SUMMARY
The receptor tyrosine kinase KIT plays an important role in development of germ cells, hematopoietic cells and interstitial pacemaker cells. Oncogenic KIT mutations play an important `driver' role in gastrointestinal stromal tumors, acute myeloid leukemias and melanoma among other cancers. Here we describe the crystal structure of a recurring, somatic oncogenic mutation located in the C-terminal Ig-like domain (D5) of the ectodomain rendering KIT tyrosine kinase activity constitutively activated. The structural analysis together with biochemical and biophysical experiments and detailed analyzes of the activities of a variety of oncogenic KIT mutations reveal that the strength of homotypic contacts and the cooperativety in the action of D4D5 regions determines whether KIT is normally regulated or constitutively activated in cancers. We propose that cooperative interactions mediated by multiple weak homotypic contacts between receptor molecules are responsible for regulating normal ligand-dependent or oncogenic RTK activation via a `zipper-like' mechanism for receptor activation.
INTRODUCTION
Stem cell factor (SCF) is a cytokine that functions as a ligand of the receptor tyrosine kinase KIT (Broudy, 1997). SCF is encoded by mouse Steel locus (Sl) and it is expressed in the form of a soluble or a membrane-anchored isoform; two isoforms generated by alternative RNA splicing and subsequent proteolytic processing of the SCF gene product (Anderson et al., 1991). It is now well established that SCF and its receptor KIT play an important role in the differentiation, proliferation and survival of hematopoietic stem cells, germ cells, and interstitial pacemaker cells of Cajal (Ashman, 1999).
KIT was initially discovered as cellular homolog of the feline sarcoma viral oncogene v-Kit (Besmer et al., 1986). KIT is a member of type-III receptor tyrosine kinase (RTK) family, which also includes platelet-derived growth factor receptors α, and β (PDGFRα, PDGFRβ), macrophage colony stimulating factor receptor (CSF1R/ Fms) and the Fms related tyrosine kinase 3 (Flt3/Flk2) (reviewed in Blume-Jensen and Hunter, 2001; Ullrich and Schlessinger, 1990). KIT is encoded by the murine White locus (W) and has two or four isoforms that are generated by alternative splicing in mouse and human, respectively (Rönnstrand, 2004). Like other members of type III RTK family, the extracellular (EC) domain of KIT is composed of five Ig-like domains, designated D1, D2, D3, D4 and D5, a single transmembrane domain (TM) and a cytoplasmic region (Figure S1A). The cytoplasmic region includes a regulatory juxtamembrane segment (JM), a catalytic tyrosine kinase domain (PTK) split by a kinase insert region, and a C-terminal tail.
Like other RTKs, KIT is stimulated by ligand-induced receptor dimerization (Schlessinger, 2000). It was shown that an SCF dimer binds to the first three membrane distal Ig-like domains of KIT (D1, D2 and D3) to form 2:2 KIT-SCF complex (Lemmon et al., 1997; Liu et al., 2007 Yuzawa et al., 2007). SCF binding induces KIT dimerization which increases the local concentration of the two membrane proximal Ig-like domains D4 and D5 resulting in conformational rearrangements and formation of homotypic contacts between D4 and D5 of neighboring KIT molecules (Mol et al., 2003; DiNitto et al., 2010; Liu et al., 2007; Yuzawa et al., 2007; Lemmon and Schlessinger 2010). KIT dimerization is followed by trans-autophoshorylation of tyrosine residues located in the juxtamembrane domain and in the activation loop of the kinase domain to release an autoinhibitory constraint and to stimulate KIT kinase activity, respectively. Tyrosine autophosphorylation sites located in the kinase insert region and in the C-terminal tail function as specific binding sites for the SH2 domains of signaling molecules that are recruited and activated in response to KIT activation (reviewed in Lemmon and Schlessinger, 2010).
Aberrant activation or KIT mutations were shown to be responsible for and are the main cause of a variety of human pathologies. Loss-of-function mutations of KIT in humans lead to piebaldism syndrome, which is characterized by hair and skin pigmentation defect, deafness, and constipation (Fleischman et al., 1991). Gain-of-function mutations of KIT are associated with various types of cancers including gastrointestinal stromal tumors (GIST), melanoma, acute myeloid leukemias (AML) and mastocytoma. Most somatic activating mutations are located in the JM, in D5 or in the kinase domain of KIT (Ashman and Griffith, 2012; Pittoni et al., 2011). Biochemical and structural analysis has shown that oncogenic mutations in JM region relieve an autoinhibitory constraint while mutations in the kinase domain enhance KIT enzymatic activity (Gajiwala et al., 2009). However it is not clear how oncogenic mutations in the extracellular domain of KIT that are primarily confined to D5, lead to ligand independent kinase activation and cell transformation. Moreover, the molecular mechanism through which D4-D4 and D5-D5 homotypic interactions mediate ligand dependent activation of WT KIT is also not understood. Here we present the crystal structure of a recurring somatic KIT mutation in D5 designated T417I,ΔD418,419 that was identified in AML patients. The structure reveals the molecular mechanism underlying oncogenic KIT activation in a ligand independent manner. Moreover, using biochemical and biophysical approaches as well as analyzing cells expressing a variety of oncogenic KIT mutations the mechanism of ligand stimulated and ligand independent KIT activation is unveiled. On the basis of these experiments, we propose that cooperative interactions mediated by multiple weak homotypic contacts between the extracellular, TM and cytoplasmic regions of KIT are responsible for stimulation of autophosphorylation, tyrosine kinase activation and cell signaling.
RESULTS
In order to elucidate the mechanism of action of oncogenic mutations in KIT extracellular region by structural and biophysical approaches, KIT fragments composed of the two membrane proximal Ig-like domains (D4D5) harboring several different oncogenic mutations in the D5, were expressed in baculovirus induced insect cells (SF9) (Figure S1B). In order to obtain diffraction quality crystals and to improve homogeneity of the protein preparations, the D4D5 fragments were deglycosylated by treatment with endoglycosidase F1. Crystallization hits were obtained for two oncogenic D4D5 mutants, the duplication of A502,Y503 (Dup A502,Y503), and the T417I, Δ418,419 mutant (Fig S1 C and D). After thorough refinement and optimization of the initial crystallization condition, large single crystals (100×80×80 μm) of the Dup A502,Y503 D4D5 mutant were obtained, but unfortunately their diffraction was not of sufficient quality to build a reliable model for the D5 domain (see Supporting Material for details).
