Abstract
The possible contribution of HLA-DRB3, -DRB4, and -DRB5 alleles to type 1 diabetes risk and to insulin autoantibody (IAA), GAD65 (GAD autoantibody [GADA]), IA-2 antigen (IA-2A), or ZnT8 against either of the three amino acid variants R, W, or Q at position 325 (ZnT8RA, ZnT8WA, and ZnT8QA, respectively) at clinical diagnosis is unclear. Next-generation sequencing (NGS) was used to determine all DRB alleles in consecutively diagnosed patients ages 1–18 years with islet autoantibody–positive type 1 diabetes (n = 970) and control subjects (n = 448). DRB3, DRB4, or DRB5 alleles were tested for an association with the risk of DRB1 for autoantibodies, type 1 diabetes, or both. The association between type 1 diabetes and DRB1*03:01:01 was affected by DRB3*01:01:02 and DRB3*02:02:01. These DRB3 alleles were associated positively with GADA but negatively with ZnT8WA, IA-2A, and IAA. The negative association between type 1 diabetes and DRB1*13:01:01 was affected by DRB3*01:01:02 to increase the risk and by DRB3*02:02:01 to maintain a negative association. DRB4*01:03:01 was strongly associated with type 1 diabetes (P = 10−36), yet its association was extensively affected by DRB1 alleles from protective (DRB1*04:03:01) to high (DRB1*04:01:01) risk, but its association with DRB1*04:05:01 decreased the risk. HLA-DRB3, -DRB4, and -DRB5 affect type 1 diabetes risk and islet autoantibodies. HLA typing with NGS should prove useful to select participants for prevention or intervention trials.
Introduction
Genome-wide association studies through the Type 1 Diabetes Genetics Consortium (1–3) not only have confirmed the strongest association to single nucleotide polymorphisms in the DR-DQ region on chromosome 6 but also have reported an additional 50 non-HLA genetic loci of lesser association (1,4–6). Numerous studies confirmed the contribution of class II DR and DQ to type 1 diabetes risk and attempted to dissect the relative risk confirmed by these two closely located loci (7–10) and to extend it to the HLA-DP locus as well (11). Other genetic factors in linkage disequilibrium with DR and DQ may contribute to either enhancing or decreasing the risk through a possible association with islet autoantibodies at the time of not only clinical diagnosis (12–16) but also seroconversion (17,18).
DRB1 is present in all individuals. Allelic variants of DRB1 are in linkage disequilibrium with either none or one of the genes DRB3, DRB4, and DRB5. In addition, there are several related pseudogenes (DRB2, DRB6, DRB7, DRB8, and DRB9). Through next-generation sequencing (NGS), an integrated genotyping system of exons 1–4 was developed to type all alleles of DRB1, DRB3, DRB4, and DRB5 (19,20). Among individuals with the DRB1*03:01 allele, the DRB3*02:02 allele has been reported to show an independent risk for type 1 diabetes compared with the DRB3*01:01 allele (21). The study concluded that the DRB3*02:02 allele not only was a marker of high-risk DRB1-DRB3 haplotypes but also increased the risk for DRB1*03:01 haplotypes, particularly in individuals homozygous for DRB1*03:01 (21).
We previously reported that HLA-DR-DQ genotypes may define various autoimmune phenotypes indicating strong differential associations with GAD65 (GAD antibody [GADA]), insulin autoantibody (IAA), IA-2 antigen (IA-2A), or the three variants (amino acids R, W, or Q on position 325) of ZnT8A (ZnT8RA, ZnT8WA, ZnT8QA, respectively) (14,22,23). In these studies, the analyses were limited to HLA-DQ alleles detected by allele-specific probes (24,25). A major limitation was that the DRB3, DRB4, and DRB5 alleles located between the DRA and DRB1 loci were not typed (26,27). In the current study, we used NGS of all DRB alleles (19,20) in consecutively diagnosed patients with type 1 diabetes with all islet autoantibodies analyzed (14,28,29) and in geographically matched control subjects (30). The main goal was to test the hypothesis that DRB3, DRB4, and DRB5 alleles modify the risk conferred by DRB1 for islet autoantibodies and type 1 diabetes.
Research Design and Methods
Study Design
We used a case-control study design to calculate odd ratios (ORs) for HLA alleles, haplotypes, and genotypes. Patients were from the nationwide Swedish Better Diabetes Diagnosis (BDD) study (14,22,28), which has involved ongoing participation since 2005 of all 42 pediatric clinics in Sweden. American Diabetes Association and World Health Organization criteria were used for the diagnosis of diabetes and to classify the disease (31). However, we included only patients who at the time of clinical diagnosis had one or several of the following autoantibodies against insulin: IAA; GADA; IA-2A; and ZnT8RA, ZnT8WA, or ZnT8QA (14,22,28). The Karolinska Institutet ethics board approved the BDD study (2004/1:9).
