Abstract
Mutations in the ALS2 gene result in early-onset amyotrophic lateral sclerosis, infantile-onset ascending hereditary spastic paraplegia and juvenile primary lateral sclerosis, suggesting prominent upper motor neuron involvement. However, the importance of alsin function for corticospinal motor neuron (CSMN) health and stability remains unknown. To date, four separate alsin knockout (AlsinKO) mouse models have been generated, and despite hopes of mimicking human pathology, none displayed profound motor function defects. This, however, does not rule out the possibility of neuronal defects within CSMN, which is not easy to detect in these mice. Detailed cellular analysis of CSMN has been hampered due to their limited numbers and the complex and heterogeneous structure of the cerebral cortex. In an effort to visualize CSMN in vivo and to investigate precise aspects of neuronal abnormalities in the absence of alsin function, we generated AlsinKO-UeGFP mice, by crossing AlsinKO and UCHL1-eGFP mice, a CSMN reporter line. We find that CSMN display vacuolated apical dendrites with increased autophagy, shrinkage of soma size and axonal pathology even in the pons region. Immunocytochemistry coupled with electron microscopy reveal that alsin is important for maintaining cellular cytoarchitecture and integrity of cellular organelles. In its absence, CSMN displays selective defects both in mitochondria and Golgi apparatus. UCHL1-eGFP mice help understand the underlying cellular factors that lead to CSMN vulnerability in diseases, and our findings reveal unique importance of alsin function for CSMN health and stability.
Introduction
To date, numerous mutations have been identified to amyotrophic lateral sclerosis (ALS) and other related motor neuron diseases. However, mutations in the Alsin2 (ALS2) gene are particularly important because, unlike other mutations, it mainly affects young people. Mutations in this gene are detected in juvenile cases of ALS, patients with juvenile primary lateral sclerosis (PLS) (1), infantile-onset ascending hereditary spastic paraplegia (IAHSP) (2) and are the most common cause of autosomal recessive juvenile ALS (1).
Alsin is a ubiquitous protein, expressed mainly in the central nervous system, and is encoded by the ALS2 gene. ALS2 is located on chromosome 2q33 contains 33 introns and 34 exons, and encodes two splice variant of alsin protein: a long form of 1657 amino acids and a short form of 396 amino acids (3). To date, 14 different mutations have been identified in the ALS2 gene in 24 ALS patients (4). In addition, a novel c.2761C > T mutation was found to cause hereditary spastic paraplegia (HSP) with bulbar involvement (5). Most of these are homozygous mutations leading to truncation of long and/or short form of alsin protein and loss of function (1).
Alsin protein is proposed to have several guanine nucleotide exchange factor (GEF) domains and can activate Rho, Rac1 and Rab5 GTPases, which play important roles in endocytosis, cytoskeleton maintenance, protein transport, cell signaling and membrane trafficking (1,3,6). There are three main domains of alsin protein: (1) an N-terminal regulator of chromatin condensation (RCC1)-like domain, which functions as GEF for the Ran family of GTPases (7); (2) a middle Dbl-homology/pleckstrin-homology (DH/PH) domain, which is similar to GEFs for Rho, Rac, Cdc42 (8,9), and was shown to acts as a GEF for Rac2 to stimulate neurite outgrowth (10) and (3) the C-terminus that has eight membrane occupation and recognition nexus motif (9) and vacuolar protein-sorting-9 (Vps9p) domain, which has GEF activity specifically for Rab5 (11). Rab5 is important for regulating organelle tethering, fusion and microtubule-dependent motility during endocytosis (12). Fibroblasts derived from AlsinKO mice have been shown to delay epidermal growth factor and brain derived neurotrophic factor receptor mediated endocytosis (13,14). Additionally, cultured hippocampal neurons from AlsinKO mice showed accumulation of enlarged Rab5-associated endosomes, leading to impairment of endosomal activity and augmentation of endosomal conversion to lysosomes (15).
Even though diverse functions of alsin in many different cells and model systems are beginning to emerge (6,8,10,11), very little is known about its role in upper motor neurons. To date, four different ALS2 knockout mouse models were independently generated to further reveal its function (13,16–18). They were generated by targeted deletion of either exon 3 (16), exons 3 and 4 (13), exon 4 (18), and exon 4 and part of exon 3 (17), using different gene targeting strategies. Alsin protein was not detected in any of these mouse lines, and alsin function was completely abolished. However, none of the mouse models recapitulated the clinical presentation of juvenile ALS, IAHSP and juvenile PLS observed in patients. Mouse models appeared healthy, without a major motor dysfunction, and displayed normal life expectancy. Minor motor coordination impairment was observed in aged animals, but spinal motor neurons were still intact with no signs of degeneration (17–19). In contrast, marked silver deposition within the descending axons of the corticospinal tract (CST) in the cervical region of the spinal cord suggested potential CSMN defects (17), a pathophysiology that was prominent in patients.
Corticospinal motor neurons (CSMN) are located in layer V of motor cortex together with thousands of other neurons and they are very few in numbers. However, they have unique ability to collect, integrate and convey cerebral cortex's input toward spinal cord targets. Due to lack of defining morphological properties and the complexity of the cerebral cortex, it is not easy to distinguish CSMN from other cortical neurons in vivo. Thus, despite their importance, investigation of CSMN pathology remains challenging. Bringing effective treatment strategies to neurodegenerative diseases requires understanding the cellular basis of their selective vulnerability. Especially in the cases of alsin mutations, which lead to diseases with prominent upper motor neuron defects, there is an urgent need to reveal the cellular events leading to CSMN vulnerability and potential degeneration.
We recently developed a novel reporter line, the UCHL1-UeGFP mice, in which eGFP expression is restricted to CSMN in the motor cortex and is stable even at later age in life (20). We generated AlsinKO-UeGFP mice, a CSMN reporter line in the AlsinKO background, enabling visualization and study of CSMN even in P500 (∼1.5 year). Here, by taking advantage of this CSMN reporter line of AlsinKO, we reveal that apical dendrite degeneration is a common cellular pathology for CSMN that become diseased for different causes. In addition, we identify mitochondria and the Golgi apparatus of CSMN as a target of cellular dysfunction in the absence of alsin, thus revealing a novel importance of alsin function for CSMN health and stability.
Results
Generation of a CSMN reporter line of AlsinKO mice
Because visualization and detailed cellular analysis of CSMN has been a challenge in mouse models, we used UCHL1-eGFP mice as a tool to generate reporter lines of motor neuron diseases in which CSMN express eGFP that is stable even at postnatal day (P) 800—2.5 years old—in vivo (20). AlsinKO mice, which lack alsin function and display very minor motor neuron circuitry defects by P300 have been generated and characterized (17). Here, CSMN reporter line of AlsinKO mice, AlsinKO-UeGFP were generated by two consecutive cross-breedings: AlsinHTmice were bred with UCHL1-eGFP to get AlsinHT-UeGFP, which were crossed back to AlsinHT mice to generate AlsinKO-UeGFP mice (Fig. 1A and B). All mice resulting from these cross-breedings were viable and fertile, without obvious developmental defects, and were born according to Mendelian ratios.
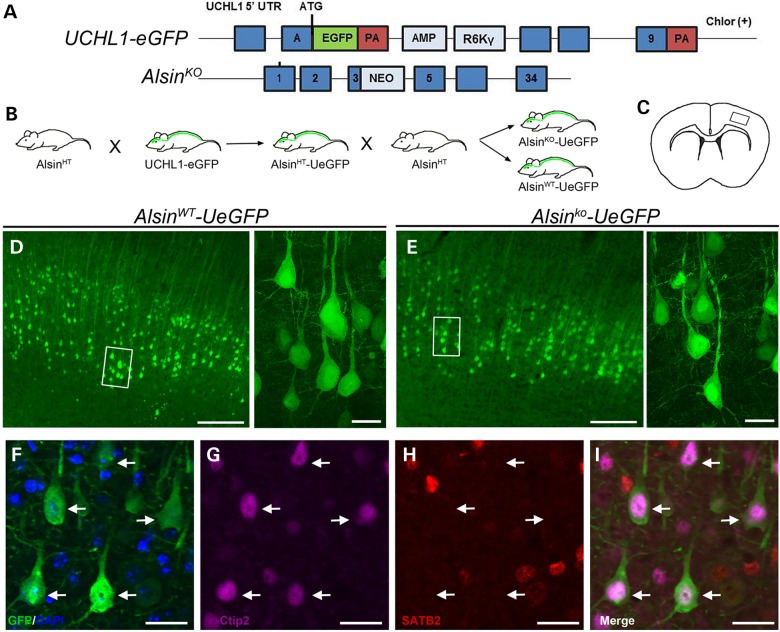
Figure 1.