Structure of the oncogenic T417I,Δ418,419 KIT D4D5 mutant
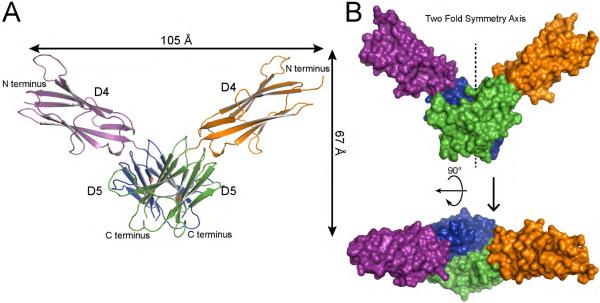
The structure of T417I,Δ418,419 mutant was determined to 2.4 Å resolution with 3 molecules in the asymmetric unit (Table S1); the first two molecules form a dimer in the asymmetric unit, and the third molecule forms a dimer with the symmetry mate (see Figure S2). A more complete trace was obtained for the two molecules forming a dimer in the asymmetric unit. The dimer interface for the third molecule was more blurred, but visible trace was the same as for the other two molecules. Dimer of T417I,Δ418,419 mutant reveals a “V” shaped 2-fold symmetric structure with approximate dimensions of 105 × 67 × 36 Å in which the two mutated D5 lie at the bottom of the “V” shaped structure forming strong contacts and the two D4 moieties constitute the two arms of the “V” (Figure 1). The overall structure is consistent with the notion that ligand-independent KIT activation induced by the T417I,Δ418,419 mutation is driven by homotypic contacts between mutated D5 species of neighboring KIT molecules. It is of note that the two salt bridges formed between Arg381 and Glu386, shown to play a critical role in ligand stimulated activation of WT KIT (Yuzawa et al., 2007), were not observed in the T417I,Δ418,419 structure.
Figure 1. Crystal structure of T417I,Δ418,419 KIT D4D5 fragment.
(A) Ribbon diagram of T417I,Δ418,419 KIT D4D5 dimer. D4 and D5 of one protomer are colored in purple and blue, and D4, D5 of another protomer are in orange and green respectively. N and C termini are labeled. Ile 417, an oncogenic mutation, is marked with a red balls. (B) Surface representation of T417I,Δ418,419 KIT D4D5 dimer. Upper panel shows a view following 90° rotation along the horizontal axis of the view shown in the bottom panel. Color coding is the same as in panel A. 2 fold symmetry axis is shown as vertical line passing through the middle of “V”.
Both KIT D4 and D5 are typical Ig-like domains composed of eight β strands (ABCC'DEFG), forming a β sheet sandwich like structure (Yuzawa et al., 2007). The overall structures of D4 or D5 of the T417I,Δ418,419 mutant are similar to those of the KIT ectodomain monomer or those of the KIT:SCF dimeric structure (Figure S3). A superposition of Cα atoms of individual D4 and D5 from KIT ectodomain (PDB code: 2EC8) to those of the T417I,Δ418,419 mutant gave a root mean square deviation (RMSD) of 0.4 Å and 1.8 Å for D4 and D5 over 80 and 64 carbon alphas, respectively. The higher RMSD value of D5 is likely caused by a deletion in β strand A and difference in DE loop folding (see Figure S3 and S4).
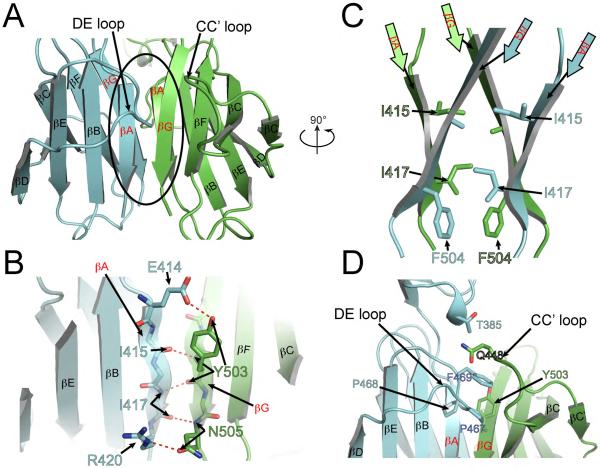
The structure presented in Figure 1 demonstrates that dimerization of the T417I,Δ418,419 mutant is solely driven by interactions mediated by homotypic D5 contacts. The solvent accessible buried surface area of the interface formed between the two protomers is approximately 1000 Å2. Majority of the contacts take place between strand A of one protomer and strand G of the second protomer (Figure 2 A, B and C). In addition, the DE loop of one protomer mediates interactions with the G strand and CC' loop of the other protomer (Figure 2A, 2D). Dimerization of two Ig-like domains of D5 results in formation of what looks like a single “elongated Ig-like domain” - each of the β sheets of one protomer are extended by a β sheet from a second protomer, resulting in a single continuous β sheet forming a β sandwich out of two “elongated” β sheets (Figure 2A). In other words, A and G strands link together two β sheets from different protomers to generate a continuous β sheets. In contrast to classical anti-parallel β sheet folding, the A and G strands are parallel to each other (Figure 2A). The interactions between A and G strands involve hydrogen bonds between backbone and side chains, as well as van der Waals contacts (Figure 2).
Figure 2. D5:D5 dimerization interface in the crystal structure of the oncogenic T417I,Δ418,419 KIT D4D5 mutant.
Ribbon representation of two D5 domains forming dimer. D5 from one protomer is colored with blue and D5 from another is in green. Secondary structure elements are labeled according to IgSF nomenclature. βA and βG – the hot spots for oncogenic mutation are highlighted in red. D4D5 dimer is rotated ~90° around vertical axis relative to the orientation of D4D5 dimer in Figure 1A. (A) Overview of D5:D5 dimerization interface. (B) Detailed view of contacts between βA of one protomer and βG of another protomer. Hydrogen bonds are shown with red dotted lines. Side chains are shown only for those residues, which form hydrogen bonds. (C) The same orientation as D4D5 dimer in Figure 1A and 90° rotated relative to orientation in A. Only strands A and G from each protomer are represented. Side chains involved in van der Waals interactions are represented with sticks model. (D) Contacts between DE loop of one protomer and strand G and CC' loop are depicted. Side chains involved in van der Waals interactions are shown with sticks and labeled.
Figure 2D shows that DE loop of one protomer makes additional van der Waals contacts with strand G and CC' loop of another protomer. Superposition of WT D5 (PDB code: 2EC8) with D5 of the T417I,Δ418,419 mutant shows a significant movement of the DE loop towards the dimerization interface. The movement of DE loop is possible because of the deletion that took place in strand A. Superposition of individual D5 moieties shows that the DE loop of the T417I,Δ418,419 mutant would clash with strand A of D5 of WT KIT (Figure S4). The overall structure of T417I,Δ418,419 mutant shows that deletion of Y418 and D419 together with a T417I mutation renders complimentary between β strand A of one protomer with β strand G of a second protomer with additional contacts with the DE loop.
