Abstract
Over 100 associated genetic loci have been robustly associated with schizophrenia. Gene prioritization and pathway analysis have focused on a priori hypotheses and thus may have been unduly influenced by prior assumptions and missed important causal genes and pathways. Using a data-driven approach, we show that genes in associated loci: (1) are highly expressed in cortical brain areas; (2) are enriched for ion channel pathways (false discovery rates <0.05); and (3) contain 62 genes that are functionally related to each other and hence represent promising candidates for experimental follow up. We validate the relevance of the prioritized genes by showing that they are enriched for rare disruptive variants and de novo variants from schizophrenia sequencing studies (odds ratio 1.67, P = 0.039), and are enriched for genes encoding members of mouse and human postsynaptic density proteomes (odds ratio 4.56, P = 5.00 × 10−4; odds ratio 2.60, P = 0.049).The authors wish it to be known that, in their opinion, the first 2 authors should be regarded as joint First Author.
Introduction
Despite recent successes in identifying rare and common genetic variants that associate with schizophrenia, the etiology of the disorder still remains unclear (1). Large-scale studies of de novo variants, rare variants and common variants have implicated the postsynaptic density (PSD) (2–4), calcium channels (4–6), targets of the fragile X mental retardation protein (FMRP, product of FMR1) (3,4,7), targets of micro-RNA mir-137 (8), glutamate pathways (9), and processes related to neurogenesis and synaptic integrity (7,9–11). However, our knowledge of the genes involved in the etiology of schizophrenia is far from complete, and in many instances, the evidence implicating putative pathogenic pathways is not decisive.
We and others have recently shown that well-powered genome-wide association studies (GWAS) of complex traits, combined with data-driven, mechanism-agnostic pathway analyses, enable identification of likely causal genes and pathways and thus provide a solid foundation for understanding causal biology (12–16). In schizophrenia, common variants are estimated to account for at least a third of genetic risk (5,17) and recent GWAS has identified more than 100 robustly associated loci (6).
Compared to GWAS-based pathway analysis of anthropometric traits, efforts to prioritize likely causal genes and enriched pathways for schizophrenia have until recently been limited by the lack of genome-wide significant associations and incomplete defined pathways (18). Moreover, except for a few studies (19–23), these analyses have been limited to either preselected pathways with a priori evidence for schizophrenia or have only identified pathways that are shared across psychiatric disorders (Supplementary Material, see (18) for a review). Furthermore, few of these analyses are followed up with independent validation. Consequently, there is a need for a comprehensive gene prioritization and pathway analysis approach for schizophrenia that includes unbiased validation and does not rely on pre-specified hypotheses.
Spatial and temporal expression patterns of implicated genes are also important to identifying promising drug targets and understanding etiology. Results from previous exome sequencing studies of de novo mutations in schizophrenia have suggested that likely causal schizophrenia genes are highly expressed during fetal development (10,11,24), but failed to replicate in the to-date largest schizophrenia exome sequencing study (3). Consequently further work is needed to investigate whether likely causal schizophrenia genes have a prenatal or postnatal expression bias.
To obtain a more comprehensive portrait of likely causal genes and biological pathways underlying schizophrenia, we applied our framework (25) for interpretation of genetic association studies (called DEPICT) to 128 genome-wide significant associations with schizophrenia recently reported by the Psychiatric Genomics Consortium (PGC) (6). DEPICT uses improved biological pathway and gene set definitions (henceforth ‘reconstituted gene sets’) to systematically prioritize the most likely causal gene(s) at associated GWAS loci and to assess whether genes in associated GWAS loci enrich for reconstituted gene sets and/or are highly expressed in specific tissues, brain areas or cell types (25). The use of reconstituted gene sets to prioritize genes and pathways is a key difference between DEPICT and other pathway methods (25) and DEPICT has been successfully applied to a wide range of polygenic traits (13,14,16,26,27). We validated the prioritized schizophrenia genes using previously published exome sequencing data (3,4) and postsynaptic density proteomics data (2). Finally, we show that prioritized genes tend to be higher expressed postnatally than prenatally.
Results
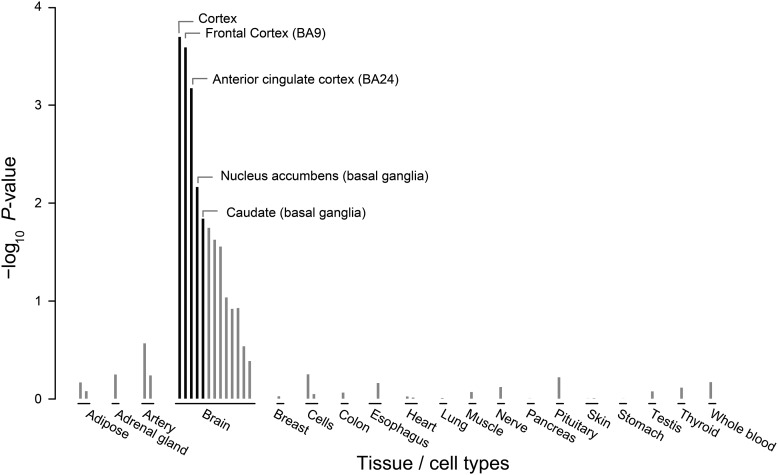
To decipher the biology implicated by GWAS loci associated with schizophrenia, we applied DEPICT to genome-wide significant associations (P < 5 × 10−8) recently identified by the PGC (6). Using 123 autosomal, non-human leukocyte antigen associated SNPs as the input to DEPICT, we first generated 109 independent loci (this differs slightly from the PGC which used a different procedure to define associated loci). We first tested whether genes in these associated loci displayed higher expression levels in particular tissues and brain areas than genes outside associated loci using RNA sequencing (RNA-Seq) data from 37 tissues evaluated in the Genotype-Tissue Expression Project (GTEx) (28). We identified five significant tissues (false discovery rates, FDR < 5%), all representing areas in the brain: cortex, frontal cortex (BA9), anterior cingulate cortex (BA24), nucleus accumbens, caudate (Fig. 1; Supplementary Material, Table S2). These findings are consistent with previous data showing that schizophrenia-associated variants co-localize with genes highly expressed in the brain (6,10,11,24) and in particular structures in the cortex (11).
