Abstract
Epidemiological studies suggest that levels of n-3 and n-6 long-chain polyunsaturated fatty acids are associated with risk of cardio-metabolic outcomes across different ethnic groups. Recent genome-wide association studies in populations of European ancestry have identified several loci associated with plasma and/or erythrocyte polyunsaturated fatty acids. To identify additional novel loci, we carried out a genome-wide association study in two population-based cohorts consisting of 3521 Chinese participants, followed by a trans-ethnic meta-analysis with meta-analysis results from 8962 participants of European ancestry. Four novel loci (MYB, AGPAT4, DGAT2 and PPT2) reached genome-wide significance in the trans-ethnic meta-analysis (log10(Bayes Factor) ≥ 6). Of them, associations of MYB and AGPAT4 with docosatetraenoic acid (log10(Bayes Factor) = 11.5 and 8.69, respectively) also reached genome-wide significance in the Chinese-specific genome-wide association analyses (P = 4.15 × 10−14 and 4.30 × 10−12, respectively), while associations of DGAT2 with gamma-linolenic acid (log10(Bayes Factor) = 6.16) and of PPT2 with docosapentaenoic acid (log10(Bayes Factor) = 6.24) were nominally significant in both Chinese- and European-specific genome-wide association analyses (P ≤ 0.003). We also confirmed previously reported loci including FADS1, NTAN1, NRBF2, ELOVL2 and GCKR. Different effect sizes in FADS1 and independent association signals in ELOVL2 were observed. These results provide novel insight into the genetic background of polyunsaturated fatty acids and their differences between Chinese and European populations.
Introduction
It is well documented that levels of circulating n-3 polyunsaturated fatty acids (PUFAs) and their dietary intake are associated with decreased risk of cardiovascular disease (1,2), type 2 diabetes (3–5), metabolic syndrome (6,7), breast cancer (8,9) and depression (10,11) in different ethnic groups. It has also been suggested that circulating n-6 PUFA levels have beneficial effect on cardio-metabolic outcomes (12–15), although controversial results remained (16,17).
Alpha-linolenic acid (ALA, 18:3n-3) and linoleic acid (LA, 18:2n-6) are two essential PUFAs that cannot be synthesized in the body (18,19). ALA serves as the substrate for production of other n-3 PUFAs, including eicosapentaenoic acid (EPA, 20:5n-3), docosapentaenoic acid (DPA, 22:5n-3), and docosahexaenoic acid (DHA, 22:6n-3), while LA serves as the substrate for production of other n-6 PUFAs, including gamma-linolenic acid (GLA, 18:3n-6), dihomo-gamma-linolenic acid (DGLA, 20:3n-6), arachidonic acid (AA, 20:4n-6) and docosatetraenoic acid (DTA, 22:4n-6) through desaturation and chain elongation (20). Therefore, genetic variants that affect uptake, desaturation and elongation of PUFAs may contribute to variation in circulating PUFA levels.
Recently, genome-wide association studies (GWAS) have identified multiple loci that were significantly associated with plasma and/or erythrocyte PUFA levels in populations of European ancestry (21–24). FADS1/2 and ELOVL2 that encode the desaturases (Δ-5 and Δ-6) and the elongase, respectively, in the PUFA metabolism pathway have been repeatedly reported to be associated with selected PUFA levels (21–24). Single nucleotide polymorphisms (SNPs) at NTAN1, NRBF2, and GCKR and SLC26A10 were associated with specific plasma PUFA levels within the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (21–23), and LPCAT3 and PCOLCE2 were also identified to be loci for association with circulating levels of specific red blood cell n-3 and n-6 PUFAs in the Framingham Offspring Study (24).
However, Chinese have different PUFA composition, with higher levels of LA, EPA, and DPA and lower levels of GLA, DGLA and AA (25), as well as lower FADS1/2 gene expression levels compared with other ethnic populations (26), suggesting a possible ethnic difference in genetic architecture. Previous GWAS for PUFA levels within CHARGE Consortium also demonstrated different allele frequencies of FADS1 and EVOVL2 variants between Chinese and European participants (22). Therefore, genetic studies in Chinese individuals may facilitate the identification of novel PUFA-associated loci. Furthermore, trans-ethnic meta-analysis that combines the GWAS data from Chinese and European populations may contribute to the identification of additional novel loci and test for the heterogeneity between these two ethnic groups. In this study, we aimed to identify novel loci for PUFA levels through a GWAS meta-analysis in two Chinese cohorts with 3521 individuals, and a trans-ethnic meta-analysis that combined the GWAS data from the Chinese populations and publically available meta-analysis results from populations of European ancestry with 8962 participants.
Results
Chinese GWAS meta-analysis
The basic characteristics of the Nutrition and Health of Aging Population in China (NHAPC) and the Multi-Ethnic Study of Atherosclerosis (MESA) were provided in Table 1. LA is the most abundant PUFA (14.2% in NHAPC and 23.2% in MESA Chinese) while GLA is the least abundant one (0.11% in NHAPC and 0.09% in MESA Chinese) in these two cohorts. The different PUFA levels may reflect the distinct diet patterns and/or differences of erythrocyte verses plasma fatty acid levels between these two studies.
Table 1.
Basic characteristics of NHAPC and MESA cohortsa
| NHAPC | MESA Chinese | |
|---|---|---|
| N | 2865 | 656 |
| Age, yearsb | 58.6 (6.0) | 62.5 (10.3) |
| Female, % | 56.6 | 51.2 |
| Fatty acids (%)b | ||
| LA | 14.2 (2.8) | 23.2 (3.5) |
| GLA | 0.11 (0.07) | 0.09 (0.05) |
| DGLA | 1.33 (0.30) | 2.77 (0.77) |
| AA | 12.9 (1.9) | 10.5 (2.1) |
| DTA | 2.63 (0.65) | NA |
| ALA | 0.28 (0.17) | 0.18 (0.07) |
| EPA | 0.44 (0.19) | 1.15 (0.85) |
| DPA | 1.73 (0.30) | 0.97 (0.22) |
| DHA | 4.42 (1.02) | 5.15 (1.40) |
aNA, not measured.
bValues are mean (SD) except where indicated otherwise.
Genetic variants at multiple loci, including MYB, AGPAT4, FADS1 and ELOVL2, were associated with selected PUFA levels at genome-wide significance (P ≤ 5 × 10−8) (Supplementary Material, Table S1 and Fig. S1).