The structure of D5 oncogenic dimer fits into the EM structure of ligand induced WT KIT dimer
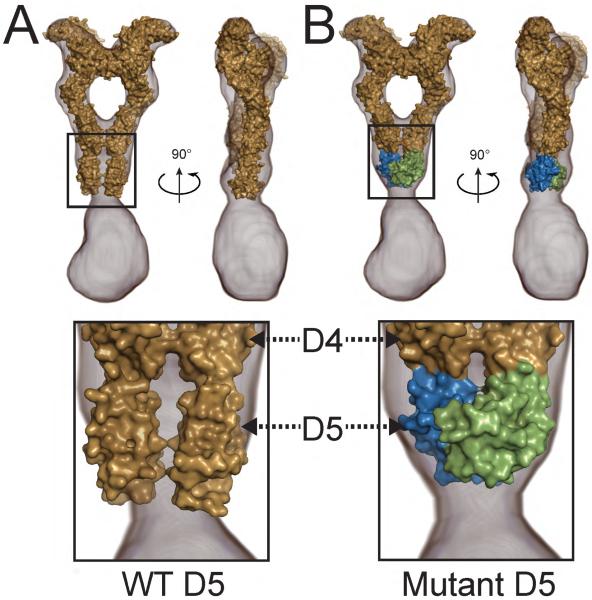
We have recently shown that the extracellular region of the EM 3D reconstruction of intact KIT dimer (EMD-2648, Opatowsky et al., 2014) is very similar to the crystal structure of the extracellular regions of SCF stimulated KIT dimers (PDB ID: 2E9W; Yuzawa et al. 2007) except for the dimerization interface of D5 contacts. Comparison of the two structures shows that D5 contacts in the crystal structure (PDB ID: 2E9W) do not fit well to the EM reconstruction of the same region in the ligand simulated intact KIT dimer (Figure 3A and Opatowsky et al., 2014). We surmise that the weak affinities of individual D4 and D5 towards homotypic interactions and the flexibility of the linker connecting these two membrane proximal Ig-like domains, together with increased dimensionality and absence of additional interactions mediated by the TM and cytoplasmic domains which hold the ligand stimulated dimers of WT KIT together, are lost in the ectodomain structure resulting in segmental flexibility of D5 relative to the rest of the isolated KIT ectodomain (Opatowsky et al., 2014). Interestingly, comparison of the crystal structure of the dimeric D5 of T417I,Δ418,419 mutant to the corresponding region in the EM envelope of SCF stimulated KIT dimer shows a very good fit between the model of D5:D5 dimer and the volume of the EM reconstruction in this region. Figure 3B demonstrates an excellent fit between the model of the extracellular domain of KIT that includes a D5:D5 dimer from T417I,Δ418,419 crystal structure and the volume of EM reconstruction in this region of ligand stimulated intact KIT receptor (EMD-2648). Based on this model, we propose that D5:D5 interface identified in D5 dimeric T417I,Δ418,419 exhibits hallmark resembling those of the fully activated membrane proximal region of WT KIT although the oncogenic mutation utilizes an interface that mediates significantly tighter homotypic D5 association.
Figure 3. Fitting of KIT ectodomain crystal structure into electron microscopy EM 3D reconstruction.
(A) Crystal structure of KIT ectodomain in complex with SCF (PDB ID 2E9W, Yuzawa et al., 2007) was fitted into the volume of the 3D EM reconstruction (EMD-2648, Opatowsky et al, 2014). KIT:SCF crystal structure is surface represented and colored with beige; 3D EM reconstruction is represented with gray mesh (EMD-2648, Opatowsky et al, 2014). Right panel is 90° rotated along vertical axis relative to the left panel. The homotypic D5 contacts seen in the crystal structure do not fit into the volume of the 3D EM reconstruction (depicted in the lower panel).
(B) D1–D4 domains from KIT:SCF complex (PDB ID 2E9W, Yuzawa et al., 2007) and D5 dimer from our current structure of T417I,Δ418,419 KIT mutant are grafted together and fitted into 3D EM reconstruction (EMD-2648, Opatowsky et al, 2014). Color code for D1–D4 of KIT and SCF is the same as in panel A. D5:D5 dimer from T417I,Δ418,419 structure are colored with blue (one protomer) and green (second protomer).
Restoration of ligand dependency by mutation of residues in the interface of dimeric D5 oncogenic KIT mutant
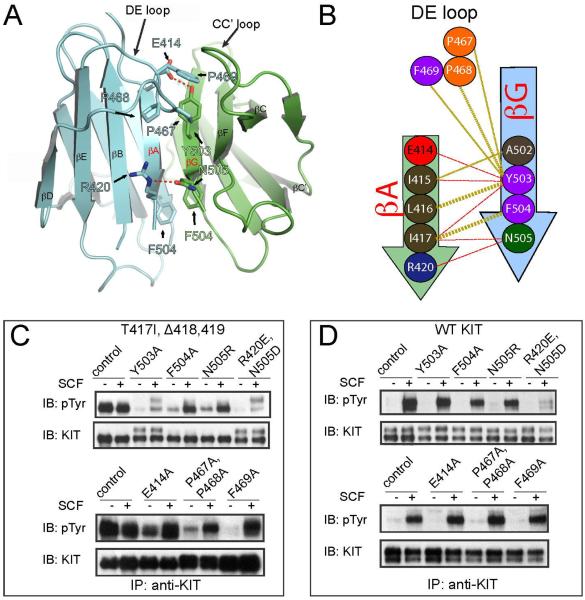
To explore the biological significance of the newly identified homotypic D5 contacts in the T417I,Δ418,419 mutant (Figure 4A and B) and to address the question of whether this interface is responsible for ligand-independent dimerization and activation of KIT, we generated and characterized the tyrosine kinase activities of full size KIT harboring the T417I,Δ418,419 mutations (Figure 4C) in comparison to WT KIT (Figure 4D). Moreover, we also examined the possibility of whether it is possible to restore ligand dependency in the T417I,Δ418,419 oncogenic mutant by generating additional mutations in amino acids critical for maintaining the interface of the D5 oncogenic mutant (Figure 4C) and as a control the same mutations were introduced in the background of WT KIT. WT or the KIT mutants were transiently expressed in HEK293 cells matched for expression level (Figure 4C and D) and their tyrosine kinase activities were compared for SCF stimulated (25 ng/ml) or unstimulated cells. (Figure 4C and D). The level of receptor tyrosine phosphorylation was determined by subjecting cell lysates to immunoprecipitation with anti-KIT antibodies followed by immunoblotting with anti-pTyr antibodies.
