Abstract
Giardia lamblia is a protistan parasite that infects and colonizes the small intestine of mammals. It is a widespread and particularly endemic in the developing world. Here we present a detailed structural study by 3-D negative staining and cryo-electron tomography of a unique Giardia organelle, the ventral disc. The disc is composed of a regular array of microtubules and associated sheets, called microribbons that form a large spiral, held together by a myriad of mostly unknown associated proteins. In a previous study we analyzed by cryo-electron tomography the central microtubule portion (here called disc body) of the ventral disc and found a large portion of microtubule associated inner (MIPs) and outer proteins (MAPs) that render these microtubules hyper-stable. With this follow-up study we expanded our 3-D analysis to different parts of the disc such as the ventral and dorsal areas of the overlap zone, as well as the outer disc margin. There are intrinsic location-specific characteristics in the composition of microtubule-associated proteins between these regions, as well as large differences between the overall architecture of microtubules and microribbons. The lateral packing of microtubule-microribbon complexes varies substantially, and closer packing often comes with contracted lateral tethers that seem to hold the disc together. It appears that the marginal microtubule-microribbon complexes function as outer, laterally contractible lids that may help the cell to clamp onto the intestinal microvilli. Furthermore, we analyzed length, quantity, curvature and distribution between different zones of the disc, which we found to differ from previous publications.
Keywords: Giardia Lamblia Ventral Disc, Cryo-Electron Microscopy, Cryo-Electron Tomography, Microtubules, Microtubule-associated Proteins (MAPS), Volume Averaging
Introduction
Giardia lamblia is a widespread zoonotic intestinal parasite that was first discovered in 1681 by Anton van Leeuwenhoek (reviewed in Boreham et al., 1990; Adam, 2001). One of the ten major parasites of humans, Giardia chronically infects over one billion people worldwide, primarily in developing countries (Savioli et al., 2006; Troeger et al., 2007) and has been included in the World Health Organization (WHO) Neglected Diseases Initiative as one of a group of diseases of global importance that are linked with poverty and limit development and socio-economic improvements (Savioli et al., 2006; Thompson, 2000). Giardia has two life cycle stages: a swimming, flagellated trophozoite form that attaches to the intestinal microvilli causing giardiasis, and an infectious cyst form that persists in the environment (Gillin et al., 1996; Adam, 2001). Cysts are ingested from contaminated water or food, then excyst and colonize the small intestine of the animal host. Following excystation, trophozoites attach to the intestinal villi using a unique microtubule (MT) structure termed the ventral disc (Holberton, 1973 & 1981; Adam, 2001; Elmendorf et al., 2003; Schwartz et al., 2012). Trophozoites that reach the colon encyst based on environmental cues and are released to infect new hosts (Roxström-Lindquist et al., 2006). Proper attachment is a viable process to target for drug development because attachment is critical for Giardia cell division and pathogenesis (Adam, 2001). Many proteins localizing to the ventral disc are novel Giardia proteins that lack homology to genes found in any other eukaryotic genome (Hagen et al., 2011). Given attachment via the ventral disc is critical for Giardia cell division and pathogenesis (Adam, 2001), these novel Giardia proteins are good candidate drug targets because they can allow for specific targeting of Giardia while avoiding damaging off-target effects to human cells.
Giardia has an elaborate microtubule cytoskeleton critical for parasite growth, motility, attachment, and development in the trophozoite and cyst stages (Adam, 2001; Elmendorf et al., 2003). The microtubule cytoskeleton in Giardia consists of eight canonical flagella and several unique microtubule structures: the median body, the funis, and the ventral disc (Elmendorf et al., 2003). The eight flagella have a motile (“9+2”) structure and each axoneme possesses elaborate ancillary structures (Holberton, 1974) that may modulate motility or attachment. Giardia uses the ventral disc – a right-handed spiral of cross-linked microtubule/microribbon complexes and associated structural elements – to attach to the villi using an undefined molecular mechanism (Holberton, 1973 & 1974; Elmendorf et al., 2003). Cytoskeletal dynamics are also important for chromosome segregation (Sagolla et al., 2006; Dawson et al., 2007), flagellar duplication (Nohynková et al., 2006), disc division (Tůmová et al., 2007), and likely encystation and excystation (Feely et al., 1982).
Here, we investigated the 3-D structure and composition of the Giardia lamblia ventral disc (see Figs. 1–3; Holberton, 1973; Schwartz et al., 2012) and closely associated structures (see Fig. 2) by negative stain and cryo-electron tomography (see Fig. 5). We analyzed different regions of the disc and found striking structural variations that suggest distinct functions amongst different regions of the disc. We produced tomographic 3-D structures of the dorsal and ventral face of the ventral disc overlap zone as well as the margins and compared them to a structure we calculated earlier from the disc body (Schwartz et al., 2012). The overall architecture of the disc shows variable lateral repeats or microtubule curvature, as well as the composition of inner and outer associated proteins, of which many are still unidentified. We also measured length and number of microtubules in the disc and came up with different values than reported earlier (Holberton, 1981).