Neurons expressing eGFP in AlsinWT-UeGFP and AlsinKO-UeGFP mice retain CSMN identity. (A) Schematic representation of the transgenic constructs of UCHL1-eGFP and AlsinKO gene used to generate AlsinWT-UeGFP and AlsinKO-UeGFP mice, respectively. (B) Breeding strategy to generate AlsinWT-UeGFP and AlsinKO-UeGFP mice, reporter line of CSMN in the AlsinKO background. (C) A cross-section area of the brain. Boxed area represents layer V of motor cortex. (D and E) eGFP+ neurons reside in layer V of the motor cortex and display pyramidal neuron morphology with prominent apical dendrites in both AlsinWT-UeGFP (D) and AlsinKO-UeGFP (E), inset are enlarged to the right. (F–I) eGFP+ neurons express Ctip2 (G) but not Satb2 (H). Scale bar: 500 µm (D and E), 20 µm (insets, F–I).
Neurons expressing eGFP (eGFP+) in AlsinWT-UeGFP and AlsinKO-UeGFP mice were located in layer V of the motor cortex, displayed pyramidal neuron morphology and a prominent apical dendrite (Fig. 1C–E). In addition, eGFP+ neurons showed co-localization with Ctip2 (subcerebral projection neuron marker (21), 95%; n = 246 neurons), but not with Satb2 (callosal projection neuron marker (22), 5%; n = 192 neurons). eGFP+ neurons retained CSMN identity in AlsinKO-UeGFP mice, even at P500 (Fig. 1F–I, arrows). CST axons, labeled with eGFP, were detected in ventral pons as previously documented in UCHL1-eGFP mice (20).
CST axons display the signs of degeneration
Silver staining revealed CST axon fiber degeneration in AlsinKO mice, by P300 (17). To further investigate whether AlsinKO-UeGFP mice recapitulate the CST axon pathology, we next studied the health of the CST axons at the level of pons (Fig. 2A). Because eGFP expression in CSMN allows visualization of single axon fibers and overall health of the CST tract, details of axonal pathology were revealed. The average fiber density was comparable between AlsinWT-UeGFP: (120.7 ± 9/unit area) and AlsinKO-UeGFP: (99.6 ± 2.2/unit area) at P300 (Fig. 2D), but there was a significant reduction by P500 AlsinWT-UeGFP: (92.1 ± 9.9/unit area); AlsinKO-UeGFP: (50.1 ± 6.8/unit area, P < 0.002) (Fig. 2D). The average axon fiber cross-sectional area was reduced both at P300 (AlsinWT-UeGFP: 3.1 ± 0.4 µm2, n = 31 axons; AlsinKO-UeGFP: 1.5 ± 0.2 µm2, n = 29 axons; P < 0.03) and P500 (AlsinWT-UeGFP: 2.5 ± 0.1 µm2, n = 28 axons; AlsinKO-UeGFP: 1.4 ± 0.1 µm2, n = 30 axons; P < 0.004; Fig. 2E).
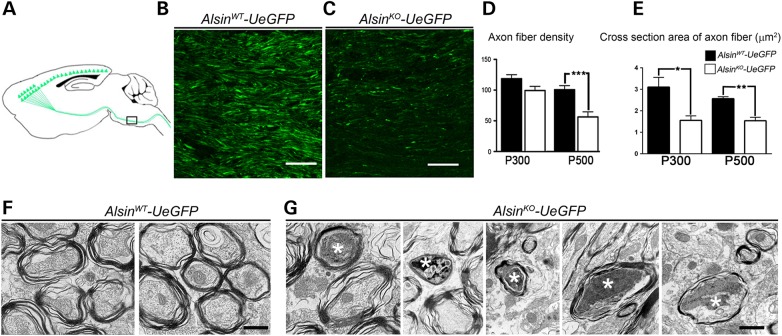
Figure 2.
Corticospinal tract axons degenerate in AlsinKO-UeGFP mice. (A) Schematic drawing of sagittal section showing CST axons. Pons, (the boxed area), is used for quantification of axon fiber density and thickness. (B and C) Representative images of eGFP+ CST axons in pons of AlsinWT-UeGFP (B) and AlsinKO-UeGFP (C) mice. (D) The average fiber density is reduced in AlsinKO-UeGFP mice at P500, but not at P300. (E) The average cross-section area of axon fiber is gradually reduced in AlsinKO-UeGFP mice at P300 and P500. (F) Electron micrographs of CST axons in ventral pons of AlsinWT-UeGFP mice show healthy axon fiber bundles. (G) There are numerous degenerating axon fibers within CST of AlsinKO-UeGFP mice at P500 displaying a wide variety of morphological defects such as: membranous debris, mitochondria aggregation, collapsed synaptic vesicles and membrane debris, electrodense material, crenated mitochondria and final stages of degeneration (denoted by stars). Bar graph values are mean ± SEM. Student's t-test, ***P < 0.0001. Scale bar: 50 µm (B and C); 500 nm (F–G).
GFP immunocytochemistry coupled with electron microscopic (EM) analysis further delineated the presence of many degenerating axon fibers only in AlsinKO-UeGFP mice. We observed healthy axon fibers in WT CSMN (Fig. 2F), and different stages of degenerating axon fibers in diseased CSMN (Fig. 2G). At early stages, large fields of membranous debris were observed (Fig. 2G), most likely representing fragments of mitochondrial membranes. In later stages, these fields become condensed, appeared darker and contained collapsed synaptic vesicles and membrane debris (Fig. 2G). Axons filled with dark electrodense material which contain clumps of crenated mitochondria and synaptic vesicles were present (Fig. 2G). Besides the ongoing pathology, signs of final stages of degeneration were also present (Fig. 2G). Because eGFP expression is restricted to CSMN, these detailed axonal analyses revealed CSMN-specific defects in the pons regions, and suggested that CSMN reporter line of AlsinKO recapitulates previously reported axon defects (17), and would be suitable to study CSMN biology in the absence of alsin function.
Lack of alsin causes subtle cellular defects but no overt cell loss
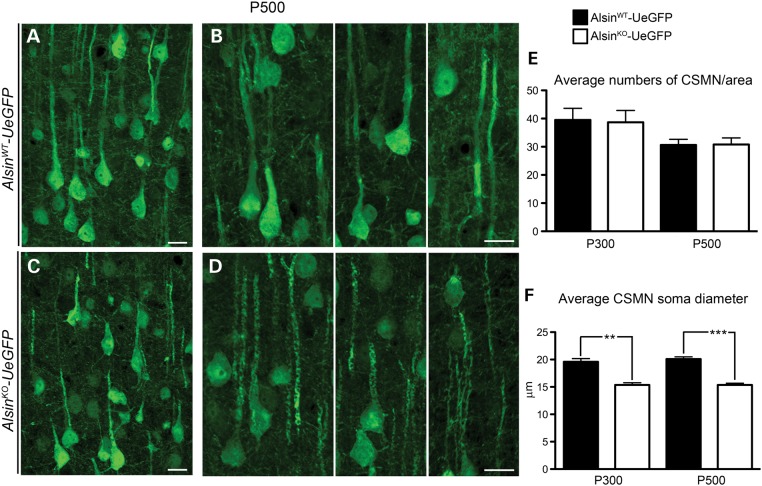
The motor cortex appeared normal and healthy with Nissl staining and NeuN immunocytochemistry in the AlsinKO mice (Supplementary Material, Fig. S1). In addition, visualization and detailed cellular analysis of eGFP+ CSMN in the absence of alsin function (Fig. 3A–D) revealed that the average number of CSMN was comparable between WT and AlsinKO mice both at P300 (AlsinWT-UeGFP: 39 ± 3, n = 464 neurons; and AlsinKO-UeGFP: 37 ± 4, n = 463 neurons); and at P500 (AlsinWT-UeGFP: 31 ± 2, n = 368 neurons; AlsinKO-UeGFP: 30 ± 2, n = 339 neurons) (Fig. 3E). These results corroborated previous reports, suggesting lack of major CSMN loss in mouse models of alsin loss of function (14,17–19). However, even though CSMN soma diameters were comparable at P50 (AlsinWT-UeGFP: 19.3 µm, n = 2 mice, n = 68 neurons; AlsinKO-UeGFP: 20.3 µm, n = 2 mice, n = 65 neurons), there was progressive reduction with age (at P300: AlsinWT-UeGFP: 19.5 ± 0.5 µm, n = 131 neurons; AlsinKO-UeGFP: 15.3 ± 0.4 µm, n = 154 neurons; P < 0.003 and by P500: AlsinWT-UeGFP: 20 ± 0.3 µm, n = 122 neurons; AlsinKO-UeGFP: 15.3 ± 0.3 µm n = 135 neurons; P < 0.0007) (Fig. 3F).
