Abstract
Chemotherapy is the mainstay of systemic treatment for triple negative breast cancer (TNBC); however, the development of drug resistance limits its effectiveness. Therefore, we investigated the underlying mechanism for drug resistance and potential approaches to overcome it for a more effective treatment for TNBCs. Using a pulse-stimulated selection strategy to mimic chemotherapy administration in the clinic, we developed a new paclitaxel-resistant MDA-MB-231 cell line and analyzed these cells for changes in autophagy activity, and the role and mechanisms of the increased autophagy in promoting drug resistance were determined. We found that the pulse-stimulated selection strategy with paclitaxel resulted in MDA-MB-231 variant cells with enhanced resistance to paclitaxel. These resistant cells were found to have enhanced basal autophagy activity, which confers a cytoprotective function under paclitaxel treatment stress. Inhibition of autophagy enhanced paclitaxel-induced cell death in these paclitaxel-resistant cells. We further revealed that up-regulated autophagy in resistant cells enhanced the clearance of damaged mitochondria. Last, we showed that the paclitaxel-resistant cancer cells acquired cross resistance to epirubicin and cisplatin. Together, these results suggest that combining autophagy inhibition with chemotherapy may be an effective strategy to improve treatment outcome in paclitaxel-resistant TNBC patients.
Keywords: Breast cancer, Autophagy, Drug resistance
Introduction
Breast cancer is one of the most common cancers in women worldwide [1–4]. With encouraging developments in systemic therapies such as chemotherapy, hormone therapy, and targeted therapy, the outcomes of breast cancer patients have improved significantly [5]. Triple-negative breast cancer (TNBC) is a subgroup of breast cancers that lacks the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) [6, 7]. This subgroup of breast cancer responds poorly to hormone therapy and HER2-targeted therapy. Therefore, chemotherapy is still the mainstay of systemic treatment for TNBC [6]. Paclitaxel is a mitotic inhibitor-based chemotherapeutic drug [8, 9], due to its high efficiency, and is widely used to treat TNBC. Unfortunately, drug resistance is still a major hurdle in the use of paclitaxel and limits its effectiveness. Accordingly, it is crucial to understand the mechanisms that drive the resistance of TNBC cancer cells to paclitaxel, in order to develop strategies that can limit resistance and improve the outcomes of TNBC patients.
Autophagy is an essential cellular process that can clear and recycle damaged cellular organelles and components and provide macromolecules in times of scarce nutrient availability [10–12]. In the autophagic process, cytoplasmic regions are sequestrated within double-membrane vesicles known as autophagosomes and delivered to lysosomes for degradation [13, 14]. Recently, the functions of autophagy have been widely studied in a variety of human diseases, including cancer [15]. Increasing evidence indicates a cytoprotective role of autophagy during breast cancer development and anticancer therapy. Dysfunctional autophagy has been shown to impair breast cancer tumorigenesis and growth [16, 17]. It was also reported that autophagy inhibition synergized with chemotherapy drug epirubicin to induce apoptosis of breast cancer cells selected for resistance to the drug by a stepwise treatment strategy [18, 19]. Nevertheless, the underlying mechanisms for autophagy inhibition to augment the anticancer effect of epirubicin are not well understood. Also importantly, the more commonly used stepwise selection used in these previous studies might not be the best option for studying the development of chemotherapy resistance [20]. This is partly because in the clinic, patients receive several cycles of treatment with chemotherapy at the same concentration each time, rather than in increasing amounts as the stepwise selection strategy.
In this study, we used a pulse stimulated strategy to mimic the clinical effects of chemotherapy to generate triple negative MDA-MB-231 cells that are resistant to paclitaxel relative to parental cells. We showed that the increased basal autophagy and ability to remove damaged mitochondria in these cells contributed to their resistance and that genetic inhibition of autophagy re-sensitized the resistant MDA-MB-231 cells to paclitaxel. These findings provide significant new insights into the mechanism of TNBC resistance to paclitaxel and suggest that combining autophagy inhibition with chemotherapy may enhance the effectiveness of treatment in paclitaxel-resistant TNBCs.
Results
MDA-MB-231 cells acquired resistance to paclitaxel after pulse-stimulated selection
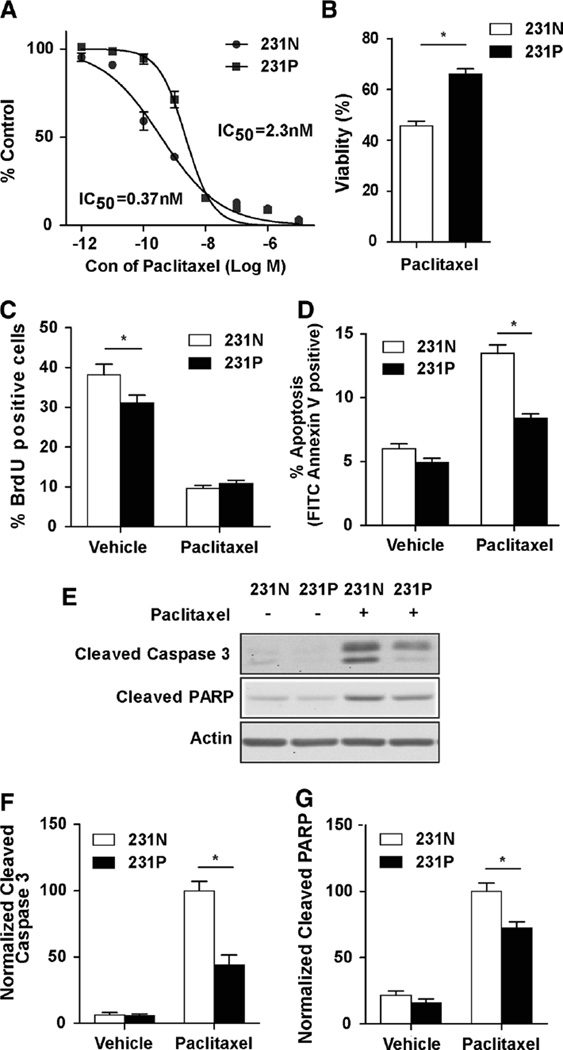
To study the mechanism of TNBC resistance to paclitaxel treatment, we performed a pulse-stimulated selection with fixed doses of paclitaxel to generate paclitaxel-resistant MDA-MB-231 cells. To mimic chemotherapy administration in the clinic, MDA-MB-231 cells were treated with only 10 cycles of paclitaxel treatment. The resulting cells are termed MDA-MB-231-P cells (231P cells), and their acquired resistance was tested by SRB assay to determine the IC50 relative to parental MDA-MB-231 cells (231N cells). 231P cells showed about six times higher IC50 compared with 231N cells (Fig. 1a). Next, cell viabilities were tested by trypan blue viability assay after both 231N and 231P cells were treated with 25 nM paclitaxel for 24 h. 231P cells showed significantly increased viability when compared with the parental cells (Fig. 1b). Apart from the increased viability in the 231P cells, we also investigated whether there was a difference in the growth inhibitory effect by paclitaxel between 231P and 231N cells. We found that 231P cells had slightly lower proliferation than 231N cells without treatment with paclitaxel, and paclitaxel treatment significantly reduced proliferation of both cells to comparable levels (Fig. 1c), suggesting that the increased paclitaxel resistance of 231P cells was not caused by altered proliferation.