Figure 4. Rescue of ligand dependency of the oncogenic T417I,Δ418,419 KIT mutant.
(A) Cartoon representation of D5D5 interface. Residues, which were mutated in (C and D) are shown with sticks and labeled. Color code is the same as in Figure 2.
(B) Schematic representation of D5:D5 interface contacts for T417I,Δ418,419 mutant. One protomer is colored by blue and another protomer by green. Beta strands are represented with arrows. Hydrogen bonds are shown with red dashes and hydrophobic interactions with golden dashes. Residues involved into the dimer interface formation are shown with circles and numbered according to WT KIT. Interface contacts were calculated using the PDBsum server (Laskowski, 2009). Only β strands A and G, and DE loop are highlighted, a full scheme of 2D secondary structure of T417I,Δ418,419 mutant is represented in Figure S5.
(C) Ligand dependent activity of T417I,Δ418,419 oncogenic KIT mutant can be fully or partially restored by point mutations of residues involved in the D5:D5 dimerization interface. (D) The same point mutations in the context of wild-type KIT (WT KIT) receptor do not interfere with tyrosine kinase activities except for the double mutation of R420E and N505D. 3T3 cells transiently expressing full length KIT receptor variants (as indicated in the upper panel) were stimulated with 25 ng/ml SCF for 6 min at 37°C. (C) Residues indicated in the upper panel were mutated in the context of T417I,Δ418,419 oncogenic KIT mutant. (D) Residues indicated in the upper panel were mutated in the context of WT KIT.
The experiment presented in Figure 4C shows that several mutations in the interface responsible for mediating dimerization of the T417I,Δ418,419 KIT oncogenic mutant resulted in full or partial restoration of ligand-dependency of this oncogenic KIT receptor. These mutations were classified into three major groups: 1. The first group included the single F504A, N505R, E414A mutations or the double P467A,P468A mutation; these four mutants exhibited different levels of basal tyrosine kinase activities that can be further enhanced in response to SCF stimulation; 2. The second group included the single Y503A and a double R420E, N505D mutations; these two mutants exhibited reduced SCF stimulated tyrosine kinase activities, and 3. The third F469A mutation exhibited WT-like characteristics, namely fully restored SCF stimulation of tyrosine kinase activity. The experiment presented in Figure 4D also shows that all the mutations, except for the R420E,N505D mutant, did not affect either basal or ligand stimulated activation of WT KIT suggesting that certain elements of T417I,Δ418,419 dimeric interface do not play a role in D5 homotypic contacts required for WT KIT ligand stimulation. The partial loss of the kinase activity of the R420E, N505D double mutation may indicate that R420 and N505 play a role in mediating D5 homotypic contacts of WT KIT or that these mutations adversely affect domain folding.
In vitro dimerization of isolated KIT D4D5 variants
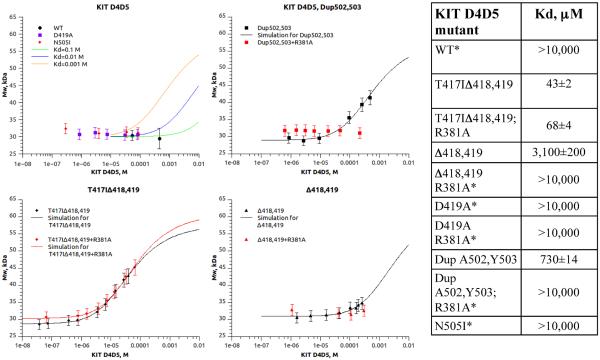
In order to further dissect mechanism of ligand independent activation of oncogenic KIT D5 mutants, we compared the capacity of different variants of KIT D4D5 fragments to form dimers in solution (Figure 5). Soluble variants of WT or oncogenic D4D5 mutants were expressed in Sf9 insect cells and purified. Different concentrations of KIT D4D5 variants were subjected to size exclusion chromatography and dimerization was monitored using multiangle laser light scattering (MALS) and refractometer index (RI) detectors to measure weight average molar mass. Dimerization constants (Kd) were determined by fitting weight average molar masses (see equation 1 in SI Experimental Procedures). The results presented in Figure 5 demonstrate that WT KIT D4D5 fragment remains monomeric in solution up to a concentration of ~0.5 mM. These data are consistent with the results of similar experiments performed with VEGFR D7, an Ig-like domain homologous to KIT D4, demonstrating that D7 of VEGFR remains monomeric in solution at similar concentrations (Yang et al., 2010). These experiments demonstrate that homotypic associations between WT D5 are very weak and likely have a secondary role in mediating lateral association between KIT membrane proximal Ig-like domains. To obtain an estimate of the dimerization constants for WT D4D5, we simulated dimerization curves with different Kd values and compared these curves with our experimental data (see equation 1 in SI Experimental Procedures section). Simulation of dimerization constants suggests that the dimerization Kd of WT D4D5 is very weak and does not exceed 10 mM (Figure 5). Dimerization experiments with KIT D4D5 fragments bearing oncogenic mutations revealed increased binding affinity between mutated D5 molecules. The experiment presented in Figure 5 demonstrates that oncogenic T417I,Δ418,419; Dup A502Y503 and Δ418,419 mutations significantly increased the dimerization affinity of D4D5 fragments (Figure 5) as compared to the dimerization of WT D4D5 which was too low to be measured using MALS. Interestingly, dimerization constants of two other oncogenic KIT mutants, D419A and N505I, were also too low to be determined at similar protein concentrations (Figure 5).
Figure 5. Multiangle light scattering (MALS) experiment demonstrating dimerization state of KIT D4D5 fragments (WT and oncogenic mutants).
Different concentrations of KIT D4D5 fragments were injected on to HPLC system equipped with light scattering (LS) detector and differential refractometer (RI). Weight-average molar mass was calculated based on LS and RI signals. The weight average molar masses were plotted as a function of concentration (dots on the graph). Solid line represents simulation of dimerization curves (top left panel) or nonlinear regression fit of the data to the model (Equation 1). Dimerization constants calculated by nonlinear regression fit are presented in the table (right panel).
