 | MG1655 ΔlacU169 (TX3772) | Lab collection |
| WM1115 | WM1074 ftsA12 | Lab collection |
| WM1125 | WM1074 ftsZ84 | Lab collection |
| WM1659 | WM1074 ftsA* | (Geissler et al., 2003) |
| WM2004 | WM1074 ΔzapA::kan | Lab collection |
| WM1281 | CH2 (recA::Tn10 ftsA0)/pDB280 (repAts ftsA+) | (Hale and de Boer, 1999) |
| WM2417 | W3110 ftsA (E124A) ΔftsN::cat | (Bernard et al., 2007) |
| DHM1 | F− glnV44(AS) recA1 endA gyrA96 thi-1 hsdR17 spoT1 rfbD1 cya-854 | (Karimova et al., 2005) |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F’ proAB lacIqZAM15 Tn10 | Stratagene |
| Plasmid | Description | Source/Reference |
| pDSW209 | Ptrc-gfp pBR322 derivative | (Weiss et al., 1999) |
| pDSW210 | Ptrc-gfp pBR322 derivative | (Weiss et al., 1999) |
| pBAD33 | pACYC184 derivative containing the araBAD promoter | (Guzman et al., 1995) |
| pKG110 | pACYC184 derivative containing the nahG promoter | J. S. Parkinson |
| pKG116 | pACYC184 derivative containing the nahG promoter | J. S. Parkinson |
| pRR48 | Ptac pBR322 derivative | J. S. Parkinson |
| pWM1260 | pET-ftsA | (Geissler et al., 2003) |
| pWM2784 | FLAG inserted between EcoRI and SacI sites of pDSW210 | (Shiomi and Margolin, 2007) |
| pWM2785 | FLAG-ftsA in pWM2784 | This study |
| pWM2786 | FLAG-ftsA-M71A in pWM2784 | This study |
| pWM2787 | FLAG-ftsA*(R286W) in pWM2784 | This study |
| pWM2788 | FLAG-ftsA-M71A+A* in pWM2784 | This study |
| pWM2789 | FLAG-ftsA-Δ1C in pWM2784 | This study |
| pWM3087 | FLAG-ftsA-D66I in pWM2784 | This study |
| pWM3088 | FLAG-ftsA-D66L in pWM2784 | This study |
| pWM3089 | FLAG-ftsA-D66R in pWM2784 | This study |
| pWM3090 | FLAG-ftsA-D66G in pWM2784 | This study |
| pWM3091 | FLAG-ftsA-D66Ain pWM2784 | This study |
| pWM3092 | FLAG-ftsA-Q67D in pWM2784 | This study |
| pWM3093 | FLAG-ftsA-Q67W in pWM2784 | This study |
| pWM3094 | FLAG-ftsA-Q67G in pWM2784 | This study |
| pWM3095 | FLAG-ftsA-E69P in pWM2784 | This study |
| pWM3096 | FLAG-ftsA-E69V in pWM2784 | This study |
| pWM3097 | FLAG-ftsA-E69R in pWM2784 | This study |
| pWM3098 | FLAG-ftsA-M71L in pWM2784 | This study |
| pWM2780 | pRR48 carrying a multi-cloning site of pKG110 | This study |
| pWM2669 | FLAG-ftsA in pWM2780 | This study |
| pWM2671 | FLAG-ftsA-M71A in pWM2780 | This study |
| pWM2781 | FLAG-ftsA* in pWM2780 | This study |
| pWM2782 | FLAG-ftsA-M71A+A* in pWM2780 | This study |
| pWM2783 | FLAG-ftsA-Δ1C in pWM2780 | This study |
| pWM2702 | FLAG-ftsA-E124A in pWM2780 | This study |
| pWM3076 | FLAG-ftsA-M71A+E124A in pWM2780 | This study |
| pWM3077 | FLAG-ftsA-I143L in pWM2780 | This study |
| pWM3078 | FLAG-ftsA-M71A+I143L in pWM2780 | This study |
| pWM3055 | FLAG-ftsA-E69P in pWM2780 | This study |
| pWM3079 | FLAG-ftsA-ftsA fusion (AA) in pWM2780 | This study |
| pWM3080 | FLAG-WT-M71A fusion in pWM2780 | This study |
| pWM3081 | FLAG-M71A-WT fusion in pWM2780 | This study |
| pWM3082 | FLAG-M71A-M71A fusion (71-71) in pWM2780 | This study |
| pWM3083 | FLAG-Δ1C-71 fusion in pWM2780 | This study |
| pWM3084 | FLAG-Δ1C-Δ1C fusion in pWM2780 | This study |
| pWM3085 | FLAG-E69P-E69P fusion (69-69) in pWM2780 | This study |