Figure 3.
Reduced soma diameter suggests potential neuronal defects, without major cell loss. (A and B) Representative images of CSMN in AlsinWT-UeGFP and (C and D) AlsinKO-UeGFP mice at P500. (E) Average numbers of CSMN were comparable between AlsinWT-UeGFP and AlsinKO-UeGFP mice. (F) The average CSMN soma diameter significantly decreased in AlsinKO-UeGFP mice at P300 and at P500. Bar graph values are mean ± SEM. Student's t-test, ** P < 0.003; *** P < 0.0007. Scale bar: 20 µm (A–D).
Increased autophagy in apical dendrites of diseased CSMN
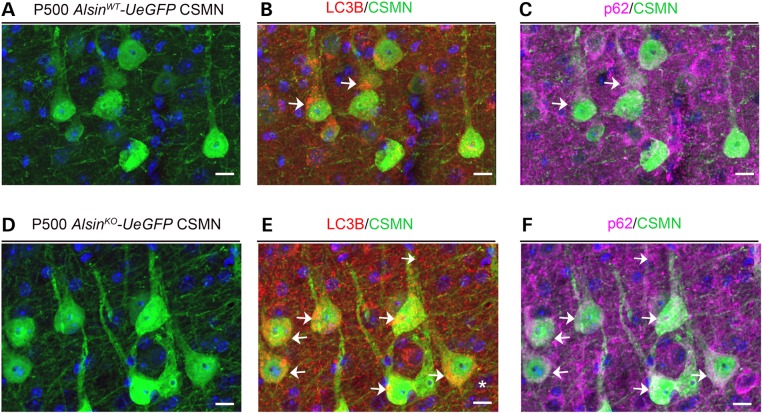
We previously reported apical dendrite degeneration in CSMN of hSOD1G93A (23) and UCHL1−/– mice (22), which suggests that this could be a common phenomenon for diseased CSMN. Interestingly, CSMN in the AlsinKO-UeGFP mice also displayed major defects in their apical dendrites (Fig. 4A–F). The percentage of CSMN with autophagic vesicles in their apical dendrites was increased in AlsinKO-UeGFP mice (at P300: AlsinWT-UeGFP: 5 ± 1%, n = 128 apical dendrites; AlsinKO-UeGFP: 4 ± 1%, n = 107 apical dendrites and P500: AlsinWT-UeGFP: 6 ± 3%, n = 74 apical dendrites; AlsinKO-UeGFP: 22 ± 2%, n = 85 apical dendrites; P < 0.02) (Fig. 4G). Interestingly, the percentages of CSMN with large somatic autophagic vesicles were comparable between WT and AlsinKO mice at both P300 (AlsinWT-UeGFP: 39 ± 4%, n = 157 cell bodies and AlsinKO-UeGFP: 33 ± 5%, n = 101 cell bodies) and P500 (AlsinWT-UeGFP: 33 ± 2%, n = 60 cell bodies; AlsinKO-UeGFP: 32 ± 7%, n = 72 cell bodies) (Fig. 4H), suggesting that increased autophagy, selectively appeared in the apical dendrites of CSMN that lacked alsin function.
Figure 4.
CSMN display increased autophagy in the absence of alsin function. (A–F) Representative images of CSMN expressing LC3B, a prominent autophagy marker, in AlsinWT-UeGFP (A) and AlsinKO-UeGFP (B–F) mice. (G and H) Quantitative analysis of percent CSMN with autophagy within their apical dendrites (G) and soma (H). Bar graph values are mean ± SEM. Student's t-test, *P < 0.02. Scale bar: 20 µm (A and B).
p62 is a selective autophagy receptor that is pivotal for mediating and modulating autophagy of ubiquitinated proteins and defective organelles (24), and its accumulation inside the neuron indicate problems with the ubiquitin proteosome system and the autophagy pathways (25). We thus investigated p62 expression together with LC3B in diseased CSMN at later ages. p62 was present in many neurons in the motor cortex, especially at P500 (Fig. 5A–F). However, the percentage of CSMN expressing p62 was higher in the absence of alsin function (AlsinWT-UeGFP: 84 ± 1%, n = 92 neurons; AlsinKO-UeGFP: 90 ± 0.5%, n = 139 neurons; P < 0.01; Fig. 5). Interestingly, diseased CSMN that include high levels of LC3B also displayed increased presence of p62 (Fig. 5D–F, arrows), suggesting defects in the autophagy pathways in the absence of alsin function.
Figure 5.
p62 is present in CSMN that expresses LC3B in AlsinKO-UeGFP mice (A–C) Representative images of CSMN (A) that express LC3B (B) and p62 (C) in the AlsinWT-UeGFP mice. (D–F) Representative images of CSMN (D) that express LC3B (E) and p62 (F) in the AlsinKO-UeGFP mice. Arrows indicate CSMN that express both LC3B and p62. Scale Bar: 10 µm.
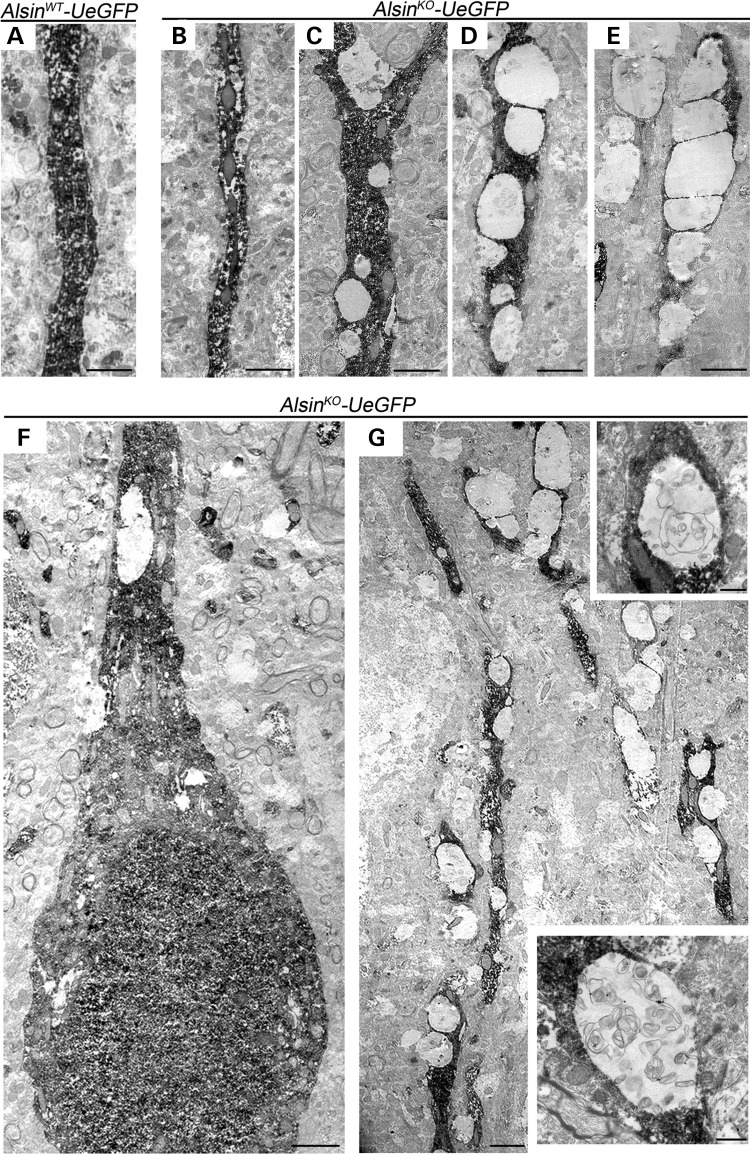
To further investigate apical dendrites of diseased CSMN, eGFP immunocytochemistry was coupled with EM analysis. eGFP expression was restricted to CSMN in the motor cortex and this helped identify CSMN among many different neurons (Fig. 6A–G), enabling visualization of ultrastructural defects selectively in CSMN. There were numerous vacuoles along the apical dendrites of CSMN (Fig. 6G), ranging from small to large. However, other neurons and CSMN in WT mice appeared healthy (Fig. 6A), suggesting that apical dendrite defects were mainly restricted to CSMN in the AlsinKO mice. Various stages of vacuolization in apical dendrites of AlsinKO-UeGFP mice were observed at P500, increasing in severity (Fig. 6B–E). Some of the vacuoles were empty (Fig. 5D and E), whereas some included pieces of cellular debris (Fig. 5G insets), suggesting an ongoing autophagy inside apical dendrites. A closer look suggested that this cellular debris could include broken mitochondria, as pieces of mitochondria were present inside these vacuoles. In contrast, WT CSMN or apical dendrites of other neurons that are not CSMN did not display such profound pathology, suggesting that these observed defects are not related to age, but rather CSMN-specific and is observed in the absence of alsin function.