Figure 1.
Genes in genome-wide significant schizophrenia meta-analysis loci enrich for cortical structures of the brain. We used DEPICT and RNA-Seq data from the GTEx Project to assess whether genes in genome-wide significant schizophrenia loci were highly expressed in any of 37 tissue/brain area annotations. Genes within the associated loci were highly expressed in 5 brain areas, most predominantly in the frontal cortex. Enrichments are grouped according to tissue-type annotations and significance, and annotations colored in black exhibited false discovery rates below 5%.
Having confirmed that genes within genome-wide significant schizophrenia loci are highly expressed in the brain, we next used DEPICT to systematically assess for enrichment of reconstituted gene sets representing a wide range of biological pathways, molecular processes, cellular compartments/structures, protein complexes and mouse phenotypes. We identified 104 significantly enriched reconstituted gene sets (FDR < 0.05, Supplementary Material, Fig. S1 and Table S3), comprising 15 canonical pathways (29,30) including ion channels, glutamate, serotonin/dopamine and neuronal growth factor signaling (P = 8.08 × 10−5) and long term potentiation (P < 6.44 × 10−4; Table 1); and 89 additional gene sets representing molecular function and biological process Gene Ontology terms (31), morphological and physiological mouse phenotypes (32) and protein complexes (33).
Table 1.
Canonical pathway gene set enrichment analysis results
| Reconstituted gene set name | Enrichment P-value | False discovery rate | Implicated in generic brain loci analysis | Top 5 genes in reconstituted gene set overlapping with associated loci |
||||
|---|---|---|---|---|---|---|---|---|
| Platelet homeostasis | 3.48 × 10−5 | 0.007 | Yes | CACNB2 (6.53) | NRGN (6.46) | RIMS1 (6.23) | ESAM (5.16) | KCNB1 (4.95) |
| Neuronal system | 6.15 × 10−5 | 0.009 | Yes | KCNB1 (8.81) | GRIN2A (8.43) | HCN1 (7.71) | RIMS1 (7.35) | CACNB2 (7.08) |
| Signalling by NGF | 8.08 × 10−5 | 0.011 | Yes | MAPK3 (6.27) | NRGN (5.5) | NAB2 (5.29) | ARHGAP1 (4.88) | GATAD2A (4.51) |
| MAPK signaling pathway | 1.04 × 10−4 | 0.011 | Yes | NAB2 (4.74) | NRGN (4.49) | KLC1 (4.32) | ZSWIM6 (3.97) | DGKZ (3.36) |
| Voltage gated potassium channels | 1.08 × 10−4 | 0.011 | No | GRIN2A (8.39) | HCN1 (8.33) | KCNB1 (8.12) | KCNV1 (7.19) | CNTN4 (6.95) |
| Calcium signaling pathway | 2.48 × 10−4 | 0.022 | No | NRGN (7.35) | GRIN2A (7.25) | KCNB1 (5.81) | CACNB2 (5.55) | RIMS1 (5.33) |
| Potassium channels | 3.19 × 10−4 | 0.03 | No | KCNB1 (8.76) | CACNB2 (7.9) | GRIN2A (7.9) | HCN1 (7.14) | CACNA1C (6.64) |
| Phosphatidylinositol signaling system | 4.69 × 10−4 | 0.041 | No | DGKZ (6.24) | HCN1 (3.91) | RIMS1 (3.7) | KCNV1 (3.69) | SRPK2 (3.55) |
| Glutamate neurotransmitter release cycle | 5.12 × 10−4 | 0.043 | Yes | INA (7.25) | RIMS1 (7.14) | SNAP91 (6.79) | HCN1 (6.5) | SEPT3 (6.03) |
| Ion channel transport | 5.23 × 10−4 | 0.044 | No | RIMS1 (7.18) | FUT9 (5.08) | HCN1 (5.05) | KCNV1 (4.86) | C11orf87 (4.82) |
| Transmission across chemical synapses | 5.49 × 10−4 | 0.045 | Yes | RIMS1 (7.97) | GRIA1 (7.57) | NRGN (7.05) | KCNB1 (6.87) | GRIN2A (6.86) |
| Serotonin neurotransmitter release cycle | 5.90 × 10−4 | 0.045 | Yes | CHRNA3 (8.83) | INA (8.07) | SNAP91 (7.69) | SEPT3 (5.94) | DOC2A (5.59) |
| Dopamine neurotransmitter release cycle | 5.90 × 10−4 | 0.045 | Yes | CHRNA3 (8.83) | INA (8.07) | SNAP91 (7.69) | SEPT3 (5.94) | DOC2A (5.59) |
| Opioid signalling | 6.15 × 10−4 | 0.045 | Yes | DRD2 (8.17) | NRGN (7.7) | MAPK3 (4.94) | DGKI (4.23) | RIMS1 (4.12) |
| Long-term potentiation | 6.44 × 10−4 | 0.046 | Yes | KCNV1 (5.04) | DGKZ (4.91) | NRGN (4.65) | RIMS1 (4.57) | GRIN2A (4.23) |
Enrichment results for reconstituted gene sets representing canonical pathways from the REACTOME (30) and KEGG (29) databases. Note, that reconstituted gene sets differ from their pre-defined (original) counterparts. The first column lists the identifiers of the original gene set, which are either prefixed by REACTOME or KEGG depending on where the original gene set was downloaded from. The second and third columns list the DEPICT gene set enrichment P-values and FDRs. The fourth column indicates whether the reconstituted gene set was implicated in the generic brain loci analysis. The last five columns list the top five genes annotated to a given reconstituted gene sets and being within associated schizophrenia loci along with the genes’ strength of association (as Z score in brackets) with the given reconstituted gene set.