| pWM2777 | FLAG-ftsA-ftsA fusion (AA) in pWM2784 | This study |
| pWM3034 | FLAG-Δ1C-WT fusion in pWM2784 | This study |
| pWM3036 | FLAG-Δ1C-Δ1C fusion in pWM2784 | This study |
| pWM3086 | FLAG-E69P-E69P fusion (69-69) in pWM2784 | This study |
| pWM3121 | FLAG-ftsA-R300E in pWM2784 | This study |
| pWM3122 | FLAG-R300E-R300E fusion (300-300) in pWM2784 | This study |
| pWM3123 | FLAG-R300E-WT fusion (300-WT) in pWM2784 | This study |
| pWM3124 | FLAG-R300E-Δ1C fusion (300-Δ1C) in pWM2784 | This study |
| pWM3125 | FLAG-(R300E+Δ1C)-WT fusion [(300+Δ1C)-WT] in pWM2784 | This study |
| pWM2760 | gfp-ftsA in pDSW209 | (Geissler et al., 2007) |
| pWM3191 | gfp-ftsA-M71A in pDSW209 | This study |
| pWM3195 | gfp-ftsA-E69P in pDSW209 | This study |
| pWM3196 | gfp-ftsA in pKG110 | This study |
| pWM3197 | gfp-ftsA-M71A in pKG110 | This study |
| pWM3200 | gfp-ftsA-E69P in pKG110 | This study |
| pKT25 | T25 fragment of B. pertussis CyaA (residues 1-224) in pSU40 | (Karimova et al., 2005) |
| pUT18C | T18 fragment of B. pertussis CyaA (residues 225-399) in pUC19 | (Karimova et al., 2005) |
| pWM3014 | T25-ftsA in pKT25 | This study |
| pWM3015 | T25-ftsA-M71A in pKT25 | This study |
| pWM3016 | T25-ftsA* in pKT25 | This study |
| pWM3017 | T25-ftsA-M71A+A* in pKT25 | This study |
| pWM3018 | T25-ftsA-Δ1C in pKT25 | This study |
| pWM3044 | T25-ftsA-E69P in pKT25 | This study |
| pWM3021 | T18-ftsA in pUT18C | This study |
| pWM3022 | T18-ftsA-M71A in pUT18C | This study |
| pWM3023 | T18-ftsA* in pUT18C | This study |
| pWM3024 | T18-ftsA-M71A+A* in pUT18C | This study |
| pWM3025 | T18-ftsA-Δ1C in pUT18C | This study |
| pWM3040 | T18-ftsA-E69P in pUT18C | This study |
| pWM3183 | T25-ftsN in pKT25 | This study |
| pKNT25-ftsZ | ftsZ-T25 in pKNT25 | (Karimova et al., 2005) |
| pWM2765 | ftsZ in pKG110 | This study |
| pWM3073 | zipA in pKG110 | This study |
| pWM3074 | zapA in pKG116 | This study |
| pWM1152 | gfp-ftsN in pDSW209 | (Corbin et al., 2004) |
| pWM3075 | gfp-ftsN in pKG116 | This study |
| pWM1806 | divIVA-ftsA in pBAD33 | (Corbin et al., 2004) |
| pWM3070 | divIVA-ftsA-M71A in pBAD33 | This study |
| pWM3071 | divIVA-ftsA-Δ1C in pBAD33 | This study |
| pWM3072 | divIVA-ftsA-E69P in pBAD33 | This study |
Plasmid constructions
Oligonucleotides for plasmid constructions and mutagenesis are listed in Table S1. Plasmids pKG110, pKG116 and pRR48 were provided by Dr. J.S. Parkinson (University of Utah). Plasmids pKG110 and pKG116 are salicylate-inducible expression vectors that confer chloramphenicol resistance. The ribosome-binding sequence of pKG110 was converted to that of E. coli cheY, which causes an increase in expression level at all inducer concentrations, to yield pKG116. Plasmid pRR48 is an IPTG-inducible expression vector that confers ampicillin resistance and is compatible with pKG110 and pKG116. To introduce a multiple cloning site into pKG110, the NdeI-SalI fragment of pRR48 was replaced with the NdeI-SalI fragment of pKG110 to yield pWM2780. Plasmid pWM2784 encodes the FLAG epitope on an EcoRI-SacI fragment (Shiomi and Margolin, 2007).
