Abstract
We investigated which neurotrophic factors may contribute to the divergence of two peripheral nerves emanating from the geniculate ganglion. We compared receptor mRNA profiles of the neurons that supply the nerves, and also the growth of their neurites in response to neurotrophic factors in culture. Three mRNAs, Gfra2, TrkA, and TrkC, were differentially expressed. Only one ligand, Neurturin, promoted substantially different nerve regrowth from the nerves, and therefore may contribute to nerve divergence. Three receptor mRNAs were expressed in 100% of the neurons: TrkB, TrkB.T2 (kinaselacking isoform), and NCAM-140. Ligands for these Trks and FRα-1 promoted more outgrowth than ligands for the other receptors. NT-3 and BDNF synergistically promoted outgrowth. Finally, receptors are coexpressed at random rates, arguing against the existence of neuronal subtypes defined by a combinatorial code of these receptors.
Keywords: Development, Guidance, Axon, Mouse, Rat, Neurotrophic factor receptor, Trk, GDNF, Geniculate ganglion
Introduction
The divergence of axonal populations at choice points offers an attractive system for studying axon guidance mechanisms. If two populations of axons encounter the same environment at the same time and react in dramatically different ways, they must be expressing different receptors or different downstream signaling molecules. Given that there are many molecules that have a minimal influence in vivo, but a notable influence on axon growth in culture, focusing on molecules that affect axonal populations in different ways may facilitate the identification of guidance cues that have a prominent role in pathfinding in vivo. An excellent system for studying nerve divergence is provided by the geniculate ganglion. Unlike most sensory ganglia, the peripheral axons of geniculate neurons diverge at the ganglion to generate two prominent nerves that exit the ganglion at a right angle to one another: the greater superficial petrosal nerve (GSP) and the combined posterior auricular nerve and chorda tympani nerve (PA/CT). The GSP grows anteriorly and medially, ultimately innervating taste buds in the palate. The PA/CT grows ventrally into the hyoid arch [1], bifurcating into the CT, which will innervate taste buds in the anterior two thirds of the tongue, and the PA, which will innervate somatosensory structures in the skin of the outer ear.
As a first step toward identifying ligand/receptor interactions that are relevant to the divergence of the PA/CT and GSP, we have characterized the expression patterns of two classes of neurotrophic factor receptors in the neurons that supply these two nerves. In addition, we compared the influence of the ligands for these receptors on the regrowth of axons that had extended into the PA/CT or GSP in vivo. We began with neurotrophic factors and their receptors for two reasons. First, no matter what guidance cues are ultimately implicated in the divergence of these nerves, it is likely that functional screening for those factors will entail promoting the outgrowth of the axons with at least one of the growth factors that we used in this study. It is therefore essential to know if the growth factors differentially affect nerve growth from the two nerves. Second, it is possible that the neurotrophic factors themselves could contribute to the divergence of the two nerves, although initial nerve growth occurs independently of several neurotrophic factors [2].
To determine which neurotrophic factor receptor mRNAs are expressed in the PA/CT and GSP neurons, we coupled retrograde labeling with single cell dissection followed by the Eberwine antisense RNA amplification method and PCR [3]. This has the advantage over in situ hybridization that several receptor mRNAs can be detected in a single specimen, enabling one to determine the coexpression patterns of these mRNAs, potentially revealing subclasses of neurons. These subclasses may differ in axon pathfinding properties, survival needs, function, or developmental stage and are therefore of broad interest.
Methods
Animal Husbandry
All aspects of animal care and treatment were carried out according to the local guidelines of the animal care committee. Pregnant dams were sacrificed by CO2 asphyxiation. Receptor expression studies were conducted on geniculate ganglia dissected from C57Bl\6 mouse embryos at embryonic day 11 (E11), Theiler stage 18 [4]. For outgrowth studies in culture, we used rat geniculate ganglia (E13, stage 18) because mouse ganglia grew poorly.
Neurotrophic Factor Receptor mRNA Expression Analysis
Dissection, Labeling, and Sectioning
Mouse embryos were decapitated and the heads were fixed overnight in 4% paraformaldehyde (EM Sciences) in phosphate-buffered saline (PBS, pH 7.4) at 4°C. The specimens were washed in PBS and further dissected to expose either the GSP or PA/CT. Crystals of Bromo-DiI (Molecular Probes) were applied to the exposed nerve and the specimens were placed in PBS containing 100 U/ml penicillin-streptomycin (Invitrogen) at 36°C for 24–48 h to permit transport back to the ganglion. Ganglia were then dissected.
Photoconversion was carried out essentially as previously described using 1.5 mg/ml diaminobenzidine (Kirkegaard & Perry) in Tris buffer (pH 8.2) [5], however, we carried out our photoconversion on intact ganglia (prior to cryosectioning). DAB precipitation did not occur in unlabeled ganglia. Photoconverted ganglia were cryoprotected with 30% sucrose, embedded in OCT (Ted Pella, Redding, Calif., USA), and 10 μm cryosections were mounted on Superfrost®/Plus slides (Fisher Scientific).
In situ Priming, Transcription, and Microdissection
Details of this procedure and that of antisense mRNA amplification, including the recipes for all solutions below, were thoroughly described previously [6, 7], and are only briefly outlined below. We will provide detailed methods on request. In overview, sections are subjected to in situ hybridization with oligo(dT)-T7 primer (Ambion) followed by AMV-Reverse Transcriptase (AMV-RT, Promega) treatment to generate cDNA. Individual cells were microdissected and transferred to a 1.5 ml Eppendorf tube and subjected to antisense RNA amplification. To determine if amplified samples contained a sufficient amount of nucleic acid to detect expression of mRNAs encoding receptors, 1μl of sample was combined with 1μl of 0.25% ethidium bromide and illuminated on an ultraviolet light box. Only samples yielding positive test drops were further processed.
Two rounds of PCR were used to test for the presence of receptor cDNAs. The reaction tube contained 2.5 U AmpliTaq DNA polymerase, 2 μl of the cDNA, 10 pmol of forward and reverse PCR primers, and 2 mmol of dNTPs, in DNA polymerase buffer (Promega). We used the same cycle durations for all of the genes we tested, altering annealing temperatures to accommodate differences in the primer sets (table 1): 94°C for 9 min to dissociate the DNA strands, then 94°C for 1 min, ∼60°C for 2 min, 72°C for 40 s. After 35 cycles as described, the last 72°C step was extended for 4 min. The reaction was then stopped at 10°C and fresh DNA polymerase and primers were added, and the process was repeated for 35 more cycles. The products were then subjected to electrophoresis in a 2% agarose gel.