We considered that this analysis could simply highlight generic pathways enriched in genes expressed in the brain. To assess this possibility, we tested whether the gene sets highlighted by DEPICT from the schizophrenia-associated loci differed from gene sets enriched in analyses based on randomly selected ‘brain loci,’ each containing a gene highly expressed in the brain (see Methods). We ran DEPICT on 100 such control sets of randomly selected brain loci and found that 20 out of the 104 gene sets remained significantly enriched when compared with these 100 control analyses (empirical P < 0.05). These gene sets clustered in five groups related to ‘channel activity’, ‘central nervous system projection neuron axonogenesis’, the ‘HOMER1 protein complex’ (HOMER1 constitutes a major part of the postsynaptic density), ‘cell recognition’, and ‘phosphatidylinositol signaling system’ (all with P < 6.2 × 10−4; Supplementary Material, Table S4).
Finally, to directly test for enrichment of gene sets that were implicated in recent exome sequencing studies (3,4), namely targets of the FMRP (34), glutamatergic postsynaptic proteins comprising activity-regulated cytoskeleton-associated protein (ARC), and N-methyl-D-aspartate receptor (NMDAR) complexes, we reconstituted these gene sets and computed their enrichment (see Methods). The only enriched reconstituted gene set was targets of FMRP (P = 4.16 × 10−6). Furthermore, to assess whether processes active in the pre-synapse and synaptic vesicles also were enriched in the genome-wide significant loci, we reconstituted gene sets encoding proteins isolated in proteomics experiments of rodent pre-synaptic active zones (35,36), pre-synaptic docking complexes (37), and synaptic vesicles (38). However, none of these four gene sets were significantly enriched (Supplementary Material, Table S5). Jointly, these results implicate processes related to dendritic spines, synaptic plasticity, and cognition/abnormal behavior but do not support key roles of presynaptic processes, the immune system or histone-related processes.
To identify likely casual schizophrenia genes, we used DEPICT to prioritize genes within the genome-wide significant loci, the majority of which contained more than one gene. In DEPICT, a gene within an associated locus is prioritized if it exhibits higher than expected pairwise similarities to genes from other associated loci (across 14,461 functional predictions). DEPICT prioritized 62 genes at 51 genome-wide significant loci (Supplementary Material, Table S6), including genes that have not been specifically hypothesized for schizophrenia before (e.g. PITPNM2, prioritization P = 4.13 × 10−8; DGKI, prioritization P = 1.03 × 10−7 and DGKZ, prioritization P = 4.95 × 10−6). To validate the relevance of the prioritized genes, we used previously published exome sequencing and PSD proteomics data that were not part of the DEPICT framework to compute the following four benchmarks. We then tested whether genes prioritized by DEPICT were more likely to harbor rare (MAF < 0.1%) disruptive variants (nonsense, essential splice site and frameshift mutations) in exome sequences of schizophrenia cases compared to controls, and whether prioritized genes were more likely than non-prioritized genes to contain de novo mutations in schizophrenia probands (3). Our results suggest that the prioritized genes enrich for these two previously identified groups of likely schizophrenia genes (odds ratio, OR, 1.67; one-sided Cochran Mantel-Haenszel statistic test P = 0.039; See Methods and Supplementary Material, Table S7). Finally, we tested whether prioritized genes are enriched for genes encoding members of either human or mouse cortex PSD proteomes (39). Prioritized genes strongly enriched for gene products from the human PSD (OR 2.60, one-sided P = 0.049) and mouse PSD (OR 4.56, one-sided P = 5.00 × 10−4) when compared to non-prioritized genes at associated loci.