Figure 6.
eGFP immunocytochemistry coupled with EM reveal major defects within apical dendrites of diseased CSMN. (A) Apical dendrite of CSMN in AlsinWT-UeGFP mice are devoid of any vacuoles and display healthy morphology. (B–E) Apical dendrites of CSMN in AlsinKO-UeGFP mice have vacuoles of different sizes and various stages. (F) A GFP labeled CSMN soma with vacuolated proximal apical dendrite and (G) distal apical dendrite with many vacuoles only in diseased CSMN. Insets display magnified view of representative vacuoles with numerous disintegrating cellular debris. Scale bar: 2 µm (A–G); 500 nm (insets).
Alsin function is important for the integrity of mitochondria and Golgi
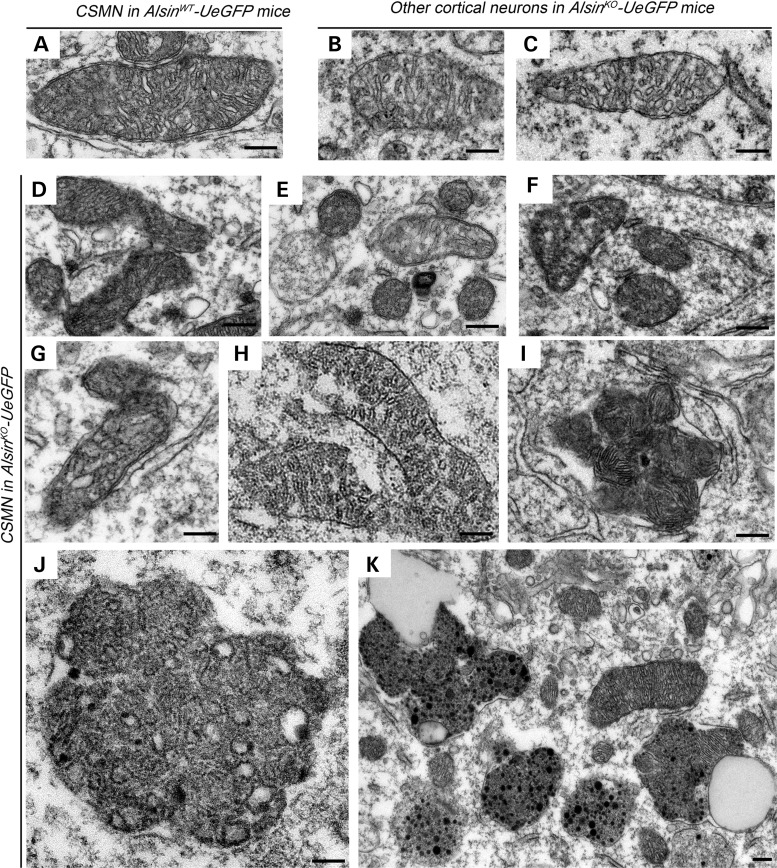
Ctip2 immunoctytochemistry coupled with EM (Supplementary Material, Fig. S2) allowed cellular analysis of CSMN with high definition in both AlsinWT-UeGFP and AlsinKO-UeGFP mice. In the absence of alsin function, CSMN displayed many cellular defects. Among cellular organelles, mitochondria (Fig. 7) and Golgi apparatus (Fig. 8) were the most affected. In diseased CSMN, mitochondria had defective inner membranes, expanded core and were disintegrated with broken cristae (AlsinWT-UeGFP: n = 30 neurons and AlsinKO-UeGFP: n = 26 neurons). Many mitochondria were found fused with each other, forming rosette-like structures (Fig. 7D–I). Fused mitochondria were engulfed in large vesicles, forming massive aggresomes (Fig. 7J and K). In contrast, mitochondria of CSMN in AlsinWT-UeGFP mice were healthy with proper membrane and cristae structure (Fig. 7A). Likewise, other cortical neurons that are not CSMN in AlsinKO-UeGFP mice (n = 17 neurons), (Fig. 7B and C) appeared normal, suggesting that this cellular pathology was mostly specific to CSMN in the absence of alsin function.
Figure 7.
Absence of alsin function leads to mitochondrial defects in CSMN. (A–C) Representative images of mitochondria present in the CSMN of AlsinWT-UeGFP mice (A) and other cortical neurons of AlsinKO-UeGFP mice (B and C). (D–K) Representative images of mitochondria present in the CSMN of AlsinKO-UeGFP mice. They display numerous structural defects, such as disintegration of inner membranes (D–G), broken cristae (H), fusion with other mitochondria (I), aggregation (J) and fusion of these aggregates with endolysosome-like structures (K). Scale bar: 200 nm (A–J); 1 µm (K).
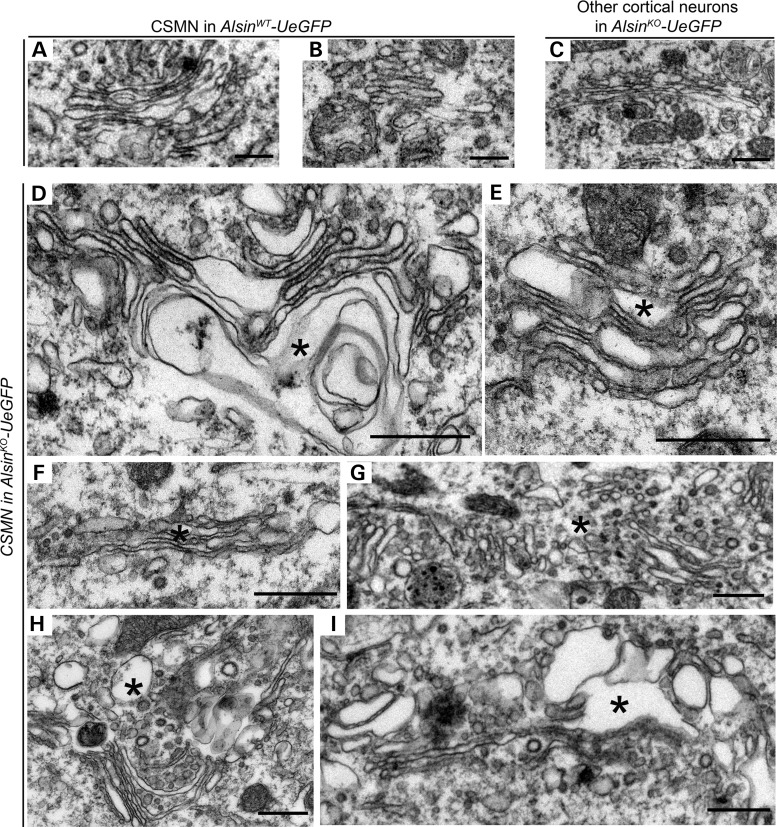
Figure 8.
Alsin function is required for the stability of Golgi apparatus in CSMN. (A–C) Representative images of Golgi apparatus present in the CSMN of AlsinWT-UeGFP mice (A) and other cortical neurons of AlsinKO-UeGFP mice (B and C). (D–I) Representative images of Golgi apparatus present in the CSMN of AlsinKO-UeGFP mice. They display numerous structural defects, such as vacuolated cisternae (denoted by asterisk) (D and E), cisternae filled with dark material (F and G) and dark vesicles and vacuoles disrupting their ultrastructure (H and I). Scale bar: 500 nm (A–I).
Parkin protein has been reported to play significant roles in the tagging and clearing of damaged mitochondria in many cells and neurons (26). We next investigated whether Parkin levels were augmented especially in diseased CSMN. Interestingly, unlike p62 (Fig. 5), the percentage of CSMN showing increased Parkin expression was low and comparable between WT and AlsinKO mice at P500 when most striking mitochondrial defects are observed (AlsinWT-UeGFP: 8 ± 2% n = 84 neurons and AlsinKO-UeGFP: 13 ± 3%, n = 78 neurons) (Supplementary Material, Fig. S3).
