Abstract
Taste receptors function as one of the interfaces between internal and external milieus. Taste receptors for sweet and umami (T1R [taste receptor, type 1]), bitter (T2R [taste receptor, type 2]), and salty (ENaC [epithelial sodium channel]) have been discovered in the recent years, but transduction mechanisms of sour taste and ENaC-independent salt taste are still poorly understood. In addition to these five main taste qualities, the taste system detects such noncanonical “tastes” as water, fat, and complex carbohydrates, but their reception mechanisms require further research. Variations in taste receptor genes between and within vertebrate species contribute to individual and species differences in taste-related behaviors. These variations are shaped by evolutionary forces and reflect species adaptations to their chemical environments and feeding ecology. Principles of drug discovery can be applied to taste receptors as targets in order to develop novel taste compounds to satisfy demand in better artificial sweeteners, enhancers of sugar and sodium taste, and blockers of bitterness of food ingredients and oral medications.
Keywords: Gustatory, sweet, bitter, umami, salty, sour, receptor, gene
INTRODUCTION
This review focuses on genetics of taste receptors in vertebrate animals, specifically, how gene structure and variation influence expression and function of taste receptors, and how this affects taste function. We describe the different taste qualities and discuss two main types of genetic variation: variation among orthologous genes in different vertebrate species, and variation of alleles within species. Although environment can also affect expression of taste receptor genes through physiological and epigenetic mechanisms, this is outside the scope of this review. For information on invertebrate taste receptors, see, e.g., [1, 2]; for other aspects of taste genetics, including genetics of taste perception, see, e.g., [3–7]).
Taste System and Taste Receptors
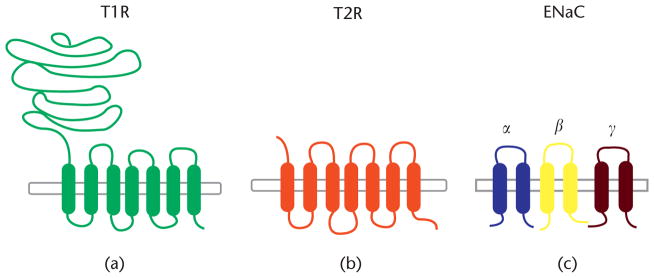
In mammals, the gustatory system comprises taste cells, afferent gustatory nerves, and brain structures involved in central processing of taste. The taste cascade begins with taste receptor cells organized in taste buds, most of which are located within gustatory papillae in the tongue. Taste bud cells come in four types: type I, II, and III cells and basal cells. Types I–III are mature taste receptor cells exposed to the oral cavity to interact with taste stimuli via taste receptor proteins. This interaction results in excitation that is transmitted via afferent gustatory nerves to the brain to evoke taste perception. Taste perception has several aspects: intensity, hedonics (pleasantness or unpleasantness), and quality. Five taste qualities are known to be perceived by humans and many nonhuman mammals: sweet, umami, bitter, salty, and sour. The existence of distinct taste qualities implies that each has a specific coding mechanism mediated by specialized taste receptors. Discovery of receptors for bitter, sweet, umami, and salty taste (Fig. 1) has supported this notion.
Fig. (1).
Known taste receptors. The T1R (a; sweet and umami) and T2R (b; bitter) proteins are G protein-coupled receptors, while ENaC (c; salty) is an ion channel. Reproduced from [6] by permission of John Wiley & Sons, Ltd.
Although several proteins have been suggested to function as taste receptors, not all of them have been unanimously accepted as such. Several requirements need to be met to affirm that a molecule functions as a taste receptor: (1) known molecular identity (DNA, RNA, and protein sequences); (2) expression in taste receptor cells; (3) interaction with appropriate ligands; and (4) changes in taste responsiveness as a result of experimentally altered receptors. All these criteria have been met for taste receptors for sweet and umami (T1R [taste receptor, type 1]), bitter (T2R [taste receptor, type 2]), and salty (ENaC [epithelial sodium channel]) taste (Fig. 1), but sour taste receptors are still unknown (although several candidates have been proposed).
Between-species Variation and Evolution of Taste Receptors
T1Rs, T2Rs, and ENaCs are conserved among vertebrates. Their respective genes and proteins are considered orthologs, because their sequence similarity suggests that they originated from common ancestral genes. The order of chromosomal location of orthologous taste receptor genes is also conserved among different species (providing an example of conserved synteny). Despite this conservation, vertebrate species differ in both sequence and numbers of receptor genes. Gene duplications, deletions, and pseudogenization contribute to species variation in gene number.
This interspecies variation has been shaped by evolutionary forces, likely reflecting adaptation to differences in their diets. The survival of all animals depends on consumption of nutrients, and taste guides animals in choosing food that is safe to consume and appropriate for bodily needs. Many of the same nutrients (e.g., sugars, amino acids, salts) are consumed by different species, yet many species have very different diets (e.g., herbivores and carnivores). Sources of nutrients may contain toxic substances, and each species may be exposed to different sets of toxins in their diets. Covariation between taste receptors and diet suggests that during the evolution the taste system adjusted to support the dietary needs of individual species.
Within-species Variation of Taste Receptors
Allelic variation of taste receptor genes is most documented in humans and mice. Naturally occurring polymorphisms contribute to individual variation in taste responses in both species. Such allelic variation can affect food perception, choice, and consumption and thus can influence nutrition and predispose to certain diseases. In mice, in addition to naturally occurring polymorphisms, mutant alleles of taste receptor genes have been produced by targeted mutagenesis. Associations of taste receptor variants with changes in responsiveness to taste stimuli provide a tool to study receptor-ligand interactions [8].
Pharmaceutical Design and Taste Receptors
There is a growing interest in developing novel taste stimuli and taste modifiers for humans and other animals. For humans, areas of interest include making foods and drinks healthier without sacrificing their palatability, and making oral medications more acceptable to patients. There is a demand for artificial sweet and umami compounds; enhancers of salty, sweet, and umami taste; blockers of bitter taste; and pharmaceutical compounds with improved sensory properties. There is also a demand for improving palatability of food for companion and farm animals and for developing nonlethal repellents of wild animals (e.g., nontoxic chemicals with aversive taste). Development of such products has been hampered by lack of knowledge of molecular identity of the taste receptors.
Discovery of most of the mammalian taste receptors in the last decade allowed scientists to apply principles of drug discovery and to use taste receptors as targets to develop novel taste stimuli, enhancers, and blockers (see, e.g., [9–11]). Growing knowledge about within- and between-species variation in taste receptors will help to tailor these products to target consumers. Recent findings of expression of taste receptors in internal organs (e.g., airway epithelia, gut, pancreas, and testes) and accumulating data that they play a role in internal chemoreception (e.g., [12–15]) also open possibilities for using taste receptors to develop drugs for treating diseases associated with these organs.
SWEET AND UMAMI TASTE
Sugars are the most common natural taste stimuli that humans describe as sweet and are innately attractive to many animals [16–18]. A wide range of other chemicals taste sweet to humans (e.g., some sugar alcohols, glycosides, amino acids, and proteins [19, 20]). Many of these compounds also evoke sucrose-like qualitative taste sensation and appetitive consummatory behavior in nonhuman mammals. In contrast to sweet taste, which humans universally appreciate, umami is lesser known. Most languages have only four words for basic taste qualities: sweet, salty, sour, and bitter. The word umami was first used around 1908 by a Japanese chemist, Dr. Kikunae Ikeda [21]. He discovered that glutamic acid and its salts evoke a taste sensation distinct from the four known taste qualities. To describe this additional basic taste quality, he combined Japanese words umai (
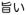 ; “delicious” or “savory”) and mi (
; “delicious” or “savory”) and mi (
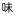 ; “taste”) to derive a new word, umami (
; “taste”) to derive a new word, umami (
 in kanji, or
in kanji, or
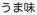 in a mixed hiragana-kanji spelling). In English, umami taste sometimes is described as “savory taste” or “glutamate taste” [22].
in a mixed hiragana-kanji spelling). In English, umami taste sometimes is described as “savory taste” or “glutamate taste” [22].
Umami-tasting compounds include some L-amino acids (e.g., glutamate and aspartate), purine 5′-ribonucleotides, theogallin, theanine, ibotenic, tricholomic, succinic, and gallic acids, and several peptides. The 5′-ribonucleotides potentiate taste of L-glutamate. Many of these compounds have other chemosensory components, in addition to umami taste. For example, in monosodium glutamate (MSG), the anion (L-glutamate) generates an umami taste, and the cation (Na+) contributes a salty taste. A good way to experience umami is to compare tastes of equimolar solutions of MSG and NaCl. Both solutions have saltiness, which is attributed to sodium, but MSG also has another taste component not present in NaCl solutions—umami. There is also strong evidence that umami taste stimuli evoke a unique (glutamate-like) taste in nonhuman animals [23–25].
Sweet and Umami Taste Receptors
G protein-coupled receptor (GPCR) proteins from the T1R family (Fig. 1a) play a central role in reception of sweet and umami taste in humans (and sucrose- and glutamate-like taste in nonhuman animals). A T1R2+3 heterodimer functions as a sweet taste receptor [26, 27] responding to a broad variety of ligands in both humans and rodents. T1R3 alone may function as a receptor for high sucrose concentrations [28]. A T1R1+3 heterodimer functions as an umami taste receptor in humans [27] and is a more broadly tuned L-amino acid taste receptor in mice and fish [29, 262], although multiple combinations of T1R2 with T1R3 also function as L-amino acid receptors in fish [262].
Depending on GPCR classification system, T1Rs belong to class C (metabotropic glutamate/pheromone) family [30–32] or the glutamate family [33]. In humans and many mammalian species, this family includes three proteins: T1R1, T1R2 and T1R3. Their corresponding gene names are taste receptor, type 1, members 1, 2, and 3, respectively, with gene symbols TAS1R1, TAS1R2, and TAS1R3 in humans and Tas1r1, Tas1r2, and Tas1r3 in other species. Numbers of T1R genes vary in some species (described below). The three human TAS1R genes are located in the short arm of human chromosome 1 (1p36) in the order TAS1R2—TAS1R1—TAS1R3. Their mouse orthologs reside in a region of conserved synteny in distal chromosome 4 in the same order: Tas1r2—Tas1r1—Tas1r3. The mouse Tas1r genes contain six coding exons that are translated into proteins of 842–858 amino acids. The T1R proteins have a predicted secondary structure that includes seven transmembrane helices forming a heptahelical domain, and a large extracellular N-terminus composed of a Venus flytrap module and a cysteine-rich domain connected to the heptahelical domain (see Fig. 1a).
T1Rs are expressed in type II taste bud cells. T1R3 is co-expressed in the same taste cells with either T1R1 or T1R2 [26, 34, 35, 63], but some taste cells express only T1R3 in mammal [26] and only T1R2s in fish [63]. This co-expression of T1Rs is consistent with their function as heterodimers (T1R1+3 and T1R2+3). Mice with targeted mutations of the T1R genes have diminished taste responses to sweet and/or umami taste stimuli [28, 36].
Data on T1R ligand specificity in mammals suggest that T1R1+3 and T1R2+3 are the main umami and sweet receptors, respectively, although T1R1 was recently demonstrated to be involved in sweet reception [263]. However, there is evidence for additional reception mechanisms for sweet or umami taste. They include glucose transporters and metabolic sensors, which may be involved in sugar tasting [37], and the ability of some sweeteners to penetrate the membrane of the taste bud cells and interact with in-tracellular targets [38–40]. Splice variants of metabotropic glutamate receptors mGluR4 and mGluR1 and the N-methyl-D-aspartate (NMDA)-type glutamate ion channel receptor (all of which are involved in glutamatergic synaptic transmission in the brain) have been proposed as candidate mammalian taste receptors for umami or glutamate taste [41–48]. Purine 5′-nucleotides may activate different taste receptors than glutamate in rats [49]. Catfish (Ictalurus punctatus) taste receptors for some L-amino acids behave as ligand-gated ion channel receptors [50–53], and combinations of T1R2+3 function as L-amino acid receptors in medaka fish (Olyzias latipes) and zebrafish (Danio rerio) [262].
Between-Species Variation of Sweet and Umami Taste Receptors
Two main processes have shaped repertoires of T1Rs in different species and their responsiveness to sweet and umami taste stimuli. First, pseudogenization and duplication changed numbers of T1R family members; pseudogenization is responsible for sweet or umami taste “blindness” in some species. Second, sequence variants within functional T1Rs resulted in species-specific differences in receptor-ligand interactions. Although ligands for the T1R receptors have been experimentally confirmed for only a few species (mostly humans and rodents, but fish T1Rs were shown to function as amino acid receptors [262]), it is likely that their orthologs in other species have similar ligand specificities. Therefore, species differences in sweet and umami taste preferences [54–62] are expected to be due to variation in the T1R genes.
Although many mammals have three T1R genes coding T1R1, T1R2, and T1R3, as described above, numbers of functional T1R genes in other vertebrate species range from complete absence in the western clawed frog, vampire bats, sea lion, and bottlenose dolphin to five in some species of fish (Table 1). Several fish species have two or three T1R2 genes, suggesting that T1R expansion in fish is probably due to duplication of Tas1r2 [63, 64].
Table 1.