Among the prioritized genes were two genes, GRIN2A and RIMS1, that exhibited a surplus of disruptive variants in schizophrenic cases compared to controls (4), de novo variants (3), and encoded members of the PSD. However, DEPICT also prioritized 14 genes with a surplus of disruptive variants or a de novo variant that did not encode PSD members (see Supplementary Material, Table S8). Predicted functions of these genes were related to diacylglycerol kinase activity (PITPNM2), hormone levels (KIAA1324L, GRAMD1B), neurotransmitter signaling (HCN1, CSMD1, KCNV1, SLC45A1, PCDHAC2), hippocampal pyramidal cell morphology and GTPase binding/activation (AKT3, PPP1R13B), ion channels (CNTN4, MBD5) and the innate immune system (CALHM2, PLCB2). Finally, DEPICT-prioritized genes included DRD2, which encodes the main target of effective antipsychotic drugs (prioritization P = 0.002), although this gene was not present in any of the four benchmark lists. Thus, while the sequencing and proteomics data provide validation of the prioritization by DEPICT, they are also clearly incomplete and imperfect benchmarks.
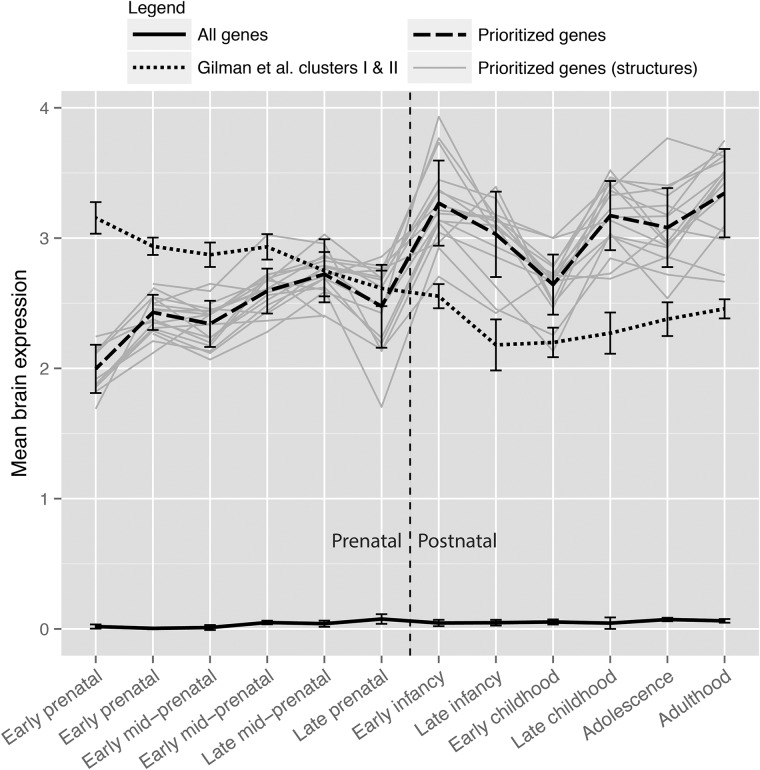
Having shown that prioritized genes enrich for rare variants in relevant exome sequencing studies and encode proteins that co-localize in the human and mouse PSDs, we investigated their temporal expression pattern using RNA-Seq-based postmortem brain expression data from the BrainSpan database (40), which covers stages from early prenatal developmental through infancy, adolescence and adulthood. In contrast to the previous studies reporting higher expression of likely causal schizophrenia genes during prenatal development (10,11,24), we observed higher expression levels in the postnatal period (fold-change 1.29; one-sided paired t-test P = 0.005; Fig. 2 and Supplementary Material, Fig. S2). We replicated these results using BrainSpan Developmental Transcriptome microarray-based gene expression data (fold-change 1.06, one-sided paired t-test P = 0.002; Supplementary Material, Fig. S3). We performed negative control analyses and found that methodological biases were unlikely to explain the tendency toward increased postnatal expression of prioritized genes (Supplementary Note). Together these results demonstrate that likely schizophrenia genes implicated by GWAS studies may act later in the life course.
Figure 2.
Genes prioritized for schizophrenia exhibited higher expression during postnatal development compared to prenatal development. The 62 genes prioritized in genome-wide significant schizophrenia loci (dashed line) exhibit higher expression during postnatal stages (early infancy through adulthood) compared to prenatal stages (early to late prenatal development; fold-change 1.29, one-sided paired t-test P = 0.005) across the different brain structures (gray lines). These findings contrast previous findings primarily based on exome sequencing data by Gilman et al. (24) showing higher expression of likely causal schizophrenia genes during prenatal stages (dotted line). These analyses were based on log2 transformed BrainSpan Developmental Transcriptome (40) RNA-Seq gene expression data. Error bars represent standard deviation across brain structures.
Discussion
Recent genome-scale studies have substantially advanced our understanding of schizophrenia (3,4,6,9–11). Here, we used previously published schizophrenia GWAS data in conjunction with a data-driven integrative approach to prioritize 62 genes and 104 reconstituted gene sets for schizophrenia. Our work differs from previous pathway analyses in an important respect: it is not based on a priori selection of candidate genes and gene sets but rather treats each gene set and the genes in associated loci equally. As a result, this work has the potential to validate the importance of previous hypotheses and also to prioritize as yet unidentified biological processes/structures, protein complexes, mouse phenotypes and genes. Importantly, we validate the relevance of the genes prioritized using our approach by replicating them using exome sequencing data and PSD proteomics data.