Study Population
Nine hundred seventy patients given a diagnosis of diabetes between 9 months and 18 years of age were sequentially enrolled in the BDD study (14,22,28). Four hundred forty-eight control subjects matched for age (1–18 years), sex, and place of residence were analyzed at the same time (30).
DNA Extraction
The Plasmid Maxiprep Kit (QIAGEN) was used to isolate DNA according to the manufacturer’s instructions from frozen whole-blood samples of patients and control subjects.
HLA NGS Analysis
The NGS HLA typing approach used PCR-based amplification of HLA and sequencing with Illumina MiSeq technology as previously described in detail (19,20). Briefly, the laboratory steps comprise consecutive PCR reactions with bar coding incorporated into the PCRs for individual sample tracking followed by application to the MiSeq system. Robust assays for each target loci of all HLA-DR alleles were developed. The depth of genotyping was extended to HLA-DRB3, -DRB4, and -DRB5 to include exons 2 and 3 for all DR alleles. The analytical tools to define haplotypes and genotypes were developed in collaboration with Scisco Genetics (Seattle, WA). To date, these tools have been tested with 100% accuracy on >2,000 control samples genotyped with the present NGS approach (19,20).
Islet Autoantibodies
GADA, IA-2A, IAA, and the three variants of ZnT8A (ZnT8RA, ZnT8WA, or ZnT8QA) were determined in quantitative radiobinding assays by using in-house standards to determine levels as previously described in detail (14,32).
Statistical Analysis
Because patients and control subjects were enrolled from a larger population study of type 1 diabetes, we used Epi Info 7.1.3.0 (http://wwwn.cdc.gov/epiinfo) for managing all epidemiological data, tracking all biological samples, and performing quality control analyses. For descriptive statistical data analysis, we used SPSS version 22 software (IBM Corporation, Chicago, IL) to calculate frequency distributions, visualizations, and two-group comparison tests of type 1 diabetes associations with pertinent epidemiological variables.
We performed allelic association analysis of multiallelic genes of HLA-DRB1, -DRB3, -DRB4, and -DRB5. Under Hardy-Weinberg equilibrium, the allelic analysis computes allelic frequencies among patients only, control subjects only, and the combination of both. Given excessive polymorphism, we chose to test allele-specific associations with type 1 diabetes by score test (33). The score test evaluates the allele-specific score under the null hypothesis with no allelic associations and is referred to here as H-score because it was developed specifically for testing haplotype associations. Under the null hypothesis, H-score has an asymptotic normal distribution, which is then used to compute the P value. To estimate allele-specific magnitudes of the type 1 diabetes association, we chose the reference allele that has comparable allelic frequencies between patients and control subjects and has relatively high allelic frequency. In comparison with this reference allele, we calculated the OR for every allele. The OR of the reference allele is 1, an OR <1 implies a protective allele, and an OR >1 is a risk allele. For all computations, we used the R function haplo.cc (http://cran.r-project.org/web/packages/haplo.stats/index.html). The function haplo.cc computes the haplotype-based association used for the haplotype analysis described next and assesses the allelic association by introducing a monomorphic locus as the second locus.
In the HLA-DR locus, alleles of HLA-DRB1 are in linkage disequilibrium with alleles of either HLA-DRB3 or -DRB4 or -DRB5. Hence, by treating these subunits and their allelic variations as different alleles, analysis of HLA-DRB1 and -DRB3, -DRB4, or -DRB5 by haplo.cc produces estimates of frequencies for all haplotypes provided that expectation numbers of corresponding haplotypes are five or more copies in both patients and control subjects. Similarly, the function haplo.cc produces H-scores and P values for all included haplotypes. By following the same principle of choosing the reference haplotype, we computed haplotype-specific ORs. To facilitate interpretation of estimated haplotype-specific ORs, we created a two-dimensional table showing H-scores for specific pairs of alleles at HLA-DRB1 and -DRB3, -DRB4, or -DRB5.
Results
DRB1 Alleles
The NGS for typing HLA-DRB1 in Swedish patients with type 1 diabetes and control subjects identified 25 different DRB1 alleles. By using DRB1*13:02:01 as the reference (equal frequency in patients and control subjects), only seven other alleles showed no association with type 1 diabetes (Table 1). The H-score of the five DRB1 alleles that were positively associated with type 1 diabetes could be ranked as DRB1*04:01:01 > *03:01:01 > *04:05:01 > *04:04:01 > *04:02:01 (Table 1). Significant negative associations were observed for a total of 13 different HLA-DRB1 alleles. The H-score ranked the top six negatively associated alleles as *15:01:01 > *07:01:01 > *13:01:01 > *11:01:01 > *11:04:01 > *14:54:01. An unadjusted P value of 0.01 for multiple comparisons was used as a threshold value for marking positive or negative associations, even though multiple alleles at this gene locus are tested on their associations with type 1 diabetes. Correcting multiple comparisons by number of alleles would have led to excessively conservative results because alleles are functional, and estimated log ORs are highly correlated. Correction was not attempted for results interpretation.