Numbers of T1R genes in vertebrates.
| Species | T1R1 | T1R2 | T1R3 |
|---|---|---|---|
| Mammals | |||
| Human (Homo sapiens) | 1 | 1 | 1 |
| Mouse (Mus musculus) | 1 | 1 | 1 |
| Rat (Rattus norvegicus) | 1 | 1 | 1 |
| Dog (Canis familiaris) | 1 | 1 | 1 |
| Opossum (Monodelphis domestica) | 1 | 1 | 1 |
| Pig (Sus scrofa) | 1 | 1 | |
| Horse (Equus caballus) | 0 | ||
| Giant panda (Ailuropoda melanoleuca) | 0 (1) | ||
| Bats* | 0 (1) | 1 | 1 |
| Vampire bats** | 0 (1) | 0 (1) | 0 (1) |
| Spotted seal (Phoca largha) | 0 (1) | ||
| Harbor seal (Phoca vitulina) | 0 (1) | ||
| Caspian seal (Pusa caspica) | 0 (1) | ||
| Northern elephant seal (Mirounga angustirostris) | 0 (1) | ||
| Australian sea lion (Neophoca cinerea) | 0 (1) | ||
| South American sea lion (Otaria byronia) | 0 (1) | ||
| Domestic cat (Felis silvestris catus) | 1 | 0 (1) | 1 |
| Tiger (Panthera tigris) | 0 (1) | ||
| Cheetah (Acinonyx jubatus) | 0 (1) | ||
| Asiatic lion (Panthera leo persica) | 0 (1) | ||
| Southern fur seal (Arctocephalus forsteri) | 0 (1) | ||
| Pacific harbor seal (Phoca vitulina richardii) | 0 (1) | ||
| Asian small-clawed otter (Amblonyx cinereus) | 0 (1) | ||
| Spotted hyena (Crocuta crocuta) | 0 (1) | ||
| Fossa (Cryptoprocta ferox) | 0 (1) | ||
| Banded linsang (Prionodon linsang) | 0 (1) | ||
| Bottlenose dolphin (Tursiops truncates) | 0 (1) | 0 (1) | 0 (1) |
| California sea lion (Zalophus californianus californianus) | 0 (1) | 0 (1) | 0 (1) |
| Birds | |||
| Zebra finch (Taeniopygia guttata) | 0 | ||
| Chicken (Gallus gallus) | 1 | 0 | 1 |
| Amphibian | |||
| Western clawed frog (Xenopus tropicalis) | 0 | 0 | 0 |
| Fish | |||
| Fugu (Takifugu rubripes) | 1 | 2 | 1 |
| Pufferfish (Tetraodon nigroviridis) | 1 | 3 | 1 |
| Zebrafish (Danio rerio) | 1 | 2 | 1 |
| Medaka (Oryzias latipes) | 1 | 3 | 1 |
Numbers reflect functional and putatively functional genes (pseudogenes). Partial genes are considered putatively functional. When different sources indicate different numbers of genes, the highest estimate is shown. Empty cells indicate lack of data.
Multiple (n=28) species, excluding vampire bats [68].
Three species of vampire bats [68].
Several species have one or more pseudogenized T1R genes. Tas1r1, which codes part of the umami heterodimer, is a pseudogene in the giant panda [65–67] and six pinnipeds (suborder Caniformia): spotted seal, harbor seal, Caspian seal, northern elephant seal, Australian sea lion, and South American sea lion [67]. Tas1r1 is absent, unamplifiable, or pseudogenized in all 31 species of bats examined [68]. These mutations deem the umami/amino acid taste receptor dimer T1R1+3 nonfunctional in these species. Most species of the order Carnivora have intact Tas1r1 and have either carnivorous or omnivorous feeding strategies. While the giant panda is also a carnivoran, unlike the other species of this order its diet is almost exclusively vegetarian (bamboo). The Tas1r1 pseudogenization was likely associated with evolution of feeding behavior in the giant panda because the estimated date of its dietary switch to bamboo coincides with the date of its Tas1r1 pseudogenization [65]. The six pinniped species examined are strictly carnivorous [67], and bats examined include fruit, insect, and blood feeders [68], so widespread Tas1r1 pseudogenization in these clades is puzzling.
The Tas1r2 gene, which codes part of the sweet heterodimer, is absent in the genomes of the chicken [64, 69, 70], zebra finch [70], and horse [70] (but see [71]). Tas1r2 is a pseudogene in four Felidae species (domestic cat, tiger, cheetah, and Asiatic lion [59, 72]) and six nonfeline species from suborders Feliformia (spotted hyena, fossa, and banded linsang) and Caniformia (southern fur seal, Pacific harbor seal, and Asian small-clawed otter) [73]. These mutations result in a nonfunctional T1R2+3 heterodimeric sweet taste receptor and explain the lack of attraction to sugars and other sweeteners documented at least in some of these species [54–59, 73]. All these animals are either obligate carnivores or, in case of the chicken, granivores and do not seek sugars in their food. Thus, loss of the Tas1r2 gene and lack of sweet taste responsiveness in these animals may be a consequence of their feeding behavior, which does not require a sweet taste receptor for proper food choice, leading to a lack of selective advantage of having a functional sweet taste receptor that recognizes sugars. Many other species of the order of Carnivora have a functional Tas1r2 structure [59, 64, 72, 73] and are attracted to sugars [59, 73–75], including dogs, lesser panda, domestic ferret, Haussa genet, meerkat, yellow mongoose, aardwolf, Canadian otter, spectacled bear, raccoon, and red wolf. Some birds also recognize sugar taste [76–78], suggesting that they may have a functional T1R2, although this has not been confirmed experimentally.
All three T1R genes are lost in the tongueless western clawed frog [64], sea lion, bottlenose dolphin [73], and vampire bats [68, 70]. Consistent with this, common vampire bats (Desmodus rotundus) are indifferent to sweet stimuli but avoid salty, sour, and bitter tastes (umami taste was not evaluated) [79]. Taste function has not been evaluated in the other species, but lack of taste receptors may be related to their feeding behavior, which involves swallowing food whole without chewing (e.g., sea lion, dolphin) [73] or feeding exclusively on blood (vampire bats) [68]. Thus, pseudogenization and duplication of T1R genes occurred independently multiple times in different vertebrate lineages during evolution.
In addition to changes in number of T1R genes, their sequence variants are also responsible for species differences in sweet and umami taste. For example, several sweeteners (aspartame, cyclamate, neohesperidin dihydrochalcone, neotame, and sweet proteins) are perceived as sweet by humans but not by rodents (e.g., [80–82]). Correspondingly, human but not rodent T1R2+3 responds to these sweeteners. Heterologously expressed T1R1+3 functions as a broadly tuned L-amino acid receptor in mice and as a more narrowly tuned umami receptor in humans. The role that sequence variation between T1R orthologs plays in species differences in sweet and umami taste has been shown in studies using heterologously expressed interspecies T1R chimeras or receptors with species-specific site-directed mutations, and in genetically engineered mice expressing human receptors [26–29, 83–89]. One well-studied example of species differences in sweet taste is sensitivity to aspartame. Humans, apes, and old-world monkeys perceive aspartame as sweet, but other primate species and most nonprimate species do not [60]. Recent studies identified sequence variants of T1R2 and T1R3 that are associated with ability to taste aspartame and are predicted to influence aspartame binding to the T1R2+3 receptor [90, 91]. Results of all these studies suggest that evolutionary changes of T1R receptors can affect their ligand-binding properties.
Within-Species Variation of Sweet and Umami Taste Receptors
Within-species polymorphisms of the T1R genes were found in humans [92], rats [93], and mice [94], but only in humans and mice have they been found to be associated with sweet or umami taste responsiveness.
Humans differ in perception of sweet (reviewed in [95–100]) and umami [101] tastes. Non-amino acid-coding single-nucleotide polymorphisms (SNPs) in the TAS1R3 promoter were found to be associated with taste sensitivity to sucrose in humans and to influence TAS1R3 promoter activity in vitro [102]. Another study found association of a missense variant in TAS1R2 with habitual consumption of sugars [103]. Polymorphisms in TAS1R1 and TAS1R3, as well as a candidate umami receptor gene, mGluR1, partially explain variation in umami taste [104–106]
In mice, variation in sweetener preference is associated with the Sac (saccharin preference) locus [107–110]. Chromosomal mapping and positional cloning of the Sac locus have identified it as the Tas1r3 gene [94, 111, 112], which was instrumental in discovery of the T1R taste receptors. Analyses of the Tas1r3 sequence variants across multiple inbred strains identified a missense polymorphism (I60T) in the extracellular N-terminus of the T1R3 protein as a candidate causative variant for ligand binding and phenotypical variation in sweet taste [94]. The effect of this polymorphism on T1R3 ligand binding was confirmed in vitro [113]. Tas1r3 polymorphisms affect behavioral and neural taste responses to many different sweeteners [114, 115], indicating that these sweeteners activate a taste receptor involving T1R3. However, Tas1r3 genotype did not affect taste responses to several sweet-tasting amino acids (L-glutamine, L-threonine, L-alanine, glycine), glucose polymers (Polycose, maltooligosaccharide), or umami, salty, sour, or bitter taste stimuli [114, 115]. The T1R3 protein is involved in transduction of both sweet and umami tastes, and disruption of the Tas1r3 gene diminishes behavioral and neural responses to both sweet and umami taste stimuli [28, 36]. Therefore, lack of effect of Tas1r3 polymorphisms on taste responses to sweet amino acids and umami taste stimuli suggests that they bind to a different site than do sweeteners, responses to which are affected by the I60T polymorphism in Tas1r3. Taste responses to glucose polymers were not affected by natural Tas1r3 polymorphisms or Tas1r3 gene knockout [116, 117], which suggests that taste of complex carbohydrates [118] is mediated by a receptor other than T1R3 (see below). Although T1R2 is a part of the sweet taste receptor and T1R1 is a part of umami/amino acid taste receptor, there is no evidence that Tas1r2 or Tas1r1 polymorphisms are associated with mouse strain differences in responses to sweet or umami taste stimuli.
BITTER TASTE
Bitter taste quality likely evolved as a mechanism for avoiding toxic foods. Bitter compounds evoke innate aversive behavior in many animal species. A large number of chemically diverse compounds elicit bitter taste in humans and taste aversion in nonhuman animals [119]. Toxins contained in plants and produced by micro-organisms are probably the stimuli that shaped species-specific repertoires of bitter taste receptors. Many oral medications have bitter taste, which may interfere with treatment compliance in human and veterinary medicine.
Bitter Taste Receptors
GPCRs from the T2R family function as mammalian bitter taste receptors (Fig. 1b) [120–122]. Depending on the GPCR classification system, T2Rs are described as distantly related to class A (rhodopsin-like) GPCRs [120], as belonging to a separate putative family [30–32], or as forming a distinct cluster within the frizzled/taste2 family [33].
Most vertebrate species have multiple T2R genes, and their numbers differ widely among species (Table 2). Many species have pseudogenes in addition to functional T2R genes. Symbols for genes encoding T2R proteins begin with TAS2R (in humans) or Tas2r (in other species), with the genes named taste receptor, type 2, member n. Besides differences in symbol letters (upper- or lowercase), human and mouse T2R genes can also be distinguished by member numbers: less than 100 for human genes (e.g., TAS2R1-TAS2R65), versus higher than 100 for mouse genes (e.g., Tas2r102-Tas2r146). Genes and proteins of other species can be distinguished by adding a lowercase letter indicating species, for example, rT2R9 for rat. In many species, T2R genes cluster in a few chromosomal locations. For example, human T2R genes map to chromosomes 5, 7, and 12, and mouse T2R genes map to chromosomes 2, 6, and 15. The T2R genes are intronless, at least in their coding region, and encode GPCR proteins that consist of ~300–330 amino acids and have a short extracellular N-terminus (Fig. 1b).
Table 2.
Numbers of T2R genes in vertebrates
| Species | Functional and putatively functional genes | Pseudogenes |
|---|---|---|
| Mammals | ||
| Human (Homo sapiens) | 28 | 16 |
| Macaque (Macaca mulatta) | 27 | 11 |
| Mouse (Mus musculus) | 36 | 7 |
| Rat (Rattus norvegicus) | 37 | 7 |
| Cow (Bos taurus) | 19 | 15 |
| Horse (Equus caballus) | 19 | 36 |
| Dog (Canis familiaris) | 16 | 5 |
| Little brown bat (Myotis lucifugus) | 17 | 9 |
| Bottlenose dolphin (Tursiops truncates) | 0 | 10 |
| Opossum (Monodelphis domestica) | 29 | 7 |
| Platypus (Ornithorhynchus anatinus) | 5 | 1 |
| Birds | ||
| White-throated sparrow (Zonotrichia albicollis) | 19 | 1 |
| Zebra finch (Taeniopygia guttata) | 7 | |
| Chicken (Gallus gallus) | 3 | 0 |
| Reptile | ||
| Lizard (Anolis carolinensis) | 39 | 10 |
| Amphibian | ||
| Western clawed frog (Xenopus tropicals) | 52 | 14 |
| Fish | ||
| Fugu (Takifugu rubripes) | 4 | 0 |
| Pufferfish (Tetraodon nigroviridis) | 6 | 0 |
| Zebrafish (Danio rerio) | 5 | 0 |
| Medaka (Oryzias latipes) | 1 | 0 |
| Stickleback (Gasterosteus aculeatus) | 3 | 0 |
T2Rs are expressed in type II taste bud cells, and their expression does not overlap with T1Rs (with the exception of zebrafish, in which a small number of taste cells co-expresses T2Rs with T1Rs [264]). It was suggested that multiple T2Rs are co-expressed in the same taste bud cells, and possibly nearly all T2Rs are expressed in each T2R-positive cell [120, 123, 124]. However, other studies suggest that different taste bud cells may express different sets of T2Rs [122, 125]. Both possibilities find support in behavioral and neurophysiological studies, some of which suggest that different bitter compounds have identical taste quality [126–129], while others suggest that the taste system can discriminate among different bitter taste stimuli [130–132].