Our work has several main findings. Using strict significance criteria, we implicate numerous gene sets and genes; in particular, we show that genes in associated schizophrenia loci enrich for reconstituted gene sets related to biological processes and mouse phenotypes including dendritic spine development, calcium, glutamate and neuronal growth factor signaling, and synaptic transmission, but also less well-established processes such as diacylglycerol kinase activity (DGKI, DGKZ, and PITPNM2). Thus our work provides further evidence for the importance of the PSD and dendritic spines in schizophrenia (41). Four of the top five most enriched gene sets nominally significant in (22) are significant in our work (postsynaptic density, postsynaptic membrane, dendritic spine, axon part). However, we did not find any support for gene sets related to histone H3-K4 methylation, nor do our analyses, unlike previous epidemiological (42) and genetic (6,22) studies, support a link between the immune system and schizophrenia. We note that the HLA locus is strongly associated with schizophrenia (6). Many immune-related genes reside in the large regions of linkage disequilibrium within HLA, so strong genetic associations in this region often lead pathway-based methods to spuriously implicate immune-related pathways; for this reason, DEPICT omits the HLA locus from analysis.
Genes expressed during adolescence and adulthood, the ages at which schizophrenia is typically diagnosed, might represent more appealing drug targets than developmental genes that act prenatally. We show that likely causal genes in schizophrenia GWAS loci exhibit higher expression during postnatal stages compared to prenatal stages. Similarly, Purcell et al. (4) showed that schizophrenia genes selected based on a broad basis of prior evidence for schizophrenia, exhibit postnatal- rather than prenatal bias in their expression (in the hippocampus and dorsolateral prefrontal cortex) (4). Moreover, using BrainSpan microarray expression data, the PGC recently showed that expression of genes encoding members of immunological and neuronal pathways jointly associated with schizophrenia, major depression and bipolar disorder, increased during childhood and plateaued around adolescence (22). These and our findings contrast previous studies reporting genes with higher expression in prenatal developmental stages (10,11,24).
A benchmark is only as good as the data used to evaluate it. Schizophrenia sequencing studies are still underpowered to identify susceptibility genes and it is likely that additional etiologic biological processes and structures besides the PSD will emerge (43). DEPICT prioritized several genes outside the PSD (e.g. DGKI, NRGN, CACNA1I, and DRD2), which are not included in any of the benchmarks used in this work, but are nevertheless strong candidates for causal contributors to schizophrenia. Future genetic studies will provide more complete data sets with which to benchmark prioritization methods and to identify additional causal genes and biological mechanisms.
In summary, by taking a data-driven approach that is not biased toward preformed hypotheses about the etiology of schizophrenia, and applying this approach to large-scale schizophrenia GWAS data, we have prioritized genes and gene sets as potentially causal for schizophrenia. We show that prioritized genes are enriched for disruptive variants in schizophrenia cases, often encode proteins localized in the PSD and are more likely to be expressed postnatally. Our work provides an example of combining large-scale neurogenetics data with data-driven methodologies to understand biological processes underlying neuropsychiatric traits.
Materials and Methods
Genome-wide association study data for schizophrenia
Genome-wide significant association (P < 5 × 10−8) from the PGC schizophrenia GWAS (6) were used as input for DEPICT. We omitted three X chromosome SNPs, one human leukocyte antigen SNP (rs115329265), and variant chr10_104957618 (which could not be mapped, but was covered by rs12887734) and used the resulting 123 autosomal SNPs as input to the DEPICT analysis (Supplementary Material, Table S9). The 123 SNPs grouped into 109 independent loci (using linkage disequilibrium r2> 0.5 and a maximum physical distance between lead SNPs and locus boundaries of 1 Mb to define loci). All genomic coordinates were defined using genome build hg19.
DEPICT overview
The DEPICT tool (Data-driven Expression Prioritized Integration for Complex Traits) performs gene set enrichment and gene prioritization based on predicted gene functions (25). For 19 987 genes, we computed the likelihood of membership in each of 14 461 pre-defined gene sets (based on similarities across in large-scale gene expression data) resulting in 14 461 reconstituted gene sets. The pre-defined gene sets included manually curated pathways from the KEGG (29), REACTOME (30), and Gene Ontology databases (31); molecular pathways derived from protein-protein interaction screens from the InWeb database (33); and phenotypic gene sets derived from mouse gene knock-out studies from the Mouse Genetics Initiative database (44), which jointly represent a wide spectrum of biological annotations. The reconstituted gene sets are used to (1) facilitate systematic prioritization of the most likely causal gene(s) at a given associated locus without limiting the analysis to genes with well-established functional annotations, and (2) to assess whether genes in associated loci enrich for particular gene sets (including biological pathways). Prior to the analyses, DEPICT constructs lists of genes at associated loci by mapping genes to loci if they reside within, or are overlapping with, boundaries defined by the most distal SNPs in either direction with LD r2 > 0.5 to the strongest associated SNP at each locus. DEPICT was run using default settings, that is using 500 permutations for bias adjustment, 20 replications for false discovery rate estimation, exclusion of the extended major histocompatibility complex region (chr. 6: 25–35 Mb) and using normalized expression data from 77 840 Affymetrix microarrays for gene set reconstitution (see (45) for details). For a complete description of DEPICT, please refer to (25). The code used to run DEPICT and other analysis conducted in this paper can be downloaded from https://github.com/perslab/ (last accessed January 19, 2016).