The ftsA gene was amplified from pWM1260 (pET-FtsA) (Geissler et al., 2003) using primers 822 and 823 cloned as an XbaI-PstI fragment into pWM2784 (pDSW-FLAG vector) (Shiomi and Margolin, 2007), yielding pWM2785 (FLAG-FtsA). The double mutations, M71A + R286W (FtsA* mutation), or M71A + FtsA*, were introduced by two-step PCR. Primers 822/907 and 823/906 were used for M71A, 822/952 and 823/951 for R286W, and pWM2785 as a template for the first PCR; the resulting products were amplified with primers 822/823. The second PCR products were cloned as XbaI-PstI fragments into pWM2784 to yield pWM2786 (pDSW-FLAG-FtsA-M71A), pWM2787 (pDSW-FLAG-FtsA*), or pWM2788 (pDSW-FLAG-FtsA-M71A+A*), respectively. The resulting plasmids including the WT construct were amplified using primers 918 and 528, and the PCR products were cloned as NdeI-XhoI fragments into pWM2780 (pRR vector) to construct pWM2669 (pRR-FLAG-FtsA), pWM2671 (pRR-FLAG-FtsA-M71A), pWM2781 (pRR-FLAG-FtsA*), or pWM2782 (pRR-FLAG-FtsA-M71A-A*), respectively. To construct pWM2789 [pDSW-FLAG-FtsA-Δ1C (ΔVal127-Gln133)] and pWM3121 (pDSW-FLAG-FtsA-R300E), we amplified ftsA using primers 822/990 and 989/823 for the Δ1C deletion, and 822/1101 and 823/1100 for the R300E mutation. The resulting PCR products were then amplified using primers 822 and 823, and cloned as an XbaI-PstI fragments into pWM2784 to yield pWM2789 and pWM3121, respectively. To construct pWM2783 [pRR-FLAG-FtsA-Δ1C(127-133)], the ftsA gene from pWM2789 was amplified using primers 918 and 528, and the PCR product was cloned as NdeI-XhoI fragment into pWM2780, yielding pWM2783. The ftsA-E124A mutant was amplified from WM2417 genomic DNA as template with primers 823 and 824, and cloned as an XbaI-PstI fragment into pWM2784. The resulting plasmid was amplified using primers 918 and 528 and the PCR product was cloned as an NdeI-XhoI fragment into pWM2780, yielding pWM2702 (pRR-FLAG-FtsA-E124A). The E124A+M71A, I143L, or I143L+M71A mutations were introduced by two-step PCR using pWM2669 as a template to yield pWM3076 (pRR-FLAG-FtsA-M71A+E124A), pWM3077 (pRR-FLAG-FtsA-I143L), or pWM3078 (pRR-FLAG-FtsA-M71A+I143L).
To construct GFP fusions to FtsA mutants, the XbaI-PstI fragments of pWM2786 (FtsA-M71A), or pWM3095 (FtsA-E69P) were cloned into pDSW209, yielding pWM3191 (GFP-FtsA-M71A) or pWM3195 (GFP-FtsA-E69P). The resulting plasmids, along with pWM2760 (GFP-FtsA), were amplified using primers 975 and 528. The PCR products were cloned as NsiI-XhoI fragments into pKG110, yielding pWM3196 (GFP-FtsA), pWM3197 (GFP-FtsA-M71A), and pWM3200 (GFP-FtsA-E69P), respectively.
To construct DivIVA fusions to FtsA mutants, the XbaI-PstI fragments of pWM2786 (FtsA-M71A), pWM2789 (FtsA-Δ1C), or pWM3095 (FtsA-E69P) were replaced with the XbaI-PstI fragment of pWM1806 (pBAD33-DivIVA-FtsA) (Corbin et al., 2004) to yield pWM3070 (pBAD33-DivIVA-FtsA-M71A), pWM3071 (pBAD33-DivIVA-FtsA-Δ1C), or pWM3072 (pBAD33-DivIVA-FtsA-E69P), respectively.