Table 1. Mouse neurotrophic factor receptor and primer information.
| Gene | Accession number | Primer sequences, forward and reverse | Product size | Annealing temperature |
|---|---|---|---|---|
| TrkA | XM_283871 | 5′-agg gcc aca tca tgg aga ac-3′ (F) 5′-gtg cag act cca aag aag c-3′ (R) | 279 bp | 59.0°C |
| TrkB | NM_008745 | 5′-gcg cgg ctc tgg ggc tta tg-3′ (F) 5′-cct gag tgt cgg ggc tgg att tag-3′ (R) |
481 bp | 58.0°C |
| TrkB.T2 | NM_008745.1 | 5′-ctg ttg cct atc cca gga ag -3′ (F) 5′-gag agg cac aat cca atg ag -3′ (R) |
232 bp | 57.0°C |
| TrkC | NM_008746 | 5′-tgc cca gcc aag tgt agt ttc t-3′ (F) 5′-gcg cct ccc cct gtt cct-3′ (R) |
526 bp | 58.5°C |
| Gfra1 | NM 010279 | 5′-gca cag cta cgg gat gct ctt ctg-3′ (F) 5′-gta gtt agg agt cat gac tgt gcc aat c-3′ (R) |
286 bp | 69.6°C |
| Gfra2 | NM 008115 | 5′-tgg cat gat tgg gtt tga ta-3′ (F) 5′-ctt tgg agt tgt tgg cct tc-3′ (R) |
356 bp | 63.5°C |
| Gfra3 | NM 010280 | 5′-ctt ggg act tgt gca act ga-3′ (F) 5′-gtg gtg gag agg aca aag ga-3′ (R) |
408 bp | 59.0°C |
| Ret | NM 009050 | 5′-taa agc agg cta cgg cat ct-3′ (F) 5′-gcc ttc tcc cag agt ttt cc-3′ (R) |
391 bp | 58.7°C |
| NCAM | X15052 | 5′-cat tgt ggg cat cct cat tgt cat ctt c-3′ (F) 5′-agt tgg cgc tgg ctt ggg ctt ctg-3′ (R) |
350 bp 1,150 bp |
59.7°C |
To insure that we were not amplifying genomic DNA, we carried out the procedure on tissue that had been mock-hybridized without oligo(dT)-T7 primer. The dot test was negative for these samples. Furthermore, PCR failed to detect TrkB or TtrkB in these samples although both of these amplicons were detected in every cell that was processed with oligo dT. In addition, we pooled 7–8 samples of PCR products from adult brain (see below) for sequencing. In every case, the sequence matched only the neurotrophic factor that we intended to detect.
Whole adult mouse brains were homogenized and total RNA was collected and reverse-transcribed to generate cDNA (protocol available on request). Each primer pair was tested on this cDNA. Negative controls were prepared similarly but during PCR, DEPC water was substituted for the sample.
Cell Culture
For these studies, we used E13 rat embryos. We tried to conduct this work using mouse embryos, but we did not succeed in getting an interpretable level of outgrowth from the mouse geniculate ganglia. Although we interpret our observations of rat geniculate ganglion outgrowth in light of the results we obtained with the mouse neurotrophic factor receptor mRNA profiling, the reader should bear in mind that there are differences between rat and mouse. Many axon guidance studies use mice for genetic studies and rat tissues for in vitro assessment of neurite growth, e.g., Charron et al. [8].
E13 rat heads were pinned in Sylgard-coated dishes. Crystals of DiI were applied to either the PA/CT in arch II or the GSP as it courses toward the internal carotid artery. DiI crystals were prevented from contacting the unintended peripheral nerves or the exposed hindbrain target region of geniculate axons. Specimens were incubated for ∼10 h at 36°C in BGJb media (Invitrogen) supplemented with 0.1 mg/ml ascorbic acid, 100 U/ml penicillin/streptomycin, and 10 ng/ml of the growth factor(s) in which the ganglia would be cultured the following day. Geniculate ganglia were dissected and up to 3 ganglia were plated on laminin-polylysine-coated coverglasses in 0.6 ml of C10-2 media [9]. After ∼8 h of culture, 0.4 ml of C10-2 media plus neurotrophic factor was added to the dish to completely submerge the ganglia. The following growth factors were tested: BDNF, NT4, NT3: recombinant human (Sigma); NGF: 2.5S mouse (Harlan, Indianapolis, Ind., USA), GDNF: recombinant rat (Sigma), Neurturin and Artemin: recombinant mouse (R&D Systems, Minneapolis, Minn., USA).
Assessment of Neurite Density and Length
At 20 h after plating, cultures were fixed as previously described [9]. Briefly, cultures were treated with 37°C 4% paraformaldehyde in 0.1 M cacodylate (pH 7.4) containing 10 mM MgCl2 and 5 mM CaCl2 for 15 min. Cultures were washed with 0.1 M cacodylate buffer followed by PBS. Cultures were mounted in PBS containing 15% Vectashield (Vector Laboratories) and imaged with a Spot RT monochrome CCD. The staining of regrowing neurites by Br-DiI is necessarily weak, as only as much dye as was retained by the somas and proximal axon segments is available to diffuse into the regrowing neurites. Therefore, in addition to subtracting background images, the contrast of the images was adjusted. The density of the labeled axons was ranked on an integral scale from 0 to 3 as described in figure 3. In the absence of growth factors, there was no outgrowth (n = 6).
Fig. 3.
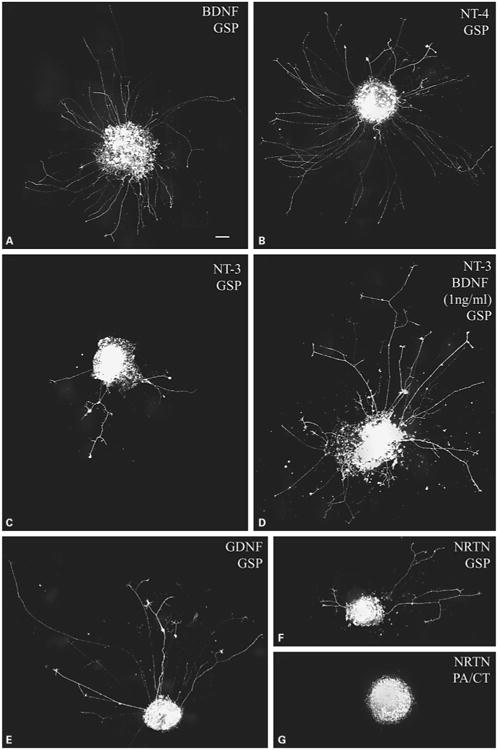
Neurotrophic factors differ in their ability to stimulate regenerative axon growth from geniculate neurons on laminin/polylysinecoated coverglasses. GSP regeneration in the presence of the indicated growth factor at the concentration (ng/ml) shown in parentheses. The low intensity staining is typical of this technique, in which DiI retained in the somas during dissection diffuses along the regrowing axon. A, B BDNF (A) and NT-4 (B) both stimulated robust regrowth of GSP axons, earning a density rating of 3 on our scale. C, D NT-3 (C) stimulated a detectable level of outgrowth (1 on our scale). When NT-3 (25 ng/ml) and a low level of BDNF (1 ng/ml) were combined (D), the outgrowth increased to level 2. E–G GDNF (E) stimulated a moderate level of regrowth of GSP axons (level 2). Neurturin promoted a low level of regrowth of GSP axons (F, level 1), but none from PA/CT axons (G, level 0). Calibration bar = 100 μm.