Gene set enrichment control analysis
We defined a set of genes with elevated expression in the human brain using the RNA-seq BrainSpan data (see the BrainSpan Developmental Transcriptome-based expression analyses section below). To derive a measure of a gene's expression in the brain, we computed for each gene the mean of the brain structure-specific and developmental stage-specific median expression levels. Next, we sampled 100 sets each comprising around 112 loci (the number of DEPICT schizophrenia loci) by sampling SNPs upstream of transcription start sites of genes with mean expression levels above the 95th percentile. We then ran DEPICT on each set of these 100 sets of input loci and summarized these results by counting how often a significantly enriched gene set (from the schizophrenia DEPICT analysis) was in observed with at least the same false discovery rate across the 100 ‘generic brain’ DEPICT runs and computed empirical P-values. Reconstituted gene sets were clustered using Affinity Propagation R software (46). The script for clustering DEPICT gene set enrichment results can be found at https://github.com/perslab/DEPICT (last accessed January 19, 2016).
Reconstitution of additional gene sets potentially relevant to schizophrenia
From Genebook (http://atgu.mgh.harvard.edu/~spurcell/genebook/genebook.cgi) we downloaded the ARC, FMRP (originally derived from 34), NMDAR, rat pre-synapse active zone (originally derived from 35) and synaptic vesicle (originally derived from 38) gene sets used in (3,4). In addition we downloaded proteomics-derived gene lists for pre-synaptic docking complexes (37) and mouse pre-synapse active zone (36). The number of genes in the original gene list and the number of genes after mapping to Ensembl gene identifiers, performing orthology mapping and limiting to genes in DEPICT can be found in Supplementary Material, Table S5. Subsequently we reconstituted the gene sets using our previously described approach (25,45). The area under the receiver operating characteristics curve estimates were high (>0.85; Supplementary Material, Table S5), indicating that the expression of the genes in each of these gene sets are strongly co-regulated. Finally, we ran DEPICT on the reconstituted gene sets.
Filtering of reconstituted gene sets and visualization
To ensure that significantly enriched reconstituted DEPICT gene sets represented biology indicated by their identifiers (reconstituted gene sets are referred to by their original gene name), we omitted reconstituted gene sets, which did not enrich for the genes in the original gene set. We used a Spearman rank-sum test to assess whether genes in the original gene set were overrepresented in the reconstituted gene set and conservatively omitted reconstituted gene sets for which the Spearman rank-sum P > 3.5 × 10−6 (∼0.05/14 461). Among 143 significantly enriched reconstituted gene sets 39 were omitted (Supplementary Material, Table S10) resulting in 104 significant reconstituted gene sets reported in this paper. The Cytoscape tool was used to visualize gene sets and their discretized overlap (47).
Genotype-Tissue Expression Project-based tissue enrichment analysis
We downloaded normalized RNA-Seq gene expression data from the GTEx Project (28) (www.gtexportal.org; release, pilot 01/31/2013, patch 1; last accessed January 19, 2016). We further processed the RNA-Seq data by Winsorizing values larger than 50 reads per kilobase of transcript per million reads mapped (RPKM) to 50 (as previously done in 4) and transforming all values to log2(1+RPKM) values. After limiting genes to genes part of DEPICT and discarding genes with no variance across all tissues, we ended up with 19 414 genes. All prioritized genes were part of the processed RNA-Seq data. We discarded tissues with less than 10 samples and computed median gene expression levels (19 682 genes) for the remaining 37 tissues (see Supplementary Material, Table S11 for the number of samples for each tissue). The resulting tissue expression data set was used in DEPICT instead of the microarray-based tissue expression matrix derived from the 37 427 microarray samples used in the standard DEPICT version. DEPICT was used to test whether genes in the genome-wide significant loci were higher expressed in a given tissue than all other genes covered in the analysis (two-sided t-test).
Schizophrenia exome sequencing study data used to validate prioritized genes
We used Swedish exome sequencing data (4) to validate genes prioritized by DEPICT. We downloaded all genes containing at least one disruptive variants (frameshift, nonsense and essential splice site variants) in cases and/or controls with minor allele frequency (MAF) <0.1% (8702 genes; download date: May 2, 2014) from the Genebook database. Next, we mapped gene symbols to Ensembl identifiers, which left us with 8,684 genes. A total of 150 genes overlapped with the 362 genes in the 109 associated schizophrenia loci. We also used Bulgarian trio exome sequencing data (3) to validate prioritized genes. We downloaded all genes with de novo variants (1368 variants in 616 genes) from the Genebook database (download date: May 2, 2014), discarded genes with de novo variants that were not replicated in (3) (referred to as ‘silent’ in the database) and mapped the remaining gene symbols to Ensembl identifiers, which left us with 613 genes, of which 16 genes overlapped with the 362 in the 109 associated schizophrenia loci. The ORs were calculated as OR = oddsprioritized/oddsnon-prioritized. For the rare variant benchmark we defined oddsprioritized = cases' count of disruptive variants in prioritized genes/controls' count of disruptive variants in prioritized genes, and oddsnon-prioritized = cases' count of disruptive variants in non-prioritized genes/controls' count of disruptive variants in non-prioritized genes. For the de novo variant benchmark we defined oddsprioritized = number of prioritized de novo variant genes/number of prioritized genes with no de novo variants, and oddsnon-prioritized = number of non-prioritized de novo variant genes/number of non-prioritized genes with no de novo variants.