To construct tandem FtsA repeats, ftsA, ftsA-M71A, ftsA-Δ1C, ftsA-E69P, and ftsA-R300E were amplified using primers 826 and 827. The resulting products were cloned as SacI-XbaI fragments into pWM2785 (FtsA), pWM2786 (FtsA-M71A), pWM2789 (FtsA-Δ1C), pWM3095 (FtsA-E69P), or pWM3121 (FtsA-R300E), yielding pWM2777 (pDSW-FLAG-AA), pWM3031 (pDSW-FLAG-WT-71), pWM3032 (pDSW-FLAG-71-WT), pWM3033 (pDSW-FLAG-71-71), pWM3035 (pDSW-FLAG-Δ1C-71), pWM3036 (pDSW-FLAG-Δ1C-Δ1C), pWM3086 (pDSW-FLAG-69-69), pWM3122 (pDSW-FLAG-300-300), pWM3123 (pDSW-FLAG-300-WT), pWM3124 (pDSW-FLAG-300-Δ1C), or pWM3125 [pDSW-FLAG-(300+Δ1C)-WT]. The construction resulted in a Ser-Arg dipeptide between two FtsA subunits. To express the tandem repeats in the pRR48 vector (pWM2780), these plasmids were further amplified using primers 918 and 696 and the PCR products were cloned as NdeI-PstI fragments into pWM2780 to yield pWM3079 (pRR-FLAG-AA), pWM3080 (pRR-FLAG-WT-71), pWM3081 (pRR-FLAG-71-WT), pWM3082 (pRR-FLAG-71-71), pWM3083 (pRR-FLAG-Δ1C-71), pWM3084 (pRR-FLAG-Δ1C-Δ1C), or pWM3085 (pRR-FLAG-69-69).
For bacterial two-hybrid assays, we used pKNT25-ftsZ kindly provided by Dr. D. Ladant (Institut Pasteur). To construct plasmids for bacterial two-hybrid assays, ftsA and its derivatives were amplified using primers 650/876 and 968/876, and the PCR products were cloned as PstI-EcoRI fragments into pKT25 and pUT18C (Karimova et al., 2005) to yield pWM3014 (T25-FtsA), pWM3015 (T25-FtsA-M71A), pWM3016 (T25-FtsA*), pWM3017 (T25-FtsA-M71A+A*), pWM3018 (T25-FtsA-Δ1C), pWM3021 (T18-FtsA), pWM3022 (T18-FtsA-M71A), pWM3023 (T18-FtsA*), pWM3024 (T18-FtsA-M71A+A*), or pWM3025 (T18-FtsA-Δ1C). These constructs had ftsA genes fused with the C terminus of either the T25 or T18 fragment, respectively, of the catalytic domain of Bordetella pertussis adenylate cyclase.
For extragenic suppression studies, the ftsZ gene was amplified using primers 965 and 967 and the PCR product was cloned as a PstI-XhoI fragment into pKG110, yielding pWM2765. The zipA and zapA genes were amplified using primers 971/972 and 973/974, respectively, and each PCR product was cloned as an NdeI-BamHI fragment into pKG110 and pKG116, yielding pWM3073 (pKG110-ZipA) and pWM3074 (pKG116-ZapA), respectively. The different salicylate-inducible plasmids were used to achieve optimal levels of ZipA or ZapA and to avoid the toxicity of excess ZipA. A gfp-ftsN fusion was amplified from pWM1152 (Corbin et al., 2004) with primers 975 and 977, and the PCR product was cloned as an NsiI-XhoI fragment into pKG116 to construct pWM3075 (pKG116-GFP-FtsN).
Site-directed mutagenesis of alpha-helix H1
To randomly mutagenize specific residues in alpha-helix H1, ftsA was amplified from pWM2669 with primer pairs of 822 + 1030, 1032, 1034, or 1036 and 823 + 1029, 1031, 1033, or 1035. The resulting PCR products were further amplified using primers 918 and 528, and the secondary products were cloned as XbaI-PstI fragments into pWM2784. WM1115 (ftsA12ts) was transformed with the resulting plasmids and plated in the presence or absence of 1 mM IPTG at 30 or 42°C. We classified plasmid-borne ftsA mutants as follows: (i) complemented WM1115 at 42°C and toxic in the presence of IPTG (wild-type FtsA-like clones), (ii) did not complement WM1115 at 42°C but toxic in the presence of IPTG (FtsA-M71A-like clones), or (iii) complemented WM1115 at 42°C but not toxic in the presence of IPTG (FtsA*-like clones). The cloned ftsA genes were then sequenced to verify the mutations. The ftsA-E69P mutant gene was further amplified using primers 918 and 528 and cloned as an NdeI-XhoI fragment into pWM2780 to yield pWM3055 (pRR-FLAG-FtsA-E69P).
Microscopy and immunoblotting
Microscopic examination of immobilized live cells, anti-FtsZ immunoflourescence microscopy, cell fixation, and staining with DAPI were done as described previously (Geissler and Margolin, 2005). Immunoblotting was performed with blots containing equal amounts of cell protein per lane using anti-FLAG (M2 antibody, Sigma), anti-DivIVA (Corbin et al., 2004), or anti-FtsZ as described previously (Shiomi and Margolin, 2007).