Fig. 1.
Development of paclitaxel-resistant MDA-MB-231 cells with pulse stimulated strategy. a Dose-dependent growth impairment of 231N and 231P cells was identified by sulphorhodamine B assay. The cells were treated in six replicates with pacltaxel for 96 h. b Cell viabilities after 24 h 25 nM paclitaxel treatment compared with vehicle in 231N and 231P cells by trypan blue viability assay (Mean ± SE are shown, n = 3). *p < 0.05. c Cell proliferations after 24 h 25 nM paclitaxel or vehicle treatment in 231N and 231P cells by BrdU incorporation assay (Mean ± SE are shown, n = 3). *p < 0.05. d Cell apoptosis after treatment with 25 nM paclitaxel for 24 h was compared with vehicle in 231N and 231P cells by Annexin V assay (Mean ± SE are shown, n = 3). *p < 0.05. e–g expression of proapoptotic markers were analyzed after 24 h 25 nM paclitaxel treatment compared with vehicle in 231N and 231P cells by Western blot. Representative results are shown in (e). Intensity of the protein bands were determined from three independent experiments by densitometry. Mean ± SE of the relative protein level (normalized to actin) are shown in (f) and (g). *p < 0.05
Paclitaxel acts as a mitotic inhibitor, inducing mitotic arrest and triggering cell apoptosis [21]. Therefore, we next tested the apoptotic levels in 231N and 231P cells after paclitaxel treatment by annexin V staining followed by flow cytometric analysis (Fig. 1d). We found that paclitaxel treatment induced apoptosis in 231P cells at a reduced level compared to 231N cells. The reduced extent of apoptosis induction by paclitaxel in 231P cells was further confirmed by Western blot analysis of cleaved caspase 3 and cleaved poly(ADP-ribose) polymerase (PARP) in these cells (Figs. 1e–g). Collectively, these results indicate that 231P cells have developed resistance to paclitaxel by reducing treatment-triggered apoptosis.
Basal autophagy is enhanced after cycles of paclitaxel treatment in MDA-MB-231 cells
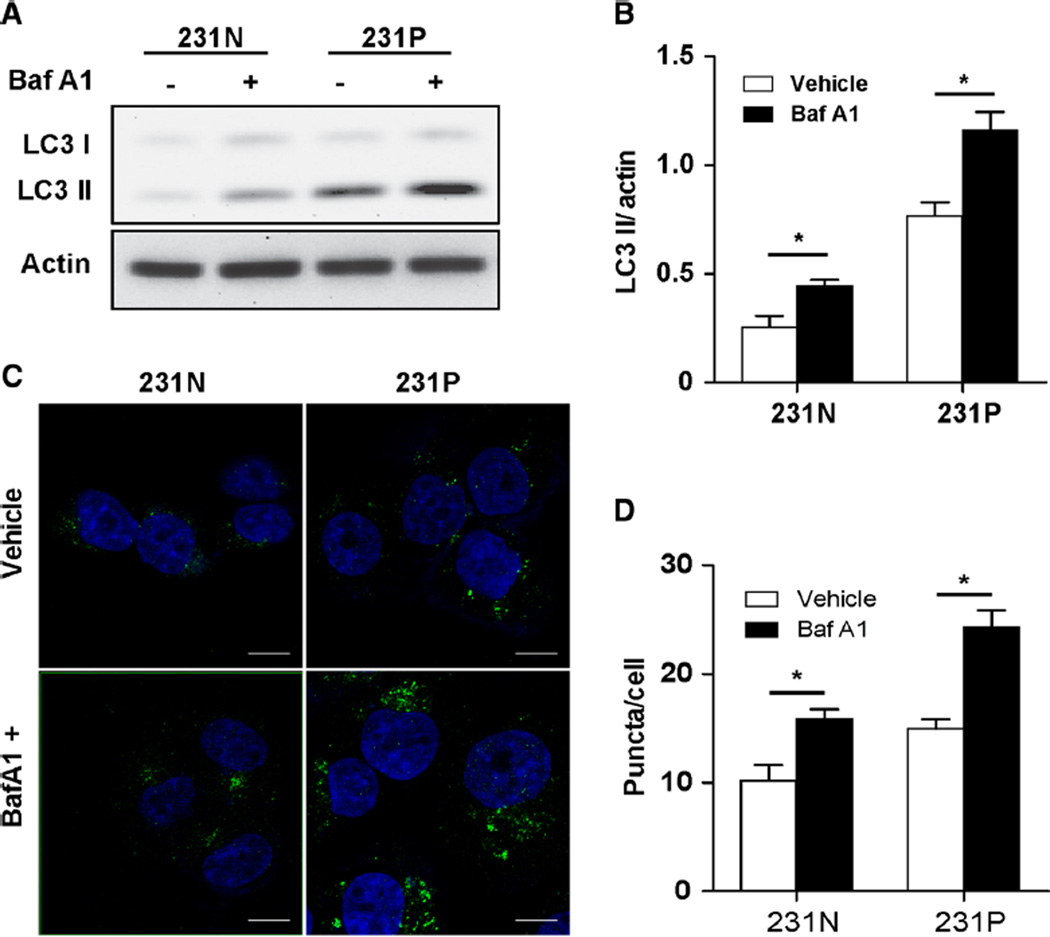
A previous report suggested increased levels of autophagy in chemotherapy-treated breast cancer patient samples [22]. To determine whether our pulse stimulation of paclitaxel affected autophagy, we compared basal autophagy levels of 231N and 231P cells. Western blot analysis showed that the autophagy marker protein LC3 II levels were increased in 231P cells compared with 231N cells (Fig. 2a). After treatment with autophagy inhibitor bafilomycin A1, we observed increased LC3 II levels in both 231N and 231P cells, indicating that autophagy flux was not blocked. Quantitation of LC3 II/actin showed a greater increase of LC3 II in 231P cells compared with 231N cells after bafilomycin A1 treatment, indicating that 231P cells had a higher basal autophagy level (Fig. 2b). To confirm this observation, LC3 II puncta in both 231N and 231P cells were analyzed by immunofluorescence (Fig. 2c). We observed increased numbers of LC3 II puncta in 231P cells in the absence of bafilomycin A1 treatment. After bafilomycin A1 treatment, 231P cells showed a greater increase of LC3 II puncta compared to 231N cells (Fig. 2d), which is in agreement with Western blot analysis. Together, these results suggest that the basal autophagy level was increased in 231P cells compared to parental 231N cells, after being subjected to paclitaxel pulse-stimulation treatment.