Statistical Analysis of Density Rankings
Nonparametric statistical analysis was carried out to determine if differences in density rankings were significant. For comparisons between the PA/CT and GSP, the Mann-Whitney U test was used with a significance threshold of p < 0.05. To compare the effects of different growth factors on PA/CT and GSP regrowth, measurements were pooled. This reduces our chances of finding a significant difference, providing a more conservative assessment of significance. In two cases, we did not combine PA/CT and GSP values: Neurturin only stimulated outgrowth from the GSP, so the PA/CT zero values were not averaged in, and we only measured the influence of 1 ng/ml BDNF on PA/CT regrowth. A Kruskal-Wallis test was performed and demonstrated significant (p < 0.05) differences among the density distributions for the different growth factors. We then conducted pairwise analysis of statistical differences using the Mann-Whitney U test followed by a sequential Bonferroni correction. The correction led to a significance threshold of 0.0015. There were 32 significant differences out of 66 possible pairwise comparisons, but we discuss only the subset of comparisons that are essential.
Results
Neurotrophic Factor Receptor mRNA Expression in Single Geniculate Ganglion Neurons That Project to the PA/CT or GSP
As described in the ‘Methods’, geniculate neurons were retrogradely labeled by applying DiI to either the PA/CT or the GSP (fig. 1A) and then photoconverted under rhodamine optics (fig. 1B). After cryosectioning, individual cells were microdissected and subjected to antisense RNA amplification. This procedure yielded sufficient cDNA to enable us to test for the presence of all of the receptor mRNAs that we studied in each cell.
Fig. 1.
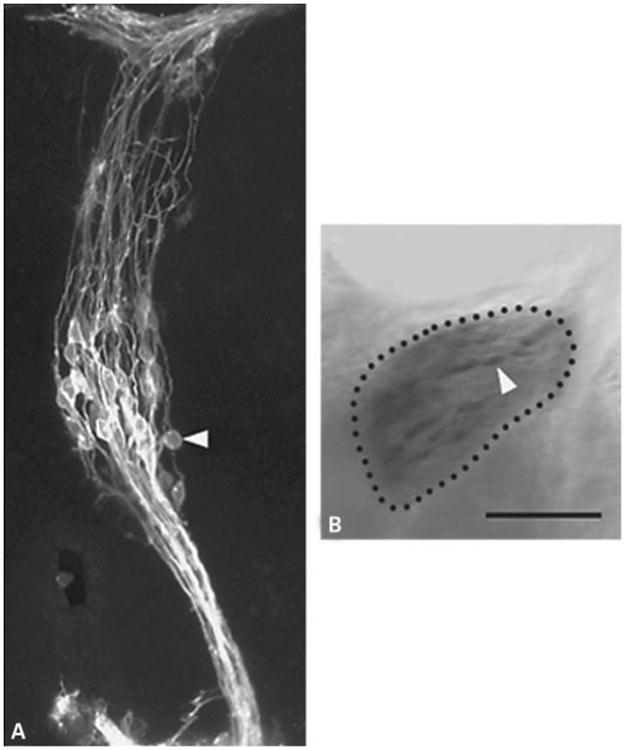
Geniculate neurons labeled by DiI and following photoconversion of DAB. A A projection of optical sections obtained from a Zeiss 410 confocal microscope showing individual neurons labeled 24 h after application of DiI to the PA/CT of a fixed E11 mouse geniculate ganglion. B A different ganglion from that shown in A following photoconversion imaged on a conventional Zeiss Axiovert 100 microscope. Bar = 100 μm.
Neurotrophin Receptor mRNAs
We tested for the selective, high-affinity neurotrophin receptors TrkA, TrkB, TrkC, and a truncated isoform of TrkB. Both TrkB and TrkC are expressed in truncated forms. The truncated forms may act as dominant negative regulators of the full-length forms [10], may bind and present ligands to full-length forms [11], and they also initiate signaling cascades distinct from those activated by the full-length isoforms [12]. We selected a truncated isoform of TrkB for several reasons. TrkB is known to be necessary for survival for over 90% of the geniculate neurons [13], whereas TrkC signaling is required for at most 11% [14], and because BDNF and NT-4, but not NT-3, promote robust outgrowth of geniculate ganglion neurons in collagen gels [1]. Among the truncated TrkB isoforms, we selected TrkB.T2.
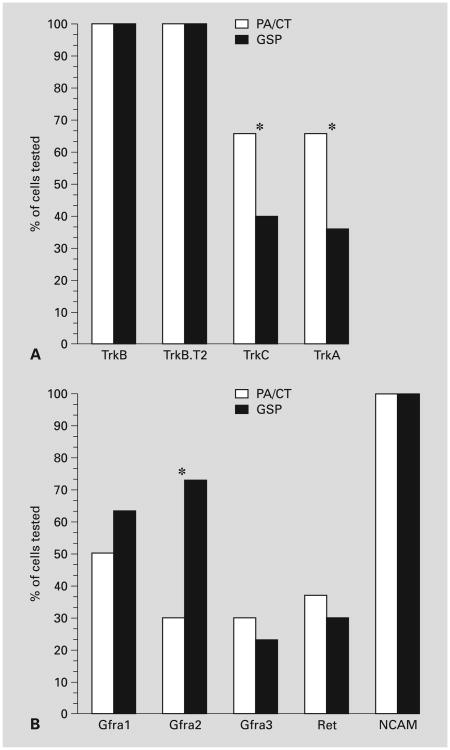
Trk mRNAs were differentially expressed in geniculate neurons at E11 (fig. 2A). TrkB and TrkB.T2 mRNAs were expressed in all PA/CT and GSP geniculate neurons (30/30). TrkC and TrkA mRNAs were expressed in only a subset of PA/CT and GSP neurons, and the size of the subpopulation that expressed these Trks differed for neurons that projected to the two nerves. TrkC and TrkA mRNA were expressed in ∼70% of the PA/CT neurons, but in only ∼40% of the GSP neurons, and these differences were significant (p < 0.05, chi-square).