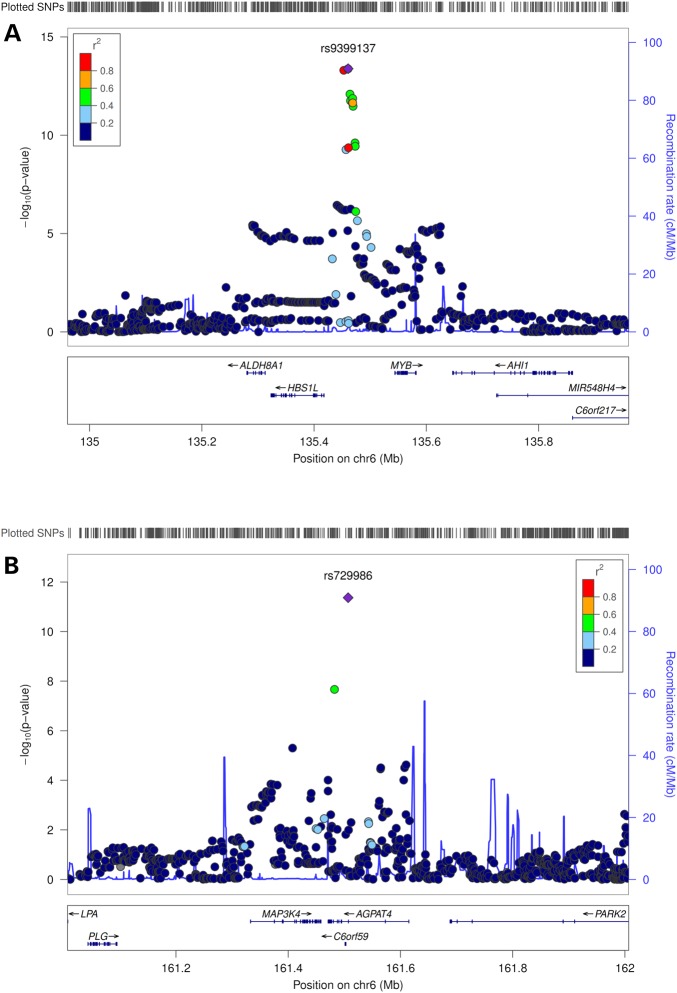
We observed novel associations of variants in MYB and AGPAT4 with DTA level (Table 2). The minor allele C of MYB-rs9399137 was significantly associated with higher level of DTA (β = 0.108, P = 4.15 × 10−14), while minor allele T of AGPAT4-rs729986 was associated with lower level of DTA (β = −0.099, P = 4.30 × 10−12) (Fig. 1). These two SNPs explained 1.18% (rs9399137) and 1.01% (rs729986) of variance in DTA, respectively. Conditional analysis by adding either of these two SNPs in the linear regression model showed that the P values remained almost unchanged (data not shown), suggesting that they were independent of each other.
Table 2.
Genome-wide significant loci identified in trans-ethnic MANTRA analysis
| SNP | Fatty acids | Chr | Position (Build36) | Gene | Effect Allele | EAF (Chinese/European) | Chinese |
European |
MANTRA |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P | β (SE) | P | log10(BF) | Phet | |||||||
| Novel loci | ||||||||||||
| rs9399137 | DTA | 6 | 135460711 | MYB | C | 30.8/26.1 | 0.108 (0.014) | 4.15 × 10−14 | −0.000 (0.002) | 0.995 | 11.5 | 1 |
| rs729986 | DTA | 6 | 161506830 | AGPAT4 | T | 32.3/9.31 | −0.099 (0.014) | 4.30 × 10−12 | 0.000 (0.003) | 0.917 | 8.69 | 1 |
| rs10899123 | GLA | 11 | 75178855 | DGAT2 | C | 79.6/90.6 | 0.007 (0.002) | 1.64 × 10−4 | 0.006 (0.001) | 9.97 × 10−5 | 6.16 | 0.004 |
| rs3134603 | DPA | 6 | 32233980 | PPT2 | A | 16.2/14.2 | 0.027 (0.009) | 0.003 | 0.021 (0.005) | 2.38 × 10−6 | 6.24 | 0.019 |
| Previously reported loci | ||||||||||||
| rs174547 | LA | 11 | 61327359 | FADS1 | C | 40.0/32.3 | 1.046 (0.070) | 4.81 × 10−50 | 1.474 (0.042) | 4.98 × 10−274 | 318 | 1 |
| GLA | 40.1/32.6 | −0.027 (0.001) | 4.59 × 10−81 | −0.016 (0.001) | 2.29 × 10−72 | 147 | 1 | |||||
| DGLA | 40.0/32.6 | 0.189 (0.007) | 8.70 × 10−146 | 0.355 (0.014) | 2.63 × 10−151 | 289 | 1 | |||||
| AA | 40.0/32.0 | −1.099 (0.039) | 1.57 × 10−174 | −1.691 (0.025) | 3.30 × 10−971 | 1141 | 1 | |||||
| DTA | 35.6/32.6 | −0.169 (0.013) | 2.18 × 10−38 | −0.048 (0.002) | 6.26 × 10−140 | 175 | 1 | |||||
| ALA | 39.9/32.5 | 0.024 (0.003) | 1.85 × 10−18 | 0.016 (0.001) | 3.47 × 10−64 | 82.7 | 0.223 | |||||
| EPA | 39.9/31.1 | −0.043 (0.004) | 9.33 × 10−24 | −0.082 (0.005) | 1.83 × 10−57 | 76.6 | 1 | |||||
| DPA | 39.9/32.6 | −0.071 (0.006) | 1.42 × 10−28 | −0.075 (0.003) | 3.79 × 10−154 | 180 | 0.007 | |||||
| rs16966952 | LA | 16 | 15043444 | NTAN1 | A | 34.8/30.9 | 0.683 (0.185) | 2.26 × 10−4 | 0.351 (0.044) | 1.23 × 10−15 | 15.8 | 0.334 |
| GLA | 35.0/31.1 | 0.000 (0.003) | 0.966 | −0.006 (0.001) | 5.05 × 10−11 | 8.36 | 0.058 | |||||
| DGLA | 35.0/31.1 | −0.069 (0.042) | 0.101 | −0.220 (0.013) | 7.55 × 10−65 | 60.8 | 0.938 | |||||
| AA | 34.8/30.8 | −0.271 (0.121) | 0.025 | −0.199 (0.031) | 2.43 × 10−10 | 9.23 | 0.104 | |||||
| rs10740118 | LA | 10 | 64771213 | NRBF2 | C | 32.3/44.0 | −0.130 (0.074) | 0.078 | −0.248 (0.043) | 8.08 × 10−9 | 6.80 | 0.146 |
| rs2281591 | DPA | 6 | 11098479 | ELOVL2 | G | 24.7/24.3 | 0.044 (0.008) | 9.19 × 10−9 | 0.044 (0.003) | 1.58 × 10−38 | 42.6 | 0.013 |
| rs3734398 | EPA | 6 | 11090959 | ELOVL2 | C | 91.0/42.7 | 0.006 (0.007) | 0.348 | 0.035 (0.005) | 3.99 × 10−12 | 6.74 | 0.813 |
| DHA | 6 | 91.0/42.8 | −0.086 (0.038) | 0.025 | −0.114 (0.014) | 1.65 × 10−15 | 14.1 | 0.068 | ||||
| rs780094 | DPA | 2 | 27594741 | GCKR | T | 52.7/41.0 | 0.018 (0.007) | 0.005 | 0.017 (0.003) | 9.04 × 10−9 | 8.43 | 0.010 |
SNP, single nucleotide polymorphism; Chr, chromosome; EAF, effect allele frequency; log10(BF), Log10(Bayers factor) calculated from MANTRA analyses; Phet, the posterior probability of heterogeneity between different ethnics.