Table 1.
Estimated allelic frequencies in pooled samples, control subjects only, and patient cases only and computed H-scores and their associated P values and estimated ORs for HLA-DRB1 among 970 patients and 448 control subjects
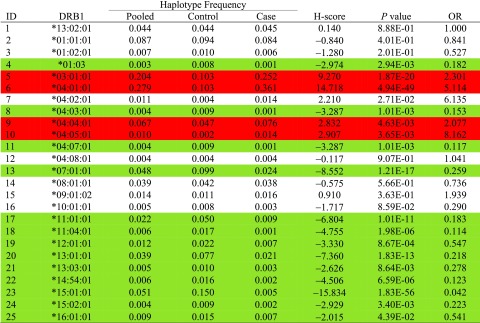 |
Red is a positive and green a negative association of statistical significance.
DRB3, DRB4, and DRB5 Alleles
The NGS for typing the HLA-DRB3, -DRB4, and -DRB5 alleles revealed eight alleles (Table 2). Subjects lacking any HLA-DRB3, -DRB4, and -DRB5 alleles (nonamplified, indicating the presence of a pseudogene) are also indicated as ID9. Using DRB3*03:01:01 as the reference (equal frequency in patients and control subjects), significant risk was conferred by DRB4*01:03:01 and DRB3*01:01:02 (Table 2). The remaining alleles, except DRB5*02:02 (neutral), were negatively associated with type 1 diabetes: DRB5*01:01:01 > DRB3*02:02:01 > DRB5*01:02 > DRB4*01:01:01.
Table 2.
Estimated allelic frequencies in pooled samples, control subjects only, and patient cases only and computed H-scores and their associated P values and estimated ORs for HLA-DRB3, -DRB4, and -DRB5 (DRB345) among 970 patients and 448 control subjects
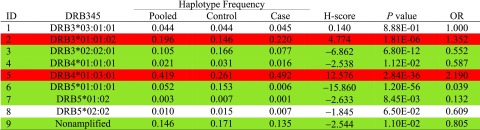 |
Red is a positive and green a negative association of statistical significance.
HLA-DRB1, -DRB3, -DRB4, and -DRB5 Haplotypes
Twenty-eight HLA-DRB1, -DRB3, -DRB4, and -DRB5 haplotypes were identified in the 1,418 subjects (Table 3). Nonamplified DRB3, DRB4, and DRB5 were found on the DRB1*01:01:01-, DRB1*01:03-, DRB1*01:02:01-, DRB1*08:01:01-, and DRB1*10:01:01-containing haplotypes. Only the DRB1*01:03-containing haplotype showed a negative association (OR 0.174, P = 0.00293) in relation to the DRB1*13:02:01-DRB3*03:01:01 reference haplotype (OR 1.0).
Table 3.
Estimated haplotypic frequencies in pooled samples, control subjects only, and patient cases only and computed H-scores and their associated P values and estimated ORs for HLA-DRB1 and DRB3, DRB4, and DRB5 (DRB345) among 970 patient and 448 control subjects
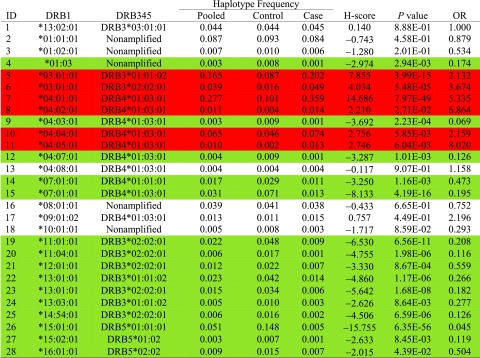 |
Red is a positive and green a negative association of statistical significance.
Positive associations were found in six haplotypes, with DRB1*04:01:01-DRB4*01:03:01 showing the highest H-score (Table 3). DRB4*01:03:01 may be present in six different haplotypes containing either DRB1*04:01:01 (risk, haplotype 7), DRB1*04:02:01 (risk, haplotype 8), DRB1*04:04:01 (risk, haplotype 10), DRB1*04:05:01 (risk, haplotype 11), DRB1*04:03:01 (protection, haplotype 9), or DRB1*04:07:01 (protection, haplotype 12). Taken together, DRB4*01:03:01 may be present on four haplotypes conferring risk and two haplotypes showing a negative association, therefore having a potential protective effect.