Most human T2Rs have been de-orphanized, mainly through the use of heterologous cell assays, and in all cases ligands were bitter-tasting compounds [133]. The number of compounds perceived by humans as bitter [119] is much larger than the number of human TAS2R genes, implying that each human T2R responds to more than one bitter ligand [134]. Consistent with this, several T2Rs are broadly tuned to detect stimuli of different chemical classes, while others appear to be more specific, activated by one or a few agonists [133]. It has been suggested that different T2R alleles may have different profiles of ligand specificity [99, 135–137]. Thus, the repertoire of bitter taste receptors may be not limited by the number of the TAS2R genes but instead may involve as many receptors as there are TAS2R alleles [137]. Compared with data in humans, few ligands for the T2R receptors have been experimentally confirmed for other species (reviewed in [148]). One non-human T2R that has been de-orphanized is the mouse receptor encoded by the Tas2r105 gene, which responds to cycloheximide in vitro [121]. Consistent with this, Tas2r105-knockout mice have selective impairment in taste responses to cycloheximide but not to other bitter or nonbitter taste stimuli [123].
In addition to activating T2R receptor proteins, some bitter compounds can interact with ion channels in the cell membrane or with intracellular targets [38–40, 138–141], which thus also function as receptors of these compounds.
Between-Species Variation of Bitter Taste Receptors
Bitter taste functions to prevent consumption of toxins with food. Because animal species differ in their diets, it is reasonable to expect that different species have unique repertoires of bitter taste receptors, shaped by natural selection.
Because many plants are bitter to humans, it has been proposed that high tolerance for bitterness would be adaptive for herbivores, which was supported by observations of lower taste sensitivity to some bitter compounds in some herbivorous species compared with omnivores or carnivores [142–144]. However, recent studies have shown that this relationship is more complex. For example, some bitter compounds (salicin and protein hydrolysates) evoked stronger avoidance in herbivores than in omnivores [145, 146]. Thus, bitter taste avoidance is species and stimulus dependent, and the premise that herbivores have a generalized reduced bitter sensitivity is not accurate.
Other examples of species differences in bitter taste include phenylthiocarbamide (PTC) and phenyl-β-D-glucopyranoside, which taste bitter to humans but are not aversive to mice [123, 147]. Compared with humans, mice are more sensitive to cycloheximide and less sensitive to denatonium (A. Bachmanov, unpublished data).
Although ligands for the T2R receptors have been experimentally confirmed for only a few species (reviewed in [148]), it is likely that T2Rs function as bitter taste receptors across vertebrates. For example, fish T2Rs are involved in the taste-evoked aversive behavior [262]. Available data for a few orthologous pairs of T2Rs suggest that receptor sequence similarity is predictive of the receptor ligand specificity. Rat and mouse Tas2r105 orthologs respond to cycloheximide [121, 149]. Orthologous human TAS2R4 and mouse Tas2r108 respond to denatonium and 6-n-propyl-2-thiouracil (PROP) [121]. Sensitive alleles of human TAS2R38 and chimpanzee Tas2r38 orthologs respond to PTC [150, 151]. On the other hand, non-orthologous T2Rs in different species may be activated by the same agonists. For example, some fish T2Rs are activated by denatonium, but they are not orthologs of the mammalian denatonium receptors [262].
As mentioned above, numbers of T2R genes in vertebrate species differ widely (Table 2). Many species have T2R pseudogenes in addition to functional T2R genes, although birds and fishes seem to lack or have small numbers of pseudogenes compared with mammals, reptiles, and amphibians. The largest number of functional or putatively functional T2Rs was predicted in the western clawed frog (n=52), while the bottlenose dolphin appears to completely lack functional T2Rs. Numbers of functional or putatively functional T2Rs also differ widely within each characterized class of animals: from 0 to 37 in mammals, from 3 to 19 in birds, and from 0 to 6 in fish.
Analyses of relatedness of the T2R genes in different species suggest a complex evolution of this gene family. Local and inter-chromosomal duplications, deletions, pseudogenization, and positive selection drove expansions and contractions of T2R repertoires in different lineages [64, 152–155]. However, little is known about how this variation in T2R repertoires affects species differences in bitter taste.
Within-Species Variation of Bitter Taste Receptors
Humans
Human TAS2R genes have substantial diversity of coding sequence, including segregating pseudogenes [136, 137, 156, 157]. This suggests that TAS2R polymorphisms may be responsible for individual differences in bitter taste (reviewed in [98–100, 158]). However, this relationship has been demonstrated for only a few genes.
TAS2R38 was identified in a positional cloning study [159] as a gene identical to a human PTC bitter taste sensitivity locus on chromosome 7q [160]. Allelic variants of TAS2R38 are associated with human perception of PTC and PROP bitterness [150, 159, 161–163]. PTC and PROP responses of cells heterologously expressing different alleles of TAS2R38 correlate with psychophysical responses of individuals carrying these alleles [150]. While wild-type mice do not show strong aversion of PTC in brief-access tests [147], mice with a taster allele of human TAS2R38 transgenically expressed in bitter-sensing cells under the control of a mouse Tas2r promoter show strong aversion to PTC [123].
Sensitive alleles of T2R38 respond to PTC, PROP, and related compounds that contain a thiourea (N-C=S) moiety. Some plants consumed by humans contain glucosinolates, which also contain the thiourea moiety, and TAS2R38 genotype can affect perception of bitterness of these plants, such as broccoli, turnips, and horseradish [164].
Although a PTC “nontaster” allele of TAS2R38 is expressed in taste buds, it does not respond to taste stimuli in vitro [150]. Because taster and nontaster alleles of TAS2R38 are maintained by balanced selection [165], it has been suggested that the “nontaster” allele may serve as a functional receptor for toxic bitter substances other than PTC that have not yet been identified [99, 135, 136, 165].
Signs of positive selection were also found for TAS2R16 encoding a receptor for β-glucopyranosides. Its ancestral allele associated with lower sensitivity to β-glucopyranosides is under positive selection only in central Africa, while the evolutionarily derived allele associated with an increased sensitivity is under positive selection in the rest of the world. It is suggested that the global pattern of allelic variation of the TAS2R16 gene depends on selective pressures of protection against malaria in Africa and protection against toxins in malaria-free zones [166].
T2R31 is a receptor for bitterness of artificial sweeteners, saccharin, and acesulfame K. Polymorphisms in the TAS2R31 gene were associated with perception of bitterness of these sweeteners in both human subjects and in vitro assays [167]. Taste perception of quinine was associated with a cluster of T2R and salivary proline-rich protein genes on chromosome 12, but tight linkage among these genes precluded the identification of a single causal genetic variant [168].
Chimpanzee
Allelic variants of chimpanzee Tas2r38, an ortholog of human TAS2R38, are also associated with taste sensitivity to PTC in individual animals. A taster allele of chimpanzee Tas2r38 responds to PTC in vitro [151]. However, variation in PTC taste sensitivity in chimpanzees depends on Tas2r38 alleles different from those in humans. While taster and nontaster alleles of the human TAS2R38 gene are defined by missense SNPs [159, 165], the PTC-insensitive allele of chimpanzee Tas2r38 encodes a truncated receptor variant [151]. Thus, nontaster Tas2r38 alleles have been independently derived in humans and chimpanzees [151].
Mice
Mouse strains differ in taste responses to bitter taste stimuli. Several genetic loci are responsible for variation in aversion to bitter-tasting quinine (Qui), cycloheximide (Cyx), copper glycinate (Glb), and the acetylated sugars sucrose octaacetate and raffinose undecaacetate (Soa/Rua), all of which map to the mouse chromosome 6, in a T2R gene cluster region (reviewed in [5, 7]). There is also considerable strain variation in sequences of the mouse T2R genes [121, 147, 169], suggesting that the genetic variation in bitter taste is due to polymorphisms of the T2R genes, as predicted by Lush [109]. However, this relationship has been demonstrated for only the Tas2r105 gene corresponding to the Cyx locus [121, 123, 147], although with some inconsistencies (see [3, 170]).
SALTY TASTE
Hedonic responses (attraction or avoidance) to salty taste depend on species, genotype, stimulus concentration, and physiological state of individuals. At lower concentrations, salty taste is often attractive, but at high concentrations it is typically aversive. Sodium balance of the body strongly influences hedonics of salty taste, with sodium depletion known to induce salt appetite in many species [171, 172].
A prototypical salty taste stimulus is sodium chloride (NaCl). LiCl also has predominantly salty taste. Many other sodium and nonsodium salts taste salty, but their saltiness is usually accompanied by additional taste quality components, most frequently described as bitterness [173].
Salty Taste Receptors
Earlier studies suggested that there are at least two transduction pathways for salty taste. This hypothesis was based on findings that amiloride, a potassium-sparing diuretic, partially suppresses taste responses to sodium and lithium salts [174–176]. Because amiloride blocks the epithelial sodium channel (ENaC), the portion of taste responses suppressed by amiloride (amiloride sensitive) is proposed to be mediated by ENaC. The portion of taste response that is not inhibited by amiloride is considered amiloride insensitive. Amiloride-sensitive and -insensitive transduction pathways of salty taste have distinct properties. The amiloride-sensitive mechanism is cation (Na+ and Li+) selective; the amiloride-insensitive mechanism is cation nonselective and can be activated by both sodium and nonsodium salts, such as KCl and NH4Cl.
ENaC is a member of the degenerin/ENaC superfamily of ion channels. The ENaC channel is a heteromer consisting of several different subunits: α, β, γ, and/or δ. Each ENaC subunit has two transmembrane domains and is encoded by a separate gene (Fig. 1c). Humans have four ENaC channel subunits, α, β, γ, and δ, encoded by the non-voltage-gated sodium channel 1 genes SCNN1A, SCNN1B, SCNN1G, and SCNN1D, respectively. Mice and rats lack the ENaCδ subunit [177] and therefore have only three ENaC subunits encoded by Scnn1a, Scnn1b, and Scnn1g. The genes for ENaC β and γ subunits are closely linked and located in humans on chromosome 16 and in mice in a region of conserved synteny on chromosome 7. The ENaCα gene is on human chromosome 12, and in the mouse it is on chromosome 6. Human ENaCδ is on chromosome 1, and in the mouse it is a pseudogene located on chromosome 4 [178].
ENaC is involved in transepithelial ion transport in many tissues (e.g., kidney, lung). Correspondingly, ENaC subunits are expressed in many epithelial tissues, including taste and nontaste lingual epithelial cells [179–182]. Based on suppression of taste responses to sodium by amiloride in some species, and based on ENaC expression in taste tissues, ENaC was proposed as a candidate component of salty taste transduction system (reviewed in [183–185]). Recent work with mice genetically engineered to lack ENaC in taste cells [186, 187] has conclusively established the importance of this channel in mediating sodium taste in mice. However, it is still not clear what other components are involved in the amiloride-sensitive salt taste transduction pathway or how they interact with ENaC.
Mechanisms for transduction of amiloride-insensitive cation-nonselective salt taste are even less understood. Earlier studies suggested that amiloride-insensitive salty taste can be explained by diffusion of sodium through tight junctions that are impermeable to amiloride; as a result, Na+ would stimulate taste bud cells through basolateral ENaC channels, which are not accessible to, and therefore are not blocked by, amiloride [139, 188, 189]. However, this paracellular pathway hypothesis is not consistent with existence of taste bud cells, in which responses to apical application of NaCl are not inhibited by amiloride [190]. The Cl− anion was also proposed to be responsible for amiloride-insensitive NaCl taste [191], but experiments with known mechanisms of transmembrane transport of Cl− did not confirm their involvement in salty taste [192, 193]. A recent study showed that concentrated solutions of sodium and nonsodium salts activate two populations of taste bud cells: type II cells expressing T2R receptors, which are also activated by bitter compounds, and type III cells, some of which are also activated by acids. Activation of these cells by salts triggers aversive behavioral response [194]. However, the biological molecules mediating amiloride-insensitive salt taste response have not yet been conclusively identified, although candidates have been proposed.