Fig. 2.
Neurotrophic factor receptor expression in PA/CT and GSP neurons (n = 30/nerve). A Neurotrophin receptors. Note that TrkB. T2 mRNA, like the full-length TrkB, was expressed in all retrogradely labeled mouse geniculate neurons at E11. B GDNF family receptor components. An asterisk indicates that PA/CT and GSP values are significantly different (p < 0.05, χ2).
GDNF Family Receptor mRNAs
We determined the frequency of mRNA expression of Gfra1, 2, and 3, and their obligate coreceptors Ret and NCAM. We did not screen for the mRNA encoding GFR1-4 because mouse nervous tissue only expresses a secreted isoform [15] and its ligand, Persephin, does not support sensory neuron survival [16]. In contrast to trk mRNA expression, none of the Gfra receptor mRNAs were expressed in all neurons at E11 (fig. 2B). Gfra1 mRNA was expressed in 50% (15/30) of PA/CT neurons and in 63% (19/30) of GSP neurons. Gfra2 mRNA was expressed in 30% (9/30) of the PA/CT neurons and 73% (19/30) of the GSP neurons, and this difference was significant (p < 0.05). Gfra3 mRNA had the lowest frequency of expression at ∼27% for both PA/CT and GSP neurons.
GFRα subtypes do not have transmembrane domains and must signal in combination with coreceptors, such as RET and NCAM [17, 18]. Ret mRNA was expressed in approximately one third of the 30 neurons that we tested for each nerve in both nerves (fig. 2B). Neuronal NCAM is expressed in multiple splice variants, including a 140-kD and a 180-kD isoform [19, 20]. The 140-kD isoform has been implicated in GDNF signaling directly and is the isoform that is expressed on growth cones, whereas NCAM-180 is thought to serve a structural role along axon shafts and at synapses, and its role in GDNF signaling has not been established. We used primers that span the region of NCAM cDNA that encodes the 800-bp insert that distinguishes these two isoforms, allowing us to determine if either or both NCAM-140 and NCAM-180 mRNA are expressed in geniculate neurons. NCAM-140 mRNA was expressed by all of the neurons in both nerves (n = 30 neurons/nerve). We found only the mRNA encoding the 140-kD isoform in these embryonic geniculate neurons, although we detected both isoforms in adult mouse brain. Thus, NCAM-140 is the predominant isoform of NCAM that is expressed by mouse geniculate neurons at E11.
Receptor Coexpression Analysis
Because we were able to conduct receptor profiling for all of the above receptors on each of the cells we dissected, we conducted coexpression analysis. We compared the observed frequency of coexpression of receptors to the coexpression frequencies that would occur if receptors were coexpressed randomly. If coexpression rates occur significantly more or significantly less than the random rate, this would likely indicate coordinated regulation of expression. This in turn would suggest that neurotrophic factor coexpression defines subclasses of neurons within the PA/CT and GSP classes. We found no evidence of nonrandom coexpression, i.e., the observed values closely followed those expected of a random distribution of the receptors. Table 2 shows the results of a subset of our analysis. Coexpression occurred at random rates in PA/CT, GSP, and combined PA/CT and GSP neuronal populations, suggesting that these receptors are expressed independently of one another.
Table 2. Observed (obs.) vs. expected (expd.) percent of geniculate neurons that express the indicated neurotrophic factor mRNAs.
| mRNA | PA/CT obs. vs. expd. | GSP obs. vs. expd. | Both obs. vs. expd. |
|---|---|---|---|
| TrkB | 7 vs. 11% | 40 vs. 38% | 23 vs. 23% |
| TrkB + A | 23 vs. 22% | 20 vs. 22% | 22 vs. 24% |
| TrkB + C | 23 vs. 22% | 23 vs. 25% | 23 vs. 26% |
| TrkB + A + C | 43 vs. 44% | 16 vs. 15% | 30 vs. 28% |
|
| |||
| Ret + Gfra1 | 17 vs. 9% | 0 vs. 4% | 8 vs. 7% |
| Ret + Gfra2 | 3 vs. 4% | 0 vs. 6% | 2 vs. 5% |
| Ret + Gfra3 | 7 vs. 4% | 0 vs. 1% | 3 vs. 2% |
| Ret + Gfra1 + 2 | 0 vs. 4% | 27 vs. 11% | 13 vs. 8% |
| Ret + Gfra1 + 3 | 0 vs. 4% | 3 vs. 12% | 2 vs. 2% |
| Ret + Gfra2 + 3 | 0 vs. 2% | 0 vs. 2% | 0 vs. 2% |
| Ret + Gfra1 + 2 + 3 | 3 vs. 2% | 0 vs. 3% | 2 vs. 3% |
Note that the ‘mRNA’ column represents cells that express the indicated mRNA(s) of a particular receptor class only. Thus, ‘TrkB’ is ‘TrkB but not TrkC or TrkA’, and does not take into account GFRa or Ret mRNA expression and ‘Ret + Gfra1’ is ‘Ret + Gfra1 but not Gfra2 and Gfra3 and disregards Trk’ mRNA expression. To calculate the percentage of neurons that would express the indicated mRNAs, and not other mRNAs of the same class if each of the mRNAs were distributed randomly, we computed the product of the percentages of cells that expressed each of the indicated mRNAs times the product of the percentages of cells that did not express the other mRNAs of that class. For example, to calculate the percentage of geniculate neurons that are expected to express Ret, Gfra1, and Gfra2 (but not Gfra3), we compute: (33.3% Ret) × (56.7% Gfra1) × (51.7% Gfra2) × (73.3% not expressing Gfra3) = 8%. Note that coexpression occurred at random rates regardless of whether it was assessed as described above, or without regard for subclasses for each neurotrophic factor receptor family. n = 30 for both PA/CT and GSP neurons.
Regenerative Outgrowth from Rat PA/CT and GSP Neurons in the Presence of Neurotrophins and GDNF Family Ligands
There are no markers to distinguish between PA/CT and GSP neurites in vitro, so we applied DiI to either the PA/CT or GSP to retrogradely label the somata, cultured the labeled ganglia for 20 h in 25 ng/ml neurotrophic factor (except where otherwise noted), and assessed the length and density of labeled neurites semiquantitatively.