Haplotypic Interactions Between DRB1 and DRB3, DRB4, and DRB5
To gain an intuitive insight into haplotypic modification of DRB1 by DRB3, DRB4, and DRB5 alleles, all H-scores (rounded to the integer for easy visualization) are shown in a two-way diagram with rows of DRB1 alleles and columns of DRB3, DRB4, and DRB5 alleles (Table 4). As shown, the presence of a single horizontal and vertical cross (i.e., DRB1*01:03 with nonamplified pseudogenes DRB1*15:02:01-DRB5*01:02 and DRB1*16:01:01-DRB5*02:02) implies that the association is haplotype specific; hence, one cannot differentiate effects of two alleles. The association between type 1 diabetes and DRB1*03:01:01 (H-score 9) was found to be affected by DRB3*01:01:02 (H-score 8) and DRB3*02:02:01 (H-score 4). Similarly, the associations between type 1 diabetes and DRB1*07:01:01 (H-score −9) and DRB1*13:01:01 (H-score −7) were modified by DRB3, DRB4, and DRB5.DRB1*07:01:01 was modified by both DRB4*01:01:01 (H-score −3) and DRB4*01:03:01 (H-score 13); hence, two DRB4 alleles with opposite risks when analyzed individually results in a negative association on the DRB1*07:01:01 haplotype. DRB1*13:01:01 was modified by both DRB3*01:01:02 (H-score 5) and DRB3*02:02:01 (H-score −7), which also showed opposite risks when analyzed individually, but when present in the DRB1*13:01:01 haplotype, the H-score of −7 indicated protection.
Table 4.
Estimated H-scores for all haplotypes between HLA-DRB1 and -DRB3, -DRB4, and -DRB5, with their marginal H-scores listed by rows (HLA-DRB1) and by columns (HLA-DRB3, -DRB4, and -DRB5)
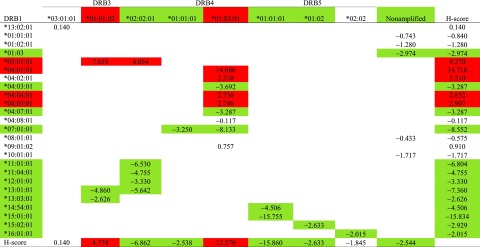 |
All H-scores are rounded to their integers, and their absolute values >2 are deemed significant. Red is a positive and green a negative association of statistical significance.
On the other hand, marginal associations of DRB3, DRB4, and DRB5 alleles are extensively modified by DRB1 alleles (Table 4). For example, DRB4*01:03:01 was marginally associated with type 1 diabetes (H-score 13), yet its association was extensively modified by DRB1 alleles from protective (H-score −8) to high risk (H-score 15).
Haplotypic Interactions Between DRB1, DRB3, DRB4, and DRB5 and Islet Autoantibodies in Patients
Levels of islet autoantibodies were determined at the time of clinical diagnosis. Levels, expressed in arbitrary units using in-house reference sera (14) or the World Health Organization standard for GADA and IA-2A (34), were transformed with cubic root to correct the excessive skewed distributions (Table 5). The most common haplotype, DRB1*04:01:01-DRB4*01:03:01 (in 36% of the patients) was used as the reference. To ease interpretation, Table 6 lists the integer part of z scores, indicating a positive association (red), negative association (green), or no statistical association (no color) (absolute values of z scores <2). At the significance level of 0.05, all haplotypes of HLA-DRB1, -DRB3, -DRB4, and -DRB5 have various patterns of associations with the six different autoantibodies. In particular, DRB3*01:01:02 and DRB3*02:02:01 regulate autoantibody association of DRB1*03:01:01 through specific autoantibodies ZnT8QA and ZnT8RA, respectively. On the other hand, DRB4*01:01:01 and DRB4*01:03:01 affected autoantibody associations of DRB1*07:01:01 through a positive regulation with GADA, while weakly regulating IA-2A with a marginal H-score of −1.60.
Table 5.
Haplotypic association analysis of DRB1 and DRB3 or DRB4 (DRB345) haplotypes with five islet autoantibodies (IAA, GADA, IA-2A, ZnT8RA, ZnT8WA, ZnT8QA) among all 448 patients, with the most common haplotype DRB1*04:01:01-DRB4*01:03:01 as the reference
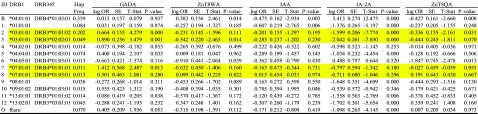 |
The yellow highlights indicate differences in haplotypic associations for ZnT8RA and ZnT8QA whether the DRB3 is either *01:01:02 or *02:02:01 on the DRB1*03:01:01 haplotype. Similarly, GADA and IA-2A vary dependent on the DRB4 subtype on the DRB1*07:01:01 haplotype. Association statistics are coefficients, SEs, z scores, and P values. Hap freq, haplotype frequency; t stat, t statistic.