We have previously demonstrated that ligand stimulated activation of KIT or PDGF-receptor (PDGFR) was strongly compromised by point mutations that prevent formation of homotypic D4 contacts mediated by salt bridges between Arg381 and Glu386 residues on D4 of neighboring receptors (Yuzawa et al., 2007; Yang et al., 2008). Although the pair of salt bridges that mediates homotypic D4 contacts are critical for KIT or PDGFR activation, these contacts are not required for KIT or PDGFR ligand stimulated dimerization (Yang et al., 2008). In order to examine potential cooperativity between D4 and D5 domains in the process of homotypic association, we mutated Arg 381 of D4 to Ala (R381A) in KIT D4D5 oncogenic variants and compared dimerization affinity of D4D5 fragments in the presence or absence of the salt bridge (contacts between Arg381 and Glu386) in D4 (Figure 5). These experiments revealed that the salt bridge that mediate D4 homotypic association are crucial for dimerization of the DupA502,Y503 as well as for Δ418,419 mutants; no dimerization was detected in these two mutants upon mutation of Arg381. In contrast, mutation of the salt bridge in D4 had virtually no effect on dimerization of the T417I,Δ418,419 oncogenic mutant. Our results show that cooperativity between D4 and D5 is crucial for dimerization of membrane proximal Ig-like domains of oncogenic mutants harboring weak homotypic D5 associations but a mutant harboring strong homotypic D5 contacts such as T417I,Δ418,419 does not require contribution from D4 for mediating ligand independent dimerization of membrane proximal Ig-like domains.
Homotypic contacts in the membrane proximal Ig-like domains influence the tyrosine kinase activity of oncogenic KIT mutants
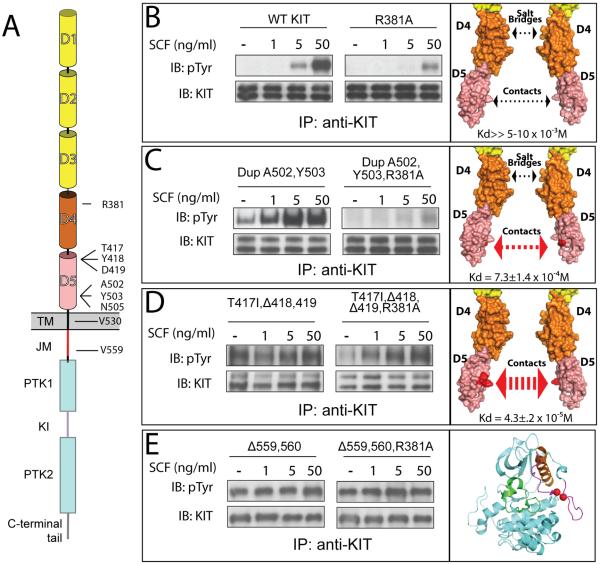
In order to gain further insight concerning the role played by D4 or D5 homotypic contacts in KIT activation, we compared tyrosine kinase activity of WT and oncogenic mutants of the full length KIT receptor in the cell based assay. We explored the possibility of whether the tyrosine kinase activity of oncogenic KIT mutants is controlled by homotypic D4 contacts between Arg381 and Glu386, a salt bridge known to play an important role in SCF induced WT KIT activation (Figure 6 and Yuzawa et al., 2007).
Figure 6. In vivo autophosphorylation of WT or oncogenic KIT mutants.
(A) Schematic representation of full length KIT receptor. D1 – D5 are Ig-like domains. D1–D3 represented with yellow, D4 and D5 with orange and pink cylinders respectively. TM is transmembrane (black), JM - juxtamembrane (red), PTK - protein tyrosine kinase (blue), KI - kinase insert (purple) domains. Oncogenic mutations and Arg381 are labeled.
Tyrosine kinase activity was compared for WT – wild type KIT receptor (B); DupA502,Y503 – duplication of Ala 502 and Tyr 503 (C); T417I ΔY418D419 – substitution of Thr 417 to Ile and deletion of Tyr 418 and Asp 419 (D), and Δ559,560 – deletion of Val 559 and 560 (E). WT or oncogenic KIT mutants (left panel) were compared to those harboring an additional mutation of an Arg 381 (middle panel) - residue responsible for D4:D4 homotypic contacts. NIH-3T3 cells stably expressing WT KIT or oncogenic mutants were stimulated with increasing concentration of SCF (as indicated in the upper panel) for 6 minutes at 37°C. Lysates of unstimulated or SCF-stimulated cells were subject for immunoprecipitation (IP) using anti-KIT antibodies followed by SDS-PAGE and immunoblotting (IB) with anti-pTyr (IB: pTyr) or anti-KIT (IB: KIT) antibodies (as indicated). Right panel – surface representation of membrane proximal Ig-like domains (B–D) or cartoon representation of the kinase domain (E), oncogenic mutations are highlighted with red. The strength of D5:D5 contacts and impact of D4 salt bridge schematically represented with arrows. Dimerization constants for D4D5 fragment are shown in the bottom of the right panel.
NIH-3T3 cells stably expressing WT or oncogenic KIT mutants were matched for similar levels of KIT receptor expression (Figure 6 bottom panels in B, C, D and E section) were stimulated with 1, 5, or 50 ng/ml of SCF or with buffer alone for six minutes at 37°C. Lysates of SCF stimulated or unstimulated cells were subjected to immunoprecipitation (IP) with anti-KIT antibodies followed by SDS-PAGE and immunoblotting (IB) with anti-p-Tyr antibodies. The experiment presented in Figure 6 (top panels in B, C, D and E section) shows SCF dependent activation of KIT autophosphorylation for WT KIT, oncogenic mutants in D5: Dup A502,Y503; T417I,Δ418,419, and deletion in the intracellular JM Δ559,560. Analysis of tyrosine kinase activity of oncogenic and WT full length KIT, and analysis of in vitro dimerization of D4D5 variants allowed us to group WT and oncogenic KIT mutants into four distinct groups based on how D4 and D5 homotypic contacts influence ligand dependency of KIT receptor variants:
Ligand-dependent KIT activation. We have shown that the binding affinity of isolated WT D4D5 towards each other is very weak with a Kd that does not exceed 10 mM. Nevertheless, binding of an SCF dimer to KIT receptor in the cell membrane leads to efficient ligand induced KIT activation driven by cooperative association between the membrane proximal D4 and D5 that is enhanced by a dimensionality effect. SCF induced KIT stimulation is dependent on salt bridge-mediated D4 homotypic contacts (Yuzawa et al., 2007) and dimerization of isolated D4D5 is prevented by compromising salt bridge formation between neighboring D4 species. (Figure 6 B).
Ligand-sensitized oncogenic KIT mutation. The dimerization constant of isolated D4D5 of the oncogenic Dup A502,Y503 is increased by at least 10–15 fold as compared to WT D5 to a Kd of approximately 730 μM (Figure 5) resulting in elevated tyrosine autophosphorylation of unoccupied KIT (basal phosphorylation, Figure 6 C). Nevertheless, KIT receptor with enhanced D5 homotypic contacts relies on D4 salt bridges formation for dimerization; dimer formation of Dup A502,Y50 D4D5 with an additional mutation in D4 (R381A) was prevented. It is notworthy that three additional oncogenic mutations in the extracellular domain (D419A; Δ418,419, and N505I) and a mutation in the transmembrane (V530I) region belong to the same category of ligand sensitized oncogenic KIT mutations (see Figure S6 for the details). Full length KIT harboring these mutations exhibited elevated basal tyrosine kinase activity, which can be further stimulated by SCF binding.