Figure 1.
Regional plots of novel loci identified in the Chinese-specific GWAS meta-analysis. Genetic coordinates are displayed along the x-axis (NCBI Build 36) and genome-wide association significance level is plotted against the y-axis as -log10(P value). LD is indicated by color scale in relationship to the most significant SNP in each association, with red for strong LD (r2 ≥ 0.8) and fading color for lower LD. (A) DTA, MYB-rs9399137; (B) DTA, AGPAT4-rs729986.
We observed a novel locus in ELOVL2 associated with DPA. ELOVL2-rs2281591, independent of the previously reported SNP rs3734398 in European-ancestry populations (22) (r2 = 0.02), was significantly associated with DPA level (β = 0.044, P = 9.19 × 10−9) (Supplementary Material, Table S1), and explained 0.68 and 2.30% of the estimated variance of DPA in the NHAPC and the MESA cohorts, respectively.
Consistent with findings from the previous GWAS studies, genetic variants in or near the FADS1/2 gene showed association signals with individual PUFA tested in the analysis (Supplementary Material, Table S1 and Fig. S1). FADS1-rs174547 minor allele C was associated with higher levels of LA (β = 1.046, P = 4.81 × 10−50), DGLA (β = 0.189, P = 8.70 × 10−146) and ALA (β = 0.024, P = 1.85 × 10−18), and lower levels of GLA (β = −0.027, P = 4.59 × 10−81), AA (β = −1.099, P = 1.57 × 10−174), DTA (β = −0.169, P = 2.18 × 10−38), EPA (β = −0.043, P = 9.33 × 10−24) and DPA (β = −0.071, P = 1.42 × 10−28). In addition, SNP rs174547 exhibited significant heterogeneity in effects on LA, DGLA and AA comparing across the NHAPC and the MESA cohorts (Pfor heterogeneity ≤ 6.18 × 10−7). To test whether the heterogeneity were introduced by differences in PUFA levels between the NHAPC and the MESA cohorts, we re-analyzed the data by implementing log-transformation and z-score normalization before cohort-specific GWAS and then combined the results by meta-analysis. The heterogeneity in association of FADS1 variants with LA, DGLA and AA remained between the two cohorts (Pfor heterogeneity ≤ 2.53 × 10−4, Supplementary Material, Table S1), suggesting that differences in PUFA levels may not be the main source of the observed heterogeneity. We estimated that the most significant SNPs on chromosome 11 explained 4.68 and 13.6% of variance in LA, 7.57 and 12.9% of GLA, 19.5 and 0.04% of DGLA, 6.33 and 19.1% of AA, 1.28 and 4.90% of ALA, 2.49 and 0.06% of EPA, 2.95 and 3.98% of DPA in NHAPC and MESA, respectively, and 3.19% of DTA in NHAPC.
We further performed association analyses using 1000 Genomes Phase I imputed data to examine whether there were multiple signals existing at the associated loci (FADS1, ELOVL2, MYB and AGPAT4). However, no additional independent association signals were observed (Supplementary Material, Table S1 and Fig. S2). The most significant SNPs at FADS1/2 and ELOVL2 are highly correlated with the ones observed in the analyses using HapMap Phase II data as the reference panel (r2 ≥ 0.62), and the leading association signals at MYB and AGPAT4 remained the same (r2 = 1).
For previously reported loci including NTAN1, NRBF2, ELOVL2, GCKR, PCOLCE2, LPCAT3 and SLC26A10 in European populations (21–24), associations of NTAN1-rs16966952 with LA and AA, ELOVL2-rs3734398 with DHA, and GCKR-rs780094 with DPA were confirmed in Chinese populations (P ≤ 0.025, Supplementary Material, Table S2).