Table 6.
Patterns of haplotypic associations with autoantibodies (with z scores)
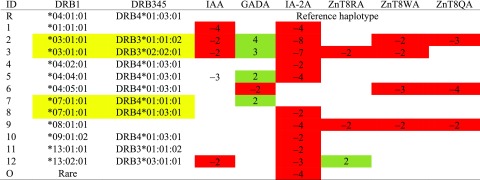 |
The yellow highlights indicate differences in haplotypic associations for ZnT8RA and ZnT8QA whether the DRB3 is either *01:01:02 or *02:02:01 on the DRB1*03:01:01 haplotype. Similarly, GADA and IA-2A vary dependent on the DRB4 subtype on the DRB1*07:01:01 haplotype.
Green indicates negative associations; red, positive associations; and blank, null associations.
Discussion
Use of the NGS to type patients with newly diagnosed type 1 diabetes and control subjects resulted in the following principal findings. First, among the 25 HLA-DRB1 alleles, only 4 (DRB1*03:01:01, DRB1*04:01:01, DRB1*04:04:01, and DRB1*04:05:01) were positively associated with type 1 diabetes. The H-score was used as a way to rank the relative degree of protection. Second, NGS detected nine alleles of DRB3, DRB4, and DRB5, including chromosomes with only nonamplified loci. Only DRB4*01:03:01 and DRB3*01:01:02 were positively associated; the remaining five alleles were negatively associated with the disease. Most importantly, DRB4 was dichotomized in that DRB4*01:03:01 was positively but DRB4*01:01:01 was negatively associated with type 1 diabetes. Similarly, DRB3*01:01:02 was positively but DRB3*02:02:01 was negatively associated with the disease. Because either one of these two alleles may be present on a haplotype containing DRB1*03:01:01, it cannot be excluded that the risk of this allele for type 1 diabetes is affected by the DRB3 alleles to either increase or decrease the risk. Third, in dissecting the extended DRB1-DRB3-DRB4-DRB5 haplotypes (28 haplotypes were identified), a major finding was that the two DRB1*03:01:01-containing haplotypes remained positively associated with diabetes whether either DRB3*01:01:02 or DRB3*02:02:01 was present (although the positively associated DRB3*01:01:02 showed a P value for risk that was three times higher than for the DRB3*02:02:01-containing haplotype).
More importantly, the marginal associations of the DRB3, DRB4, and DRB5 alleles were extensively affected by DRB1 alleles. The analyses of the different extended DRB1-DRB3-DRB4-DRB5 haplotypes strongly suggest that the risk for type 1 diabetes cannot be assigned to a single DRB1 allele but that DRB3, DRB4, and DRB5 in linkage disequilibrium have to be taken into account when dissecting the role of HLA-DR in type 1 diabetes.
The NGS offered considerable strength to the study. The method used allows extended DRB1-DRB3-DRB4-DRB5 haplotypes to be computed without information on descent. The sequencing of coding regions of exon 1–4 of both DRB1 and DRB3, DRB4, and DRB5 allowed the detection of all functional genes, whereas pseudogenes were not amplified. Despite the presence of pseudogenes in the DRB3-DRB4-DRB5 region, it was possible to compute complete haplotypes also in subjects with pseudogenes such as DRB1*01:01:01, DRB1*01:02:01, DRB1*08:01:01, and DRB1*10:01:01, respectively. The methods for NGS of HLA alleles vary between approaches and methodologies; however, the present method is a strength because it allows high-resolution typing of alleles, which until now have been understudied. In addition, due to reduced costs of PCR, novel instrumentations and approaches in bioinformatics make NGS HLA typing affordable and precise.
The current study population is unique because it represents consecutive patients with newly diagnosed type 1 diabetes in Sweden (22,28). The patients in the current study were all born to parents who were born in Sweden, as was the case for their grandparents (28). According to Swedish pediatric diabetes guidelines, all patients with type 1 diabetes <18 years old are seen by diabetes specialists in one of the pediatric diabetes clinics in Sweden. A potential weakness to this study is that we were not able for reasons of funding to analyze a control group of equal number as the patients. However, the current control subjects were selected to represent the geographical location of the patients (30).