The Golgi complex is one of the most important cellular organelles for post-translational modification of proteins and controlling secretory pathways and transportation of proteins to proper targets within the cell. We found major defects within the Golgi complex of CSMN that lacked alsin function (AlsinKO-UeGFP: n = 36 neurons; Fig. 8). Their cisternae were enlarged and some were broken, indicating severe structural defects (Fig. 8D–I, asterisk). In addition, there were many vesicles in different sizes and content, especially along the Golgi apparatus (Fig. 8F and G). The membranes of these vesicles were thick and appeared dark suggesting potential problems with fusion and recycling. These modifications were only observed in the CSMN of AlsinKO-UeGFP mice; WT CSMN (AlsinWT-UeGFP: n = 24 neurons), and other neurons that were not CSMN in the AlsinKO-UeGFP mice (n = 16) did not display similar severe pathology (Fig. 8A–C), and appeared healthy and normal.
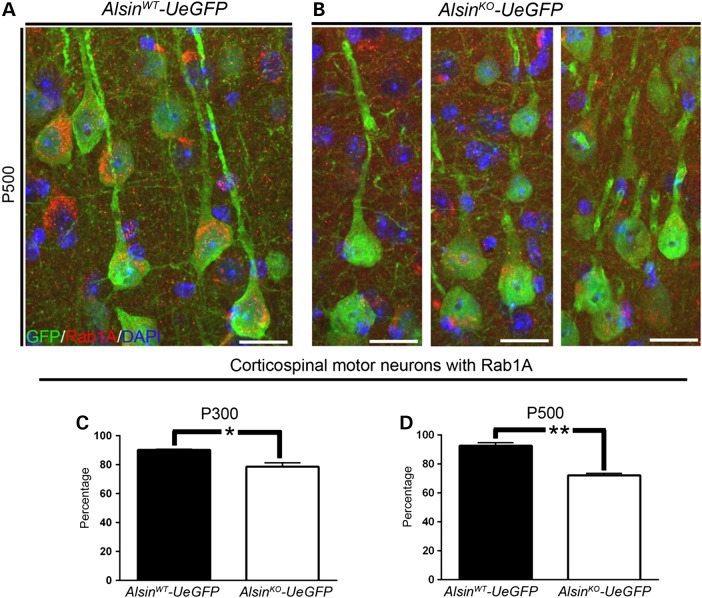
Rab1A is a member of small GTPase family that plays an important role in maintaining integrity of the Golgi apparatus (27). Rab1A expression in AlsinWT CSMN remained unchanged with age and CSMN expressed high levels of Rab1A both at P300 (AlsinWT-UeGFP: 92 ± 2%, n = 229 neurons) and P500 (AlsinWT-UeGFP: 93 ± 1%, n = 109 neurons; Fig. 9A–D). However, there was significant reduction in the number of CSMN that expressed Rab1A in the AlsinKO-UeGFP mice (P300: AlsinKO-UeGFP: 79 ± 3%, n = 163 neurons; P < 0.01 and P500: AlsinKO-UeGFP: 72 ± 1%, n = 299 neurons; P < 0.001), suggesting a progressive decline in Rab1A levels especially in the absence of alsin function (Fig. 9). Similarly, AlsinWT and AlsinKO CSMN were comparable and both displayed high levels of Rab1A by P50 (AlsinWT-UeGFP: 93%, n = 147 neurons, n = 2 mice and AlsinKO-UeGFP: 94%, n = 153 neurons, n = 2 mice).
Figure 9.
Rab1A expression is reduced in diseased CSMN. (A and B) Representative images of Rab1A expression in CSMN of AlsinWT-UeGFP (A) and AlsinKO-UeGFP mice (B). (C and D) Percentage of CSMN with Rab1A is progressively reduced in the absence of alsin function at P300 (C) and P500 (D). Bar graph values are mean ± SEM. Student's t-test, *P < 0.01, **P < 0.001. Scale bar: 20 µm (A and B).
Discussion
CSMN have a unique importance for initiation and modulation of voluntary movement by receiving, integrating, translating and transmitting cerebral cortex's input toward spinal cord targets. Therefore, they are very important components of the motor neuron circuitry, which degenerates in many motor neuron diseases (28,29). As expected, CSMN cellular degeneration is a hallmark of many diseases, such as PLS, HSP and IAHSP (28–31). They also undergo progressive degeneration together with spinal motor neurons in ALS patients (29). However, our knowledge of the cellular and molecular basis of CSMN degeneration is limited.
Mutations identified in patients with motor neuron diseases suggest many important cellular events that may lead to motor neuron death. For example, mutations in the ALS2 gene are exceptionally important as they are present in patients with the juvenile form of ALS, juvenile PLS, HSP and IAHSP (2,3). Because upper motor neurons are the primary motor neuron population that is affected in these diseases, it has been suggested that alsin function may be particularly important for upper motor neurons (1).
Mounting evidence suggests that alsin is central to multiple important cellular functions, such as endocytosis, micropinocytosis, membrane trafficking and maintaining cytoskeleton integrity (9,32,33). Alsin protein has been shown to modulate activities of Rab5 and Rac1 (9,33). Loss of alsin protein function leads to disruption of fusion between endosomes and autophagosomes obstructing endolysosomal-mediated protein degradation (34). However, how motor neurons, and especially upper motor neurons, are affected in the absence of alsin function remained unknown.
Humans heavily depend on their upper motor neurons, and the CST is the major axonal path connecting cortex and spinal cord. However, rodents do not fully recapitulate human anatomy, and other subcerebral paths such as rubrospinal tract and reticulospinal tracts are prominent in mice (35). It is possible that CSMN may indeed undergo specific degeneration in mouse models without obvious motor dysfunction. Thus, developing disease models with selective CSMN vulnerability is a challenge, because mouse models may not display overt disease phenotype even though upper motor neurons become vulnerable. In addition, transgenic mice that express multiple copy numbers of human mutations may facilitate disease pathology, and even though models that have a single mutated gene may take a longer time to develop the disease, they may better represent the cellular or neuronal pathology that occurs in human patients (36). Therefore, investigation of vulnerable neurons at later ages of well-defined models may reveal the underlying causes of vulnerability in humans.
To date, many mouse models that either lack or overexpress the mutant form of the genes identified in ALS patients have been generated, and only very few display motor function defects and recapitulate human pathology. This was interpreted as lack of translation from mouse to human; consequently, questions were raised concerning the relevance of the results in mouse studies, especially for drug discovery studies (37). We, however, propose that if comparisons were made at a cellular level, the translation would be more apparent and informative. In line with this reasoning, even yeast cells helped reveal novel disease pathways and cellular events that are responsible for neuronal degeneration in different neurodegenerative diseases (38). We believe that by moving our focus of attention from mice to specific neurons that display vulnerability, it is possible to obtain a better understanding for the underlying causes that lead to their selective vulnerability, which could indeed be translational.
In an effort to make CSMN visible in the absence of alsin function and to study the cellular defects that give rise to their selective vulnerability, we generated AlsinKO-UeGFP mice. Our detailed cellular analysis suggested novel disease targets in CSMN that lacked alsin function. We found major defects in mitochondria with broken cristae and vacuolated cisternae. Defective mitochondria were found to cluster together in diseased CSMN, whereas other cortical neurons, even in AlsinKO-UeGFP mice, remained unaffected. These findings are supported by previous reports showing interaction of alsin with neurocalcin alpha (39) and spartin (40), two proteins that are critically important for modulating Ca+ influx and uptake at the site of mitochondria. Parkin is critically important for mitophagy (41,42). However, we did not observe high levels of Parkin expression in CSMN that lack alsin function, albeit major mitochondrial defects were present. In contrast, p62 expression was increased in diseased CSMN. p62 is recruited to defective mitochondria for their clearance (43), and it is implicated in degradation of misfolded proteins and defective cellular organelles through autophagy–lysosome pathway (44). In addition, recent studies also suggest that p62 can lead to mitophagy in a Parkin-independent manner (45).
Even though mitochondrial defects in CSMN constitute a novel finding within the context of alsin function, such mitochondria abnormalities in motor neurons of other mouse models of ALS such as hSOD1G93A (46) and TDP-43 (47) have been previously shown. Our findings, together with previous reports, strengthen the hypothesis that mitochondrial pathology, which results in dysregulation of metabolism, oxidative stress and insufficient energy production, may be a common mechanism for both upper and lower motor neuron vulnerability.