One candidate is TRPV1 (transient receptor potential cation channel, subfamily V, member 1; formerly named vanilloid receptor subtype 1, or capsaicin receptor). TRPV1 is a transducer of painful thermal stimuli and is also activated by capsaicin, a pungent ingredient in “hot” chili peppers [195]. Based on taste nerve recording experiments using TRPV1 agonists and antagonists and Trpv1-knockout mice, a TRPV1 variant was proposed to function as an amiloride-insensitive salt taste receptor in rodents [196]. However, Trpv1-knockout mice do not have deficiencies in behavioral taste responses to salt [197 198]. Moreover, recent studies did not confirm initial findings of effects of TRPV1 antagonists and Trpv1 gene knockout on taste nerve responses [199, 200]. In addition, data on expression of TRPV1 in the lingual tissue are also inconsistent with its role in salt taste reception. Some immunohistochemical studies detected its expression in rat trigeminal afferent neurons innervating the oral cavity but not in taste bud cells [201, 202]. In another study, TRPV1 immunoreactivity has been detected in rat taste bud cells expressing T1R and T2R receptors [203]. If TRPV1 functions as a salt taste receptor, then co-expression of TRPV1 with T1Rs would suggest that salts should activate T1R-expressing cells, which contradicts some findings [194]. It is likely that TRPV1 contributes to oral chemosensory responses to salts through its expression in trigeminal (somatosensory) nerve endings [200, 204], but not in taste bud cells.
A second candidate for amiloride-insensitive salt taste receptor is TRPML3 (MCOLN3, mucolipin 3), which, like TRPV1, belongs to the transient receptor potential (TRP) family of cation channels [205]. However, mouse taste receptor cells do not appear to express physiologically relevant levels of TRPML3 [206].
Between-Species Variation of Salty Taste Receptors
Animal species differ in their avidity for salt. For example, licking crystallized salt is common among wild animals, particularly herbivores, whose food contains little sodium. On the other hand, meat, the diet of strict carnivores, has sufficient sodium to satisfy their bodily needs [172, 173]. Species differences in need-induced salt appetite may have central rather than peripheral origin. Nevertheless, species differences in peripheral taste mechanisms may also exist.
The best-known example of possible species differences in salty taste reception is sensitivity to amiloride. Neural and/or behavioral taste responses to NaCl are attenuated by amiloride in the mouse, rat, gerbil, hamster, dog, rhesus monkey, and frog [184]. However, amiloride generally does not change perception of saltiness in humans but reduces total intensity and sourness of NaCl and LiCl [184 207]. These species differences may be attributed to presence of the ENaCδ subunit in humans but not rodents. Because ENaCδ is expressed in human taste bud cells [208, 209], it was proposed that the substitution of ENaCα with ENaCδ in the human salty taste receptor deems it less amiloride sensitive [210], consistent with known substantially reduced amiloride sensitivity of δβγ ENaC channel compared with αβγ ENaC [211]. It was proposed that the amiloride-sensitive gustatory mechanisms may be of minor importance for human taste [184]. However, this does not mean that ENaC is not involved in human salty taste, because differences in subunit composition of the ENaC channel may be responsible for species differences in amiloride sensitivity of salty taste.
If ENaCδ explains reduced amiloride sensitivity of salty taste in humans, other species with an intact ENaCδ gene should also have diminished amiloride sensitivity of salty taste. ENaCδ is annotated in genomes of several species. In addition to several primate species, it is present in the dog, cat, cow, sheep, horse, guinea pig, Tasmanian devil, and chicken ([212] and http://www.ncbi.nlm.nih.gov). (The gene annotated as Scnn1d in the African clawed frog, Xenopus laevis, encodes the ENaCε subunit [212]). Amiloride-sensitive NaCl taste responses have been described in dogs and some primates, species with intact ENaCδ [184, 213, 214]. This brings into question whether presence or absence of ENaCδ can explain amiloride sensitivity or insensitivity across all animal species. However, it is not known whether ENaCδ is expressed in taste bud cells of these animals, as it is in humans. Furthermore, amiloride sensitivity in these animals was characterized using taste nerve recordings, but such data are not available for humans. In mice, amiloride sensitivity to NaCl does not always correlate well between behavioral and neural responses (e.g., [215]). Similarly, the contribution of ENaCδ may be different for amiloride sensitivity of behavioral and neural responses to NaCl.
It is interesting that salt taste responses in Drosophila involve degenerin/ENaC channels PPK11 and PPK19 [216], suggesting that salt-sensing mechanisms by degenerin/ENaC channels may have evolved in a common ancestor of vertebrate and invertebrate animals.
Within-Species Variation of Salty Taste Receptors
Previous studies reported individual variation in human salty taste perception and differences among mouse and rat strains in salt preferences and amiloride sensitivity of chorda tympani responses to NaCl (reviewed in [5–7]). Inbred mouse strains also differ in NaCl taste thresholds [217]. Polymorphisms of ENaC subunit genes have been associated with amiloride sensitivity of NaCl taste responses in mice [218] and humans [219].
SOUR TASTE
Sour taste is evoked by acids. Protons are the critical stimulus, although sourness can be affected by the anion. For example, sourness of organic acids is stronger than sourness of inorganic acids at the same pH. Acids can stimulate not only gustatory but also oral somatosensory chemoreceptors. However, gustatory and somato-sensory responses to acids can be distinguished based on acid concentrations. Typically, responses to dilute acid solutions are predominantly gustatory, and a somatosensory component requires higher acid concentrations (discussed in [220]).
Sour Taste Receptors
A sour taste receptor is presumably an ion channel. Several candidate sour (acid) taste receptors have been proposed, but none has been definitely proven (reviewed in [3]). Recent studies suggested that ion channels encoded by the genes Pkd1l3 and Pkd2l1 (polycystic kidney disease-1- and -2-like) function as sour taste receptors [221–223]. However, targeted mutations in these genes have no or only modest effect on taste responses to acids [220, 224].
Between-Species Variation in Sour Taste
Animals of many species detect, and typically avoid, sour taste. We are not aware of any published data on species differences in sour taste. It is possible that all species have similar needs for orosensory detection of acids, and thus sour taste may be conserved across species.
Within-Species Variation in Sour Taste
Genetics likely contributes to within-species differences in sour taste: heritable differences in sour taste were reported in human twins [225] and among inbred mouse strains [226]. These genetic studies may assist with identification of the still elusive sour taste receptor through the positional cloning approach, which facilitated identification of T1R and T2R receptors (e.g., [159, 178]).
TASTE RECEPTORS FOR NONCANONICAL AND COMPLEX TASTE STIMULI
Water
Water consumption is crucial for animals’ survival and is regulated by thirst, a specialized water appetite [171]. Therefore, animals must be able to detect water or hypo-osmotic fluids through oral chemosensation. Consistent with this, water can evoke taste responses (reviewed in [139, 227]). Aquaporins are expressed in taste receptor cells and were proposed as sensors for hypo-osmotic stimuli [227, 228].
Taste perception of water by humans depends on preceding adaptation of the oral cavity to different taste solutions (and probably to saliva) [229–232]. Water elicits a strong sweet taste when it is applied to the oral cavity after exposure to sweet taste blockers. This phenomenon has been labeled “sweet water aftertaste” [233]. This adaptation-dependent perception of water taste is mediated by the T1R2+3 sweet taste receptor: washing out a sweet taste inhibitor with water shifts the receptor from an inactive to an active state, which initiates transduction events evoking perception of sweetness [234].
Ethanol
Ethanol evokes sweet and bitter taste (reviewed in [235]). Consistent with this, alcohol consumption is associated with allelic variants of the Tas1r3 sweet taste receptor gene in mice and TAS2R bitter taste receptor genes in humans. A more sensitive Tas1r3 allele makes the pleasant sweet taste component of ethanol stronger and facilitates consumption of ethanol by mice [236]. More sensitive alleles of the TAS2R genes make the unpleasant bitter taste component of ethanol stronger and suppress alcohol consumption in humans [163, 237].
Kokumi and Calcium
Kokumi is derived from the Japanese word koku (
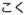 ; “body”) in the same way as umami is derived from the word umai. Kokumi describes oral sensations of continuity, mouthfulness and thickness, and enhancement of basic taste sensations. Examples of kokumi substances are glutathione and some other γ-glutamyl peptides contained in foodstuffs [238, 239]. Kokumi compounds activate the extracellular calcium-sensing receptor (CaSR) [240] that is expressed in taste bud cells [241]. Activation of CaSR-expressing taste bud cells [242] by kokumi substances suggests that kokumi can be categorized as a taste sensation, although the definition of kokumi as a taste quality remains debatable.
; “body”) in the same way as umami is derived from the word umai. Kokumi describes oral sensations of continuity, mouthfulness and thickness, and enhancement of basic taste sensations. Examples of kokumi substances are glutathione and some other γ-glutamyl peptides contained in foodstuffs [238, 239]. Kokumi compounds activate the extracellular calcium-sensing receptor (CaSR) [240] that is expressed in taste bud cells [241]. Activation of CaSR-expressing taste bud cells [242] by kokumi substances suggests that kokumi can be categorized as a taste sensation, although the definition of kokumi as a taste quality remains debatable.
Calcium salts stimulate taste [243], which can be mediated by the T1R3 receptor [244, 245] in addition to the CaSR.
Fat
The fatty acid transporter CD36 and GPCRs GPR40 and GPR120 are expressed in taste bud cells and have been proposed to be involved in oral detection of fatty acids [246–254]. Polymorphisms of the human CD36 gene have been associated with oral fat perception [255, 256].
Complex Carbohydrates
Rats and some other species perceive taste of polysaccharides as qualitatively distinct from taste of sugars [118]. However, a molecular mechanism for perception of complex carbohydrates is unknown.
CONCLUSION
This review demonstrates how gene structure and variation influence expression and function of taste receptors, which affects how the taste system functions. The two main types of gene variation, orthologous genes in different species and alleles within species, account for variations in behaviors toward tastes between and within species. Although taste receptors for sweet and umami (T1R), bitter (T2R), and salty (ENaC) are known, we know little about their across-species variations, and sour taste and ENaC-independent salt taste are still poorly understood. Other, noncanonical taste receptors have been proposed, including those that detect water, fat, and complex carbohydrates, though much research remains to confirm them and uncover their mechanisms. These and other as yet discovered taste receptors can be targets for screening novel taste compounds. Thus, further research in these areas holds the promise of improved nutrition, health, and well-being.
Acknowledgments
Supported by NIH grant R01DC00882 and an Ajinomoto Amino Acid Research Program grant (AAB).