Trans-ethnic meta-analysis in Chinese and European-ancestry populations
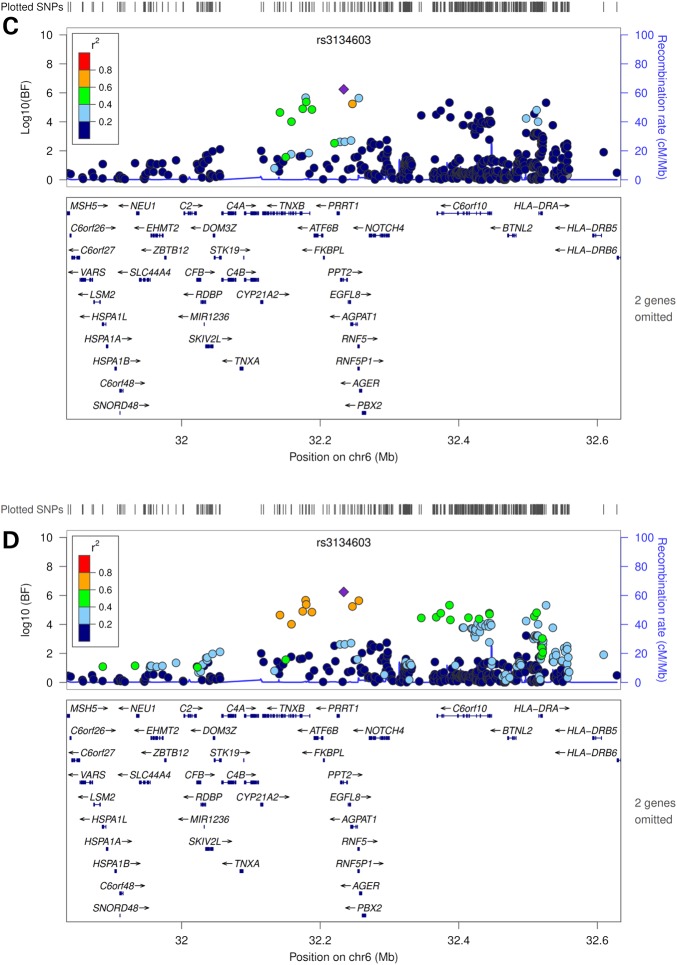
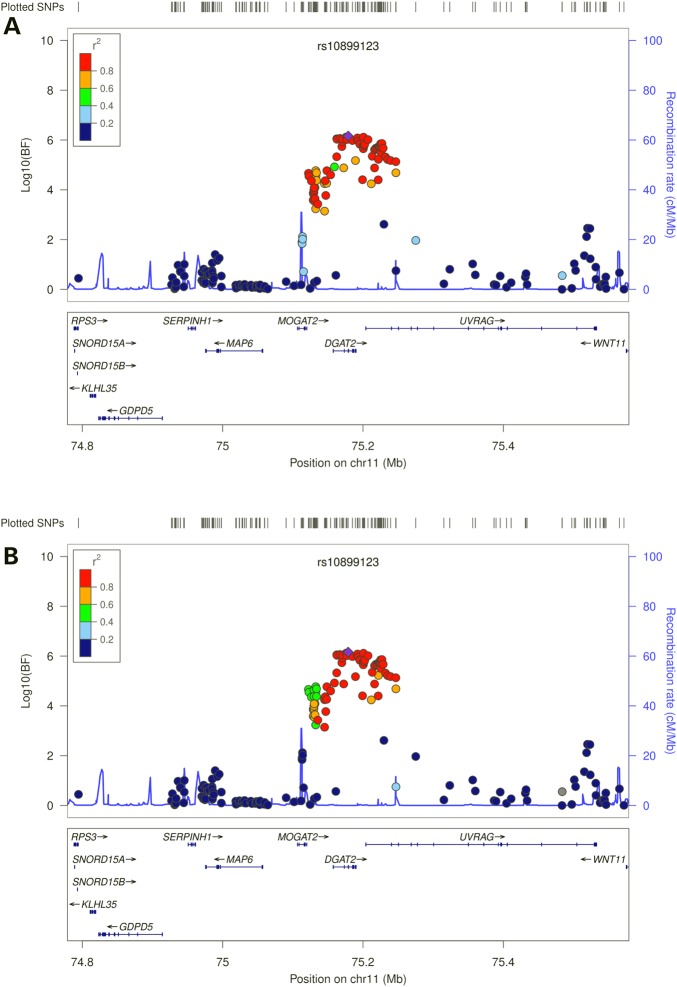
In the trans-ethnic meta-analysis that combined the GWAS data from Chinese and European-ancestry populations, we not only confirmed the two novel loci (MYB and AGPAT4) identified in the Chinese-specific GWAS meta-analyses (log10(Bayes Factor) (BF) ≥ 8.69), but also identified two additional novel loci (DGAT2 and PPT2) at genome-wide significance (log10(BF) ≥ 6.16) (Table 2 and Fig. 2). DGAT2-rs10899123 and PPT2-rs3134603 were also significantly associated with GLA and DPA, respectively, in the meta-analysis that combined results from the Chinese- and European-specific GWAS using METAL (P = 4.04 × 10−8 for DGAT2-rs10899123 and P = 3.28 × 10−8 for PPT2-rs3134603, respectively, data not shown), and nominally significant in either of the ethnic-specific GWAS meta-analyses, with consistent directions of associations and comparable effect sizes across the two ethnic groups (Table 2). However, MYB-rs9399137 and AGPAT4-rs729986 showed significant heterogeneity between Chinese and European-ancestry populations (Posterior probability of heterogeneity (Phet) = 1, Table 2).
Figure 2.
Continued.
Figure 2.
Regional plots of novel loci identified in the trans-ethnic MANTRA analysis. Genetic coordinates are displayed along the x-axis (NCBI Build 36) and genome-wide association significance level is plotted against the y-axis as log10 (BF value). LD is indicated by color scale in relationship to the most significant SNP in each association, with red for strong LD (r2 ≥ 0.8) and fading color for lower LD. (A) GLA, DGAT2-rs10899123, based on HapMap Phase II JPT+CHB data; (B) GLA, DGAT2-rs10899123, based on HapMap Phase II CEU data; (C) DPA, PPT2-rs3134603, based on HapMap Phase II JPT+CHB data; (D) DPA, PPT2-rs3134603, based on HapMap Phase II CEU data.
We also confirmed the previously reported associations of FADS1 with LA (log10(BF) = 318), GLA (log10(BF) = 147), DGLA (log10(BF) = 289), AA (log10(BF) = 1141), DTA (log10(BF) = 175), ALA (log10(BF) = 82.7), EPA (log10(BF) = 76.6) and DPA (log10(BF) = 180), of NTAN1 with LA (log10(BF) = 15.8), GLA (log10(BF) = 8.36), DGLA (log10(BF) = 60.8) and AA (log10(BF) = 9.23), of NRBF2 with LA (log10(BF) = 6.80), of ELOVL2 with EPA (log10(BF) = 6.74) and DHA (log10(BF) = 14.1), and of GCKR with DPA (log10(BF) = 8.43) (Table 2), but failed to replicate previously reported associations of LPCAT3-rs2110073 with LA, PCOLCE2-rs6778966 with AA (24), and SLC26A10-rs2277324 with DHA (21) (log10(BF) ≤ 0.881, data not shown). Notably, the effect sizes of rs174547 on PUFA levels exhibited significant difference between Chinese and European-ancestry populations (Phet = 1), except for ALA and DPA (Phet ≤ 0.223), and ELOVL2-rs3734398 associated with EPA and DHA exhibited different allele frequencies between Chinese and European-ancestry populations (Table 2). The newly identified independent signal ELOVL2-rs2281591 was significantly associated with DPA level (log10(BF) = 42.6), with consistent directions and comparable effect sizes between the two ethnic groups (Table 2).
The most significant SNP at each locus is listed in Supplementary Material, Table S3. Comparing the most associated SNPs identified in the Chinese-specific GWAS, European-specific GWAS and the trans-ethnic meta-analysis, SNPs at FADS1 and DGAT2 are correlated, while SNPs at ELOVL2, MYB, AGPAT4 and PPT2 are population-specific (Supplementary Material, Table S4). Some variants in or near MYB and AGPAT4 showed nominal evidence of association with DTA in European populations (P < 0.05). However, these SNPs are not in linkage disequilibrium (LD) with the leading signals (MYB-rs9399137 and AGPAT4-rs729986) identified in the Chinese populations (r2 ≤ 0.05).