Fig. 2.
MDA-MB-231 paclitaxel-resistant cells show up-regulated basal autophagy. a LC3 II levels were evaluated by Western blot. 231N and 231P cells were cultured under normal condition treated with vehicle (−), 200 nM bafilomycin A1 (+) for 2 h. b Based on above Western blot results, normalized LC3 II was evaluated by densitometry quantitation of LC3 II/actin in the presence of bafilomycin A1 compared with vehicle in 231N and 231P cells (Mean ± SE are shown, n = 3). *p < 0.05. c Cells were treated with vehicle, 200 nM bafilomycin A1 for 2 h and analyzed by immunofluorescence using LC3B antibody and DAPI to stain nuclei. Scale bars = 10 µm. d Autophagy level (measured by green puncta) was quantified (Mean ± SE are shown, n = 3). *p < 0.05
Up-regulated basal autophagy confers a cytoprotective function under paclitaxel stress
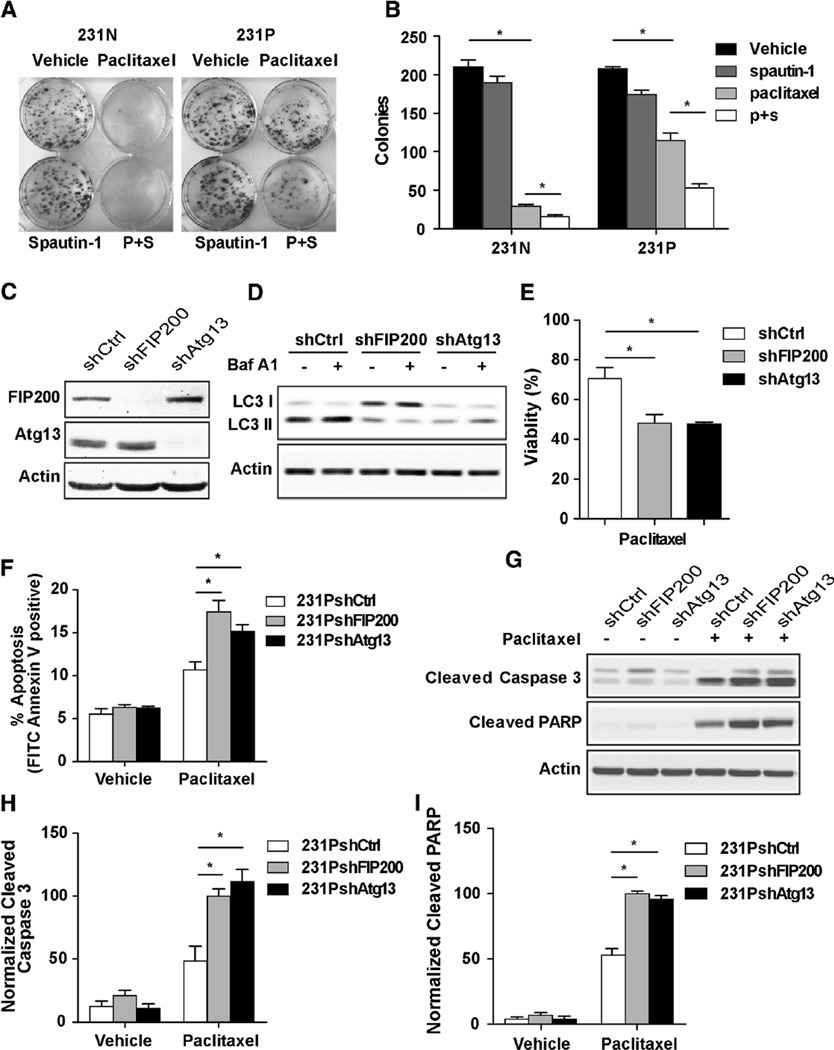
To determine whether the up-regulated basal autophagy plays a cytoprotective role and facilitates the resistance to paclitaxel in 231P cells, we examined the effect of autophagy inhibition on these cells in response to paclitaxel. Spautin-1 is a recently described autophagy inhibitor which acts through promoting the degradation of Vps34 required for autophagy initiation [23]. We performed clonogenic assays for both 231N and 231P cells with paclitaxel treatment in the absence or presence of spautin-1. We found that while paclitaxel treatment suppressed colony formation in both 231N and 231P cells, the 231P cells showed only a moderate decrease, consistent with their resistance to the drug (Figs. 3a and b). Treatment with spautin-1 alone had little effect on colony formation of either 231N or 231P cells. However, the presence of spautin-1 significantly reduced the remaining colony formation ability of 231P cells in the presence of paclitaxel, suggesting that the increased autophagy in these cells is at least partially responsible for their resistance to paclitaxel.
Fig. 3.