PA/CT and GSP Lengths Were the Same
If the outgrowth was sufficiently dense (averaging over 1.5), then the lengths of 5 or more neurites representing the extent of the halo of outgrowth were measured and averaged. The following treatments (25 ng/ml) induced sufficient outgrowth for analysis: BDNF, NT-4, NT-3 + 1 ng/ml BDNF, and GDNF. In none of these cases did we find that the lengths of regenerating PA/CT neurites differed significantly from those of GSP neurites (Student's t test, p > 0.05, n values as in fig. 4A, data not shown).
Fig. 4.
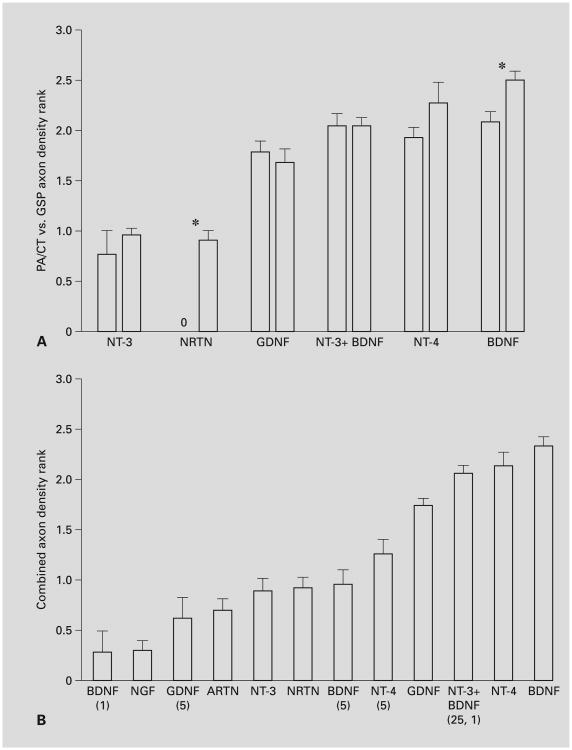
The density of PA/CT or GSP axon regrowth induced by neurotrophic factors. Except where noted otherwise in parentheses, the growth factor concentration was 25 ng/ml. The regrowth density of each ganglion was rated by 3 observers who did not know the treatments. Ratings were as follows: 0 = no regrowth (fig. 3G); 1 = low regrowth (3–6 axons, fig. 3C); 2 = moderate regrowth (e.g., fig. 3E); 3 = high (e.g., fig. 3A, B). The three measurements were averaged for each ganglion, and these averages were averaged for each trophic factor. Statistical analysis and significant differences are discussed in the ‘Results’. A Comparison of PA/CT (left bar of each pair) and GSP (right bar of each pair) outgrowth density for growth factor treatments that resulted in a low level of outgrowth (∼1 or higher). The axon regrowth density was significantly different for the PA/CT and GSP when BDNF (n = 11, 10 for PA/CT and GSP, respectively) or Neurturin (n = 10, 12) were used (*p < 0.05). No significant difference was observed when the following neurotrophic factors were used: NT-3 (n = 7, 8) or NT-3 in combination with 1 ng/ml BDNF (n = 7, 6), GDNF (n = 9, 9), or NT-4 (n = 5, 6). B Comparison of the combined PA/CT and GSP regrowth data for each growth factor (with the exception of Neurturin, for which only the GSP regrowth is plotted, and 1 ng/ml BDNF, for which only PA/CT regeneration was tested). N-values for each treatment are as follows: BDNF 1 ng/ml (n = 6), NGF 25 ng/ml (n = 14), GDNF 5 ng/ml (n = 6), ARTN 25 ng/ml (n = 12), NT-3 25 ng/ml (n = 15), NRTN 25 ng/ml (n = 12), BDNF 5 ng/ml (n = 14), NT-4 5 ng/ml (n = 10), GDNF 25 ng/ml (n = 18), NT-3 + BDNF (n = 13), NT-4 25 ng/ml (n = 11), BDNF 25 ng/ml (n = 21).
Neurotrophin-Stimulated Regrowth
BDNF and NT-4 induced the most regrowth of all of the growth factors that we tested, averaging moderate to high (2–2.5) densities from the GSP (fig. 3A, B) and PA/CT (fig. 4). Curiously, both of these factors induced slightly more regrowth from GSP than from PA/CT neurons (fig. 4A), and in the case of BDNF, the difference between the regrowth densities was significant. This was not expected because TrkB mRNA is expressed by all mouse geniculate neurons at this stage, and there may be more PA/CT neurons than GSP neurons. The amount of regrowth promoted by BDNF (25, 5, and 1 ng/ml) and NT-4 (25 and 5 ng/ml) was dose-dependent (fig. 4B, all pair-wise differences were significant except 5 vs. 1 ng/ml BDNF). NT-3 induced a low level of regrowth (fig. 3C, 4B), that was significantly less than that induced by BDNF and NT-4. The density of NT-3-stimulated regrowth was comparably low for both nerves (fig. 4A). When a low concentration of BDNF (1 ng/ml) was combined with 25 ng/ml NT-3, the density of regrowing neurites from both nerves was significantly more robust than with NT-3 alone and with 5 ng/ml BDNF alone (fig. 3D, 4B). It was not statistically different from the regrowth in 25 ng/ml BDNF. Given that 1 ng/ml BDNF stimulated virtually no outgrowth (fig. 4B), this finding suggests that BDNF and NT-3 interact synergistically. As with NT-3 alone, NT-3 plus BDNF-stimulated PA/CT and GSP regrowth were not significantly different (fig. 4A). NGF typically induced no regrowth (fig. 4B). Thus, despite the fact that approximately one third of the mouse geniculate neurons express TrkC and TrkA mRNA, their ligands induced little regrowth of PA/CT or GSP neurites. There was no apparent synergistic interaction when 25 ng/ml NGF plus 1 ng/ml BDNF were applied (n = 6, not shown).