To narrow association signals to potentially causative variants, we calculated 95 and 99% credible regions surrounding the top hits at the MANTRA loci (Supplementary Material, Table S5). We observed decreased numbers of SNPs contained within the 99% credible sets at loci including FADS1, NTAN1, NRBF2 and ELOVL2. We then examined variants in the 99% credible sets at each locus for sequence overlap with potential regulatory sites by search them in the HaploReg V3 database (27). The results showed that most of the variants in the 99% credible sets or their proxies (r2 > 0.8 in 1000 Genomes Pilot I) lie at strong enhancers or alter protein binding sites in multiple cell lines (Supplementary Material, Tables S6 and S7).
Cis-eQTL analysis and metabolic traits associated with the novel loci
To evaluate whether the significant variants at each novel loci affect gene transcription levels as cis-effect expression quantitative traits (cis-eQTLs), we performed cis-eQTL analysis by searching the publicly available Genotype-Tissue Expression (GTEx, http://gtexportal.org/home/) and Blood eQTL (http://genenetwork.nl/bloodeqtlbrowser/) in adipose tissue and blood. MYB-rs9399137 was significantly associated with ALDH8A1 transcript level in subcutaneous adipose tissue (P = 3.54 × 10−8, Supplementary Material, Table S8). DGAT2-rs10899123 and PPT2-rs3134603 were significantly associated with transcript levels of DGAT2 and SKIV2L, respectively, in blood (P = 1.36 × 10−13 and 1.09 × 10−6, respectively, Supplementary Material, Table S8). Whether there are relationships between gene transcription levels and fatty acid metabolism remained to be known.
We explored associations of these newly identified loci in our study with metabolic traits and outcomes in the public GWAS databases of GIANT, DIAGRAM, CARDIoGRAMplusC4D Consortiums and Metabolomics GWAS Server (http://mips.helmholtz-muenchen.de/proj/GWAS/gwas/). Interestingly, genetic variants in PPT2 were suggestively associated with type 2 diabetes and body mass index in the public databases of the GIANT and DIAGRAM Consortiums (Supplementary Material, Table S9).
Discussion
In this first trans-ethnic meta-analysis of PUFA levels in Chinese and European-ancestry populations consisting of more than 12 000 participants, we identified four novel loci (MYB, AGPAT4, DGAT2 and PPT2) at genome-wide significance. We also confirmed five previously reported loci (FADS1, NTAN1, NRBF2, ELOVL2 and GCKR) and observed an independent signal in ELOVL2-rs2281591.
MYB-rs9399137 is located in the upstream of MYB gene and AGPAT4-rs729986, DGAT2-rs10899123 and PPT2-rs3134603 are all located in the intronic region of these genes. MYB (v-myb myeloblastosis viral oncogene homolog) encodes a transcription factor and influences the regulation of hematopoiesis and may also play a role in tumorigenesis. A previous study using gene-based approach demonstrated suggestive association of MYB with body mass index in children (28). More recently, GWAS of plasma triglycerides after an n-3 PUFA supplementation also observed different allele frequencies of MYB-rs6920829 between the responders and the non-responders (29). AGPAT4 (1-acylglycerol-3-phosphate O-acyltransferase 4) encodes 1-acylglycerol-3-phosphate O-acyltransferase 4, a member of the AGPAT family. This membrane protein converts lysophosphatidic acid to phosphatidic acid in the second step of the de novo phospholipid biosynthesis pathway. Interestingly, the previous meta-analyses of GWASs in European-ancestry populations also observed suggestive evidence for the association of AGPAT3-rs7435 with DPA and significant association of AGPAT1-rs3134950 with DTA after adjustment for AA (22,23). Altogether, these finding suggest an involvement of the AGPAT family enzymes in PUFA level regulation.
Consistent with our results, previous studies in Europeans also observed suggestive associations of DGAT2 and PPT2 with GLA and DPA, respectively (22,23). DGAT2 (diacylglycerol O-acyltransferase 2) encodes an enzyme that catalyzes the transfer of long chain fatty acyl-CoAs to diacylglycerol in the synthesis of triglycerides. DGAT2-rs499974 was previously reported to be associated with lower HDL level in European populations (30). PPT2 (palmitoyl-protein thioesterase 2) encodes palmitoyl-protein thioesterase 2, a lysosomal hydrolase that catalyzes the hydrolysis of long chain fatty acyl CoAs (31). Although PPT2 is located near AGPAT1, a locus for phospholipid levels (32), PPT2-rs3134603 is in weak LD with AGPAT1-rs1061808 (the top hit for phospholipid levels, r2 = 0.279). Moreover, reciprocal conditional analyses by adding PPT2-rs3134603 or AGPAT1-rs1061808 in the main model suggested that they were largely independent of each other (Padj = 0.010 and 0.643, P = 0.003 and 0.138, respectively, data not shown). This evidence suggests for the potential roles of these four novel loci in fatty acid metabolism, but the exact mechanism underlying these associations remains to be investigated.
Genetic variants in the elongase gene ELOVL2 were associated with higher levels of DPA in the previous GWAS meta-analyses in European-ancestry populations (22,24). However, the top hit rs2281591 observed in the Chinese-specific GWAS meta-analysis was not in LD with the reported rs3734398 in European-ancestry populations (r2 = 0.02 in CHB and r2 = 0.36 in CEU). Reciprocal conditional analyses in Chinese by adding rs2281591 or rs3734398 in the main model did not materially changed the P-values of association with DPA (Padj = 8.29 × 10−8 and 0.183, P = 9.19 × 10−9 and 0.035, respectively), further indicating that they are largely independent of each other. SNP rs2281591 is in the intronic region of ELOVL2 while rs3734398 is located in the 3′-UTR of this gene. The minor allele frequency (MAF) of rs3734398 was relatively lower in CHB (MAF = 10%) than in CEU (MAF = 45%) according to HapMap data. The previously published GWAS for PUFA levels within CHARGE Consortium also observed distinct allele frequencies of ELOVL2-rs3734398 between Chinese in MESA cohort and European participants (22), which was consistent with the current findings.