To our knowledge, there is only one previous publication on NGS of DRB genes (21), which studied 143 control subjects and 337 patients of a much larger cohort and reported that both DRB3*01:01 and DRB3*02:02 alleles showed an increased risk for type 1 diabetes. In particular, the authors suggested that on DRB1*03:01 haplotypes, the DRB3*02:02 allele contributes to type 1 diabetes risk (21). The current analysis of patients with newly diagnosed type 1 diabetes and control subjects shows results consistent with this conclusion. However, the current results differ because we found that DRB3*02:02 was negatively associated with type 1 diabetes (Table 2), and when considered with DRB1*03:01:01, we found that DRB3*02:02:01 reduced the risk for type 1 diabetes compared with the DRB1*03:01:01-DRB3*01:01:02 haplotype (Table 3).
The DRB1*03:01:01-containing haplotypes may carry either the DRB3*02:02:01 or the DRB3*02:02:01 alleles. It was recently demonstrated that children homozygous for DR3/3 (DRB1*03:01:01/DRB1*03:01:01) in the TEDDY (The Environmental Determinants of Diabetes in the Young) study had an increased risk for developing GADA as their first islet autoantibody (17). Further studies are therefore needed to determine whether DRB3*02:02:01 affects the risk for GADA as the first islet autoantibody.
The negative association of DRB1*01:03 with type 1 diabetes was independent of DRB3, DRB4, and DRB5 because it resides on a haplotype unable to express any of these DR subtypes. The interpretation is that the DRB1 protein heterodimer confers protection probably by inducing immunological tolerance.
A paucity of studies of other autoimmune diseases have investigated the possible contribution of DRB1-DRB3-DRB4-DRB5 haplotypes. Thrombotic thrombocytopenic purpura was reported to be positively associated with DRB3 and negatively associated with DRB4, although the DRB3 and DRB4 subtyping was at low resolution to conclude whether the association was due to linkage disequilibrium to either DR3 or DR4 (35). The meaning of the results in the current study is that DRB3, DRB4, and DRB5 may affect not only the risk for type 1 diabetes but also the risk of having certain islet autoantibodies (Table 5). The data strongly suggest that DRB3 on the DRB1*03:01:01 haplotype affects the risk of having GADA or ZnT8RA (positive association) or IA-2A (negative association). Patients with DRB1*07:01:01-DRB4*01:01:01 have an increased risk for GADA at the time of clinical diagnosis. Therefore, determining to what extent the DRB3, DRB4, and DRB5 β-chains are able to form heterodimers with the DRA α-chains is critical. It cannot be excluded that peptide presentation on DRB3, DRB4, or DRB5 heterodimers may induce immune responses related to autoimmunity. It was found, for example, that peptides from group A streptococcal vaccine epitopes are effectively presented on DRB3, DRB4, and DRB5 heterodimers (36).
In conclusion, HLA-DRB3, -DRB4, and -DRB5 genetic factors should be taken into account when dissecting the role of HLA-DR in the risk for islet autoimmunity and progression to the clinical onset of type 1 diabetes. The contribution of these heterodimer proteins in the immune response to infectious agents may be of particular interest in studying the etiology of islet autoimmunity and the progression to the clinical onset of type 1 diabetes.
Article Information
Acknowledgments. The authors thank Anita Ramelius, Linda Faxius, Karl Moritz, Petra Moritz, Ingrid Wigheden, Ida Jönsson, and Rasmus Håkansson (Department of Clinical Sciences, Lund University/Clinical Research Centre, Malmö, Sweden) for expert technical assistance.
Funding. The study was supported by a grant from the European Foundation for the Study of Diabetes Clinical Research Grants Programme 2013 and in part by the Swedish Child Diabetes Foundation; the National Institutes of Health (DK-63861, DK-26190); the Swedish Research Council, including a Linné grant to Lund University Diabetes Centre; the Skåne County Council for Research and Development; the Swedish Association of Local Authorities and Regions; and the Knut and Alice Wallenberg Foundation.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. L.P.Z., S.A., M.Z., D.E.G., and Å.L. contributed to the data research and analysis and writing of the manuscript. A.C., H.E.L., G.F., S.A.I., J.L., C.M., M.P., U.S., and E.Ö. contributed to the design of the BDD study, data research, discussion, and review of the manuscript. I.K. contributed to the data research (control subjects) and review of the manuscript. C.-W.P. and W.C.N. contributed to the NGS, data research, and review of the manuscript. Å.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Appendix
Members of the BDD Study Group include: Bengt-Olof Samuelsson (Borås), Kalle Snellman (Eskilstuna), Anna Olivecrona (Falun), Åke Stenberg (Gällivare), Lars Skogsberg (Gävle), Nils Östen Nilsson (Halmstad), Jan Neiderud (Helsingborg), Torun Torbjörnsdotter (Huddinge), Thomas Hägg (Hudiksvall), Kristina Hemmingsson (Härnösand), Göran Lundström (Kalmar), Magnus Ljungcrantz (Karlskrona), Maria Forsberg (Karlstad), Karin Larsson (Kristianstad), Christer Gundewall (Kungsbacka), Karin Åkesson (Jönköping), Rebecka Enander (Lidköping), Ulf Samuelsson (Linköping), Helena Elding Larsson (Malmö), Agneta Brännström (Luleå), Maria Nordwall (Norrköping), Lennart Hellenberg (Nyköping), Elena Lundberg (Skellefteå), Henrik Tollig (Skövde), Britta Björsell (Sollefteå), Eva Örtqvist (Stockholm/KS), Björn Stjernstedt (Sundsvall), Nils Wramner (Trollhättan), Ragnar Hanås (Uddevalla), Ingemar Swenne (Uppsala), Margareta Blomgren (Visby), Anders Thåström (Västervik), Carl-Göran Arvidsson (Västerås), Stig Edvardsson (Växjö), Björn Jönsson (Ystad), Torsten Gadd (Ängelholm), Jan Åman (Örebro), Rein Florell (Örnsköldsvik), and Anna-Lena Fureman (Östersund).