Constitutively activated ligand-independent oncogenic KIT mutations. The dimerization constant of isolated D4D5 harboring the oncogenic T417I,Δ418,419 mutation is increased by 200–250 fold as compared to WT D5 to a Kd of approximately 43 μM. Moreover, full length KIT receptor harboring this mutation exhibited ligand-independent tyrosine kinase activity that does not depend upon salt bridge-mediated homotypic D4 contacts (Figure 6 D).
Constitutively activated ligand-independent oncogenic mutations in KIT cytoplasmic domain. Gain of function mutations in cytoplasmic domain such as the Δ559,560 mutant relieve an autoinhibitory constraint (cis autoinhibition) rendering KIT receptor constitutively activated (Figure 6 E). These mutations do not depend on dimerization of the extracellular domain and as a consequence do not rely on D4:D4 and/or D5:D5 homotypic contact formation.
DISCUSSION
Determination of the crystal structure of a somatic mutation in the extracellular domain of KIT reveals how a highly regulated ligand stimulated RTK is converted into a constitutively-activated ligand-independent oncogenic receptor. These experiments demonstrate that oncogenic mutations that alter the strength of homotypic interactions between the membrane proximal Ig-like domain D5 have a profound effect on KIT activation. Moreover, cooperativety in the interactions between WT or mutated D5 with D4 of KIT extracellular domain enable the maintenance of either normal ligand regulated WT KIT activation or the generation of several distinct forms of aberrantly activated oncogenic KIT mutants.
Analysis of the crystal structure shows that the majority of interactions between the two neighboring D5 of the oncogenic mutant T417I,Δ418,419 take place between A and G β-strands and that nearly all gain of function mutations in KIT ectodomain are located in these two β-strands (Ashman and Griffith, 2012; Pittoni et al., 2011). We have also shown that the binding affinity of homotypic contacts between D5 of WT KIT is very low, but deletions, substitutions and/or point mutations in A or G β-strands that increase the binding affinity between oncogenic D5 mutations have a strong impact on KIT tyrosine kinase activation resulting in either ligand-dependent or ligand-independent mechanisms. In addition to A and G β-strands mediated contacts in D5 dimer of T417I,Δ418,419 mutant, the DE loop also makes interactions with β-strand G and CC' loop. Experiments presented in the manuscript demonstrate that these interactions play an important role in maintaining the dimerization interface of the oncogenic T417I,Δ418,419 mutant and that disruption of these interactions results in full or partial restoration ligand dependency of tyrosine kinase activity of the D5 oncogenic KIT mutant (Figure 4 A, B and C).
It is clear that the D5 dimer in the crystal structure of the soluble extracellular KIT domain shows less compact contacts in comparison to the D5 dimer seen in the EM envelope of full size dimeric KIT (Figure 3A and Opatowsky et al, 2014, EMD-2648). The limited D5 contacts seen in the crystal structure of the extracellular domain are likely caused by the flexibility of the hinge between D4 and D5, by the weak binding affinity of homotypic D5 contacts and by severing additional homotypic interactions mediated by the TM and cytoplasmic domains of neighboring KIT molecules which are seen in the EM reconstruction of SCF stimulated dimer of intact KIT receptor. We show here that the structure of the oncogenic D5 mutant dimer fits well into the envelope of EM reconstruction of ligand stimulated intact KIT receptor dimer. The excellent fit of the oncogenic D5 dimer is caused by the substantial 100-250 fold increase in the dimerization constant of the D5 oncogenic mutant resulting in more compact contacts (Figure S7) similar to those seen in the EM envelope of corresponding region of WT KIT. It is noteworthy that most of the mutations introduced in the interface of the oncogenic D5 dimer that rescue ligand dependency of KIT do not affect ligand induced activation of WT KIT. Moreover, as the dimerization constant of oncogenic D5 dimer is several hundred fold higher then the dimerization constant of WT D5 dimer it is likely that the dimerization interface of normal D5 contacts induced by ligand stimulation differs for the most part from the oncogenic D5 dimeric interface.
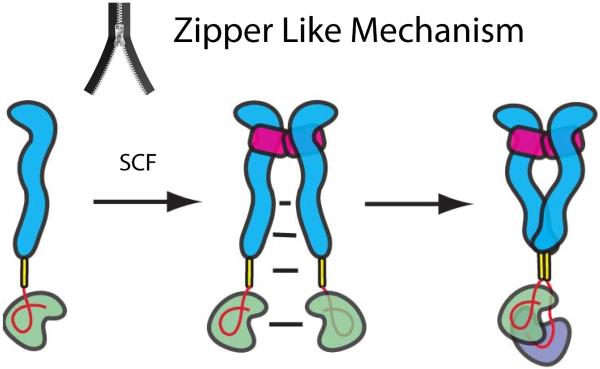
On the basis of the structural and biochemical analyses described in this report, it is possible to draw a new and more refined model for how SCF binding stimulates KIT activation and how oncogenic mutations in D5 of KIT extracellular region activate KIT via a ligand-independent (constitutively) mechanism or through a ligand sensitized manner. Prior to ligand binding, the lion's share of WT KIT molecules exist in the cell membrane as laterally mobile, inactive receptor monomers. While a small population of dimeric KIT molecules may exist prior to ligand binding, the binding affinity of KIT monomers towards each other is too low to drive the formation of a significant population of KIT dimers before SCF stimulation. The structural and biochemical experiments presented in this and earlier reports have shown that KIT dimerization is entirely driven by SCF binding, whose sole role (as a homodimeric cytokine) is to bring two KIT molecules together. The binding of SCF to the ligand binding region (i.e. D1, D2 and D3) brings the remaining parts of the two KIT molecules (D4, D5, TM and the cytoplasmic region) to a distance of 75 Å from each other (Opatowsky et al., 2014 and Yuzawa et al., 2007). At this short distance, the local concentration of KIT protomers is approximately 10−5M (Klein et al, 2004; Yang et al., 2008). This high, local concentration allows multiple weak homotypic contacts to form between D4, D5 and other parts of neighboring KIT molecules and leads to robust receptor-receptor association. We propose that the inter-receptor contacts propagate along the entire length of neighboring KIT molecules in a cooperative manner via a “zipper-like” mechanism that moves from the SCF binding region to D4, D5 to the TM domain and finally to the cytoplasmic region leading to close interactions throughout the entire length of KIT structure of each receptor protomer (Fig 7). The dimerization constant of two D4D5 fragments carrying a T417I,Δ418,419 D5 mutations towards each other (Kd = 43 μM) is 100-250 fold higher than the dimerization constant of WT D4D5. Such changes in dissociation constant have a profound effect on receptor dimerization since the apparent concentrations of the membrane proteins on the cell surface are significantly higher due to reduced dimensionality. We previously used an approach based on “average distance to nearest neighbor” calculation (Klein et al, 2004) to estimate that the apparent concentration of a receptor in the membrane of a cell expressing 20,000 receptors per cell is approximately 1–3 μM (Klein et al, 2004; Yang et al., 2008). It was similarly proposed by Grasberger et al. (1986), that the dimerization constant for membrane proteins on the cell surface is enhanced by approximately 106 times compared to the same interactions in solution. Accordingly, approximately 10% of KIT harboring the strong oncogenic T417I,Δ418,419 D5 mutation are displayed on the cell surface in the form of activated receptor dimers and less than 1% of the Dup Ala502,Tyr503 D5 mutation, which has a less tight dimerization Kd, are displayed on the cell surface as activated dimers that can be further activated by ligand stimulation. Moreover, the D419A or N505I D5 mutations which exhibit dimerization Kd in the range of 10−3 M show high basal tyrosine kinase activity and sensitized SCF stimulation in comparison to ligand stimulation of WT KIT.