In addition to observing novel genetic variants, we confirmed previously reported loci at FADS1, NTAN1, NRBF2, ELOVL2 and GCKR. The strongest associations were observed on chromosome 11 in the region which contained FADS1/2, FEN1, C11orf9 and C11orf10, as previously reported in populations of European ancestry (21–24). The previously reported association signal rs174547 at this locus showed consistent directions of associations but significantly different effect sizes on PUFA levels, except for ALA and DPA, between Chinese and European-ancestry populations (Table 2). Different PUFA measurement compartments across studies (plasma phospholipid versus erythrocyte membrane fatty acids) may be one source of heterogeneity. Significant heterogeneity of FADS1-rs174547 associated with LA, DGLA and AA between the NHAPC (erythrocyte membrane fatty acids) and the MESA (plasma phospholipid fatty acids) cohorts was also observed (Supplementary Material, Table S1). GWAS using normalized data did not eliminate evidence of heterogeneity between the NHAPC and the MESA cohorts, indicating differences in PUFA levels may not be the main source of the observed heterogeneity. Dietary patterns which influences fatty acid intake may also contribute to the discrepancy. A recent study in European populations observed interaction effects between dietary n-3 PUFA intake and genetic variants in FADS1/2 on plasma PUFA levels, but not on erythrocyte membrane fatty acid levels (33). Further study is required to determine the mechanism underlying heterogeneity of effect sizes across populations.
Calculation of 99% credible sets across the association signals at each identified locus suggested improvement in fine mapping resolution at loci FADS1, NTAN1, NRBF2 and ELOVL2. These findings may help guide decision for the future targeted sequencing studies to pinpoint the true causal variant.
In summary, GWAS of PUFA levels in Chinese and trans-ethnic meta-analyses in Chinese and European-ancestry populations revealed four novel loci (MYB, AGPAT4, DGAT2 and PPT2) and confirmed five previously reported genetic variants (FADS1/2, NTAN1, NRBF2, GCKR, ELOVL2). These results should stimulate future research of how these genetic loci and related pathways affect PUFA metabolism and establish the foundation for further genetic and functional research.
Materials and Methods
Study cohorts
The study sample consisted of 3521 unrelated Chinese participants from NHAPC and MESA. The NHAPC study is a population-based cohort study and 2865 participants with PUFA data were included in the current analysis. These participants were recruited from Beijing and Shanghai, 50–70 years old. The study design, methods and measurements of this cohort study have been described in detail elsewhere (34). The MESA study is a population-based study of 6814 men and women of diverse ethnic groups. Participants aged from 45 to 84 years old were recruited from six field centers in the United States. Of them, 656 participants of Chinese descent were included in the current analysis. The study design and data collection have been described in detail previously (35). The two cohort studies were approved by local committees and informed consents were obtained from all participants.
Meta-analyses of GWAS of PUFA levels in participants of European ancestry from the Atherosclerosis Risk in Communities Study (ARIC), the Coronary Artery Risk Development in Young Adults Study (CARDIA), the Cardiovascular Health Study (CHS), the Invecchiare in Chianti Study (InCHIANTI) and MESA were previously published (22,23). We used the publically available meta-analysis results (http://www.chargeconsortium.com/main/results), and included a total of 8866 and 8962 European-ancestry participants in the n-3 and n-6 trans-ethnic meta-analyses, respectively.
Fatty acids measurements
For the NHAPC samples, erythrocyte membrane fatty acids including LA, GLA, DGLA, AA, DTA, ALA, EPA, DPA and DHA were measured as previously described (36). After being extracted by hexane and isopropanol, erythrocyte membrane fatty acids were incubated with mixture of methanol and sulfuric acid for fatty acid methyl esters (FAME). FAMEs were then separated by gas chromatography (Agilent 6890 GC). Individual FAME was identified by positive chemical ionization using methane as reagent gas (Agilent 5975B). In MESA, the method for plasma PUFA measurement has been described in detail elsewhere (37). Briefly, lipids were extracted from fasting EDTA plasma using a chloroform/methanol extraction method and the cholesterol esters, triglyceride, phospholipids and free fatty acids were separated by thin layer chromatography. FAMEs were obtained from the phospholipids, separated using gas chromatography, and detected by gas chromatography flame ionization. DTA was not measured in MESA cohort. Individual fatty acid of both cohorts was expressed as the percentage of total fatty acids.
Genotyping, imputation and quality control
For the NHAPC and the MESA studies, genotyping was performed separately using Illumina Human660W and Affymetrix 6.0 genotyping arrays, respectively. Quality control filters were applied at both individual and SNP levels. At the individual level, samples with call rates <97% in NHAPC and call rate <95% in MESA were excluded. At the SNP level, SNPs with call rates <95%, monomorphic SNPs in MESA, or MAF <1% in NHAPC were excluded. Imputation was then done by IMPUTE (http://mathgen.stats.ox.ac.uk/impute/impute.html) (38) in both studies using HapMap Phase II data. For the significantly associated loci in chromosome 6 and chromosome 11, imputation using 1000 Genomes Phase I data was also conducted. Imputed SNPs with an estimated call rate ≤95%, or poor imputation quality, defined as the info measure ≤0.5 (NHAPC) or an observed variance divided by expected variance ≤0.8 (MESA) were excluded.
GWAS meta-analysis in Chinese populations
Associations between genotypes and PUFA levels were determined using linear regression analysis under an additive genetic model, adjusted for age, sex, region/study site and principal components (PCs) of ancestry (1 PC in MESA and 2 PCs in NHAPC). Robust variance estimators were applied to derive standard errors and to calculate P-values using ProbABEL v0.1-9c in MESA and v0.4.3 in NHAPC from the GenABEL suite of programs (http://www.genabel.org/packages/ ProABEL) (39). Association analyses were performed using log-transformed and z-score normalized fatty acids data to test whether differences in PUFA levels could introduce heterogeneity between Chinese populations from the NHAPC and the MESA cohorts. To test whether there are better candidate variants at the associated loci, 1000 Genomes Phase I imputed data were used for association analysis for the significant loci in chromosome 6 and chromosome 11 in Chinese populations.
Genome-wide association of each SNP with PUFAs from the NHAPC and the MESA cohorts were combined using inverse-variance based meta-analysis in METAL (http://www.sph.umich.edu/csg/abecasis/metal). Genomic control was conducted before meta-analysis to minimize potential confounding resulted from population stratification. Inflation factors (λ) were 1.087 (LA), 1.012 (GLA), 1.147 (ALA), 1.002 (DGLA), 1.014 (AA), 1.006 (EPA), 1.025 (DPA) and 1.014 (DHA) in NHAPC and 1.023 (LA), 1.034 (GLA), 1.025 (ALA), 1.030 (DGLA), 1.034 (AA), 1.042 (EPA), 1.018 (DPA) and 1.034 (DHA) in MESA, suggesting minimal genome-wide inflation due to population stratification. Since DTA was not measured in MESA, the results in the NHAPC study were reported directly without meta-analysis. SNPs with P ≤ 5 × 10−8 were considered genome-wide significant. The proportion of PUFA variance explained by a specific allele was calculated based on the following formula: β2 × 2 × MAF(1-MAF)/Var(Y), where β is the coefficient for one copy of the allele, MAF is the minor allele frequency and Var(Y) is the variance of the PUFA.