GDNF Family Ligand-Stimulated Regrowth
GDNF family ligands also stimulated regrowth of PA/CT and GSP neurites, and the level of regrowth more closely paralleled the frequencies of cognate receptor expression in the mouse geniculate ganglion than was observed for neurotrophin-stimulated regrowth. GDNF (fig. 3E, 4B) was significantly more effective in promoting regrowth than the other GDNF family members Neurturin (fig. 3F, G, 4B) and Artemin (fig. 4B). GDNF-stimulated regrowth density did not differ between the PA/CT and GSP (fig. 4A), in parallel with the nearly identical frequencies of Gfra1 mRNA expression observed in the mouse neurons (fig. 2B). Neurturin did stimulate a significant difference in regrowth density: GSP regrowth (fig. 3F) was low (density average of 0.92), but no regrowth was stimulated from PA/CT neurons (fig. 3G, 4A). This difference correlates with our finding that Gfra 2 mRNA was expressed in significantly more mouse GSP neurons than PA/CT neurons (fig. 2B). Artemin, a GFRα-3 ligand, usually stimulated no regrowth from either the PA/CT or GSP neurons (fig. 4B).
Discussion
We evaluated whether neurotrophic factor receptor mRNAs are differentially expressed in rodent geniculate neurons that supply divergent peripheral nerves (the PA/CT and GSP), and whether the ligands for these receptors differentially influence the growth of neurites from those neuronal populations. We found a variety of neurotrophic factor receptor mRNA expression patterns, and in three cases the expression levels of a particular receptor differed significantly between PA/CT and GSP neurons. The proportion of neurons that expressed a particular receptor mRNA ranged from 23 to 100% and correlated weakly with the robustness of regrowth induced by its cognate ligand. Our results are consistent with the possibility that neurotrophic factor signaling contributes to the divergence of PA/CT and GSP axons. In addition, mRNA coexpression analysis indicated that these receptors are expressed independently of one another, i.e., we did not detect subtypes based on receptor expression. To our knowledge, this is the first report to compare receptor expression analysis using antisense RNA amplification with in vitro outgrowth properties of neuronal populations giving rise to divergent axons. Note that caution must be exercised in comparing these results because the molecular anatomy was carried out on mouse ganglia whereas the outgrowth assays were carried out on rat ganglia. We discuss the amplification method first, followed by the results.
The antisense RNA amplification method was designed to provide a proportional amplification of mRNAs [3, 21]. Nonetheless there could be differences in the amplification of mRNAs, and therefore differences in the frequency of expression of different receptor mRNAs must be interpreted cautiously. However, differences in the frequency of expression of a particular mRNA between two populations of neurons cannot result exclusively from the technique. In addition to caveats about the amplification procedure, it should be noted that our retrograde labeling was carried out three quarters to a full day after the first axons exit and diverge from the geniculate ganglion, and it is possible that by this time receptors or downstream signaling molecules involved in divergence-related guidance decisions have been down-regulated. Attempts to retrogradely label neurons at earlier stages yielded too few labeled neurons for analysis.
Three mRNAs were differentially distributed between the PA/CT and GSP: Gfra2 (73% GSP vs. 30% PA/CT), and TrkC and TrkA (67% PA/CT vs. ∼40% GSP). In only one of these cases did the cognate ligand exert a corresponding differential effect on regrowth. Neurturin, the ligand for GFRα-2, induced a low level of outgrowth from rat GSP neurites, but none from PA/CT neurites. Additional studies are required to determine if Neurturin is differentially distributed along the paths explored by the PA/CT and GSP at the time of divergence (E10 in mouse). Experiments are underway to determine if Neurturin is more attractive to GSP neurites than PA/CT neurites using an in situ approach [22]. The percentages of GSP neurons and PA/CT neurons that express Gfra2 mRNA parallel the percentages of gustatory neurons contributing to the two nerves [23] (in adult rats), raising the possibility that GFRα-2 expression distinguishes gustatory neurons from the PA somatosensory neurons within this ganglion.
TrkC and TrkA mRNA were detected at higher frequencies in the PA/CT than in the GSP, but NT-3 induced the same low level of outgrowth from PA/CT and GSP neurons and NGF did not elicit any outgrowth. TrkC is evidently not required for divergence in vivo [24]. A robust synergy was observed when NT-3 was combined with an otherwise ineffective level of BDNF in our regrowth assay. NT-3 has been shown to interact synergistically with NGF in a chemotropism assay [25], and with BDNF in regulating survival of postnatal spiral ganglion neurons [26]. The synergistic effect on regrowth density could reflect enhanced neuronal survival or enhanced neuritogenesis. NT-3 can signal through TrkB or TrkA receptors [27] as well as TrkC. In some systems, NT-3 can substitute for BDNF, but NT-3 does not rescue BDNF-deprived geniculate neurons from death nor does it support innervation of lingual taste buds [28]. It will be important to determine if NT-3 and BDNF interact synergistically throughout development in light of the juxtaposition of BDNF mRNA in taste buds and NT-3 mRNA in surrounding nongustatory epithelium of the tongue [29] at the time of arrival by CT neurites [30]. Given that BDNF and NT-4 stimulate different outgrowth morphologies [1], it will be interesting to see if NT-3 and NT-4 interact synergistically as well.
Among the receptor mRNAs that were not differentially expressed in the PA/CT and GSP, the most abundantly expressed were those encoding TrkB, TrkB.T2, and NCAM-140, and Gfra1, all of which were expressed in 57–100% of the mouse neurons tested. NT-4, GDNF, and NT-3 + BDNF did not induce significantly different outgrowth densities between the rat PA/CT and GSP, and BDNF induced significantly (albeit slightly) more GSP neurite outgrowth. We anticipated that PA/CT neurite regrowth would be more robust than GSP regrowth because there are more PA/CT than GSP neurons in the adult [23], and the majority (60–75%) of neurons have been born at the time the labeling commences [31]. Given that at least three ligands induced comparable outgrowth from GSP and PA/CT neurons, it may be that comparable numbers of GSP and PA/CT neurons are being labeled, e.g., if similar numbers of PA/CT and GSP neurons have extended axons to the site of dye application. However, it is also possible that there are fewer GSP axons labeled but GSP neurons are heartier than PA/CT neurons. In light of the latter possibility, Neurturin's differential influence on GSP and PA/CT regrowth density must be interpreted cautiously.
There was only a weak correlation between the percentage of mouse neurons that expressed mRNA encoding a particular receptor and the density of regrowth elicited by the ligand for that receptor from rat geniculate ganglia. Ligands for TrkB promoted significantly more outgrowth than ligands of receptors that had mRNA expression frequencies of 50% or less. However there are equally strong negative correlations. Gfra2 and Gfra1 mRNA were both present in over 60% of the mouse GSP neurons, but Neurturin induced significantly less regrowth in rat GSP neurons than did GDNF. Approximately half of the neurons were TrkA or TrkC mRNA-positive but neither NT-3 nor NGF induced substantial regrowth, nor did they promote growth from unlabeled geniculate ganglia cultured in collagen gels [1]. NGF also fails to stimulate neurite outgrowth from TrkA expressing trigeminal neurons at early embryonic stages [32]. This could result from lack of neuritogenic signaling or poor survival of those neurons.