We find that in addition to mitochondrial defects, the Golgi apparatus also displays many ultrastructural abnormalities in the CSMN of AlsinKO mice. Rab1A is reported to be important for the maintenance of Golgi apparatus (27), and we find reduced levels of Rab1A in diseased CSMN. Presence of disintegrating Golgi apparatus in CSMN that lack alsin function suggests potential problems with protein modifications, processing and transportation. Even though this is the first direct evidence of defects in Golgi apparatus in CSMN, previous studies eluted to the potential involvement of alsin in the maintenance of Golgi apparatus. Rab1 promotes disassembly of Golgi bodies when mutated (48). It is possible that in the absence of alsin, Rab1 activity is dysregulated in the AlsinKO mice. Interestingly, anterior horn cells of familial ALS patients with SOD1 mutation display fragmentation of their Golgi apparatus (49), and this was also observed in the spinal motor neurons of hSOD1G93A mice (50). These reports, together with our findings, indicate a critical importance of the health of Golgi bodies, especially for upper motor neurons that lack alsin function.
Apical dendrite degeneration begins to emerge as a common phenomenon that occurs in vulnerable and degenerating CSMN. We previously reported apical dendrite defects in hSOD1G93A (23) and UCHL1−/− (22) mice. Now we show that the CSMN of AlsinKO mice display similar major ultrastructural defects in the apical dendrites. Increased LC3B together with p62 suggests active involvement of problems with autophagy, especially in CSMN that becomes vulnerable in the absence of alsin function. This represents an important problem for maintenance of CSMN function, because most cortical inputs converge onto CSMN at the site of apical dendrite (51). Disintegration of apical dendrites may imply failure of CSMN to receive the required input and modulation from cortical neurons (52).
The UCHL1-UeGFP reporter line provides a powerful tool to investigate CSMN biology in health and diseases. Our results demonstrate, for the first time, that upper motor neurons of AlsinKO mice display major cellular defects, but no cell loss. This is different from findings in hSOD1G93A transgenic ALS mouse, which display early and progressive CSMN degeneration (53). Building evidence suggest that neuron numbers do not fully inform about disease. For example, CSMN numbers do not change with disease progression in hSOD1G93A rat model of ALS; however, correction of mutated SOD1 gene in the motor cortex, using adeno associated viruses (AAV) approaches, significantly improved the health of CSMN and the connectivity of the motor neuron circuitry (54). This study suggested that re-establishment of CSMN health improves overall motor function, even independently of neuron number.
By shifting our focus from AlsinKO mice to CSMN that lack alsin function, we revealed major cellular defects that occur selectively in CSMN, which would otherwise remain unnoticed. Our findings add further evidence to the fact that important pathology is present at a cellular level even when the mouse do not display major motor function defects, and suggest that defining pathology as a function of cell number may be misleading, and that focusing our attention to the vulnerable motor neurons will begin to reveal underlying causes of selective vulnerability.
Materials and Methods
Mice
All animal procedures were performed in compliance with the standards set by National Institutes of Health and were approved by the Northwestern University Animal Care and Use committee. The following mouse strains were used in this study: wild-type (WT), AlsinKO (gift of Drs Deng and Siddique, Northwestern University), UCHL1-eGFP, AlsinKO-UeGFP and AlsinWT-UeGFP mice (generated by the Ozdinler laboratory). Hemizygous UCHL1-eGFP males were bred to hemizygous AlsinHT females to generate AlsinHT-UeGFP mice (Fig. 1A). AlsinKO-UeGFP and AlsinWT-UeGFP mice were generated on a mixed C57/BL6 and B6/SJL background and animals of both sexes were used in this study. Progeny were initially screened at birth for eGFP expression using fluorescent flashlight (BlueStar GFP, NightSea). DNA was isolated from tail biopsies by precipitation using isopropanol. Genotyping was performed by polymerase chain reaction (PCR) to detect presence of UCHL1-eGFP gene (forward 5′-CCTACGGCGTGCAGTGCTTCAGC-3′ and reverse 5′-CGGCGAGCTGCACGCTGCCGTCCTC-3′ primers) PCR cycle (94°C for 2 min (94°C for 30 s, 62°C for 45 s, 72°C for 45 s) for 30 cycles, 72°C for 10 min) and mutant allele (Neo) (forward 5′-CAGCTGTGCTCGACGTTGTCACTGA-3′ and reverse 5′-GCCTTGAGCCTGGCGAACAGTTC-3′) and WT fragment (forward 5′-GGAAACTCTGGTACTGAGCTATAG-3′ and reverse 5′-CCAGCTCACTGGCCTAACTCTAGATGTC-3′) of the Als2 gene (94°C for 2 min (94°C for 30 s; 62°C for 30 s; 72°C for 30 s) for 30 cycles, 72°C for 8 min).
Tissue processing
Mice were deeply anesthetized with intraperitoneal injection of ketamine (90 mg/kg), and xylazine (10 mg/kg; Fort Dodge Animal Health, Fort Dodge, IA, USA) prior to transcardiac perfusion with phosphate-buffered saline (PBS) and 4% paraformaldehyde (PFA) in PBS. Intact cortex and spinal cord were dissected out, post-fixed in 4% PFA overnight and stored in PBS with 0.01% sodium azide at 4°C. Left hemispheres were cryopreserved in 30% sucrose in PBS, and 20 µm frozen sections were cut using sliding microtome. Right hemispheres were sectioned at 50 µm using Leica vibratome (Leica VT1000S, Leica Inc., Nussloch, Germany).
Immunocytochemistry
Fifty micrometers thick floating sections were incubated in blocking solution (PBS, 0.05% bovine serum albumin, 2% fetal bovine serum, 1% Triton X-100 and 0.1% saponin) for 30 min prior to addition of primary antibodies: rat anti-Ctip2 antibody (1:500; Abcam), rat anti-GFAP (1:2000, Invitrogen), chicken anti-GFP (1:1000, Abcam), rabbit anti-Iba1 (1:500, Wako), rabbit anti-LC3B (1:1000, Invitrogen), mouse anti-Parkin (1:200, Abcam), mouse anti-SQSTM1/p62 (1:200, Abcam), rabbit anti-Rab1A (1:200, Proteintech) and mouse anti-Satb2 (1:1000, Millipore) at 4°C overnight. After extensive washes, sections were incubated in appropriate secondary antibodies in blocking solution: Alexa Fluor 488 (1:1000, Invitrogen), and Cy3-conjugated (1:1000, Molecular Probes) for 2 h at room temperature. Antigen retrieval was performed for rat anti-Ctip2 antibody (1:500; Abcam) as previously described (22). Sections were counterstained with DAPI.
Immunocytochemistry coupled with EM
In a subset of experiments, immunocytochemistry was followed by EM analysis. Mice were perfused with EM grade 4% PFA at P300 and P500. One hemisphere of the brain was sectioned at 150 µm, coronally on a vibrotome (Leica). The sections were post-fixed in 2% PFA and 0.5% glutaraldehyde for an hour, they were cryoprotected with glycerol–dimethylsulfoxide mixture followed by freeze–thaw at least four times and treated with 1% sodium borohydrate. They were then treated with 0.3% H2O2–10% methanol in TBS (100 mm Tris–HCl and 150 mm NaCl, pH 7.6) and 5% NGS–1% bovine serum albumin in TBS to block non-specific binding of primary antibody. They were incubated overnight with rabbit anti-GFP antibody (1:1000; Invitrogen) or rat anti-Ctip2 antibody (1:500, Invitrogen). Biotinylated goat anti-rabbit IgG (1:500) or biotinylated goat anti-rat IgG (1:500) were used as secondary antibodies, and diaminobenzidine was applied as the chromogen (ABC Elite kit, Vector Laboratories, Burlingame, CA, USA). Sections were then post-fixed in buffered 2% OsO4, rinsed with distilled water and stained in 1% uranyl acetate, again rinsed with distilled water, dehydrated in ascending grades of ethanol with transition fluid propylene oxide and embedded in resin mixture with Embed 812 and cured in a 60°C oven for 3 days. The sections in which primary motor cortex was present and visible under bright field illumination on a dissecting scope were selected. Approximately, 5 mm wide × 7 mm long piece of the motor cortex from these sections was dissected under the microscope, mounted on resin block and was sectioned on a Leica Ultracut UC6 ultramicrotome. Seventy nanometers thin sections were collected on 200 mesh copper–palladium grids. Grids were counter stained with 8% radioactive depleted uranyl acetate and 0.2% lead citrate.
Imaging
Nikon Eclipse TE2000-E (Nikon Inc., Melville, NY, USA) and Leica TCS SP5 confocal microscopes (Leica Inc., Bensheim, Germany) were used to acquire low and high magnification images of fluorescent samples, respectively. Z-stacks were processed using ImageJ (National Institutes of Health, Bethesda, MD, USA, http://imagej.nih.gov/ij). For EM imaging, grids were examined on FEI Tecnai Spirit Transmission Electron Microscope at 120 kV. Images were captured on FEI Eagle camera using FEI TIA software.