Figure 7. A “zipper-like” mechanism for ligand-dependent or oncogenic KIT activation.
SCF binding brings two KIT monomers together. The increase in local concentration of KIT upon ligand induced dimerization enables formation of multiple low affinity associations between different regions of neighboring KIT molecules by means of a “zipper-like” process that propagates in a cooperative manner from the SCF binding region to D4, D5, TM and to the cytoplasmic region leading to formation of asymmetric KIT dimers. We propose that the asymmetric kinase arrangements represent snapshots of molecular interactions taking place between two tyrosine kinase domains poised towards trans autophosphorylation of tyrosine residues located in different parts of the cytoplasmic region in an orderly manner. In this process the membrane proximal cis JM autoinhibited kinase may function as a substrate (green) and the membrane distal kinase will function as an active enzyme (purple). After trans autophosphorylation the roles of the two kinase molecules are switched. Ectodomain of KIT depicted in blue, TM in yellow, JM region is shown as a red loop and SCF in magenta.
A zipper-like mechanism was previously proposed for a surface receptor protein tyrosine phosphatase α (RPTPα), where multiple weak interactions between two monomers zipper up into dimers (Jiang et al, 2000). However, by contrast to RTK activation which is mediated by ligand induced receptor dimerization it was proposed that monomeric RPTPα functions as the active tyrosine phosphatase while dimerization RPTPα serves as a mechanism for inhibition of tyrosine kinase activity of this family of enzymes. The molecular mechanism depicted in Figure 7 for ligand stimulated or oncogenically induced KIT activation relies on efficient lateral encounters between mobile receptor molecules in the cell membrane enabled by reduced dimensionality and cooperative “zipper-like” action of several specific but weak homotypic associations between each of D4, D5, TM and/or cytoplasmic regions of neighboring KIT molecules.By introducing a R381A mutation that disrupts salt bridges essential for mediating homotypic contacts between neighboring D4 we show strong cooperativity in the action of the two membrane proximal Ig-like domains, essential for ligand induced stimulation of WT KIT as well as ligand-independent or ligand-sensitized activation of certain oncogenic KIT mutations. However, certain oncogenic mutations such as the T417I,Δ418,419 mutation in D5 are minimally affected by preventing homotypic D4 contacts and the ΔV559 oncogenic mutation in the intracellular JM region is not affected at all by preventing D4 homotypic association.
The delicate balance between ligand binding, formation of homotypic receptor contacts, trans autophosphorylation and kinase stimulation are disrupted by oncogenic mutations in different regions of KIT molecule shown to play critical regulatory roles. Comparison of the tyrosine kinase activities of oncogenic KIT mutations has shown that enhanced association between oncogenic D5 mutants or release of cis autoinhibition by oncogenic JM mutation may initiate a “zipper-like” process that leads to robust KIT contacts and tyrosine kinase activation in a ligand-independent manner. Other oncogenic mutations become more responsive to ligand stimulation by sensitizing the “zipper like” process, but still in a ligand-dependent manner. We have previously demonstrated that upon ligand stimulation the cytoplasmic domains of KIT form asymmetric complexes (Opatowsky et al, 2014). We proposed that the asymmetric kinase arrangements represent snapshots of molecular interactions that take place between two kinase molecule poised towards trans autophosphorylation of tyrosines on different parts of the cytoplasmic region with distances of 50 Å from each other in an orderly manner (Furdui et al., 2006; Lew et al., 2009; Bae et al., 2010).
How general is the zipper-like mechanism for activation of RTKs and other membrane receptors? Because of their structural similarity, all type-III and type-V RTKs are probably activated by a zipper-like mechanism similar to the one presented in Figure 7. The salient features of the mechanism presented in Figure 7 may apply for the activation of all RTK and perhaps also for other multi-domain containing surface receptor possessing a single TM irrespective of the exact mechanism by which their ligands stimulate receptor dimerization.
EXPERIMENTAL PROCEDURES
Proteins Expressions and purifications
Different DNA constructs of human KIT D4D5 fragments (aa 308-514 and 308-507) were used in this study: KIT D4D5 variants with noncleavable 6xHis tag at the C-terminus were used in the light scattering experiments, and D4D5 variants with N-terminus 6xHis tag followed by TEV cleavage site were used for crystallization (see Supplementary Material). All constructs contained the original signal sequence of human KIT. The cDNAs were cloned into pFastBac1 (Invitrogen) by NcoI and HindIII restriction sites. Site directed mutagenesis was used to generate oncogenic variants of KIT D4D5.
Soluble KIT D4D5 fragments were expressed in Sf9 insect cells according to the Bac-to-Bac instruction manual (Invitrogen). For light scattering experiments KIT D4D5 fragments were purified by Ni-affinity followed and size exclusion chromatographies. The D4D5 fragments of KIT used for crystallization was partially deglycosylated using recombinant Endoglycosidase F1 and additionally purified by anion exchange and size exclusion chromatography. For details see Supplemental Experimental Procedures.
Crystallization and Data Collection
T417/IΔ418,419 variant of KIT D4D5 was crystallized at 22°C by hanging-drop technique containing equal volumes of protein solution and reservoir buffer (10%–18% [w/v] PEG 3350, 20 mM CoCl2). All crystals were transferred to the reservoir solution supplemented with 30% glycerol, loop mounted and flash frozen in liquid nitrogen. The crystals belonged to centered monoclinic C2 space group (a=163.59, b=63.54, c=81.55 Å and β=117.47°) with three molecules in the asymmetric unit. The solvent content of these crystals was around 51%.