ABBREVIATIONS
- CaSR
Calcium-sensing receptor
- ENaC
Epithelial sodium channel
- GPCR
G protein-coupled receptor
- mGluR
Metabotropic glutamate receptor
- MSG
Monosodium glutamate
- PROP
6-n-propyl-2-thiouracil
- PTC
Phenylthiocarbamide
- SNP
Single-nucleotide polymorphism
- T1R
Taste receptor, type 1
- T2R
Taste receptor, type 2
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
LITERATURE CITED
- 1.Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–44. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montell C. A taste of the Drosophila gustatory receptors. Curr Opin Neurobiol. 2009;19:345–53. doi: 10.1016/j.conb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu Rev Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmanov AA, Bosak NP, Floriano WB, et al. Genetics of sweet taste preferences. Flavour Fragr J. 2011;26:286–294. doi: 10.1002/ffj.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boughter JD, Bachmanov AA. Genetics and evolution of taste. In: Firestein S, Beauchamp GK, editors. Olfaction and Taste. Elsevier/Academic Press; San Diego: 2008. pp. 371–390. [Google Scholar]
- 6.Bachmanov AA, Boughter JD. eLS 2012. John Wiley & Sons, Ltd; Chichester: 2012. Genetics of Taste Perception. http://www.els.net/ [DOI] [Google Scholar]
- 7.Boughter JD, Jr, Bachmanov AA. Behavioral genetics and taste. BMC Neurosci. 2007;8(Suppl 3):S3. doi: 10.1186/1471-2202-8-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachmanov AA. Genetic approach to characterize interaction of sweeteners with sweet taste receptors in vivo. Chem Senses. 2005;30(Suppl 1):i82–i83. doi: 10.1093/chemse/bjh124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slack JP, Brockhoff A, Batram C, et al. Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr Biol. 2010;20:1104–9. doi: 10.1016/j.cub.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene TA, Alarcon S, Thomas A, et al. Probenecid inhibits the human bitter taste receptor TAS2R16 and suppresses bitter perception of salicin. PLoS One. 2011;6:e20123. doi: 10.1371/journal.pone.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Servant G, Tachdjian C, Tang XQ, et al. Positive allosteric modulators of the human sweet taste receptor enhance sweet taste. Proc Natl Acad Sci USA. 2010;107:4746–51. doi: 10.1073/pnas.0911670107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolskee RF, Dyer J, Kokrashvili Z, et al. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci USA. 2007;104:15075–80. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee RJ, Xiong G, Kofonow JM, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;122:4145–59. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen S, Depoortere I. Nutrient sensing in the gut: new roads to therapeutics? Trends Endocrinol Metab. 2013;24:92–100. doi: 10.1016/j.tem.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Behrens M, Meyerhof W. Gustatory and extragustatory functions of mammalian taste receptors. Physiol Behav. 2011;105:4–13. doi: 10.1016/j.physbeh.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 16.McCaughey SA. The taste of sugars. Neurosci Biobehav Rev. 2008;32:1024–43. doi: 10.1016/j.neubiorev.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner JE, Glaser D, Hawilo ME, Berridge KC. Comparative expression of hedonic impact: affective reactions to taste by human infants and other primates. Neurosci Biobehav Rev. 2001;25:53–74. doi: 10.1016/s0149-7634(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 18.Berridge KC. Measuring hedonic impact in animals and infants: microstructure of affective taste reactivity patterns. Neurosci Biobehav Rev. 2000;24:173–98. doi: 10.1016/s0149-7634(99)00072-x. [DOI] [PubMed] [Google Scholar]
- 19.DuBois GE. New insights on the coding of the sweet taste message in chemical structure. In: Salvadori G, editor. Firmenich Jubilee Symposium 1895–1995. Allured Publishing Corp; Carol Stream, IL, USA: Geneva, Switzerland: 1995. pp. 32–95. [Google Scholar]
- 20.Schiffman SS, Gatlin CA. Sweeteners: State of knowledge review. Neurosci Biobehav Rev. 1993;17:313–345. doi: 10.1016/s0149-7634(05)80015-6. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda K. New seasonings. Chem Senses. 2002;27:847–9. doi: 10.1093/chemse/27.9.847. [DOI] [PubMed] [Google Scholar]
- 22.Bachmanov AA. Umami: Fifth taste? Flavor enhancer? Perfumer and Flavorist. 2010;35:52–57. [Google Scholar]
- 23.Ninomiya Y, Funakoshi M. Behavioural discrimination between glutamate and the four basic taste substances in mice. Comp Biochem Physiol. 1989;92:365–370. doi: 10.1016/0300-9629(89)90577-x. [DOI] [PubMed] [Google Scholar]
- 24.Ninomiya Y, Funakoshi M. Peripheral neural basis for behavioural discrimination between glutamate and the four basic taste substances in mice. Comp Biochem Physiol. 1989;92:371–376. doi: 10.1016/0300-9629(89)90578-1. [DOI] [PubMed] [Google Scholar]
- 25.Murata Y, Beauchamp GK, Bachmanov AA. Taste perception of monosodium glutamate and inosine monophosphate by 129P3/J and C57BL/6ByJ mice. Physiol Behav. 2009;98:481–8. doi: 10.1016/j.physbeh.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson G, Hoon MA, Chandrashekar J, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–90. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Staszewski L, Xu H, et al. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 2002;99:4692–6. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao GQ, Zhang Y, Hoon MA, et al. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–66. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 29.Nelson G, Chandrashekar J, Hoon MA, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 30.Kolakowski LF., Jr GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- 31.Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- 32.GPCRDB. Information system for G protein-coupled receptors (GPCRs) http://www.gpcr.org/7tm/
- 33.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 34.Max M, Shanker YG, Huang L, et al. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 35.Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–8. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 36.Damak S, Rong M, Yasumatsu K, et al. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science. 2003;301:850–3. doi: 10.1126/science.1087155. [DOI] [PubMed] [Google Scholar]
- 37.Yee KK, Sukumaran SK, Kotha R, Gilbertson TA, Margolskee RF. Glucose transporters and ATP-gated K+ (KATP) metabolic sensors are present in type 1 taste receptor 3 (T1r3)-expressing taste cells. Proc Natl Acad Sci USA. 2011;108:5431–5436. doi: 10.1073/pnas.1100495108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naim M, Nir S, Spielman AI, et al. Hypothesis of receptor-dependent and receptor-independent mechanisms for bitter and sweet taste transduction: Implications for slow taste onset and lingering aftertaste. In: Given P, Parades D, editors. Chemistry of Taste: Mechanisms, Behaviors, and Mimics. American Chemical Society; Washington, DC: 2002. pp. 2–17. ACS symposium series; 825. [Google Scholar]
- 39.Peri I, Mamrud-Brains H, Rodin S, et al. Rapid entry of bitter and sweet tastants into liposomes and taste cells: implications for signal transduction. Am J Physiol Cell Physiol. 2000;278:C17–25. doi: 10.1152/ajpcell.2000.278.1.C17. [DOI] [PubMed] [Google Scholar]
- 40.Zubare-Samuelov M, Shaul ME, Peri I, et al. Inhibition of signal termination-related kinases by membrane-permeant bitter and sweet tastants: potential role in taste signal termination. Am J Physiol Cell Physiol. 2005;289:C483–92. doi: 10.1152/ajpcell.00547.2004. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhari N, Yang H, Lamp C, et al. The taste of monosodium glutamate: membrane receptors in taste buds. J Neurosci. 1996;16:3817–3826. doi: 10.1523/JNEUROSCI.16-12-03817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhari N, Roper SD. Molecular and physiological evidence for glutamate (umami) taste transduction via a G protein-coupled receptor. Ann N Y Acad Sci. 1998;855:398–406. doi: 10.1111/j.1749-6632.1998.tb10598.x. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhari N, Landin AM, Roper SD. A metabotropic glutamate receptor variant functions as a taste receptor. Nat Neurosci. 2000;3:113–9. doi: 10.1038/72053. [DOI] [PubMed] [Google Scholar]
- 44.San Gabriel A, Uneyama H, Yoshie S, Torii K. Cloning and characterization of a novel mGluR1 variant from vallate papillae that functions as a receptor for L-glutamate stimuli. Chem Senses. 2005;30(Suppl 1):i25–i26. doi: 10.1093/chemse/bjh095. [DOI] [PubMed] [Google Scholar]
- 45.Brand JG. Receptor and transduction processes for umami taste. J Nutr. 2000;130:942S–5S. doi: 10.1093/jn/130.4.942S. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi Y, Zviman MM, Brand JG, Teeter JH, Restrepo D. Measurement of membrane potential and [Ca2+]i in cell ensembles: application to the study of glutamate taste in mice. Biophys J. 1996;71:1057–1070. doi: 10.1016/S0006-3495(96)79306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyono T, Seta Y, Kataoka S, et al. Expression of metabotropic glutamate receptor group I in rat gustatory papillae. Cell Tissue Res. 2003;313:29–35. doi: 10.1007/s00441-003-0740-2. [DOI] [PubMed] [Google Scholar]
- 48.Toyono T, Seta Y, Kataoka S, et al. Expression of the metabotropic glutamate receptor, mGluR4a, in the taste hairs of taste buds in rat gustatory papillae. Arch Histol Cytol. 2002;65:91–6. doi: 10.1679/aohc.65.91. [DOI] [PubMed] [Google Scholar]
- 49.Lin W, Ogura T, Kinnamon SC. Responses to di-sodium guanosine 5′-monophosphate and monosodium L-glutamate in taste receptor cells of rat fungiform papillae. J Neurophysiol. 2003;89:1434–9. doi: 10.1152/jn.00994.2002. [DOI] [PubMed] [Google Scholar]
- 50.Grosvenor W, Kaulin Y, Spielman AI, et al. Biochemical enrichment and biophysical characterization of a taste receptor for L-arginine from the catfish, Ictalurus puntatus. BMC Neurosci. 2004;5:25. doi: 10.1186/1471-2202-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosvenor W, Feigin AM, Spielman AI, et al. The arginine taste receptor. Physiology, biochemistry, and immunohistochemistry. Ann N Y Acad Sci. 1998;855:134–42. doi: 10.1111/j.1749-6632.1998.tb10555.x. [DOI] [PubMed] [Google Scholar]
- 52.Kumazawa T, Brand JG, Teeter JH. Amino acid-activated channels in the catfish taste system. Biophys J. 1998;75:2757–66. doi: 10.1016/S0006-3495(98)77719-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finger TE, Bryant BP, Kalinoski DL, et al. Differential localization of putative amino acid receptors in taste buds of the channel catfish, Ictalurus punctatus. J Comp Neurol. 1996;373:129–38. doi: 10.1002/(SICI)1096-9861(19960909)373:1<129::AID-CNE11>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 54.Beauchamp GK, Maller O, Rogers JG. Flavor preferences in cats (Felis catus and Panthera sp. ) J Comp Physiol Psychol. 1977;91:1118–1127. [Google Scholar]
- 55.Bartoshuk LM, Jacobs HL, Nichols TL, Hoff LA, Ryckman JJ. Taste rejection of nonnutritive sweeteners in cats. J Comp Physiol Psychol. 1975;89:971–5. doi: 10.1037/h0077172. [DOI] [PubMed] [Google Scholar]
- 56.Halpern BP. Gustatory nerve responses in the chicken. Am J Physiol. 1962;203:541–4. doi: 10.1152/ajplegacy.1962.203.3.541. [DOI] [PubMed] [Google Scholar]
- 57.Ganchrow JR, Steiner JE, Bartana A. Behavioral reactions to gustatory stimuli in young chicks (Gallus gallus domesticus) Dev Psychobiol. 1990;23:103–17. doi: 10.1002/dev.420230202. [DOI] [PubMed] [Google Scholar]
- 58.Kare MR. Comparative aspects of the sense of taste. In: Kare MR, Halpern BP, editors. Physiological and Behavioral Aspects of Taste. The University of Chicago Press; Chicago: 1961. pp. 6–15. [Google Scholar]
- 59.Li X, Glaser D, Li W, et al. Analyses of sweet receptor gene (Tas1r2) and preference for sweet stimuli in species of Carnivora. J Hered. 2009;100(Suppl 1):S90–100. doi: 10.1093/jhered/esp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glaser D, Tinti JM, Nofre C. Evolution of the sweetness receptor in primates. I. Why does alitame taste sweet in all Prosimians and Simians, and aspartame only in Old World Simians? Chem Senses. 1995;20:573–584. doi: 10.1093/chemse/20.5.573. [DOI] [PubMed] [Google Scholar]
- 61.Bachmanov AA, Tordoff MG, Beauchamp GK. Sweetener preference of C57BL/6ByJ and 129P3/J mice. Chem Senses. 2001;26:905–13. doi: 10.1093/chemse/26.7.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roura E, Humphre B, Klasingc K, Swartd M. Is the pig a good umami sensing model for humans? A comparative taste receptor study. Flavour Fragr J. 2011;26:282–285. [Google Scholar]
- 63.Ishimaru Y, Okada S, Naito H, et al. Two families of candidate taste receptors in fishes. Mech Dev. 2005;122:1310–21. doi: 10.1016/j.mod.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Shi P, Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- 65.Zhao H, Yang JR, Xu H, Zhang J. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol Biol Evol. 2010;27:2669–73. doi: 10.1093/molbev/msq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li R, Fan W, Tian G, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–7. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sato JJ, Wolsan M. Loss or major reduction of umami taste sensation in pinnipeds. Naturwissenschaften. 2012;99:655–9. doi: 10.1007/s00114-012-0939-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao H, Xu D, Zhang S, Zhang J. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol Evol. 2011;4:73–9. doi: 10.1093/gbe/evr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagerstrom MC, Hellstrom AR, Gloriam DE, et al. The G protein-coupled receptor subset of the chicken genome. PLoS Comput Biol. 2006;2:e54. doi: 10.1371/journal.pcbi.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao H, Zhou Y, Pinto CM, et al. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol Biol Evol. 2010;27:2642–50. doi: 10.1093/molbev/msq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daly K, Al-Rammahi M, Arora DK, et al. Expression of sweet receptor components in equine small intestine: relevance to intestinal glucose transport. Am J Physiol Regul Integr Comp Physiol. 2012;303:R199–208. doi: 10.1152/ajpregu.00031.2012. [DOI] [PubMed] [Google Scholar]