Up-regulated basal autophagy plays a cytoprotective function under paclitaxel stress. (a, b) 10 days clonogenic assays for 231N and 231P cells. Cells were plated in triplicate and treated with vehicle or 10 µM spautin-1 for 24 h. 25 nM paclitaxel were added into the medium for the last 4 h. Medium were replaced with fresh complete medium without drugs and cultured. Representative results are shown in (a). Colony numbers were counted and Mean ± SE of the numbers from three experiments are shown in (b). *p < 0.05. c Expressions of FIP200 and Atg13 were analyzed by Western blot in 231PshCtrl, 231PshFIP200 and 231PshAtg13 cells under normal culture condition. d LC3 II level was evaluated by Western blot. 231PshCtrl, 231PshFIP200 and 231PshAtg13 cells were cultured under normal condition treated with vehicle, 200 nM bafilomycin A1 for 2 h. e Cell viabilities after 24 h 25 nM paclitaxel treatment compared with vehicle in 231PshCtrl, 231PshFIP200 and 231PshAtg13 cells by trypan blue viability assay (Mean ± SE are shown, n = 3). *p < 0.05. f Cell apoptosis after 24 h 25 nM paclitaxel treatment compared with vehicle in 231N and 231P cells by Annexin V assay (Mean ± SE are shown, n = 3). *p < 0.05. g–i Expressions of proapoptotic markers were analyzed after 24 h 25 nM paclitaxel treatment compared with vehicle in 231PshCtrl, 231PshFIP200 and 231PshAtg13 cells by Western blot. Representative results are shown in (g). Intensity of the protein bands were determined from three independent experiments by densitometry. Mean ± SE of the relative protein level (normalized to actin) are shown in (h and i). *p < 0.05
Next, we further investigated the role of the increased autophagy of 231P cells using genetic inhibition of autophagy by shRNA against autophagy genes FIP200 and Atg13. FIP200 and Atg13 form a complex with ULK1, which is essential for the initiation of the autophagy process [13]. As shown in Fig. 3c, treatment of the cells with shRNA against FIP200 or Atg13 reduced the expression of the corresponding proteins efficiently and specifically. As expected, shRNA knockdown of FIP200 or Atg13 significantly decreased autophagy in these cells, as evidenced by the decreased LC3 II levels compared to that in cells treated with control shRNA (Fig. 3d). Consistent with results using pharmacological inhibitors, autophagy inhibition by FIP200 or Atg13 knockdown significantly decreased viability of 231P cells under paclitaxel treatment (Fig. 3e). To further validate the cytoprotective effect of autophagy in 231P cells, paclitaxel-induced apoptosis was assessed. Annexin V assay shows significantly higher levels of apoptosis in 231P cells with FIP200 or Atg13 knockdown compared to that with control shRNA (Fig. 3f). Western blot analysis further confirmed these findings, showing increased amount of cleaved caspase 3 and PARP in 231P cells with FIP200 or Atg13 knockdown (Figs. 3g–i). These results indicate that up-regulated basal autophagy in 231P cells plays a cytoprotective function under paclitaxel stress. They also indicate that autophagy inhibition can attenuate paclitaxel resistance by enhancing paclitaxel-induced apoptosis in 231P cells.
Up-regulated basal autophagy enhances the clearance of dysfunctional mitochondria in paclitaxel-resistant cancer cells
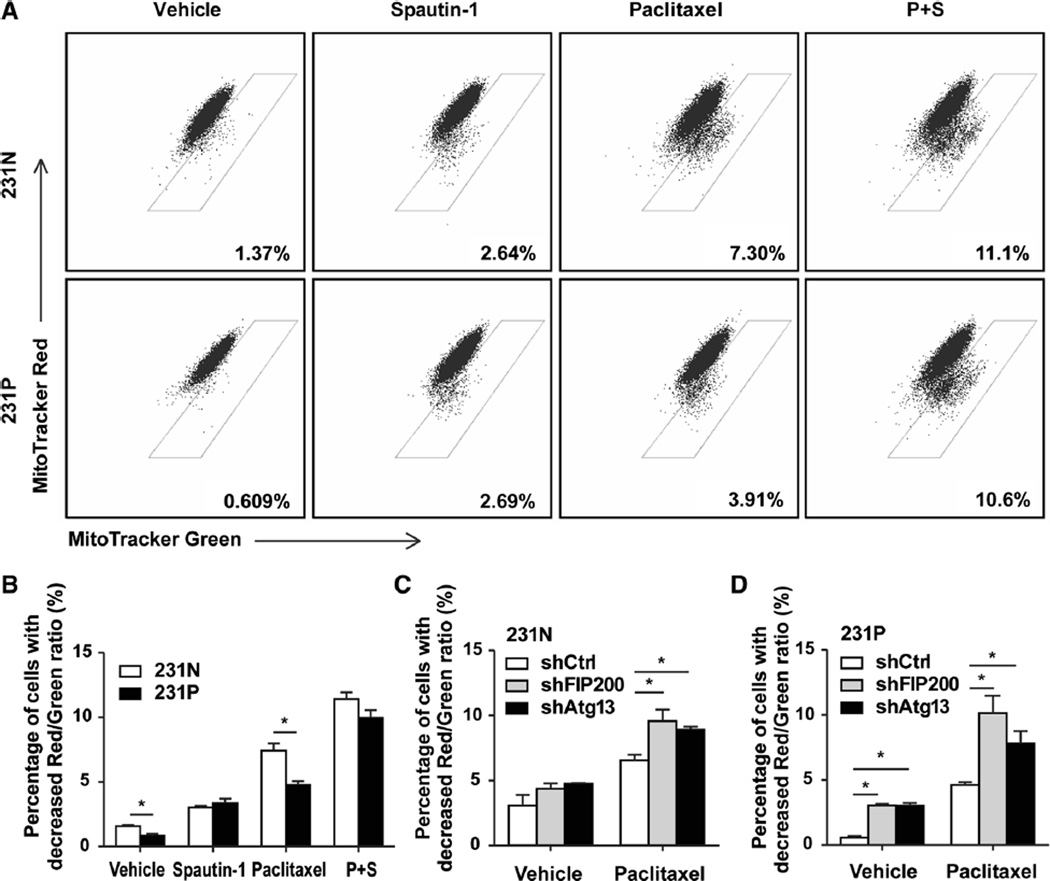
Having established that up-regulated autophagy reduces apoptosis in 231P cells treated with paclitaxel, we sought to delineate the mechanism by which autophagy plays a cytoprotective function in 231P cells. It is well established that multiple apoptotic pathways can be activated by damaged mitochondria [24, 25]. Autophagy is responsible for the clearance of damaged mitochondria [26]. Our previous research has shown that inhibition of autophagy by FIP200 ablation causes accumulation of mitochondria with abnormal morphology [16]. We posited that up-regulated autophagy in 231P cells contributes to the enhanced clearance of damaged mitochondria, protecting 231P cells from apoptosis induced by damaged mitochondria. To test this hypothesis, we used MitoTracker Green FX and Mito-Tracker Red CMXRos to analyze mitochondrial function in 231N and 231P cells. MitoTracker Green FX staining represents the total mitochondrial mass, while MitoTracker Red CMXRos indicate the amount of respiring mitochondria [27]. Therefore, a 1:1 ratio of labeling for these two indicates that all mitochondria are functional in a cell, whereas a reduced value for MitoTracker Red CMXRos staining compared to MitoTracker Green FX staining indicates cells with damaged mitochondria [28]. Flow cytometry analyses showed that 231P cells have a lower level of damaged mitochondria (i.e., fraction of cells below the 1:1 diagonal line for labeling of the two trackers) compared with 231N cells (Figs. 4a and b, vehicle treated cells), which is consistent with their increased basal autophagy activity. Treatment with the autophagy inhibitor spautin-1 increased mitochondrial damage to comparably higher levels for both 231N and 231P cells, providing further support that the reduced level of damaged mitochondria in 231P cells was due to their increased autophagy.
Fig. 4.