Plate viability spot assays
Plate viability spot assays were performed by spotting 10-fold dilutions of exponentially growing cells that were at equivalent densities, starting with a 10−1 dilution. Cells were grown overnight in LB supplemented with appropriate antibiotics at 30°C, diluted 1:100 and grown at 30°C for 4 h prior to spotting on plates with different inducer concentrations at 30 or 42°C.
Bacterial two-hybrid assays
Plasmids pKT25 and pUT18C carrying ftsA, ftsN, or pKNT25-ftsZ genes were used to co-transform the E. coli cya strain DHM1. Fresh colonies from plates were resuspended directly into 200 μl of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). 100 μl of the cell suspension was used to measure OD600 and 900 μl of Z-buffer was added to the rest (100 μl). Then, 40 μl CHCl3 and 20 μl 0.1 % SDS were added, vortexed and the reaction equilibrated to 30°C. 200 μl of a 4 mg/ml stock of o-nitrophenyl-β-d-galactopyranoside were added to the lysed cells and the mixtures were incubated at 30°C until yellow color developed. Then, 250 μl of 2 M Na2CO3 was added to stop the reaction. Finally, the OD420 was measured and specific β-galactosidase activity (Miller units) was determined.
Supplementary Material
Acknowledgments
We thank Dr. John S. Parkinson for providing plasmids pKG110, pKG116, and pRR48, Dr. Daniel Ladant for providing plasmids, strains, and antibody for the bacterial two-hybrid system, and Dr. Tushar K. Beuria for constructing the plasmid encoding T25-ftsN (pWM3183). This work was supported by National Institutes of Health grant R01-GM61074.
Abbreviations
- WT
wild-type
- IPTG
isopropyl-thiogalactoside
- DAPI
4’,6-diamidino-2-phenylindole
- DIC
differential interference contrast
REFERENCES
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Carettoni D, Gomez-Puertas P, Yim L, Mingorance J, Massidda O, Vicente M, Valencia A, Domenici E, Anderluzzi D. Phage-display and correlated mutations identify an essential region of subdomain 1C involved in homodimerization of Escherichia coli FtsA. Proteins. 2003;50:192–206. doi: 10.1002/prot.10244. [DOI] [PubMed] [Google Scholar]
- Corbin BD, Geissler B, Sadasivam M, Margolin W. A Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell SC, Anderson RE, Lowe J. Crystal structure of the bacterial cell division inhibitor MinC. Embo J. 2001;20:2454–2461. doi: 10.1093/emboj/20.10.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J. Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J. Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lallo G, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology. 2003;149:3353–3359. doi: 10.1099/mic.0.26580-0. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Shiomi D, Margolin W. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology. 2007;153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:514–526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Petrovska I, Boyd D, Beckwith J. Mutants, suppressors, and wrinkled colonies: Mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J. Bacteriol. 2007;189:633–645. doi: 10.1128/JB.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 2000;182:3965–3971. doi: 10.1128/jb.182.14.3965-3971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SO, Thompson LS, Harry EJ. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-Ring assembly. J. Bacteriol. 2005;187:6536–6544. doi: 10.1128/JB.187.18.6536-6544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, Biton J, Vicente M, Mingorance J, Massidda O. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 2005;55:699–711. doi: 10.1111/j.1365-2958.2004.04432.x. [DOI] [PubMed] [Google Scholar]
- Low HH, Moncrieffe MC, Löwe J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 2004;341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell. Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol. 2007;64:1129–1138. doi: 10.1111/j.1365-2958.2007.05735.x. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol. Microbiol. 2004;53:1359–1371. doi: 10.1111/j.1365-2958.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda S, Vicente M, Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W. The C-terminal domain of MinC inhibits assembly of the Z ring in Escherichia coli. J. Bacteriol. 2007;189:236–243. doi: 10.1128/JB.00666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Margolin W. FtsZ dynamics during the cell division cycle of live Escherichia coli. J. Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanedar S, Margolin W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Den Ent F, Löwe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Pearce KH, Payne DJ. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2000;270:387–392. doi: 10.1006/bbrc.2000.2439. [DOI] [PubMed] [Google Scholar]
- Yim L, Vandenbussche G, Mingorance J, Rueda S, Casanova M, Ruysschaert JM, Vicente M. Role of the carboxy terminus of Escherichia coli FtsA in self- interaction and cell division. J. Bacteriol. 2000;182:6366–6373. doi: 10.1128/jb.182.22.6366-6373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol. 2000;182:6203–6213. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.