- 72.Li X, Li W, Wang H, et al. Pseudogenization of a sweet-receptor gene accounts for cats’ indifference toward sugar. PLoS Genet. 2005;1:27–35. doi: 10.1371/journal.pgen.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang P, Josue J, Li X, et al. Major taste loss in carnivorous mammals. Proc Natl Acad Sci USA. 2012;109:4956–61. doi: 10.1073/pnas.1118360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grace J, Russek M. The influence of previous experience on the taste behavior of dogs toward sucrose and saccharin. Physiol Behav. 1969;4:553–558. [Google Scholar]
- 75.Ferrell F. Preference for sugars and nonnutritive sweeteners in young beagles. Neurosci Biobehav Rev. 1984;8:199–203. doi: 10.1016/0149-7634(84)90041-1. [DOI] [PubMed] [Google Scholar]
- 76.Martinez del Rio C, Stevens BR, Daneke DE, Andreadis PT. Physiological correlates of preference and aversion for sugars in three species of birds. Physiol Zool. 1988;61:222–229. [Google Scholar]
- 77.Matson KD, Millam JR, Klasing KC. Thresholds for sweet, salt, and sour taste stimuli in cockatiels (Nymphicus hollandicus) Zoo Biol. 2001;20:1–13. doi: 10.1002/zoo.1001. [DOI] [PubMed] [Google Scholar]
- 78.Stiles FG. Taste preferences, color preferences, and flower choice in humminbirds. The Condor. 1976;78:10–26. [Google Scholar]
- 79.Thompson JD, Elias DJ, Shumake SA, Gaddis SE. Taste preferences of the common vampire bat (Desmodus rotundus) J Chem Ecol. 1982;8:715–721. doi: 10.1007/BF00988313. [DOI] [PubMed] [Google Scholar]
- 80.Danilova V, Hellekant G, Tinti JM, Nofre C. Gustatory responses of the hamster Mesocricetus auratus to various compounds considered sweet by humans. J Neurophysiol. 1998;80:2102–2112. doi: 10.1152/jn.1998.80.4.2102. [DOI] [PubMed] [Google Scholar]
- 81.Bachmanov AA, Tordoff MG, Beauchamp GK. Sweetener preference of C57BL/6ByJ and 129P3/J mice. Chem Senses. 2001;26:905–913. doi: 10.1093/chemse/26.7.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Inoue M, McCaughey SA, Bachmanov AA, Beauchamp GK. Whole-nerve chorda tympani responses to sweeteners in C57BL/6ByJ and 129P3/J mice. Chem Senses. 2001;26:915–923. doi: 10.1093/chemse/26.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winnig M, Bufe B, Meyerhof W. Valine 738 and lysine 735 in the fifth transmembrane domain of rTas1r3 mediate insensitivity towards lactisole of the rat sweet taste receptor. BMC Neurosci. 2005;6:22. doi: 10.1186/1471-2202-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang P, Ji Q, Liu Z, et al. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J Biol Chem. 2004;279:45068–75. doi: 10.1074/jbc.M406779200. [DOI] [PubMed] [Google Scholar]
- 85.Xu H, Staszewski L, Tang H, et al. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA. 2004;101:14258–63. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang P, Cui M, Ji Q, et al. Molecular mechanisms of sweet receptor function. Chem Senses. 2005;30(Suppl 1):i17–i18. doi: 10.1093/chemse/bjh091. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y, Hoon MA, Chandrashekar J, et al. Coding of sweet, bitter, and umami tastes. Different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 88.Jiang P, Cui M, Zhao B, et al. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J Biol Chem. 2005;280:34296–305. doi: 10.1074/jbc.M505255200. [DOI] [PubMed] [Google Scholar]
- 89.Jiang P, Cui M, Zhao B, et al. Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J Biol Chem. 2005;280:15238–46. doi: 10.1074/jbc.M414287200. [DOI] [PubMed] [Google Scholar]
- 90.Li X, Bachmanov AA, Maehashi K, et al. Sweet Taste Receptor Gene Variation and Aspartame Taste in Primates and Other Species. Chem Senses. 2011;36:453–475. doi: 10.1093/chemse/bjq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu B, Ha M, Meng XY, et al. Molecular Mechanism of Species-Dependent Sweet Taste toward Artificial Sweeteners. J Neurosci. 2011;31:11070–6. doi: 10.1523/JNEUROSCI.0791-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim UK, Wooding S, Riaz N, Jorde LB, Drayna D. Variation in the human TAS1R taste receptor genes. Chem Senses. 2006;31:599–611. doi: 10.1093/chemse/bjj065. [DOI] [PubMed] [Google Scholar]
- 93.Lu K, McDaniel AH, Tordoff MG, et al. No relationship between sequence variation in protein coding regions of the Tas1r3 gene and saccharin preference in rats. Chem Senses. 2005;30:231–40. doi: 10.1093/chemse/bji019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reed DR, Li S, Li X, et al. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci. 2004;24:938–46. doi: 10.1523/JNEUROSCI.1374-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reed DR, McDaniel AH. The human sweet tooth. BMC Oral Health. 2006;6(Suppl 1):S17. doi: 10.1186/1472-6831-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reed DR, Li X, Bachmanov AA, Mascioli K, Beauchamp GK. The molecular basis of the mammalian sweet tooth. In: Medeiros-Neto G, Halpern A, Bouchard C, editors. Progress in Obesity Research. John Libbey Eurotext Ltd; London: 2003. pp. 304–306. [Google Scholar]
- 97.Reed DR, Bachmanov AA, Beauchamp GK, Tordoff MG, Price RA. Heritable variation in food preferences and their contribution to obesity. Behav Genet. 1997;27:373–387. doi: 10.1023/a:1025692031673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reed DR, Tanaka T, McDaniel AH. Diverse tastes: Genetics of sweet and bitter perception. Physiol Behav. 2006;88:215–26. doi: 10.1016/j.physbeh.2006.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drayna D. Human taste genetics. Annu Rev Genomics Hum Genet. 2005;6:217–35. doi: 10.1146/annurev.genom.6.080604.162340. [DOI] [PubMed] [Google Scholar]
- 100.Kim UK, Breslin PA, Reed D, Drayna D. Genetics of human taste perception. J Dent Res. 2004;83:448–53. doi: 10.1177/154405910408300603. [DOI] [PubMed] [Google Scholar]
- 101.Lugaz O, Pillias AM, Faurion A. A new specific ageusia: some humans cannot taste L-glutamate. Chem Senses. 2002;27:105–15. doi: 10.1093/chemse/27.2.105. [DOI] [PubMed] [Google Scholar]
- 102.Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr Biol. 2009;19:1288–93. doi: 10.1016/j.cub.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eny KM, Wolever TM, Corey PN, El-Sohemy A. Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. Am J Clin Nutr. 2010;92:1501–10. doi: 10.3945/ajcn.2010.29836. [DOI] [PubMed] [Google Scholar]
- 104.Chen QY, Alarcon S, Tharp A, et al. Perceptual variation in umami taste and polymorphisms in TAS1R taste receptor genes. Am J Clin Nutr. 2009;90:770S–779S. doi: 10.3945/ajcn.2009.27462N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raliou M, Boucher Y, Wiencis A, et al. Tas1R1-Tas1R3 taste receptor variants in human fungiform papillae. Neurosci Lett. 2009;451:217–21. doi: 10.1016/j.neulet.2008.12.060. [DOI] [PubMed] [Google Scholar]
- 106.Shigemura N, Shirosaki S, Sanematsu K, Yoshida R, Ninomiya Y. Genetic and molecular basis of individual differences in human umami taste perception. PLoS One. 2009;4:e6717. doi: 10.1371/journal.pone.0006717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fuller JL. Single-locus control of saccharin preference in mice. J Hered. 1974;65:33–36. doi: 10.1093/oxfordjournals.jhered.a108452. [DOI] [PubMed] [Google Scholar]
- 108.Lush IE. The genetics of tasting in mice. VI. Saccharin, acesulfame, dulcin and sucrose. Genet Res. 1989;53:95–99. doi: 10.1017/s0016672300027968. [DOI] [PubMed] [Google Scholar]
- 109.Lush IE, Hornigold N, King P, Stoye JP. The genetics of tasting in mice. VII. Glycine revisited, and the chromosomal location of Sac and Soa. Genet Res. 1995;66:167–174. doi: 10.1017/s0016672300034510. [DOI] [PubMed] [Google Scholar]
- 110.Belknap JK, Crabbe JC, Plomin R, et al. Single-locus control of saccharin intake in BXD/Ty recombinant inbred (RI) mice: Some methodological implications for RI strain analysis. Behav Genet. 1992;22:81–100. doi: 10.1007/BF01066794. [DOI] [PubMed] [Google Scholar]
- 111.Bachmanov AA, Li X, Reed DR, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li X, Bachmanov AA, Li S, et al. Genetic, physical and comparative map of the subtelomeric region of mouse chromosome 4. Mamm Genome. 2002;13:5–19. doi: 10.1007/s0033501-2109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr Biol. 2005;15:1948–52. doi: 10.1016/j.cub.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 114.Inoue M, Reed DR, Li X, et al. Allelic variation of the Tas1r3 taste receptor gene selectively affects behavioral and neural taste responses to sweeteners in the F2 hybrids between C57BL/6ByJ and 129P3/J mice. J Neurosci. 2004;24:2296–303. doi: 10.1523/JNEUROSCI.4439-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Inoue M, Glendinning JI, Theodorides ML, et al. Allelic variation of the Tas1r3 taste receptor gene selectively affects taste responses to sweeteners: evidence from 129. B6-Tas1r3 congenic mice Physiol Genomics. 2007;32:82–94. doi: 10.1152/physiolgenomics.00161.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zukerman S, Glendinning JI, Margolskee RF, Sclafani A. T1R3 taste receptor is critical for sucrose but not Polycose taste. Am J Physiol Regul Integr Comp Physiol. 2009;296:R866–76. doi: 10.1152/ajpregu.90870.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Treesukosol Y, Blonde GD, Spector AC. T1R2 and T1R3 subunits are individually unnecessary for normal affective licking responses to Polycose: implications for saccharide taste receptors in mice. Am J Physiol Regul Integr Comp Physiol. 2009;296:R855–65. doi: 10.1152/ajpregu.90869.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sclafani A. The sixth taste? Appetite. 2004;43:1–3. doi: 10.1016/j.appet.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 119.Glendinning JI. Is the bitter rejection response always adaptive? Physiol Behav. 1994;56:1217–27. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- 120.Adler E, Hoon MA, Mueller KL, et al. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 121.Chandrashekar J, Mueller KL, Hoon MA, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 122.Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 123.Mueller KL, Hoon MA, Erlenbach I, et al. The receptors and coding logic for bitter taste. Nature. 2005;434:225–9. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 124.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–94. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 125.Behrens M, Foerster S, Staehler F, Raguse JD, Meyerhof W. Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogenous population of bitter responsive taste receptor cells. J Neurosci. 2007;27:12630–40. doi: 10.1523/JNEUROSCI.1168-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spector AC, Kopka SL. Rats fail to discriminate quinine from denatonium: implications for the neural coding of bitter-tasting compounds. J Neurosci. 2002;22:1937–41. doi: 10.1523/JNEUROSCI.22-05-01937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aspen J, Gatch MB, Woods JH. Training and characterization of a quinine taste discrimination in rhesus monkeys. Psychopharmacology (Berl) 1999;141:251–7. doi: 10.1007/s002130050832. [DOI] [PubMed] [Google Scholar]
- 128.Chan CY, Yoo JE, Travers SP. Diverse bitter stimuli elicit highly similar patterns of Fos-like immunoreactivity in the nucleus of the solitary tract. Chem Senses. 2004;29:573–81. doi: 10.1093/chemse/bjh062. [DOI] [PubMed] [Google Scholar]
- 129.Scott TR, Giza BK, Yan J. Gustatory neural coding in the cortex of the alert cynomolgus macaque: the quality of bitterness. J Neurophysiol. 1999;81:60–71. doi: 10.1152/jn.1999.81.1.60. [DOI] [PubMed] [Google Scholar]
- 130.Caicedo A, Roper SD. Taste receptor cells that discriminate between bitter stimuli. Science. 2001;291:1557–60. doi: 10.1126/science.291.5508.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dahl M, Erickson RP, Simon SA. Neural responses to bitter compounds in rats. Brain Res. 1997;756:22–34. doi: 10.1016/s0006-8993(97)00131-5. [DOI] [PubMed] [Google Scholar]
- 132.Frank ME, Bouverat BP, MacKinnon BI, Hettinger TP. The distinctiveness of ionic and nonionic bitter stimuli. Physiol Behav. 2004;80:421–31. doi: 10.1016/j.physbeh.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 133.Meyerhof W, Batram C, Kuhn C, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35:157–70. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 134.Behrens M, Meyerhof W. Bitter taste receptors and human bitter taste perception. Cell Mol Life Sci. 2006;63:1501–9. doi: 10.1007/s00018-006-6113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim UK, Drayna D. Genetics of individual differences in bitter taste perception: lessons from the PTC gene. Clin Genet. 2005;67:275–80. doi: 10.1111/j.1399-0004.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 136.Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat. 2005;26:199–204. doi: 10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- 137.Meyerhof W. Elucidation of mammalian bitter taste. Rev Physiol Biochem Pharmacol. 2005;154:37–72. doi: 10.1007/s10254-005-0041-0. [DOI] [PubMed] [Google Scholar]
- 138.Sawano S, Seto E, Mori T, Hayashi Y. G-protein-dependent and -independent pathways in denatonium signal transduction. Biosci Biotechnol Biochem. 2005;69:1643–51. doi: 10.1271/bbb.69.1643. [DOI] [PubMed] [Google Scholar]
- 139.Lindemann B. Taste reception. Physiol Rev. 1996;76:719–766. doi: 10.1152/physrev.1996.76.3.719. [DOI] [PubMed] [Google Scholar]
- 140.Rosenzweig S, Yan W, Dasso M, Spielman AI. Possible novel mechanism for bitter taste mediated through cGMP. J Neurophysiol. 1999;81:1661–5. doi: 10.1152/jn.1999.81.4.1661. [DOI] [PubMed] [Google Scholar]