Up-regulated basal autophagy enhances the clearance of dysfunctional mitochondria in paclitaxel-resistant cancer cells. a Mitochondrial functions of 231N and 231P cells were analyzed by flow cytometry, stained with 25 nM Mitotracker Green and 25 nM Mitotracker Red. 231N and 231P cells were treated with vehicle, 25 nM paclitaxel, 10 µM spautin-1, or the combination for 24 h. b Mitochondria damage results were evaluated based on flow cytometry results above (Mean ± SE are shown, n = 3). *p < 0.05. c Mitochondrial damage results were evaluated based on mitochondrial function analysis by flow cytomety for 231NshCtrl, 231NshFIP200 and 231NshAtg13 cells. Cells were treated with vehicle, 25nM paclitaxel, 10 µM spautin-1, or the combination for 24 h (Mean ± SE are shown, n = 3). *p < 0.05. d Mitochondrial damage results were evaluated based on mitochondrial function analysis by flow cytomety for 231PshCtrl, 231PshFIP200 and 231PshAtg13 cells. Cells were treated with vehicle, 25 nM paclitaxel, 10 µM spautin-1, or the combination for 24 h (Mean ± SE are shown, n = 3). *p < 0.05
As expected, treatment of the cells with paclitaxel increased damaged mitochondria in both cells, but 231P cells still exhibited a lower level compared to that in 231N cells. Similar to vehicle controls, addition of spautin-1 increased the damaged mitochondria to higher and comparable levels for these two cells. The results from pharmacological inhibition of autophagy with spautin-1 were corroborated with genetic inhibition of autophagy-related genes. After knockdown of FIP200 or Atg13, mitochondrial damage was increased in both 231N and 231P cells after treatment with vehicle or paclitaxel (Figs. 4c and d). These results are consistent with that enhanced basal autophagy improved the ability of 231P cells to maintain mitochondrial homeostasis by increasing the clearance of damaged mitochondria.
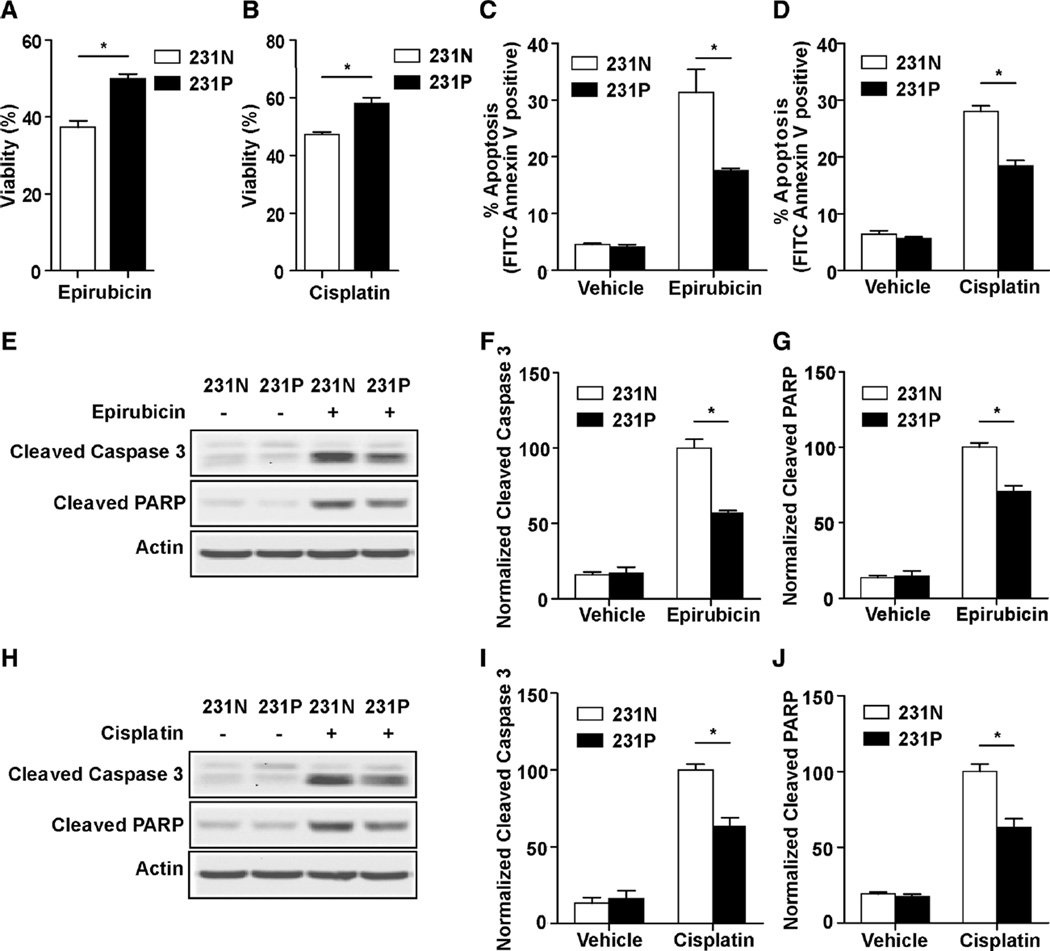
Paclitaxel-resistant cancer cells show cross resistance to epirubicin and cisplatin
Besides the increased resistance to paclitaxel, we investigated whether up-regulated autophagy in 231P cells also facilitates cross resistance to other chemotherapy drugs. Epirubicin and cisplatin are two drugs which are also widely used in breast cancer chemotherapy. When 231N and 231P cells were treated with epirubicin or cisplatin for 24 h, 231P cells show significantly increased viabilities compared to 231 N cells (Figs. 5a and b). Annexin V assay showed a significant decrease in apoptotic events in 231P cells with epirubicin or cisplatin treatment (Figs. 5c and d). Additionally, Western blot analysis showed decreased levels of cleaved caspase 3 and PARP in 231P cells compared with 231N cells after treatment with epirubicin (Figs. 5e–g) or cisplatin (Figs. 5h–j). These results suggest that paclitaxel-resistant 231P cells also develop a level of resistance to other chemotherapy drugs.
Fig. 5.