An especially appealing aspect of the single cell approach is the prospect for detecting subtypes of neurons based on their coexpression profiles. We were surprised to find that coexpression of each receptor pair occurred at approximately the rate predicted if coexpression occurred randomly. Thus, we found no evidence that any pairs of receptors form a combinatorial code [33] that is relevant to divergence or that could distinguish PA and CT neurons. Randomness of coexpression frequencies could signify that coexpression is not functionally significant, and for that matter, that the receptors or their downstream signaling systems are not functional at this stage, consistent with the lack of correlation between receptor expression and outgrowth described above. It may also indicate that expression is functionally redundant. At present there is evidence that cells that express multiple GFRα receptors respond similarly to different GDNF ligands [16], but in some systems GFRα's in the same cell regulate distinct functions [34].
Although the expression of most Trk and Gfra mRNAs has not been reported previously for early-stage embryonic mouse GSP and PA/CT neurons, our observations are in general agreement with most previous work in rodents using other techniques, e.g., in situ hybridization or genetic mutants [35–37]. Our results are in partial agreement with those of Farbman et al. [6], who used the antisense RNA amplification to assess neurotrophin receptor expression in rat geniculate neurons that had been immunostained for TrkB or TrkC. In both studies all embryonic neurons (rat E13, mouse E11) expressed TrkB isoform mRNAs. However, in the rat study only one third of the neurons expressed both the full-length and truncated isoforms whereas we found that all mouse geniculate neurons expressed both receptor mRNAs. Second, in the study on E13 rat neurons, neither TrkC nor TrkA mRNA were detected whereas approximately half of the mouse neurons were positive for these Trk mRNAs. Differences in TrkA and TrkC mRNA expression could result from the different approaches – the cells that stain most clearly for TrkB may represent a subpopulation that tends not to express the other Trk receptor mRNAs. It is also possible that it is more difficult to detect TrkA and TrkC mRNA in rat than it is in mouse. TrkC mRNA was not detected in rat geniculate neurons that stained positively for TrkC protein, consistent with this notion [but see ref. 6]. Finally, it is also possible that mouse and rat neurons differ in the neurotrophic factor receptor mRNAs that they express. We note that Farbman et al. [6] did not detect p75 in any E13 rat geniculate ganglion neuron. Our observations on neurite outgrowth by regenerating rat geniculate ganglion neurons are consistent with the receptor profiles of both studies.
Coupling expression profiling with a differential regrowth assay identified signaling systems that may be relevant to the divergence of the GSP and PA/CT. Three neurotrophic factor receptor mRNAs, Gfra2, and TrkC and TrkA, were differentially expressed in populations of neurons with divergent axons, and the remaining mRNAs were not differentially expressed. Neurturin stimulated no regrowth of PA/CT neurites, but significant (albeit low) regrowth of GSP neurites. If Neurturin were more concentrated in the path of the GSP than in the path of the PA/CT, GSP growth may be promoted more along the GSP path. However, Neurturin promoted significantly less regrowth than other neurotrophic factors expressed in the branchial arches, so it may not have a prominent role in controlling nerve growth. Although our data implicated only one signaling system as potentially contributing to divergence, the finding that the other factors are not likely to have a role in divergence is also useful, as they can be used to promote GSP and PA/CT outgrowth in future tests of candidate divergence factors. We also uncovered a synergistic interaction between BDNF and NT-3. Finally, the expression profiles of the receptor mRNAs do not support a the presence of nerve-specific, functional, or developmental stage-specific subtypes at this stage, so further studies are required to identify the molecular underpinnings of the differences between these subtypes.
Acknowledgments
We thank Dr. Al Farbman and Mitra Bhattacharyya for technical advice on molecular biology methods. We are grateful to Dr. Martin Berg for sharing his expertise in statistics. This work was supported by a research grant from the NIDCD, 5 R21 DC05253-02 to MWR.
References
- 1.Rochlin MW, O'Connor R, Giger RJ, Verhaa gen J, Farbman AI. Comparison of neuro-trophin and repellent sensitivities of early embryonic geniculate and trigeminal axons. J Comp Neurol. 2000;422:579–593. [PubMed] [Google Scholar]
- 2.Markus A, Patel TD, Snider WD. Neurotrophic factors and axonal growth. Curr Opin Neurobiol. 2002;12:523–531. doi: 10.1016/s0959-4388(02)00372-0. [DOI] [PubMed] [Google Scholar]
- 3.Phillips J, Eberwine JH. Antisense RNA amplification: A linear amplification method for analyzing the MRNA population from single living cells. Methods. 1996;10:283–288. doi: 10.1006/meth.1996.0104. [DOI] [PubMed] [Google Scholar]
- 4.Theiler K. The House Mouse: Development and Normal Stages from Fertilization to 4 Weeks of Age. Berlin: Springer; 1972. [Google Scholar]
- 5.Singleton CD, Casagrande VA. A reliable and sensitive method for fluorescent photoconversion. J Neurosci Methods. 1996;64:47–54. doi: 10.1016/0165-0270(95)00097-6. [DOI] [PubMed] [Google Scholar]
- 6.Farbman AI, Brann JH, Rozenblat A, Rochlin MW, Weiler E, Bhattacharyya M. Developmental expression of neurotrophin receptor genes in rat geniculate ganglion neurons. J Neurocytol. 2004;33:331–343. doi: 10.1023/B:NEUR.0000044194.71426.ee. [DOI] [PubMed] [Google Scholar]
- 7.Eberwine J, Crino P. Analysis of MRNA populations from single live and fixed cells of the central nervous system. Curr Protocols Neurosci. 1997;5:3.1.5–3.1.15. doi: 10.1002/0471142301.ns0503s00. [DOI] [PubMed] [Google Scholar]