Postsynaptic density proteomics data used to validate prioritized genes
Genes encoding protein members of the PSD have previously implicated in especially schizophrenia copy number variation studies (2). To validate genes prioritized by DEPICT, we retrieved purified human and mouse cortex PSD proteomics data (39) from the Genes2Cognition database (www.genes2cognition.org/db/GeneList/L00000059; last accessed January 19, 2016). We downloaded the ‘consensus’ sets of 748 human genes (see Supplementary Material, Table S12) and 1061 mouse genes (see Supplementary Material, Table S13) encoding proteins identified in each of three biological replicas in human and mouse proteomics experiments (download date: October 23, 2013). After mapping gene symbols to Ensembl identifiers we were left with 745 human PSD genes and 969 mouse PSD genes (of which 21 and 27 overlapped with the 362 genes in the 109 associated schizophrenia loci, respectively; Supplementary Material, Tables S11 and S12). For each benchmark we defined oddsprioritized = number of prioritized PSD genes/number of prioritized genes non-PSD genes, and oddsnon-prioritized = number of non-prioritized PSD genes/number of non-prioritized non-PSD genes.
Brainspan Developmental Transcriptome-based expression analyses
We downloaded normalized BrainSpan Developmental Transcriptome RNA-Seq data and microarray data (see www.brainspan.org/static/download.html; download date October 31, 2014; last accessed January 19, 2016). We further processed the RNA-Seq data as described in (4), that is values larger than 50 RPKM were Winsorized to 50 and transformed to log2(1+RPKM) values. In total 4,586 genes were affected by Winsorizing (∼2% of the expression values). No further normalization was applied to the microarray normalization. A total of eight prioritized genes were missing from the RNA-Seq data (Supplementary Material, Table S14). The developmental stages were defined using the BrainSpan Developmental Transcriptome technical white paper, release October 2013 v.5 (http://help.brain-map.org/display/devhumanbrain/Documentation; last accessed January 19, 2016). We computed median gene expression levels for all 26 structures (across all stages; see Supplementary Material, Table S15 for sample counts) and for all 12 stages (across all structures; see Supplementary Material, Table S16 for sample counts). To assess whether prioritized genes were higher expressed in postnatal compared to prenatal stages, we computed for each prioritized gene the average expression prenatal and postnatal expression value and then used a one-sided paired t-test to signify the difference in means between the two categories (prenatal group mean = 3.07; postnatal mean = 2.75). Temporal trajectories of genes identified by Gilman et al. (24) were plotted the same way as we plotted the prioritized genes identified by DEPICT, none of these genes overlapped with the prioritized genes (Supplementary Material, Fig. S4).
Negative controls analyses
To ensure that DEPICT did not bias the results toward postnatally expressed genes (gene expression data is used as part of the DEPICT gene prioritization framework), we performed 1000 null GWAS, ran DEPICT gene prioritization on each of them and recomputed the pre- versus postnatal expression test for each of them. In addition, DEPICT was used to prioritize genes for traits with at least 10 genome-wide significant associations reported in the GWAS Catalog database (48) (see Supplementary Material, Figure S5). DEPICT was run with the same settings as described above.
Supplementary Material
Funding
This work was supported by the Danish Council for Independent Research Medical Sciences (FSS) (to T.H.P.); the Alfred Benzon Foundation (to T.H.P.); the Lundbeck Foundation (to T.H.P.); the Novo Scholarship Programme (to P.T.); the National Institute of Diabetes and Digestive and Kidney Diseases (2R01DK075787 to J.N.H.); the National Institute of Mental Health (grant U01 MH094421 to P.G.C.); the Netherlands Organization for Scientific Research (NWO-VENI grant 916-10135 and NWO VIDI grant 917-14374 to L.F.); the Horizon Breakthrough grant from the Netherlands Genomics Initiative (92519031 to L.F.); the National Institute of General Medicine (T32GM074905 to S.L.); and the European Community's Health Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 259867 (to L.F.).
Supplementary Material
Acknowledgements
We thank the PGC for making schizophrenia summary statistics available.
Conflict of Interest statement. None declared.