Quantification and data analysis
Three mice were used for each genotype and time point, and three comparable sections that span the motor cortex were used for each mice for all statistical analysis. Numbers are noted in the Results section, unless otherwise.
Quantification of numbers of CSMN and soma diameter assessment
Average numbers of CSMN were counted using maximum projection images acquired from 50 µm thick coronal sections of the brains isolated from P300 and P500 AlsinKO-UeGFP and AlsinWT-UeGFP mice (n = 3). An equivalent area of the motor cortex in three serial sections (at least ∼600 µm apart) was imaged with 10× objective field per mouse that represents motor cortex area (Section 1: Bregma 1.18 mm, interaural 4.98 mm; Section 2: Bregma 0.74 mm, interaural 4.54 mm and Section 3: Bregma 0.14 mm, interaural 3.94 mm) (55). eGFP-positive (eGFP+) neurons with pyramidal morphology and an apical dendrite were counted in each area by a blinded observer. The soma diameter of eGFP+ neurons with prominent apical dendrite was assessed by measuring and averaging the length across dorsoventral and mediolateral axis of each neuron. At least 90 neurons were analyzed per genotype.
Fiber density analysis
CST in the ventral pons was imaged in three, 20 µm thick sections per mouse (AlsinWT-UeGFP and AlsinKO-UeGFP mice at P300 and P500; n = 3) using 63× objective. Z-stacks of identical volume (7 µm) were processed with ImageJ software to generate maximum intensity projections. Images with a defined area (168 × 246 µm) were converted to grayscale, and this is accepted as ‘unit area’ for quantification. Mean gray pixel value was measured in each image. At least three images were taken and analyzed per genotype.
Axon fiber cross-section area measurement
CST in the ventral pons area of the P300 and P500 AlsinKO-UeGFP and AlsinWT-UeGFP mice was imaged in 20 µm sections using 63× objective of Leica TCS SP5 confocal microscopes with z-stacks (Leica Inc., Bensheim, Germany). The resulting stacks were loaded into the open source software platform Fiji (Fiji/ImageJ http://imagej.nih.gov/ij/i, NIH) (56). Simple neurite tracer plugin of Fiji software was used to trace an axon fiber using Hessian-based analysis (57). Traced axon fibers were filled using fill volume feature using the Dijkstra's algorithm at a set intensity (determined by taking the average of adjusted thresholds from each axon). Volume of traced axon fiber was divided by its length to measure cross-section area of an individual axon fiber.
Statistical analysis
Prism software (GraphPad Software Inc., La Jolla, CA, USA) was used for all statistical analyses. D'Agostino and Pearson normality test was performed on all data sets (WT and KO) prior to all statistical tests. Student's t-test was used to determine differences between two experimental conditions, and statistically significant differences were taken at P < 0.05.
Supplementary Material
Funding
This work was supported by grant from NIH R21-NS085750-01, Les Turner ALS Foundation and Wenske Foundation (P.H.O.). We thank Drs Deng and Siddique (Northwestern University, Chicago) for providing AlsinKO mice. We thank Mr Lennell Reynolds Jr, and Center for Advanced microscopy, Northwestern University for helping with EM imaging. We also thank the Microscopy and Imaging Facility at Stanley Manne Children's Research Institute and B. Goossens for help with confocal microscopy. Funding to pay the Open Access publication charges for this article was provided by the Les Turner ALS Foundation.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
References
- 1.Yang Y., Hentati A., Deng H.X., Dabbagh O., Sasaki T., Hirano M., Hung W.Y., Ouahchi K., Yan J., Azim A.C. et al. (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat. Genet., 29, 160–165. [DOI] [PubMed] [Google Scholar]
- 2.Eymard-Pierre E., Lesca G., Dollet S., Santorelli F.M., di Capua M., Bertini E., Boespflug-Tanguy O. (2002) Infantile-onset ascending hereditary spastic paralysis is associated with mutations in the alsin gene. Am. J. Hum. Genet., 71, 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadano S., Hand C.K., Osuga H., Yanagisawa Y., Otomo A., Devon R.S., Miyamoto N., Showguchi-Miyata J., Okada Y., Singaraja R. et al. (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet., 29, 166–173. [DOI] [PubMed] [Google Scholar]
- 4.Eker H.K., Unlu S.E., Al-Salmi F., Crosby A.H. (2014) A novel homozygous mutation in ALS2 gene in four siblings with infantile-onset ascending hereditary spastic paralysis. Eur. J. Med. Genet., 57, 275–278. [DOI] [PubMed] [Google Scholar]
- 5.Wakil S.M., Ramzan K., Abuthuraya R., Hagos S., Al-Dossari H., Al-Omar R., Murad H., Chedrawi A., Al-Hassnan Z.N., Finsterer J. et al. (2014) Infantile-onset ascending hereditary spastic paraplegia with bulbar involvement due to the novel ALS2 mutation c.2761C>T. Gene, 536, 217–220. [DOI] [PubMed] [Google Scholar]
- 6.Abiko H., Fujiwara S., Ohashi K., Hiatari R., Mashiko T., Sakamoto N., Sato M., Mizuno K. (2015) Rho guanine nucleotide exchange factors involved in cyclic-stretch-induced reorientation of vascular endothelial cells. J. Cell Sci., 128, 1683–1695. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff F.R., Ponstingl H. (1991) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature, 354, 80–82. [DOI] [PubMed] [Google Scholar]
- 8.Kanekura K., Hashimoto Y., Kita Y., Sasabe J., Aiso S., Nishimoto I., Matsuoka M. (2005) A Rac1/phosphatidylinositol 3-kinase/Akt3 anti-apoptotic pathway, triggered by AlsinLF, the product of the ALS2 gene, antagonizes Cu/Zn-superoxide dismutase (SOD1) mutant-induced motoneuronal cell death. J. Biol. Chem., 280, 4532–4543. [DOI] [PubMed] [Google Scholar]
- 9.Topp J.D., Gray N.W., Gerard R.D., Horazdovsky B.F. (2004) Alsin is a Rab5 and Rac1 guanine nucleotide exchange factor. J. Biol. Chem., 279, 24612–24623. [DOI] [PubMed] [Google Scholar]
- 10.Tudor E.L., Perkinton M.S., Schmidt A., Ackerley S., Brownlees J., Jacobsen N.J., Byers H.L., Ward M., Hall A., Leigh P.N. et al. (2005) ALS2/Alsin regulates Rac-PAK signaling and neurite outgrowth. J. Biol. Chem., 280, 34735–34740. [DOI] [PubMed] [Google Scholar]
- 11.Otomo A., Hadano S., Okada T., Mizumura H., Kunita R., Nishijima H., Showguchi-Miyata J., Yanagisawa Y., Kohiki E., Suga E. et al. (2003) ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum. Mol. Genet., 12, 1671–1687. [DOI] [PubMed] [Google Scholar]
- 12.Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol., 2, 107–117. [DOI] [PubMed] [Google Scholar]
- 13.Devon R.S., Orban P.C., Gerrow K., Barbieri M.A., Schwab C., Cao L.P., Helm J.R., Bissada N., Cruz-Aguado R., Davidson T.L. et al. (2006) Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc. Natl. Acad. Sci. USA, 103, 9595–9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadano S., Benn S.C., Kakuta S., Otomo A., Sudo K., Kunita R., Suzuki-Utsunomiya K., Mizumura H., Shefner J.M., Cox G.A. et al. (2006) Mice deficient in the Rab5 guanine nucleotide exchange factor ALS2/alsin exhibit age-dependent neurological deficits and altered endosome trafficking. Hum. Mol. Genet., 15, 233–250. [DOI] [PubMed] [Google Scholar]
- 15.Lai C., Xie C., Shim H., Chandran J., Howell B.W., Cai H. (2009) Regulation of endosomal motility and degradation by amyotrophic lateral sclerosis 2/alsin. Mol. Brain, 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai H., Shim H., Lai C., Xie C., Lin X., Yang W.J., Chandran J. (2008) ALS2/alsin knockout mice and motor neuron diseases. Neurodegener. Dis., 5, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng H.X., Zhai H., Fu R., Shi Y., Gorrie G.H., Yang Y., Liu E., Dal Canto M.C., Mugnaini E., Siddique T. (2007) Distal axonopathy in an alsin-deficient mouse model. Hum. Mol. Genet., 16, 2911–2920. [DOI] [PubMed] [Google Scholar]
- 18.Yamanaka K., Miller T.M., McAlonis-Downes M., Chun S.J., Cleveland D.W. (2006) Progressive spinal axonal degeneration and slowness in ALS2-deficient mice. Ann. Neurol., 60, 95–104. [DOI] [PubMed] [Google Scholar]