- 141.Takeuchi H, Tsunenari T, Kurahashi T, Kaneko A. Physiology of morphologically identified cells of the bullfrog fungiform papilla. Neuroreport. 2001;12:2957–62. doi: 10.1097/00001756-200109170-00040. [DOI] [PubMed] [Google Scholar]
- 142.Jacobs WW. Taste responses in wild and domestic guinea pigs. Physiol Behav. 1978;20:579–588. doi: 10.1016/0031-9384(78)90250-0. [DOI] [PubMed] [Google Scholar]
- 143.Glendinning JI. Is the bitter rejection response always adaptive? Physiol Behav. 1994;56:1217–1227. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- 144.Nolte DL, Mason JR, Lewis SL. Tolerance of bitter compounds by an herbivore, Cavia porcellus. J Chem Ecol. 1994;20:303–308. doi: 10.1007/BF02064438. [DOI] [PubMed] [Google Scholar]
- 145.Field KL, Bachmanov AA, Mennella JA, Beauchamp GK, Kimball BA. Protein hydrolysates are avoided by herbivores but not by omnivores in two-choice preference tests. PLoS One. 2009;4:e4126. doi: 10.1371/journal.pone.0004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Field KL, Beauchamp GK, Kimball BA, Mennella JA, Bachmanov AA. Bitter avoidance in guinea pigs (Cavia porcellus) and mice (Mus musculus and Peromyscus leucopus) J Comp Psychol. 2010;124:455–9. doi: 10.1037/a0020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nelson TM, Munger SD, Boughter JD., Jr Taste sensitivities to PROP and PTC vary independently in mice. Chem Senses. 2003;28:695–704. doi: 10.1093/chemse/bjg062. [DOI] [PubMed] [Google Scholar]
- 148.Bachmanov AA, Beauchamp GK. Taste Receptor Genes. Annu Rev Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 150.Bufe B, Breslin PA, Kuhn C, et al. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–7. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wooding S, Bufe B, Grassi C, et al. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–4. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- 152.Go Y. Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol Biol Evol. 2006;23:964–72. doi: 10.1093/molbev/msj106. [DOI] [PubMed] [Google Scholar]
- 153.Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 2003;20:805–14. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- 154.Conte C, Ebeling M, Marcuz A, Nef P, Andres-Barquin PJ. Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol Genomics. 2003;14:73–82. doi: 10.1152/physiolgenomics.00060.2003. [DOI] [PubMed] [Google Scholar]
- 155.Liman ER. Use it or lose it: molecular evolution of sensory signaling in primates. Pflugers Arch. 2006;453:125–31. doi: 10.1007/s00424-006-0120-3. [DOI] [PubMed] [Google Scholar]
- 156.Ueda T, Ugawa S, Ishida Y, et al. Identification of coding single-nucleotide polymorphisms in human taste receptor genes involving bitter tasting. Biochem Biophys Res Commun. 2001;285:147–51. doi: 10.1006/bbrc.2001.5136. [DOI] [PubMed] [Google Scholar]
- 157.Wang X, Thomas SD, Zhang J. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 2004;13:2671–8. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- 158.Guo SW, Reed DR. The genetics of phenylthiocarbamide perception. Ann Hum Biol. 2001;28:111–42. doi: 10.1080/03014460151056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim UK, Jorgenson E, Coon H, et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–5. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 160.Drayna D, Coon H, Kim UK, et al. Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum Genet. 2003;112:567–72. doi: 10.1007/s00439-003-0911-y. [DOI] [PubMed] [Google Scholar]
- 161.Prodi DA, Drayna D, Forabosco P, et al. Bitter taste study in a sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chem Senses. 2004;29:697–702. doi: 10.1093/chemse/bjh074. [DOI] [PubMed] [Google Scholar]
- 162.Mennella JA, Pepino MY, Reed DR. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics. 2005;115:e216–22. doi: 10.1542/peds.2004-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Duffy VB, Davidson AC, Kidd JR, et al. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 2004;28:1629–37. doi: 10.1097/01.ALC.0000145789.55183.D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–4. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 165.Wooding S, Kim UK, Bamshad MJ, et al. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–46. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Soranzo N, Bufe B, Sabeti PC, et al. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 2005;15:1257–65. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 167.Roudnitzky N, Bufe B, Thalmann S, et al. Genomic, genetic and functional dissection of bitter taste responses to artificial sweeteners. Hum Mol Genet. 2011;20:3437–49. doi: 10.1093/hmg/ddr252. [DOI] [PubMed] [Google Scholar]
- 168.Reed DR, Zhu G, Breslin PA, et al. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 2010;19:4278–85. doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nelson TM, Munger SD, Boughter JD., Jr Haplotypes at the Tas2r locus on distal chromosome 6 vary with quinine taste sensitivity in inbred mice. BMC Genet. 2005;6:32. doi: 10.1186/1471-2156-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Boughter JD, Jr, Raghow S, Nelson TM, Munger SD. Inbred mouse strains C57BL/6J and DBA/2J vary in sensitivity to a subset of bitter stimuli. BMC Genet. 2005;6:36. doi: 10.1186/1471-2156-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fitzsimons JT. The Physiology of Thirst and Sodium Appetite. Cambridge University Press; Cambridge: 1979. [PubMed] [Google Scholar]
- 172.Denton D. The hunger for salt: An anthropological, physiological and medical analysis. Springer-Verlag; Berlin: 1984. [Google Scholar]
- 173.Beauchamp GK, Stein LJ. Salt Taste. In: Firestein S, Beauchamp GK, editors. Olfaction and Taste. Elsevier/Academic Press; San Diego: 2008. pp. 401–408. [Google Scholar]
- 174.Schiffman S, Lockhead E, Mars F. Amiloride reduces the taste intensity of Na+ and Li+ salts and sweeteners. Proc Natl Acad Sci USA. 1983;80:6136–6140. doi: 10.1073/pnas.80.19.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Heck GL, Mierson S, DeSimone JA. Salt taste transduction occurs through an amiloride-sensitive sodium transport pathway. Science. 1984;223:403–405. doi: 10.1126/science.6691151. [DOI] [PubMed] [Google Scholar]
- 176.Brand J, Teeter J, Silver W. Inhibition by amiloride of chorda tympani responses evoked by monovalent salts. Brain Res. 1985;334:207–214. doi: 10.1016/0006-8993(85)90212-4. [DOI] [PubMed] [Google Scholar]
- 177.Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–67. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- 178.Bachmanov AA, Li X, Reed DR, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001;26:925–33. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li XJ, Blackshaw S, Snyder SH. Expression and localization of amiloride-sensitive sodium channel indicate a role for non-taste cells in taste perception. Proc Natl Acad Sci USA. 1994;91:1814–8. doi: 10.1073/pnas.91.5.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lindemann B, Barbry P, Kretz O, Bock R. Occurrence of ENaC subunit mRNA and immunocytochemistry of the channel subunits in taste buds of the rat vallate papilla. Ann N Y Acad Sci. 1998;855:116–27. doi: 10.1111/j.1749-6632.1998.tb10553.x. [DOI] [PubMed] [Google Scholar]
- 181.Kretz O, Barbry P, Bock R, Lindemann B. Differential expression of RNA and protein of the three pore-forming subunits of the amiloride-sensitive epithelial sodium channel in taste buds of the rat. J Histochem Cytochem. 1999;47:51–64. doi: 10.1177/002215549904700106. [DOI] [PubMed] [Google Scholar]
- 182.Lin W, Finger TE, Rossier BC, Kinnamon SC. Epithelial Na+ channel subunits in rat taste cells: localization and regulation by aldosterone. J Comp Neurol. 1999;405:406–20. doi: 10.1002/(sici)1096-9861(19990315)405:3<406::aid-cne10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 183.Lindemann B. Sodium taste. Curr Opin Nephrol Hypertens. 1997;6:425–9. doi: 10.1097/00041552-199709000-00003. [DOI] [PubMed] [Google Scholar]
- 184.Halpern BP. Amiloride and vertebrate gustatory responses to NaCl. Neurosci Biobehav Rev. 1998;23:5–47. doi: 10.1016/s0149-7634(97)00063-8. [DOI] [PubMed] [Google Scholar]
- 185.Boughter JD, Jr, Gilbertson TA. From channels to behavior: an integrative model of NaCl taste. Neuron. 1999;22:213–5. doi: 10.1016/s0896-6273(00)81082-x. [DOI] [PubMed] [Google Scholar]
- 186.Bosak N, Inoue M, Nelson T, et al. Epithelial sodium channel (ENaC) is involved in reception of sodium taste: evidence from mice with a tissue-specific conditional targeted mutation of the ENaCa gene (Abstract). Chem Senses; AChemS XXXII Annual Meeting; 2010 April 21–25; St. Petersburg (FL). 2010. [DOI] [Google Scholar]
- 187.Chandrashekar J, Kuhn C, Oka Y, et al. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ye Q, Heck GL, DeSimone JA. Voltage dependence of the rat chorda tympani response to Na+ salts: implications for the functional organization of taste receptor cells. J Neurophysiol. 1993;70:167–78. doi: 10.1152/jn.1993.70.1.167. [DOI] [PubMed] [Google Scholar]
- 189.Mierson S, Olson MM, Tietz AE. Basolateral amiloride-sensitive Na+ transport pathway in rat tongue epithelium. J Neurophysiol. 1996;76:1297–309. doi: 10.1152/jn.1996.76.2.1297. [DOI] [PubMed] [Google Scholar]
- 190.Yoshida R, Horio N, Murata Y, et al. NaCl responsive taste cells in the mouse fungiform taste buds. Neuroscience. 2009;159:795–803. doi: 10.1016/j.neuroscience.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 191.Formaker BK, Hill DL. An analysis of residual NaCl taste response after amiloride. Am J Physiol. 1988;255:R1002–7. doi: 10.1152/ajpregu.1988.255.6.R1002. [DOI] [PubMed] [Google Scholar]
- 192.Elliott EJ, Simon SA. The anion in salt taste: a possible role of paracellular pathways. Brain Res. 1990;535:9–17. doi: 10.1016/0006-8993(90)91817-z. [DOI] [PubMed] [Google Scholar]
- 193.Rehnberg BG, MacKinnon BI, Hettinger TP, Frank ME. Anion modulation of taste responses in sodium-sensitive neurons of the hamster chorda tympani nerve. J Gen Physiol. 1993;101:453–65. doi: 10.1085/jgp.101.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Oka Y, Butnaru M, von Buchholtz L, Ryba NJ, Zuker CS. High salt recruits aversive taste pathways. Nature. 2013;494:472–475. doi: 10.1038/nature11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Caterina MJ, Schumacher MA, Tominaga M, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 196.Lyall V, Heck GL, Vinnikova AK, et al. The mammalian amiloride-insensitive non-specific salt taste receptor is a vanilloid receptor-1 variant. J Physiol. 2004;558:147–59. doi: 10.1113/jphysiol.2004.065656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ruiz C, Gutknecht S, Delay E, Kinnamon S. Detection of NaCl and KCl in TRPV1 knockout mice. Chem Senses. 2006;31:813–20. doi: 10.1093/chemse/bjl024. [DOI] [PubMed] [Google Scholar]
- 198.Treesukosol Y, Lyall V, Heck GL, DeSimone JA, Spector AC. A psychophysical and electrophysiological analysis of salt taste in Trpv1 null mice. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1799–809. doi: 10.1152/ajpregu.00587.2006. [DOI] [PubMed] [Google Scholar]
- 199.Breza JM, Contreras RJ. Anion size modulates salt taste in rats. J Neurophysiol. 2012;107:1632–48. doi: 10.1152/jn.00621.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Smith KR, Treesukosol Y, Paedae AB, Contreras RJ, Spector AC. Contribution of the TRPV1 channel to salt taste quality in mice as assessed by conditioned taste aversion generalization and chorda tympani nerve responses. Am J Physiol Regul Integr Comp Physiol. 2012;303:R1195–205. doi: 10.1152/ajpregu.00154.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kido MA, Muroya H, Yamaza T, Terada Y, Tanaka T. Vanilloid receptor expression in the rat tongue and palate. J Dent Res. 2003;82:393–7. doi: 10.1177/154405910308200513. [DOI] [PubMed] [Google Scholar]
- 202.Ishida Y, Ugawa S, Ueda T, Murakami S, Shimada S. Vanilloid receptor subtype-1 (VR1) is specifically localized to taste papillae. Brain Res Mol Brain Res. 2002;107:17–22. doi: 10.1016/s0169-328x(02)00441-2. [DOI] [PubMed] [Google Scholar]
- 203.Moon YW, Lee JH, Yoo SB, Jahng JW. Capsaicin receptors are colocalized with sweet/bitter receptors in the taste sensing cells of circumvallate papillae. Genes Nutr. 2010;5:251–5. doi: 10.1007/s12263-009-0164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Riera CE, Vogel H, Simon SA, le Coutre J. Artificial sweeteners and salts producing a metallic taste sensation activate TRPV1 receptors. Am J Physiol Regul Integr Comp Physiol. 2007;293:R626–34. doi: 10.1152/ajpregu.00286.2007. [DOI] [PubMed] [Google Scholar]
- 205.Moyer B, Zlotnik A, Hevezi P, et al. Identification of TRPML3 (MCOLN3) as a Salty Taste Receptor and Use in Assays for Identifying Taste (Salty) Modulators and/or Therapeutics that Modulate Sodium Transport, Absorption or Excretion and/or Aldosterone and/or Vasopressin Production or Release. WO/2009/008950. Patent Pub No. 2009
- 206.Castiglioni AJ, Remis NN, Flores EN, Garcia-Anoveros J. Expression and vesicular localization of mouse Trpml3 in stria vascularis, hair cells, and vomeronasal and olfactory receptor neurons. J Comp Neurol. 2011;519:1095–114. doi: 10.1002/cne.22554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ossebaard CA, Polet IA, Smith DV. Amiloride effects on taste quality: comparison of single and multiple response category procedures. Chem Senses. 1997;22:267–75. doi: 10.1093/chemse/22.3.267. [DOI] [PubMed] [Google Scholar]
- 208.Huque T, Cowart BJ, Dankulich-Nagrudny L, et al. Sour ageusia in two individuals implicates ion channels of the ASIC and PKD families in human sour taste perception at the anterior tongue. PLoS One. 2009;4:e7347. doi: 10.1371/journal.pone.0007347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stähler F, Riedel K, Demgensky S, et al. A role of the epithelial sodium channel in human salt taste transduction? Chemosensory Perception. 2008;1:78–90. [Google Scholar]
- 210.Brand JG, Huque T. Human salty taste receptor and methods of modulating salty taste perception. WO2008051447 A2 2008 Patent Pub No.