Trans-ethnic meta-analysis in Chinese and European-ancestry populations
Trans-ethnic meta-analysis was performed to identify additional novel loci for PUFA levels, to narrow the genomic functional regions represented by the identified association signals. Association statistics of Chinese and European-ancestry populations were combined by using the trans-ethnic meta-analysis approach implemented in MANTRA, which was specifically designed to perform trans-ethnic meta-analysis and adopted a Bayesian framework (40). Log10(BF) ≥ 6 was considered genome-wide significant and SNPs with Phet ≥ 0.5 were considered showing significant heterogeneity between Chinese and European-ancestry populations. 95 and 99% credible regions surrounding the top hits from the trans-ethnic MANTRA analysis were calculated. We estimated credible sets of SNPs by first defining a ±500 kb region surrounding the lead SNPs and ranking the SNPs within this region according to their BF values. The cumulative posterior probabilities of these ranked SNPs were then combined until 95 and 99% confidence was reached.
Cis-eQTL analysis and metabolic traits associated with the novel loci
We searched the publicly available eQTL databases, GTEx (http://gtexportal.org/home/) and Blood eQTL (http://genenetwork.nl/bloodeqtlbrowser/) for cis-eQTLs to explore the functional mechanism underlying the observed associations. Subcutaneous and visceral adipose tissue samples were selected in the GTEx browser.
We used the public GWAS databases of GIANT, DIAGRAM, CARDIoGRAMplusC4D Consortiums and Metabolomics GWAS Server (http://mips.helmholtz-muenchen.de/proj/GWAS/gwas/) to explore associations of these newly identified loci in our study with metabolic traits and outcomes.
Supplementary Material
Funding
The NHPAC study is supported by the National Basic Research Program of China (973 Program 2012CB524900), and the National Natural Science Foundation of China (30930081, 81170734, 81321062, and 81471013). We are grateful to all participants of the NHAPC, and also thank our colleagues at the laboratory and local CDC staffs of Beijing and Shanghai for their assistance with data collection. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-64278. MESA SHARe genotyping was performed as Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant HL105756. The authors thank all investigators, staff and participants from the studies of NHAPC and MESA cohorts for their contributions to this work. We appreciate the European PUFA GWAS data sharing by the CHARGE Consortium.
Supplementary Material
Acknowledgments
Conflicts of Interest statement. None declared.
References
- 1.Mozaffarian D., Wu J.H. (2011) Omega-3 fatty acids and cardiovascular disease: effects on risk factors, molecular pathways, and clinical events. J. Am. Coll. Cardiol., 58, 2047–2067. [DOI] [PubMed] [Google Scholar]
- 2.Schwab U., Lauritzen L., Tholstrup T., Haldorssoni T., Riserus U., Uusitupa M., Becker W. (2014) Effect of the amount and type of dietary fat on cardiometabolic risk factors and risk of developing type 2 diabetes, cardiovascular diseases, and cancer: a systematic review. Food. Nutr. Res., 58, doi:10.3402/fnr.v58.25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang T., Sun J., Chen Y., Xie H., Xu D., Huang J., Li D. (2014) Genetic variants in desaturase gene, erythrocyte fatty acids, and risk for type 2 diabetes in Chinese Hans. Nutrition, 30, 897–902. [DOI] [PubMed] [Google Scholar]
- 4.Brostow D.P., Odegaard A.O., Koh W.P., Duval S., Gross M.D., Yuan J.M., Pereira M.A. (2011) Omega-3 fatty acids and incident type 2 diabetes: the Singapore Chinese Health Study. Am. J. Clin. Nutr., 94, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rylander C., Sandanger T.M., Engeset D., Lund E. (2014) Consumption of lean fish reduces the risk of type 2 diabetes mellitus: a prospective population based cohort study of Norwegian women. PLoS One, 9, e89845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G., Sun Q., Hu F.B., Ye X., Yu Z., Zong G., Li H., Zhou Y., Lin X. (2012) Erythrocyte n-3 fatty acids and metabolic syndrome in middle-aged and older Chinese. J. Clin. Endocrinol. Metab., 97, E973–E977. [DOI] [PubMed] [Google Scholar]
- 7.Carpentier Y.A., Portois L., Malaisse W.J. (2006) n-3 fatty acids and the metabolic syndrome. Am. J. Clin. Nutr., 83, 1499S–1504S. [DOI] [PubMed] [Google Scholar]
- 8.Zheng J.S., Hu X.J., Zhao Y.M., Yang J., Li D. (2013) Intake of fish and marine n-3 polyunsaturated fatty acids and risk of breast cancer: meta-analysis of data from 21 independent prospective cohort studies. BMJ, 346, f3706. [DOI] [PubMed] [Google Scholar]
- 9.Shannon J., King I.B., Moshofsky R., Lampe J.W., Gao D.L., Ray R.M., Thomas D.B. (2007) Erythrocyte fatty acids and breast cancer risk: a case-control study in Shanghai, China. Am. J. Clin. Nutr., 85, 1090–1097. [DOI] [PubMed] [Google Scholar]
- 10.Assies J., Pouwer F., Lok A., Mocking R.J., Bockting C.L., Visser I., Abeling N.G., Duran M., Schene A.H. (2010) Plasma and erythrocyte fatty acid patterns in patients with recurrent depression: a matched case-control study. PLoS One, 5, e10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiemeier H., van Tuijl H.R., Hofman A., Kiliaan A.J., Breteler M.M. (2003) Plasma fatty acid composition and depression are associated in the elderly: the Rotterdam Study. Am. J. Clin. Nutr., 78, 40–46. [DOI] [PubMed] [Google Scholar]
- 12.Wu J.H., Lemaitre R.N., King I.B., Song X., Psaty B.M., Siscovick D.S., Mozaffarian D. (2014) Circulating omega-6 polyunsaturated fatty acids and total and cause-specific mortality: the Cardiovascular Health Study. Circulation, 130, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farvid M.S., Ding M., Pan A., Sun Q., Chiuve S.E., Steffen L.M., Willett W.C., Hu F.B. (2014) Dietary linoleic acid and risk of coronary heart disease: a systematic review and meta-analysis of prospective cohort studies. Circulation, 130, 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh K., Hu F.B., Manson J.E., Stampfer M.J., Willett W.C. (2005) Dietary fat intake and risk of coronary heart disease in women: 20 years of follow-up of the nurses’ health study. Am. J. Epidemiol., 161, 672–679. [DOI] [PubMed] [Google Scholar]