231P cells show cross-resistance to epirubicin and cisplatin. a and b Cell viabilities after 24 h of 25 µM epirubicin (a) or 5 µM cisplatin (b) treatment compared with vehicle in 231N and 231P cells by Trypan blue viability assay (Mean ± SE are shown, n = 3). *p < 0.05. c and d Cell apoptosis after 24 h 25 µM epirubicin (c) or 5 µM cisplatin (d) treatment compared with vehicle in 231N and 231P cells by Annexin V assay (Mean ± SE are shown, n = 3). *p < 0.05. e–j Expression of proapoptotic markers were analyzed after 24 h of 25 µM epirubicin (e–g) or 5 µM cisplatin (h–j) treatment compared with vehicle in 231N and 231P cells by Western blot. Representative results are shown in (e and h). Intensity of the protein bands were determined from three independent experiments by densitometry. Mean ± SE of the relative protein level (normalized to actin) are shown in (f and g), and (i and j), respectively. *p < 0.05
Discussion
The role of autophagy in cancer is thought to be context dependent. Although there are still conflicting data on the role of autophagy in tumor initiation, most of the currently available evidence supports a protective role for autophagy in the survival of established tumors [29, 30]. Periods of extensive growth coupled with insufficient blood supply expose cancer cells to nutritional deprivation and hypoxic stresses, which may inhibit proliferation and cause cell death. The capability of autophagy to degrade intracellular constituents provides cells with energy and recycles damaged cellular organelles to maintain intracellular homeostasis and promote survival [31–35]. In addition to the important cytoprotective role during cancer growth, autophagy also seems to impart a survival advantage to cancer cells under therapeutic stress and to facilitate drug resistance in several types of cancer, including breast cancer [36–47]. Recent studies have suggested that breast cancer patients with higher expression of the autophagy marker LC3B have a worse outcome [48–50]. Additionally, the levels of autophagy in breast cancer tumor cells can be regulated in response to drug treatment. Elevated autophagy has been reported in drug resistant breast cancers and has been shown to contribute to acquired resistance [51, 52]. Another recent study showed higher autophagy levels in epirubicin-resistant TNBCs [19]. Here, we used a different strategy than that used in many studies, namely pulse-stimulated selection that better mimics chemotherapy in human patients, and developed a new TNBC MDA-MB-231 cell line with resistance to paclitaxel. We observed elevated basal autophagy in these cells and showed that enhanced autophagy plays a cytoprotective role during paclitaxel treatment and participates in paclitaxel resistance development. Therefore, our results provide further support that combined autophagy inhibition with paclitaxel treatment can be a more effective treatment by promoting cell death in resistant TNBCs.
Based on the encouraging research results of autophagy inhibition therapy, the autophagy inhibitor chloroquine is being studied in two clinic trials for breast cancer therapy (in the Study of the Efficacy of Chloroquine in the Treatment of Ductal Carcinoma in Situ [ClinicalTrials.gov, NCT01023477], and Chloroquine With Taxane Chemotherapy for Advanced or Metastatic Breast Cancer Patients Who Have Failed an Anthracycline [NCT01446016]). Notably, a recent study, based on a panel of breast cancer cell lines, shows that TNBC cells are more autophagy addicted and more sensitive to autophagy inhibition [53]. Although various factors contribute to chemotherapy resistance in cancer cells, these results highlight the importance of autophagy in chemo-resistant TNBC, and indicate TNBC patients may receive more benefit from autophagy inhibition therapy.
Although the mechanisms causing cytotoxicity vary among the different classes of chemotherapy drugs, in general, the cytotoxic effect perturbs cellular homeostasis and induces cancer cell death through activation of multiple apoptotic pathways. There is much evidence supporting the notion that damaged mitochondria are the most significant trigger of apoptosis [24, 25]. Anticancer drugs including paclitaxel can cause mitochondria damage, leading to permeabilization of the mitochondrial outer membrane and release of cytochrome c into the cytoplasm [54, 55]. Cytochrome c in the cytoplasm activates the caspase cascade and leads to apoptosis. Autophagy is involved in recycling of damaged cellular organelles, including mitochondria. It is the primary mechanism for removing damaged mitochondria and plays an important role in maintaining mitochondrial homeostasis in cells [56–58]. Reports show that inhibition of autophagy results in accumulation of damaged mitochondria [31–33]. These findings are consistent with our previous data, which shows inhibition of autophagy by FIP200 ablation causes accumulation of mitochondria with abnormal morphology [16]. In this study, we also observe an accumulation of damaged mitochondria following pharmacological autophagy inhibition with spautin-1 or knockdown of FIP200 and Atg13 genes in MDA-MB-231 cells. Our findings also showed that there were fewer cells with damaged mitochondria in paclitaxel-resistant MDA-MB-231 cells under both normal and paclitaxel-treated conditions compared with the parental cells. Paclitaxel in combination with autophagy inhibition can increase the number of cells with damaged mitochondria to a level that becomes comparable in both the resistant and parental cells. These results indicate that up-regulated autophagy in resistant MDA-MB-231 cells enhances the clearance of damaged mitochondria caused by the cytotoxic effect of paclitaxel, and protects cancer cells from mitochondrial-mediated apoptosis.
Next, we speculated that up-regulated autophagy in paclitaxel-resistant TNBC cells may confer them with cross resistance to other types of chemotherapy drugs. Therefore, we decided to investigate the chemotherapeutic drugs epirubicin and cisplatin. Epirubicin is an anthracycline drug which acts by intercalating with DNA, promoting complex formations, and subsequently inhibiting DNA and RNA synthesis [59]. Cisplatin is a platinum-based drug which binds with DNA to form intra-strand crosslinks and adducts, leading to conformational changes and ultimately affecting replication [60]. We observe that paclitaxel-resistant MDA-MB-231 cells also show resistance to both epirubicin and cisplatin treatment, with similarly reduced levels of apoptosis. These findings indicate that the cytoprotective function of elevated autophagy is not specific to paclitaxel resistance, but also to other chemotherapy drugs. This evidence illustrates the potential for cancer cells to develop not only resistance to the chemotherapy it was exposed to, but also cross resistance to other kinds of chemotherapy drugs with the help of enhanced autophagy. Our findings suggest that the combination of autophagy inhibition therapy with chemotherapy may help to prevent cancer cells from developing resistance.
Although further studies will be required to clarify the mechanisms that elevate the basal autophagy level in resistant cancer cells, our results provide evidence and rationale for autophagy inhibition in combination with chemotherapy drugs to treat primary or resistant TNBC.
Materials and methods
Antibodies and reagents
The following antibodies were used: cleaved caspase-3 (Cell Signaling Technology), cleaved PARP (Cell Signaling Technology), LC3B (Cell Signaling Technology), β-actin (Sigma-Aldrich), FIP200 (ProteinTech Group), Atg13 (Cell Signaling Technology), BrdU (Developmental Studies Hybridoma Bank), goat anti-rabbit FITC (Jackson ImmunoResearch Laboratories). The following reagents were used: bafilomycin A1, spautin-1, paclitaxel, epirubicin, cisplatin, puromycin, BrdU, trichloroacetic acid, sulphorhodamine B, protease inhibitor cocktails, DAPI were from Sigma-Aldrich. MitoTracker Green FX (Invitrogen) and MitoTracker Red CMXRos (Invitrogen), Permount SP15-100 Toluene Solution (Fisher Scientific), DNase I (Roche).