- 211.Ji HL, Zhao RZ, Chen ZX, et al. delta ENaC: a novel divergent amiloride-inhibitable sodium channel. Am J Physiol Lung Cell Mol Physiol. 2012;303:L1013–26. doi: 10.1152/ajplung.00206.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Giraldez T, Rojas P, Jou J, Flores C, Alvarez de la Rosa D. The epithelial sodium channel delta-subunit: new notes for an old song. Am J Physiol Renal Physiol. 2012;303:F328–38. doi: 10.1152/ajprenal.00116.2012. [DOI] [PubMed] [Google Scholar]
- 213.Hellekant G, Ninomiya Y, Danilova V. Taste in chimpanzees II: single chorda tympani fibers. Physiol Behav. 1997;61:829–41. doi: 10.1016/s0031-9384(96)00562-8. [DOI] [PubMed] [Google Scholar]
- 214.Danilova V, Danilov Y, Roberts T, et al. Sense of taste in a new world monkey, the common marmoset: recordings from the chorda tympani and glossopharyngeal nerves. J Neurophysiol. 2002;88:579–94. doi: 10.1152/jn.2002.88.2.579. [DOI] [PubMed] [Google Scholar]
- 215.Eylam S, Spector AC. Oral amiloride treatment decreases taste sensitivity to sodium salts in C57BL/6J and DBA/2J mice. Chem Senses. 2003;28:447–58. doi: 10.1093/chemse/28.5.447. [DOI] [PubMed] [Google Scholar]
- 216.Liu L, Leonard AS, Motto DG, et al. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 2003;39:133–46. doi: 10.1016/s0896-6273(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 217.Ishiwatari Y, Bachmanov AA. NaCl taste thresholds in 13 inbred mouse strains. Chem Senses. 2012;37:497–508. doi: 10.1093/chemse/bjr135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Shigemura N, Ohkuri T, Sadamitsu C, et al. Amiloride-sensitive NaCl taste responses are associated with genetic variation of ENaC α-subunit in mice. Am J Physiol Regul Integr Comp Physiol. 2008;294:R66–R75. doi: 10.1152/ajpregu.00420.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Dias AG, Rousseau D, Duizer L, et al. Genetic variation in putative salt taste receptors and salt taste perception in humans. Chem Senses. 2013;38:137–45. doi: 10.1093/chemse/bjs090. [DOI] [PubMed] [Google Scholar]
- 220.Nelson TM, Lopezjimenez ND, Tessarollo L, et al. Taste function in mice with a targeted mutation of the Pkd1l3 gene. Chem Senses. 2010;35:565–77. doi: 10.1093/chemse/bjq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ishimaru Y, Inada H, Kubota M, et al. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci USA. 2006;103:12569–74. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Huang AL, Chen X, Hoon MA, et al. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–8. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.LopezJimenez ND, Sainz E, Cavenagh MM, et al. Two novel genes, Gpr113, which encodes a family 2 G-protein-coupled receptor, and Trcg1, are selectively expressed in taste receptor cells. Genomics. 2005;85:472–82. doi: 10.1016/j.ygeno.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 224.Horio N, Yoshida R, Yasumatsu K, et al. Sour taste responses in mice lacking PKD channels. PLoS One. 2011;6:e20007. doi: 10.1371/journal.pone.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wise PM, Hansen JL, Reed DR, Breslin PA. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem Senses. 2007;32:749–54. doi: 10.1093/chemse/bjm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bachmanov AA, Tordoff MG, Beauchamp GK. Acid acceptance in 28 mouse strains (Abstract) Chem Senses. 2000;25:600. [Google Scholar]
- 227.Gilbertson TA, Baquero AF, Spray-Watson KJ. Water taste: the importance of osmotic sensing in the oral cavity. J Water Health. 2006;4(Suppl 1):35–40. [PubMed] [Google Scholar]
- 228.Gilbertson TA, Kim I, Siears NL, Zhang H, Liu L. The water response in taste cells: expression of aquaporin-1, -2 and -5 and the characterization of hypoosmic-induced currents in mammalian taste cells (Abstract) Chem Senses. 1999;24:596. [Google Scholar]
- 229.Bartoshuk LM. NaCl thresholds in man: thresholds for water taste or NaCl taste? J Comp Physiol Psychol. 1974;87:310–25. doi: 10.1037/h0036788. [DOI] [PubMed] [Google Scholar]
- 230.Bartoshuk LM. Water taste in mammals. In: Weijnen JAWM, Mendelson J, editors. Drinking behavior: oral stimulation, reinforcement, and preference. Plenum Press; New York: 1977. pp. 317–339. [Google Scholar]
- 231.Bartoshuk LM, McBurney DH, Pfaffmann C. Taste of sodium chloride solutions after adaptation to sodium chloride: implications for the “water taste”. Science. 1964;143:967–8. doi: 10.1126/science.143.3609.967. [DOI] [PubMed] [Google Scholar]
- 232.McBurney DH, Bartoshuk LM. Water taste in mammals. In: Schneider D, editor. Olfaction and Taste IV. Proceedings of the Fourth International Symposium held in Starnberg; Germany. August 2–4 1971; Stuttgart: Wissenschaftlische Verlagsgesellschaft MBH; 1972. pp. 329–335. [Google Scholar]
- 233.DuBois GE. Unraveling the biochemistry of sweet and umami tastes. Proc Natl Acad Sci USA. 2004;101:13972–3. doi: 10.1073/pnas.0405991101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Galindo-Cuspinera V, Winnig M, Bufe B, Meyerhof W, Breslin PA. A TAS1R receptor-based explanation of sweet ‘water-taste’. Nature. 2006;441:354–7. doi: 10.1038/nature04765. [DOI] [PubMed] [Google Scholar]
- 235.Bachmanov AA, Kiefer SW, Molina JC, et al. Chemosensory factors influencing alcohol perception, preferences, and consumption. Alcohol Clin Exp Res. 2003;27:220–31. doi: 10.1097/01.ALC.0000051021.99641.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bachmanov AA, Reed DR, Li X, et al. Voluntary ethanol consumption by mice: genome-wide analysis of quantitative trait loci and their interactions in a C57BL/6ByJ x 129P3/J F2 intercross. Genome Res. 2002;12:1257–68. doi: 10.1101/gr.129702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hinrichs AL, Wang JC, Bufe B, et al. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78:103–11. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Dunkel A, Koster J, Hofmann T. Molecular and sensory characterization of gamma-glutamyl peptides as key contributors to the kokumi taste of edible beans (Phaseolus vulgaris L. ) J Agric Food Chem. 2007;55:6712–9. doi: 10.1021/jf071276u. [DOI] [PubMed] [Google Scholar]
- 239.Ueda Y, Yonemitsu M, Tsubuku T, Sakaguchi M, Miyajima R. Flavor characteristics of glutathione in raw and cooked foodstuffs. Biosci Biotechnol Biochem. 1997;61:1977–80. doi: 10.1271/bbb.61.1977. [DOI] [PubMed] [Google Scholar]
- 240.Ohsu T, Amino Y, Nagasaki H, et al. Involvement of the calcium-sensing receptor in human taste perception. J Biol Chem. 2010;285:1016–22. doi: 10.1074/jbc.M109.029165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.San Gabriel A, Uneyama H, Maekawa T, Torii K. The calcium-sensing receptor in taste tissue. Biochem Biophys Res Commun. 2009;378:414–8. doi: 10.1016/j.bbrc.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 242.Maruyama Y, Yasuda R, Kuroda M, Eto Y. Kokumi substances, enhancers of basic tastes, induce responses in calcium-sensing receptor expressing taste cells. PLoS One. 2012;7:e34489. doi: 10.1371/journal.pone.0034489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tordoff MG. Calcium: taste, intake, and appetite. Physiol Rev. 2001;81:1567–97. doi: 10.1152/physrev.2001.81.4.1567. [DOI] [PubMed] [Google Scholar]
- 244.Tordoff MG, Shao H, Alarcon LK, et al. Involvement of T1R3 in calcium-magnesium taste. Physiol Genomics. 2008;34:338–48. doi: 10.1152/physiolgenomics.90200.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tordoff MG, Alarcon LK, Valmeki S, Jiang P. T1R3: a human calcium taste receptor. Sci Rep. 2012;2:496. doi: 10.1038/srep00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Fukuwatari T, Kawada T, Tsuruta M, et al. Expression of the putative membrane fatty acid transporter (FAT) in taste buds of the circumvallate papillae in rats. FEBS Lett. 1997;414:461–4. doi: 10.1016/s0014-5793(97)01055-7. [DOI] [PubMed] [Google Scholar]
- 247.Zhang X, Fitzsimmons RL, Cleland LG, et al. CD36/fatty acid translocase in rats: distribution, isolation from hepatocytes, and comparison with the scavenger receptor SR-B1. Lab Invest. 2003;83:317–32. doi: 10.1097/01.lab.0000059923.67198.ba. [DOI] [PubMed] [Google Scholar]
- 248.Laugerette F, Passilly-Degrace P, Patris B, et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. 2005;115:3177–84. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cartoni C, Yasumatsu K, Ohkuri T, et al. Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci. 2010;30:8376–82. doi: 10.1523/JNEUROSCI.0496-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Matsumura S, Mizushige T, Yoneda T, et al. GPR expression in the rat taste bud relating to fatty acid sensing. Biomed Res. 2007;28:49–55. doi: 10.2220/biomedres.28.49. [DOI] [PubMed] [Google Scholar]
- 251.Gaillard D, Laugerette F, Darcel N, et al. The gustatory pathway is involved in CD36-mediated orosensory perception of long-chain fatty acids in the mouse. FASEB J. 2008;22:1458–68. doi: 10.1096/fj.07-8415com. [DOI] [PubMed] [Google Scholar]
- 252.El-Yassimi A, Hichami A, Besnard P, Khan NA. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neuro-transmitter release in mouse CD36-positive gustatory cells. J Biol Chem. 2008;283:12949–59. doi: 10.1074/jbc.M707478200. [DOI] [PubMed] [Google Scholar]
- 253.Sclafani A, Ackroff K, Abumrad NA. CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1823–32. doi: 10.1152/ajpregu.00211.2007. [DOI] [PubMed] [Google Scholar]
- 254.Galindo MM, Voigt N, Stein J, et al. G protein-coupled receptors in human fat taste perception. Chem Senses. 2012;37:123–39. doi: 10.1093/chemse/bjr069. [DOI] [PubMed] [Google Scholar]
- 255.Pepino MY, Love-Gregory L, Klein S, Abumrad NA. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J Lipid Res. 2012;53:561–6. doi: 10.1194/jlr.M021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Keller KL, Liang LC, Sakimura J, et al. Common variants in the CD36 gene are associated with oral fat perception, fat preferences, and obesity in African Americans. Obesity (Silver Spring) 2012;20:1066–73. doi: 10.1038/oby.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bachmanov AA. Genetic architecture of sweet taste. In: Weerasinghe DK, DuBois GE, editors. Sweetness and Sweeteners: Biology, Chemistry and Psychophysics. American Chemical Society; Washington, D.C: 2008. pp. 18–47. [Google Scholar]
- 258.Shi P, Zhang J. Extraordinary diversity of chemosensory receptor gene repertoires among vertebrates. Results Probl Cell Differ. 2009;47:57–75. doi: 10.1007/400_2008_4. [DOI] [PubMed] [Google Scholar]
- 259.Dong D, Jones G, Zhang S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 2009;9:12. doi: 10.1186/1471-2148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Davis JK, Lowman JJ, Thomas PJ, et al. Evolution of a bitter taste receptor gene cluster in a New World sparrow. Genome Biol Evol. 2010;2:358–70. doi: 10.1093/gbe/evq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zhou Y, Dong D, Zhang S, Zhao H. Positive selection drives the evolution of bat bitter taste receptor genes. Biochem Genet. 2009;47:207–15. doi: 10.1007/s10528-008-9218-y. [DOI] [PubMed] [Google Scholar]
- 262.Oike H, Nagai T, Furuyama A, et al. Characterization of ligands for fish taste receptors. J Neurosci. 2007;27:5584–92. doi: 10.1523/JNEUROSCI.0651-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kusuhara Y, Yoshida R, Ohkuri T, Yasumatsu K, Voigt A, Hubner S, Maeda K, Boehm U, Meyerhof W, Ninomiya Y. Taste responses in mice lacking taste receptor subunit T1R1. J Physiol. 2013;591:1967–1985. doi: 10.1113/jphysiol.2012.236604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ohmoto M, Okada S, Nakamura S, Abe K, Matsumoto I. Mutually exclusive expression of Gaia and Ga14 reveales diversification of taste receptor cells in zebrafish. J Comp Neurol. 2011;519:1616–1629. doi: 10.1002/cne.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]