References
- 1.McCarroll S.A., Feng G., Hyman S.E. (2014) Genome-scale neurogenetics: methodology and meaning. Nat. Neurosci., 17, 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirov G., Pocklingto A.J., Holmans P., Ivanov D., Ikeda M., Ruderfer D., Moran J., Chambert K., Toncheva D., Georgieva L. et al. (2012) De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry, 17, 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromer M., Pocklingto A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M. et al. (2014) De novo mutations in schizophrenia implicate synaptic networks. Nature, 506, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O'Dushlaine C., Chambert K., Bergen S.E., Kähler A. et al. (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ripke S., O'Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M. et al. (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet., 45, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.-H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H. et al. (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature. 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P.H., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E. et al. (2008) Large recurrent microdeletions associated with schizophrenia. Nature, 455, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Genome-wide association study identifies five new schizophrenia loci. (2011) Nat. Genet., 43, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A. et al. (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science, 320, 539–543. [DOI] [PubMed] [Google Scholar]
- 10.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M. (2012) De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet., 44, 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulsuner S., Walsh T., Watts A.C., Lee M.K., Thornton A.M., Casadei S., Rippey C., Shahin H., Nimgaonkar V.L., Go R.C.P. et al. (2013) Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell, 154, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arking D.E., Pulit S.L., Crotti L., van der Harst P., Munroe P.B., Koopmann T.T., Sotoodehnia N., Rossin E.J., Morley M., Wang X. et al. (2014) Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet., 46, 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z. et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E. et al. (2015) New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A. et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geller F., Feenstra B., Carstensen L., Pers T.H., van Rooij I.A.L.M., Körberg I.B., Choudhry S., Karjalainen J.M., Schnack T.H., Hollegaard M.V. et al. (2014) Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat. Genet., 10.1038/ng.3063. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan P.F., Posthuma D. (2014) Biological pathways and networks implicated in psychiatric disorders. Curr. Opin. Behav. Sci., 2, 58–68. [Google Scholar]
- 19.Jia P., Wang L., Meltzer H.Y., Zhao Z. (2010) Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr. Res., 122, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Dushlaine C., Kenny E., Heron E., Donohoe G., Gill M., Morris D., Corvin A. (2011) Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol. Psychiatry, 16, 286–292. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y.H., Kim J.-H., Song G.G. (2013) Pathway analysis of a genome-wide association study in schizophrenia. Gene, 525, 107–115. [DOI] [PubMed] [Google Scholar]
- 22.Network P. (2015) Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juraeva D., Haenisch B., Zapatka M., Frank J., Witt S.H., Mühleisen T.W., Treutlein J., Strohmaier J., Meier S., Degenhardt F. et al. (2014) Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia. PLoS Genet., 10, e1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilman S.R., Chang J., Xu B., Bawa T.S., Gogos J.A., Karayiorgou M., Vitkup D. (2012) Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat. Neurosci., 15, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pers T.H., Karjalainen J.M., Chan Y., Westra H.-J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T. et al. (2015) Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun., 6, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Valk R.J.P., Kreiner-Møller E., Kooijman M.N., Guxens M., Stergiakouli E., Sääf A., Bradfield J.P., Geller F., Hayes M.G., Cousminer D.L. et al. (2015) A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum. Mol. Genet., 24, 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J. et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. The Genotype-Tissue Expression (GTEx) project. (2013) Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res., 40, D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croft D., O'Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B. et al. (2011) Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res., 39, D691–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E. (2014) The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res., 42, D810–D817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lage K., Karlberg E.O., Størling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tümer Z., Pociot F., Tommerup N. et al. (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol., 25, 309–316. [DOI] [PubMed] [Google Scholar]
- 34.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y.S., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. et al. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morciano M., Beckhaus T., Karas M., Zimmermann H., Volknandt W. (2009) The proteome of the presynaptic active zone: from docked synaptic vesicles to adhesion molecules and maxi-channels. J. Neurochem., 108, 662–675. [DOI] [PubMed] [Google Scholar]
- 36.Weingarten J., Laßek M., Mueller B.F., Rohmer M., Lunger I., Baeumlisberger D., Dudek S., Gogesch P., Karas M., Volknandt W. (2014) The proteome of the presynaptic active zone from mouse brain. Mol. Cell. Neurosci., 59, 106–118. [DOI] [PubMed] [Google Scholar]
- 37.Boyken J., Grønborg M., Riedel D., Urlaub H., Jahn R., Chua J. (2013) Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron, 78, 285–297. [DOI] [PubMed] [Google Scholar]
- 38.Takamori S., Holt M., Stenius K., Lemke E.A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P. et al. (2006) Molecular anatomy of a trafficking organelle. Cell, 127, 831–846. [DOI] [PubMed] [Google Scholar]
- 39.Bayés A., van de Lagemaat L.N., Collins M.O., Croning M.D.R., Whittle I.R., Choudhary J.S., Grant S.G.N. (2011) Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci., 14, 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M.M., Pletikos M., Meyer K.A., Sedmak G. et al. (2011) Spatio-temporal transcriptome of the human brain. Nature, 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penzes P., Cahill M.E., Jones K.A., VanLeeuwen J.-E., Woolfrey K.M. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci., 14, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benros M.E., Mortensen P.B., Eaton W.W. (2012) Autoimmune diseases and infections as risk factors for schizophrenia. Ann. N. Y. Acad. Sci., 1262, 56–66. [DOI] [PubMed] [Google Scholar]
- 43.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. (2014) Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA, 111, E455–E464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bult C.J., Richardson J.E., Blake J.A., Kadin J.A., Ringwald M., Eppig J.T., Baldarelli R.M., Baya M., Beal J.S., Begley D.A. et al. (2000) Mouse genome informatics in a new age of biological inquiry. Proc. IEEE Int. Symp. Bio-Informatics Biomed. Eng. 10.1109/BIBE.2000.889586. [DOI] [Google Scholar]
- 45.Fehrmann R.S.N., Karjalainen J.M., Krajewska M., Westra H.-J., Maloney D., Simeonov A., Pers T.H., Hirschhorn J.N., Jansen R.C., Schultes E.A. et al. (2015) Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet., 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 46.Bodenhofer U., Kothmeier A., Hochreiter S. (2011) APCluster: an R package for affinity propagation clustering. Bioinformatics, 27, 2463–2464. [DOI] [PubMed] [Google Scholar]
- 47.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Lotia S., Pico A.R., Bader G.D., Ideker T. (2012) A travel guide to Cytoscape plugins. Nat. Methods, 9, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA, 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.