Cell culture and lentiviral transduction
MDA-MB-231 cells (from American Type Culture Collection) were cultured at 37 °C, 5 % CO2 in Gibco DMEM supplemented with 10 % fetal bovine serum (FBS). For the generation of resistant cell lines MDA-MB-231P, 25 nM paclitaxel was added to MDA-MB-231 culture medium for 4 h and then culturing continued with complete culture medium.MDA-MB-231 cells were left to recover to normal proliferation rates before starting the next cycle of treatment as mentioned above. After a total of 10 cycles of paclitaxel treatment, MDA-MB-231P cells were cultured normally with complete culture medium. For lentiviral infection, MDA-MB-231 cells were infected with lentivirus encoding FIP200 (TRCN 000000350426, Sigma-Aldrich) and Atg13 (TRCN 000000172801, Sigma-Aldrich) short hairpin RNA (shRNA) or scrambled sequence. Cells were then selected with 1 µg/ml puromycin.
Sulphorhodamine B assay
Drugs cytotoxicity was determined using the sulphorhodamine B (SRB) assay. Cells were plated with 3 × 103 cells/well with six replicates in 96-well plates. After overnight culture to allow cells to adhere, culture medium containing paclitaxel at different concentrations was added. One plate was tested at 0 h time point, and other plates were tested until 96 h. The cells were fixed with 10 % Trichloroacetic acid for 1 h at 4 °C and washed five times with water. 0.4 % SRB in 1 % acetic acid was added to stain for 30 min and washed five times with 1 % acetic acid and allowed to dry. 10 mM Tris-base was added to dissolve SRB and absorbance was measured with a plate reader at 530 nm. IC50 was presented as the drug concentration that killed 50 % of cells relative to the untreated control.
Trypan blue viability assay
Cells were plated in 6-well plates (15 × 104 cells/well) and incubated overnight. Drugs or vehicle were added for 24 h and the cells were counted with a Fuchs-Rosenthal Counting Chamber. Dead cells were excluded with Trypan blue.
BrdU incorporation assay
Cells were plated in 6-well plates (15 × 104 cells/well) and incubated overnight. The next day cells were treated with 25 nM paclitaxel for 24 h, and 10 µM BrdU was added for the last 4 h. Then cells were fixed for 1 h with 4 % paraformaldehyde and washed five times with PBS. Fixed cultures were treated for 30 min in 2 M HCl, then washed three time with PBS. They were then permeabilized with 0.3 % Triton X-100 in PBS for 15 min and blocked with 2 % BSA, 0.3 % Triton X-100 in PBS for 30 min, then incubated with primary antibody anti-BrdU at a dilution of 1:200 in 2 % BSA overnight at 4 °C. After washing with PBS for 15 min, the cells were incubated with goat anti-rabbit FITC secondary antibody (1:200) for 1 h at room temperature, and mounted with Permount SP15-100 toluene solution.
Western blot analysis
Cells were lysed in RIPA buffer (10 mM Tris Cl, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 20 mM Na4P2O7, 2 mM Na3VO4, 1 % Triton X-100, 10 % glycerol, 0.1 % sodium dodecyl sulfate, 0.5 % deoxycholate) supplemented with protease inhibitor cocktails. Cell lysates were separated by 10 or 12 % SDS-PAGE and transferred to nitrocellulose membrane. Different antibodies were used as described in the antibodies and reagents section above.
Immunofluorescence analysis
Cells were fixed in 4 % paraformaldehyde for 15 min, washed three times with PBS, and then permeabilized with 0.3 % Triton X-100 in PBS for 15 min. After blocking with 2 % BSA and 0.3 % Triton X-100 in PBS for 30 min, cells were incubated with primary antibody LC3B at a dilution of 1:200 in 2 % BSA overnight at 4 °C. After washing with PBS for 15 min, the cells were incubated with Goat Anti-Rabbit FITC secondary antibody (1:200) for 1 h at room temperature, and mounted with Permount SP15-100 toluene solution. LC3 II puncta images were acquired with a fixed thickness section on a Zeiss LSM710 confocal microscope with a Zeiss AxioObserver Z1 stand. The numbers of puncta per cell were counted manually.
Clonogenic assay
Cells were suspended in DMED supplemented with 10 % FBS and plated in 6-well plates (1 × 103 cells/well) and incubated overnight. They were then treated with 10 µM spautin-1 for 24 h. 25 nM paclitaxel was added into the medium for the last 4 h. Medium were replaced with fresh medium without drugs and cultured for 7–10 days. Colonies were stained with 0.1 % crystal violet and counted.
Mitochondrial function assay with flow cytometry
Cells were plated in 6-well plates (15 × 104 cells/well) and incubated overnight. 25 nM MitoTracker Green FX and 25 nM MitoTracker Red CMXRos were added and the cells were incubated for 15 min at 37 °C. Then cells were washed with DPBS three times and then suspended in Hank’s buffered salt solution (HBSS) supplemented with 2 % FBS and DNase I. DAPI was added to discriminate live from dead cells. Flow cytometry analysis was performed on BD FACSAria™ III.
Annexin V with propidium iodide assay with flow cytometry
Cells were plated in 6-well plates (15 × 104 cells/well) and incubated overnight. Drugs or vehicle was added for 24 h. Cells were harvested and stained with FITC Annexin V and propidium iodide using a FITC Annexin V/dead cell apoptosis kit according to the manufacture instructions (Invitrogen). Flow cytometry analysis was performed on BD FACSAria™ III.
Statistical analysis
For cell viability, statistical significance was evaluated by unpaired, 2-tailed Student t test using p < 0.05 as indicative of statistical significance. For IC50 analysis, nonlinear regression analysis was used. Statistical analysis was performed using GraphPad Prism 5.
Acknowledgments
This research was supported by NIH Grant CA163493 and CA150926 to J.-L. Guan.
Footnotes
Conflict of interest The authors declare no conflict of interest.
Contributor Information
Jian Wen, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA; Department of Breast Surgery, The Fourth Affiliated Hospital of China Medical University, Shenyang 110032, Liaoning, China; Department of Breast Surgery, The First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning, China.
Syn Yeo, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA.
Chenran Wang, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA.
Song Chen, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA.
Shaogang Sun, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA.
Michael A. Haas, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA
Wei Tu, Department of Breast Surgery, The Fourth Affiliated Hospital of China Medical University, Shenyang 110032, Liaoning, China.
Feng Jin, Department of Breast Surgery, The First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning, China.