- 15.Mahendran Y., Agren J., Uusitupa M., Cederberg H., Vangipurapu J., Stancakova A., Schwab U., Kuusisto J., Laakso M. (2014) Association of erythrocyte membrane fatty acids with changes in glycemia and risk of type 2 diabetes. Am. J. Clin. Nutr., 99, 79–85. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury R., Warnakula S., Kunutsor S., Crowe F., Ward H.A., Johnson L., Franco O.H., Butterworth A.S., Forouhi N.G., Thompson S.G. et al. (2014) Association of dietary, circulating, and supplement fatty acids with coronary risk: a systematic review and meta-analysis. Ann. Intern. Med., 160, 398–406. [DOI] [PubMed] [Google Scholar]
- 17.McGee D.L., Reed D.M., Yano K., Kagan A., Tillotson J. (1984) Ten-year incidence of coronary heart disease in the Honolulu Heart Program. Relationship to nutrient intake. Am. J. Epidemiol., 119, 667–676. [DOI] [PubMed] [Google Scholar]
- 18.Sioen I., Vyncke K., De Maeyer M., Gerichhausen M., De Henauw S. (2013) Dietary intake and food sources of total and individual polyunsaturated fatty acids in the Belgian population over 15 years old. Lipids, 48, 729–738. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q., Ma J., Campos H., Hankinson S.E., Hu F.B. (2007) Comparison between plasma and erythrocyte fatty acid content as biomarkers of fatty acid intake in US women. Am. J. Clin. Nutr., 86, 74–81. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto K., Yoshizawa A.C., Okuda S., Kuma K., Goto S., Kanehisa M. (2008) The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid. Res., 49, 183–191. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T., Shen J., Abecasis G.R., Kisialiou A., Ordovas J.M., Guralnik J.M., Singleton A., Bandinelli S., Cherubini A., Arnett D. et al. (2009) Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet., 5, e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaitre R.N., Tanaka T., Tang W., Manichaikul A., Foy M., Kabagambe E.K., Nettleton J.A., King I.B., Weng L.C., Bhattacharya S. et al. (2011) Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet., 7, e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan W., Steffen B.T., Lemaitre R.N., Wu J.H., Tanaka T., Manichaikul A., Foy M., Rich S.S., Wang L., Nettleton J.A. et al. (2014) Genome-wide association study of plasma n6 polyunsaturated Fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ. Cardiovasc. Genet., 7, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tintle N.L., Pottala J.V., Lacey S., Ramachandran V., Westra J., Rogers A., Clark J., Olthoff B., Larson M., Harris W. et al. (2015) A genome-wide association study of saturated, mono- and polyunsaturated red blood cell fatty acids in the Framingham Heart Offspring Study. Prostaglandins Leukot. Essent. Fatty Acids, 94, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffen B.T., Steffen L.M., Tracy R., Siscovick D., Jacobs D., Liu K., He K., Hanson N.Q., Nettleton J.A., Tsai M.Y. (2012) Ethnicity, plasma phospholipid fatty acid composition and inflammatory/endothelial activation biomarkers in the Multi-Ethnic Study of Atherosclerosis (MESA). Eur. J. Clin. Nutr., 66, 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang M., Rahman M.A., Ai H., Li X., Harbige L.S. (2006) Diet and gene expression: delta-5 and delta-6 desaturases in healthy Chinese and European subjects. Ann. Nutr. Metab., 50, 492–498. [DOI] [PubMed] [Google Scholar]
- 27.Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melen E., Himes B.E., Brehm J.M., Boutaoui N., Klanderman B.J., Sylvia J.S., Lasky-Su J. (2010) Analyses of shared genetic factors between asthma and obesity in children. J. Allergy Clin. Immunol., 126, 631–637 e631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudkowska I., Guenard F., Julien P., Couture P., Lemieux S., Barbier O., Calder P.C., Minihane A.M., Vohl M.C. (2014) Genome-wide association study of the plasma triglyceride response to an n-3 polyunsaturated fatty acid supplementation. J. Lipid. Res., 55, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Global Lipids Genetics Consortium Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L. et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta P., Soyombo A.A., Atashband A., Wisniewski K.E., Shelton J.M., Richardson J.A., Hammer R.E., Hofmann S.L. (2001) Disruption of PPT1 or PPT2 causes neuronal ceroid lipofuscinosis in knockout mice. Proc. Natl Acad. Sci. USA, 98, 13566–13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demirkan A., van Duijn C.M., Ugocsai P., Isaacs A., Pramstaller P.P., Liebisch G., Wilson J.F., Johansson A., Rudan I., Aulchenko Y.S. et al. (2012) Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet., 8, e1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith C.E., Follis J.L., Nettleton J.A., Foy M., Wu J.H., Ma Y., Tanaka T., Manichakul A.W., Wu H., Chu A.Y. et al. (2015) Dietary fatty acids modulate associations between genetic variants and circulating fatty acids in plasma and erythrocyte membranes: meta-analysis of nine studies in the CHARGE consortium. Mol. Nutr. Food Res., 59, 1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y., Li H., Loos R.J., Yu Z., Ye X., Chen L., Pan A., Hu F.B., Lin X. (2008) Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes, 57, 2834–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bild D.E., Bluemke D.A., Burke G.L., Detrano R., Diez Roux A.V., Folsom A.R., Greenland P., Jacob D.R. Jr., Kronmal R., Liu K. et al. (2002) Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol., 156, 871–881. [DOI] [PubMed] [Google Scholar]
- 36.Zong G., Ye X., Sun L., Li H., Yu Z., Hu F.B., Sun Q., Lin X. (2012) Associations of erythrocyte palmitoleic acid with adipokines, inflammatory markers, and the metabolic syndrome in middle-aged and older Chinese. Am. J. Clin. Nutr., 96, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao J., Schwichtenberg K.A., Hanson N.Q., Tsai M.Y. (2006) Incorporation and clearance of omega-3 fatty acids in erythrocyte membranes and plasma phospholipids. Clin. Chem., 52, 2265–2272. [DOI] [PubMed] [Google Scholar]
- 38.Marchini J., Howie B., Myers S., McVean G., Donnelly P. (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913. [DOI] [PubMed] [Google Scholar]
- 39.Aulchenko Y.S., Struchalin M.V., van Duijn C.M. (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics, 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris A.P. (2011) Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol., 35, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.