Footnotes
Members of the BDD Study Group are listed in the appendix.
References
- 1.Rich SS, Concannon P, Erlich H, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci 2006;1079:1–8 [DOI] [PubMed] [Google Scholar]
- 2.Cooper JD, Howson JM, Smyth D, et al.; Type 1 Diabetes Genetics Consortium . Confirmation of novel type 1 diabetes risk loci in families. Diabetologia 2012;55:996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013;8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med 2009;360:1646–1654 [DOI] [PubMed] [Google Scholar]
- 5.Concannon P, Erlich HA, Julier C, et al.; Type 1 Diabetes Genetics Consortium . Type 1 diabetes: evidence for susceptibility loci from four genome-wide linkage scans in 1,435 multiplex families. Diabetes 2005;54:2995–3001 [DOI] [PubMed] [Google Scholar]
- 6.Barrett JC, Clayton DG, Concannon P, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson G, Valdes AM, Noble JA, et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens 2007;70:110–127 [DOI] [PubMed] [Google Scholar]
- 8.Kockum I, Sanjeevi CB, Eastman S, Landin-Olsson M, Dahlquist G, Lernmark A Complex interaction between HLA DR and DQ in conferring risk for childhood type 1 diabetes. Eur J Immunogenet 1999;26:361–372 [DOI] [PubMed] [Google Scholar]
- 9.Graham J, Kockum I, Sanjeevi CB, et al. Negative association between type 1 diabetes and HLA DQB1*0602-DQA1*0102 is attenuated with age at onset. Swedish Childhood Diabetes Study Group. Eur J Immunogenet 1999;26:117–127 [PubMed] [Google Scholar]
- 10.Erlich H, Valdes AM, Noble J, et al.; Type 1 Diabetes Genetics Consortium . HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdes AM, Thomson G, Erlich HA, Noble JA. Association between type 1 diabetes age of onset and HLA among sibling pairs. Diabetes 1999;48:1658–1661 [DOI] [PubMed] [Google Scholar]
- 12.Graham J, Hagopian WA, Kockum I, et al.; Diabetes Incidence in Sweden Study Group; Swedish Childhood Diabetes Study Group . Genetic effects on age-dependent onset and islet cell autoantibody markers in type 1 diabetes. Diabetes 2002;51:1346–1355 [DOI] [PubMed] [Google Scholar]
- 13.Qu HQ, Polychronakos C. The effect of the MHC locus on autoantibodies in type 1 diabetes. J Med Genet 2009;46:469–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delli AJ, Vaziri-Sani F, Lindblad B, et al.; Better Diabetes Diagnosis Study Group . Zinc transporter 8 autoantibodies and their association with SLC30A8 and HLA-DQ genes differ between immigrant and Swedish patients with newly diagnosed type 1 diabetes in the Better Diabetes Diagnosis study. Diabetes 2012;61:2556–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savola K, Bonifacio E, Sabbah E, et al.; Childhood Diabetes in Finland Study Group . IA-2 antibodies--a sensitive marker of IDDM with clinical onset in childhood and adolescence. Diabetologia 1998;41:424–429 [DOI] [PubMed] [Google Scholar]
- 16.Sabbah E, Savola K, Kulmala P, et al.; The Childhood Diabetes in Finland Study Group . Disease-associated autoantibodies and HLA-DQB1 genotypes in children with newly diagnosed insulin-dependent diabetes mellitus (IDDM). Clin Exp Immunol 1999;116:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krischer JP, Lynch KF, Schatz DA, et al.; TEDDY Study Group . The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 2015;58:980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilonen J, Hammais A, Laine AP, et al. Patterns of β-cell autoantibody appearance and genetic associations during the first years of life. Diabetes 2013;62:3636–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson WC, Pyo CW, Vogan D, et al. An integrated genotyping approach for HLA and other complex genetic systems. Hum Immunol 2015;76:928–938 [DOI] [PubMed] [Google Scholar]