Jun-Lin Guan, Email: guanjl@uc.edu, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA.
References
- 1.Zeng H, Zheng R, Zhang S, Zou X, Chen W. Female breast cancer statistics of 2010 in China: estimates based on data from 145 population-based cancer registries. J thorac dis. 2014;6(5):466–470. doi: 10.3978/j.issn.2072-1439.2014.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 4.Zagouri F, Sergentanis TN, Tsigginou A, Dimitrakakis C, Zografos GC, Dimopoulos MA, Psaltopoulou T. Female breast cancer in Europe: statistics, diagnosis and treatment modalities. J thorac dis. 2014;6(6):589–590. doi: 10.3978/j.issn.2072-1439.2014.06.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 6.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. New Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 7.Stevens KN, Vachon CM, Couch FJ. Genetic susceptibility to triple-negative breast cancer. Cancer Res. 2013;73(7):2025–2030. doi: 10.1158/0008-5472.CAN-12-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs P, Csaba G, Pallinger E, Czaker R. Effects of taxol treatment on the microtubular system and mitochondria of Tetrahymena. Cell Biol Int. 2007;31(7):724–732. doi: 10.1016/j.cellbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 11.Wirth M, Joachim J, Tooze SA. Autophagosome formation–the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23(5):301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 14.Xie ZP, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 15.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. New Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 16.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25(14):1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22(2):165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun W-L, Chen J, Wang Y-P, Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7(9):1035–1044. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 19.Chittaranjan S, Bortnik S, Dragowska WH, Xu J, Abeysundara N, Leung A, Go NE, DeVorkin L, Weppler SA, Gelmon K, et al. Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clin Cancer Res. 2014;20(12):3159–3173. doi: 10.1158/1078-0432.CCR-13-2060. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3(7):502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 21.Andre N, Braguer D, Brasseur G, Goncalves A, Lemesle-Meunier D, Guise S, Jordan MA, Briand C. Paclitaxel induces release of cytochrome c from mitochondria isolated from human neuroblastoma cells’. Cancer Res. 2000;60(19):5349–5353. [PubMed] [Google Scholar]
- 22.Chen S, Jiang YZ, Huang L, Zhou RJ, Yu KD, Liu Y, Shao ZM. The residual tumor autophagy marker LC3B serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Clin Cancer Res: Off J Am Assoc Cancer Res. 2013;19(24):6853–6862. doi: 10.1158/1078-0432.CCR-13-1617. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15(22):2922–2933. [PubMed] [Google Scholar]
- 25.Gottlieb E, Armour SM, Harris MH, Thompson CB. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003;10(6):709–717. doi: 10.1038/sj.cdd.4401231. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441(2):523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pendergrass W, Wolf N, Poot M. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry Part A. J Int Soc Anal Cytol. 2004;61(2):162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 28.Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14(5):454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 29.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res: Off J Am Assoc Cancer Res. 2011;17(4):654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155(6):1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4(8):914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72(7):1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy Sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3(11):1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Investig. 2007;117(2):326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68(5):1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 39.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3(147):ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72(1):230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 41.Levy JM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, Morgan MJ, Mirsky DM, Handler MH, Foreman NK, et al. Autophagy inhibition improves chemosensitivity in BRAFV600E brain tumors. Cancer Discov. 2014;4(7):773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25(1):23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 43.Lopez G, Torres K, Liu J, Hernandez B, Young E, Belousov R, Bolshakov S, Lazar AJ, Slopis JM, McCutcheon IE, et al. Autophagic survival in resistance to histone deacetylase inhibitors: novel strategies to treat malignant peripheral nerve sheath tumors. Cancer Res. 2011;71(1):185–196. doi: 10.1158/0008-5472.CAN-10-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen HG, Yang JC, Kung HJ, Shi XB, Tilki D, Lara PN, Jr, Devere White RW, Gao AC, Evans CP. Targeting autophagy overcomes enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33(36):4521–4530. doi: 10.1038/onc.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Donovan TR, O’Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011;7(5):509–524. doi: 10.4161/auto.7.6.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112(3):389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 47.Song J, Qu Z, Guo X, Zhao Q, Zhao X, Gao L, Sun K, Shen F, Wu M, Wei L. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5(8):1131–1144. doi: 10.4161/auto.5.8.9996. [DOI] [PubMed] [Google Scholar]
- 48.Zhao H, Yang M, Zhao J, Wang J, Zhang Y, Zhang Q. High expression of LC3B is associated with progression and poor outcome in triple-negative breast cancer. Med Oncol. 2013;30(1):475. doi: 10.1007/s12032-013-0475-1. [DOI] [PubMed] [Google Scholar]
- 49.Choi J, Jung W, Koo JS. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology. 2013;62(2):275–286. doi: 10.1111/his.12002. [DOI] [PubMed] [Google Scholar]
- 50.Ladoire S, Chaba K, Martins I, Sukkurwala AQ, Adjemian S, Michaud M, Poirier-Colame V, Andreiuolo F, Galluzzi L, White E, et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy. 2012;8(8):1175–1184. doi: 10.4161/auto.20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, Browning D, Rawson J, Smith SB, Barrett JT, et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7(9):2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 52.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One. 2009;4(7):e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP, Hansen RJ, Morgan MJ, Porter CC, Gustafson DL, et al. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res. 2014;74(9):2579–2590. doi: 10.1158/0008-5472.CAN-13-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varbiro G, Veres B, Gallyas F, Jr, Sumegi B. Direct effect of Taxol on free radical formation and mitochondrial permeability transition. Free Radic Biol Med. 2001;31(4):548–558. doi: 10.1016/s0891-5849(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 55.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 56.Namba T, Takabatake Y, Kimura T, Takahashi A, Yamamoto T, Matsuda J, Kitamura H, Niimura F, Matsusaka T, Iwatani H, et al. Autophagic clearance of mitochondria in the kidney copes with metabolic acidosis. J Am Soc Nephrol. 2014;25(10):2254–2266. doi: 10.1681/ASN.2013090986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15(4):403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyer-Guittaut M, Poillet L, Liang Q, Bole-Richard E, Ouyang X, Benavides GA, Chakrama FZ, Fraichard A, Darley-Usmar VM, Despouy G, et al. The role of GABARAPL1/GEC1 in autophagic flux and mitochondrial quality control in MDA-MB-436 breast cancer cells. Autophagy. 2014;10(6):986–1003. doi: 10.4161/auto.28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wind NS, Holen I. Multidrug resistance in breast cancer: from in vitro models to clinical studies. Int J Breast Cancer. 2011;2011(967419) doi: 10.4061/2011/967419. 10.4061/2011/967419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289(24):17163–17173. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]