Structure Determination
The structure of T417I,Δ418,419 variant of KIT D4D5 was solved by molecular replacement. The program Phaser (McCoy et al., 2007) and the CCP4 suite (Winn et al., 2011) were used to locate 3 copies of individual D4 domain of KIT and 2 copies of individual D5 domain of KIT ((Yuzawa et al., 2007), PDB code: 2EC8). Part of the third copy of D5 domain was built manually. Positions of the Co atoms were determined by MR-SAD. Although strong difference density was observed in the second part of the third D5 domain, the B factor for this region was too high to allow for a reliable model to be built. Multiple rounds of the model rebuilding and refinement were carried out with Coot (Emsley et al., 2010), REFMAC5 (Murshudov et al., 2011) using a maximum-likelihood target with TLS, and Phenix (Adams et al., 2010). Water molecules were added automatically by Phenix-refine during the final rounds of the refinement. The final model of KIT D4D5 variant comprises residues 310 – 506 (numbering according to WT human KIT), excluding poorly ordered regions (see Table S1 for data collection and refinement statistics details). Molecular images were produced using Pymol molecular visualization system (www.pymol.org). Dimerization interface areas were evaluated with PISA (Krissinel and Henrick, 2007) and PDBsum (Laskowski, 2009).
Cell Culture, Immunoprecipitation, and Immunoblotting Experiments
NIH-3T3 and HEK-293 cells were cultured in the presence of DMEM containing 10% bovine serum and fetal bovine serum respectively. A retroviral vector, pBABE, containing a puromycin resistance gene was utilized to make stable NIH-3T3 cell lines expressing KIT-WT and its oncogenic mutants. For transient expression in HEK-293 cells we used pCDNA3.1 expression vector and cells were transfected with 1μg DNA using lipofectamin (Invitrogen). Prior to lysis cells were stimulated with indicated concentrations of human SCF (previously described, (Yuzawa et al., 2007) and then lysed in lysis buffer (Lax et al., 2002). Cell lysates were incubated over night with anti-KIT antibodies (polyclonal antibodies raised against the full length extracellular domain), separated on SDS-PAGE and immunoblotted with either anti-KIT antibodies or anti-pY antibodies. Procedures for cell immunoprecipitation and immunoblotting with indicated antibodies were performed as previously described (Batzer et al., 1994; Mohammadi et al., 1996).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grant from Kolltan (IL). The SEC-MALS instrumentation was supported by NIH Award Number 1S10RR023748-01 (EF). We thank the members of Schlessinger laboratory for valuable discussions and critical comments and Nathan Kucera for his help with manuscript preparation. We also thank the staff of NSLS of X25, X29A and X6A beamlines.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Williams DE, Tushinski R, Gimpel S, Eisenman J, Cannizzaro LA, Aronson M, Croce CM, Huebner K, Cosman D. Alternate splicing of mRNAs encoding human mast cell growth factor and localization of the gene to chromosome 12q22-q24. Cell Growth Differ. 1991;2:373–378. [PubMed] [Google Scholar]
- Ashman LK. The biology of stem cell factor and its receptor C-kit. Int. J. Biochem. Cell Biol. 1999;31:1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- Ashman LK, Griffith R. Therapeutic targeting of c-KIT in cancer. Expert Opin. Investig. Drugs. 2012:1–13. doi: 10.1517/13543784.2013.740010. [DOI] [PubMed] [Google Scholar]
- Bae JH, Schlessinger J. Asymmetric tyrosine kinase arrangements in activation or autophosphorylation of receptor tyrosine kinases. Mol. Cells. 2010;29:443–448. doi: 10.1007/s10059-010-0080-5. [DOI] [PubMed] [Google Scholar]
- Batzer A, Rotin D, Urena J, Skolnik E, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol. Cell. Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besmer P, Murphy JE, George PC, Qiu FH, Bergold PJ, Lederman L, Snyder HW, Brodeur D, Zuckerman EE, Hardy WD. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986;320:415–421. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Broudy VC. Stem Cell Factor and Hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- DiNitto JP, Deshmukh GD, Zhang Y, Jacques SL, Coli R, Worrall JW, Diehl W, English JM, Wu JC. Function of activation loop tyrosine phosphorylation in the mechanism of c-Kit auto-activation and its implication in sunitinib resistance. J. Biochem. 2010;147:601–609. doi: 10.1093/jb/mvq015. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman RA, Saltman DL, Stastny V, Zneimer S. Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10885–10889. doi: 10.1073/pnas.88.23.10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, English JM, Greig MJ, He Y-A, Jacques SL, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasberger B, Minton a P., DeLisi C, Metzger H. Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. U. S. A. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, den Hertog J, Hunter T. Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface. Molecular and Cellular Biology. 2000;20(16):5917–29. doi: 10.1128/mcb.20.16.5917-5929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P, Mattoon D, Lemmon M. a, Schlessinger J. A structure-based model for ligand binding and dimerization of EGF receptors. Proc. Natl. Acad. Sci. U. S. A. 2004;101:929–934. doi: 10.1073/pnas.0307285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–9. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax I, Wong A, Lamothe B, Lee A, Frost A, Hawes J, Schlessinger J. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol. Cell. 2002;10:709–719. doi: 10.1016/s1097-2765(02)00689-5. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Pinchasi D, Zhou M, Lax I, Schlessinger J. Kit receptor dimerization is driven by bivalent binding of stem cell factor. J. Biol. Chem. 1997;272:6311–6317. doi: 10.1074/jbc.272.10.6311. [DOI] [PubMed] [Google Scholar]
- Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000021. ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol. Cell. Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Lim KB, Sridhar V, Zou H, Chien EYT, Sang B-C, Nowakowski J, Kassel DB, Cronin CN, McRee DE. Structure of a c-kit product complex reveals the basis for kinase transactivation. J. Biol. Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opatowsky Y, Lax I, Tomé F, Bleichert F, Unger VM, Schlessinger J. Structure, domain organization, and different conformational states of stem cell factor-induced intact KIT dimers. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1772–1777. doi: 10.1073/pnas.1323254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittoni P, Piconese S, Tripodo C, Colombo MP. Tumor-intrinsic and -extrinsic roles of c-Kit: mast cells as the primary off-target of tyrosine kinase inhibitors. Oncogene. 2011;30:757–769. doi: 10.1038/onc.2010.494. [DOI] [PubMed] [Google Scholar]
- Rönnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell. Mol. Life Sci. 2004;61:2535–2548. doi: 10.1007/s00018-004-4189-6. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.