- 19.Cai H., Lin X., Xie C., Laird F.M., Lai C., Wen H., Chiang H.C., Shim H., Farah M.H., Hoke A. et al. (2005) Loss of ALS2 function is insufficient to trigger motor neuron degeneration in knock-out mice but predisposes neurons to oxidative stress. J. Neurosci., 25, 7567–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasvoina M.V., Genc B., Jara J.H., Sheets P.L., Quinlan K.A., Milosevic A., Shepherd G.M., Heckman C.J., Ozdinler P.H. (2013) eGFP expression under UCHL1 promoter genetically labels corticospinal motor neurons and a subpopulation of degeneration-resistant spinal motor neurons in an ALS mouse model. J. Neurosci., 33, 7890–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arlotta P., Molyneaux B.J., Chen J., Inoue J., Kominami R., Macklis J.D. (2005) Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron, 45, 207–221. [DOI] [PubMed] [Google Scholar]
- 22.Jara J.H., Genc B., Cox G.A., Bohn M.C., Roos R.P., Macklis J.D., Ulupinar E., Ozdinler P.H. (2015) Corticospinal motor neurons are susceptible to increased ER stress and display profound degeneration in the absence of UCHL1 function. Cereb. Cortex, 25, 4259–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jara J.H., Villa S.R., Khan N.A., Bohn M.C., Ozdinler P.H. (2012) AAV2 mediated retrograde transduction of corticospinal motor neurons reveals initial and selective apical dendrite degeneration in ALS. Neurobiol. Dis., 47, 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z., Yang L., Xu S., Zhang Z., Cao Y. (2015) The receptor proteins: pivotal roles in selective autophagy. Acta Biochim. Biophys. Sin., 47, 571–580. [DOI] [PubMed] [Google Scholar]
- 25.Herhaus L., Dikic I. (2015) Expanding the ubiquitin code through post-translational modification. EMBO Rep., 16, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. (2010) p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy, 6, 1090–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coune P.G., Bensadoun J.C., Aebischer P., Schneider B.L. (2011) Rab1A over-expression prevents Golgi apparatus fragmentation and partially corrects motor deficits in an alpha-synuclein based rat model of Parkinson's disease. J. Park. Dis., 4, 373–387. [DOI] [PubMed] [Google Scholar]
- 28.Brown R.H. Jr, Robberecht W. (2001) Amyotrophic lateral sclerosis: pathogenesis. Semin. Neurol., 21, 131–139. [DOI] [PubMed] [Google Scholar]
- 29.Fink J.K. (2002) Hereditary spastic paraplegia. Neurol. Clin., 20, 711–726. [DOI] [PubMed] [Google Scholar]
- 30.Fink J.K. (2006) Hereditary spastic paraplegia. Curr. Neurol. Neurosci., 6, 65–76. [DOI] [PubMed] [Google Scholar]
- 31.Rainier S., Chai J.H., Tokarz D., Nicholls R.D., Fink J.K. (2003) NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6). Am. J. Hum. Genet., 73, 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunita R., Otomo A., Mizumura H., Suzuki-Utsunomiya K., Hadano S., Ikeda J.E. (2007) The Rab5 activator ALS2/alsin acts as a novel Rac1 effector through Rac1-activated endocytosis. J. Biol. Chem., 282, 16599–16611. [DOI] [PubMed] [Google Scholar]
- 33.Otomo A., Kunita R., Suzuki-Utsunomiya K., Mizumura H., Onoe K., Osuga H., Hadano S., Ikeda J.E. (2008) ALS2/alsin deficiency in neurons leads to mild defects in macropinocytosis and axonal growth. Biochem. Biophys. Res. Commun., 370, 87–92. [DOI] [PubMed] [Google Scholar]
- 34.Hadano S., Yoshii Y., Otomo A., Kunita R., Suzuki-Utsunomiya K., Pan L., Kakuta S., Iwasaki Y., Iwakura Y., Ikeda J.E. (2010) Genetic background and gender effects on gross phenotypes in congenic lines of ALS2/alsin-deficient mice. Neurosci. Res., 68, 131–136. [DOI] [PubMed] [Google Scholar]
- 35.Lemon R.N. (2008) Descending pathways in motor control. Annu. Rev. Neurosci., 31, 195–218. [DOI] [PubMed] [Google Scholar]
- 36.Joyce P.I., McGoldrick P., Saccon R.A., Weber W., Fratta P., West S.J., Zhu N., Carter S., Phatak V., Stewart M. et al. (2015) A novel SOD1-ALS mutation separates central and peripheral effects of mutant SOD1 toxicity. Hum. Mol. Genet., 24, 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayden E.C. (2014) Misleading mouse studies waste medical resources. Nature, doi:10.1038/nature.2014.14938. [Google Scholar]
- 38.Khurana V., Lindquist S. (2010) Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat. Rev. Neurosci., 11, 436–449. [DOI] [PubMed] [Google Scholar]
- 39.Masutani T., Taguchi K., Kumanogoh H., Nakamura S., Maekawa S. (2008) Molecular interaction of neurocalcin alpha with alsin (ALS2). Neurosci. Lett., 438, 26–28. [DOI] [PubMed] [Google Scholar]
- 40.Cobanoglu G., Ozansoy M., Basak A.N. (2012) Are alsin and spartin novel interaction partners? Biochem. Biophys. Res. Commun., 427, 1–4. [DOI] [PubMed] [Google Scholar]
- 41.Narendra D., Tanaka A., Suen D.F., Youle R.J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol., 183, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka A. (2010) Parkin-mediated selective mitochondrial autophagy, mitophagy: Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett., 584, 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y., Tanaka M. (2011) p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J. Cell Sci., 124, 2692–2701. [DOI] [PubMed] [Google Scholar]
- 44.Goldberg A.L. (2003) Protein degradation and protection against misfolded or damaged proteins. Nature, 426, 895–899. [DOI] [PubMed] [Google Scholar]
- 45.Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Hoke A., Dawson V.L., Dawson T.M. et al. (2014) Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J., 33, 2798–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q., Vande Velde C., Israelson A., Xie J., Bailey A.O., Dong M.Q., Chun S.J., Roy T., Winer L., Yates J.R. et al. (2010) ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import. Proc. Natl. Acad. Sci. USA, 107, 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magrane J., Cortez C., Gan W.B., Manfredi G. (2014) Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet., 23, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson B.S., Nuoffer C., Meinkoth J.L., McCaffery M., Feramisco J.R., Balch W.E., Farquhar M.G. (1994) A Rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J. Cell Biol., 125, 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita Y., Okamoto K., Sakurai A., Gonatas N.K., Hirano A. (2000) Fragmentation of the Golgi apparatus of the anterior horn cells in patients with familial amyotrophic lateral sclerosis with SOD1 mutations and posterior column involvement. J. Neurol. Sci., 174, 137–140. [DOI] [PubMed] [Google Scholar]
- 50.Mourelatos Z., Gonatas N.K., Stieber A., Gurney M.E., Dal Canto M.C. (1996) The Golgi apparatus of spinal cord motor neurons in transgenic mice expressing mutant Cu,Zn superoxide dismutase becomes fragmented in early, preclinical stages of the disease. Proc. Natl. Acad. Sci. USA, 93, 5472–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson C.T., Sheets P.L., Kiritani T., Shepherd G.M. (2010) Sublayer-specific microcircuits of corticospinal and corticostriatal neurons in motor cortex. Nat. Neurosci., 13, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jara J.H., Genc B., Klessner J.L., Ozdinler P.H. (2014) Retrograde labeling, transduction, and genetic targeting allow cellular analysis of corticospinal motor neurons: implications in health and disease. Front. Neuroanat., 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozdinler P.H., Benn S., Yamamoto T.H., Guzel M., Brown R.H. Jr, Macklis J.D. (2011) Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G(9)(3)A transgenic ALS mice. J. Neurosci., 31, 4166–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomsen G.M., Gowing G., Latter J., Chen M., Vit J.P., Staggenborg K., Avalos P., Alkaslasi M., Ferraiuolo L., Likhite S. et al. (2014) Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J. Neurosci., 34, 15587–15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paxinos G., Franklin K.B.J. (2008) The Mouse Brain in Stereotaxic Coordinates. Academic Press/Elsevier, Amsterdam. [Google Scholar]
- 56.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Longair M.H., Baker D.A., Armstrong J.D. (2011) Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics, 27, 2453–2454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.