- 20.Smith AG, Pyo CW, Nelson W, et al. Next generation sequencing to determine HLA class II genotypes in a cohort of hematopoietic cell transplant patients and donors. Hum Immunol 2014;75:1040–1046 [DOI] [PubMed] [Google Scholar]
- 21.Erlich HA, Valdes AM, McDevitt SL, et al.; Type 1 Diabetes Genetics Consortium (T1DGC) . Next generation sequencing reveals the association of DRB3*02:02 with type 1 diabetes. Diabetes 2013;62:2618–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlsson A, Kockum I, Lindblad B, et al. ; Swedish Better Diabetes Diagnosis Study Group. Low risk HLA-DQ and increased body mass index in newly diagnosed type 1 diabetes children in the Better Diabetes Diagnosis study in Sweden. Int J Obes 2012;36:718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson C, Larsson K, Vaziri-Sani F, et al. The three ZNT8 autoantibody variants together improve the diagnostic sensitivity of childhood and adolescent type 1 diabetes. Autoimmunity 2011;44:394–405 [DOI] [PubMed] [Google Scholar]
- 24.Kiviniemi M, Hermann R, Nurmi J, et al.; TEDDY Study Group . A high-throughput population screening system for the estimation of genetic risk for type 1 diabetes: an application for the TEDDY (the Environmental Determinants of Diabetes in the Young) study. Diabetes Technol Ther 2007;9:460–472 [DOI] [PubMed] [Google Scholar]
- 25.Hagopian WA, Erlich H, Lernmark A, et al.; TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes 2011;12:733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Texier C, Pouvelle-Moratille S, Busson M, Charron D, Ménez A, Maillère B. Complementarity and redundancy of the binding specificity of HLA-DRB1, -DRB3, -DRB4 and -DRB5 molecules. Eur J Immunol 2001;31:1837–1846 [DOI] [PubMed] [Google Scholar]
- 27.Holdsworth R, Hurley CK, Marsh SG, et al. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens 2009;73:95–170 [DOI] [PubMed] [Google Scholar]
- 28.Delli AJ, Lindblad B, Carlsson A, et al.; Better Diabetes Diagnosis (BDD) Study Group . Type 1 diabetes patients born to immigrants to Sweden increase their native diabetes risk and differ from Swedish patients in HLA types and islet autoantibodies. Pediatr Diabetes 2010;11:513–520 [DOI] [PubMed] [Google Scholar]
- 29.Andersson C, Kolmodin M, Ivarsson SA, et al. Islet cell antibodies (ICA) identify autoimmunity in children with new onset diabetes mellitus negative for other islet cell antibodies. Pediatr Diabetes 2014;15:336–344 [DOI] [PubMed] [Google Scholar]
- 30.Gyllenberg A, Asad S, Piehl F, et al.; Swedish Childhood Diabetes Study Group; Diabetes Incidence in Sweden Study Group; Better Diabetes Diagnosis Study group . Age-dependent variation of genotypes in MHC II transactivator gene (CIITA) in controls and association to type 1 diabetes. Genes Immun 2012;13:632–640 [DOI] [PubMed] [Google Scholar]
- 31.American Diabetes Association . Diagnosis and classification of diabetes mellitus. Diabetes Care 2014;37(Suppl. 1):S81–S90 [DOI] [PubMed] [Google Scholar]
- 32.Vaziri-Sani F, Delli AJ, Elding-Larsson H, et al. A novel triple mix radiobinding assay for the three ZnT8 (ZnT8-RWQ) autoantibody variants in children with newly diagnosed diabetes. J Immunol Methods 2011;371:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mire-Sluis AR, Gaines Das R, Lernmark A. The World Health Organization International Collaborative Study for islet cell antibodies. Diabetologia 2000;43:1282–1292 [DOI] [PubMed] [Google Scholar]
- 35.Scully M, Brown J, Patel R, McDonald V, Brown CJ, Machin S. Human leukocyte antigen association in idiopathic thrombotic thrombocytopenic purpura: evidence for an immunogenetic link. J Thromb Haemost 2010;8:257–262 [DOI] [PubMed] [Google Scholar]
- 36.Guilherme L, Alba MP, Ferreira FM, et al. Anti-group A streptococcal vaccine epitope: structure, stability, and its ability to interact with HLA class II molecules. J Biol Chem 2011;286:6989–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]