- 8.Charron F, Stein E, Jeong J, McMahon AP, Tessier-Lavigne M. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell. 2003;113:11–23. doi: 10.1016/s0092-8674(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 9.Rochlin MW, Itoh K, Adelstein RS, Bridgman PC. Localization of myosin II A and B isoforms in cultured neurons. J Cell Sci. 1995;108:3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- 10.Eide FF, Vining ER, Eide BL, Zang K, Wang XY, Reichardt LF. Naturally occurring truncated TrkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J Neurosci. 1996;16:3123–3129. doi: 10.1523/JNEUROSCI.16-10-03123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck KD, Lamballe F, Klein R, Barbacid M, Schauwecker PE, McNeill TH, Finch CE, Hefti F, Day JR. Induction of noncatalytic TrkB neurotrophin receptors during axonal sprouting in the adult hippocampus. J Neurosci. 1993;13:4001–4014. doi: 10.1523/JNEUROSCI.13-09-04001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxter GT, Radeke MJ, Kuo RC, Makrides V, Hinkle B, Hoang R, Medina-Selby A, Coit D, Valenzuela P, Feinstein SC. Signal transduction mediated by the truncated TrkB receptor isoforms, TrkB. T1 and TrkB. T2. J Neurosci. 1997;17:2683–2690. doi: 10.1523/JNEUROSCI.17-08-02683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritzsch B, Sarai PA, Barbacid M, Silos-Santiago I. Mice with a targeted disruption of the neurotrophin receptor TrkB lose their gustatory ganglion cells early but do develop taste buds. Int J Dev Neurosci. 1997;15:563–576. doi: 10.1016/s0736-5748(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 14.Tessarollo L, Tsoulfas P, Donovan MJ, Palko ME, Blair-Flynn J, Hempstead BL, Parada LF. Targeted deletion of all isoforms of the TrkC gene suggests the use of alternate receptors by its ligand neurotrophin-3 in neuronal development and implicates TrkC in normal cardiogenesis. Proc Natl Acad Sci USA. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl M, Timmusk T, Rossi J, Saarma M, Airaksinen MS. Expression and alternative splicing of mouse Gfra4 suggest roles in endocrine cell development. Mol Cell Neurosci. 2000;15:522–533. doi: 10.1006/mcne.2000.0845. [DOI] [PubMed] [Google Scholar]
- 16.Airaksinen MS, Saarma M. The GDNF family: Signalling, biological functions and therapeutic value (review) Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 17.Paratcha G, Ledda F, Ibanez CF. The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell. 2003;113:867–879. doi: 10.1016/s0092-8674(03)00435-5. See Comment. [DOI] [PubMed] [Google Scholar]
- 18.Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- 19.Panicker AK, Buhusi M, Thelen K, Maness PF. Cellular signalling mechanisms of neural cell adhesion molecules (review) Front Biosci. 2003;8:d900–d911. doi: 10.2741/1014. [DOI] [PubMed] [Google Scholar]
- 20.Pollerberg GE, Burridge K, Krebs KE, Goodman SR, Schachner M. The 180-kD component of the neural cell adhesion molecule NCAM is involved in cell-cell contacts and cytoskeleton-membrane interactions. Cell Tissue Res. 1987;250:227–236. doi: 10.1007/BF00214676. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Hastie T, Whitfield ML, Borresen-Dale AL, Jeffrey SS. Optimization and Evaluation of T7 Based RNA Linear Amplification Protocols for CDNA Microarray Analysis. BMC Genomics. 2002;3:31. doi: 10.1186/1471-2164-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker KL, Meyer M, Barde YA. Neurotrophins are required for nerve growth during development. Nat Neurosci. 2001;4:29–37. doi: 10.1038/82868. [DOI] [PubMed] [Google Scholar]
- 23.Miller IJ, Gomez MM, Lubarsky EH. Distribution of the facial nerve to taste receptors in the rat. Chem Senses. 1978;3:397–410. [Google Scholar]
- 24.O'Connor R, Tessier-Lavigne M. Identification of maxillary factor, a maxillary process-derived chemoattractant for developing trigeminal sensory axons. Neuron. 1999;24:165–178. doi: 10.1016/s0896-6273(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 25.Cao X, Shoichet MS. Investigating the synergistic effect of combined neurotrophic factor concentration gradients to guide axonal growth. Neuroscience. 2003;122:381–389. doi: 10.1016/j.neuroscience.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Mou K, Hunsberger CL, Cleary JM, Davis RL. Synergistic effects of BDNF and NT-3 on postnatal spiral ganglion neurons. J Comp Neurol. 1997;386:529–539. [PubMed] [Google Scholar]
- 27.Huang EJ, Reichardt LF. Trk receptors: Roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 28.Agerman K, Hjerling-Leffler J, Blanchard MP, Scarfone E, Canlon B, Nosrat C, Ernfors P. BDNF gene replacement reveals multiple mechanisms for establishing neurotrophin specificity during sensory nervous system development. Development. 2003;130:1479–1491. doi: 10.1242/dev.00378. [DOI] [PubMed] [Google Scholar]
- 29.Nosrat CA, Olson L. Brain-derived neurotrophic factor MRNA is expressed in the developing taste bud-bearing tongue papillae of rat. J Comp Neurol. 1995;360:698–704. doi: 10.1002/cne.903600413. [DOI] [PubMed] [Google Scholar]
- 30.Farbman AI, Mbiene JP. Early development and ignalingns of taste bud-bearing papillae on the rat tongue. J Comp Neurol. 1991;304:172–186. doi: 10.1002/cne.903040203. [DOI] [PubMed] [Google Scholar]
- 31.Altman J, Bayer SA. Development of the cranial nerve ganglia and related nuclei in the rat. Adv Anat Embryol Cell Biol. 1982;74:1–90. doi: 10.1007/978-3-642-68479-1. [DOI] [PubMed] [Google Scholar]
- 32.Arumae U, Pirvola U, Palgi J, Kiema TR, Palm K, Moshnyakov M, Ylikoski J, Saarma M. Neurotrophins and their receptors in rat peripheral trigeminal system during maxillary nerve growth. J Cell Biol. 1993;122:1053–1065. doi: 10.1083/jcb.122.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- 34.Stucky CL, Rossi J, Airaksinen MS, Lewin GR. GFR alpha2/neurturin ignaling regulates noxious heat transduction in isolectin B4-binding mouse sensory neurons. J Physiol. 2002;545:43–50. doi: 10.1113/jphysiol.2002.027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernfors P, Merlio JP, Persson H. Cells expressing mRNA for neurotrophins and their receptors during embryonic rat development. Eur J Neurosci. 1992;4:1140–1158. doi: 10.1111/j.1460-9568.1992.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 36.Golden JP, DeMaro JA, Osborne PA, Milbrandt J, Johnson EM., Jr Expression of neurturin, GDNF, and GDNF family-receptor MRNA in the developing and mature mouse. Exp Neurol. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- 37.Nosrat CA. Neurotrophic factors in the tongue: Expression patterns, biological activity, relation to innervation and studies of neurotrophin knockout mice. Ann NY Acad Sci. 1998;855:28–49. doi: 10.1111/j.1749-6632.1998.tb10544.x. [DOI] [PubMed] [Google Scholar]