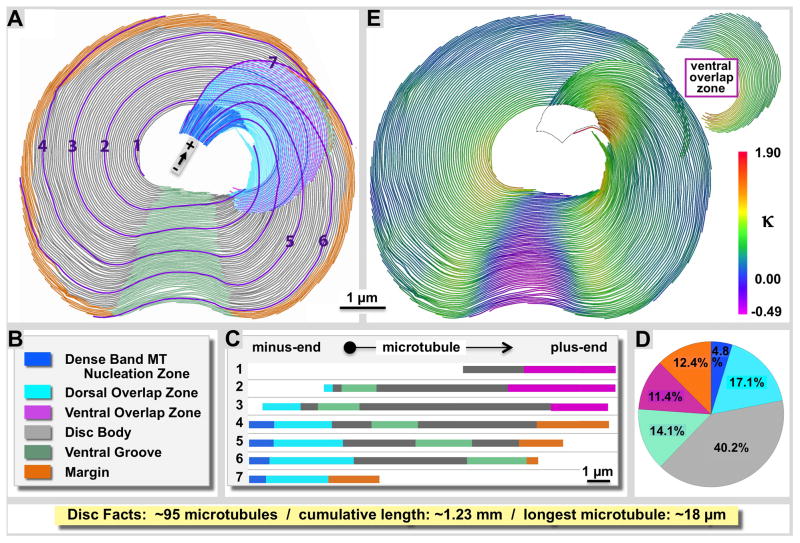
Figure 1. Structural variations between different zones of the Giardia ventral disc.
A: We have separated the ventral disc into six different regions according to the color-coded legend (B), and show seven individual microtubule traces and how they enter and exit the various regions of the disc (C). The disc forms a right-handed spiral of microtubules and regularly associated microribbons and proteins. D: Regional microtubule composition of the ventral disc. The disc body (grey) is by far the largest region with relatively little variation among lateral microtubule spacing and associated proteins. The ventral groove (green) shows an opposite curvature of the microtubule-microribbon complexes (against the spiral twist) as shown on the curvature heat map in panel E. The margin (orange), the dense band nucleation region, as well as the ventral and dorsal overlap zones (pink and cyan) feature very distinct building plans (illustrated in figures 4–7). The dense band microtubule nucleation zone ((blue; see also Figs. 3 and 4) is the origin of about 59% of the approximately 95 microtubules we counted in the disc, the others originate at the inner edge of the disc. All microtubules are parallel with the minus end towards their nucleation zone and the plus-ends at their termination zone such as margin (orange) or ventral overlap zone (pink).
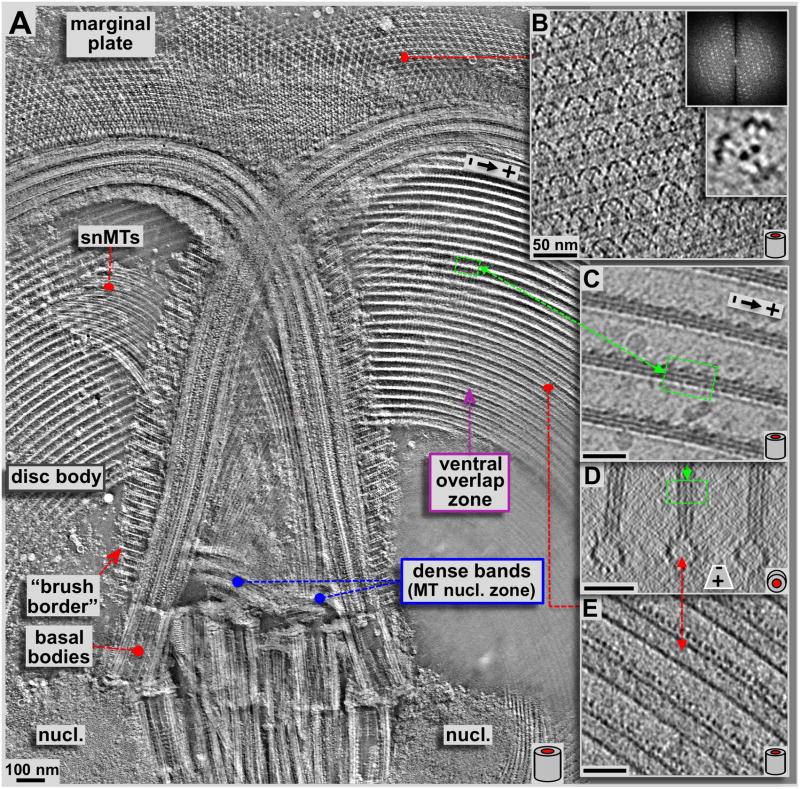
Figure 3. Detailed 3-D analysis of the upper region of the ventral disc, anterior flagella, basal bodies and the marginal plate.
A: High-resolution ~50 nm thick xy-slice (within the plane of the ventral disc) from a tomographic 3-D reconstruction of a negatively stained, isolated Giardia ventral disc. The view covered by panel A corresponds to the area marked with a yellow frame in figure 2. This slice is about 50 nm below (ventral of) the one in figure 2 and cuts through the ventral overlap zone, rather than the dorsal one as in figure 2. Here we can see some details at higher resolution, especially the crystalline pattern of the marginal plate, the so-called supernumerary microtubules (snMTs; see Fig. 4B & E), and the 3-D arrangement of the microtubule-microribbon complexes. Panels B–E are all thin tomographic slices from cryo-tomograms and therefore are not from the same tomogram as in A but represent identical regions. Panel B shows a 1.5 nm slice through the marginal plate, a paracrystalline array (see diffraction) associated with the anterior flagella. Hence, the highly regular subunits can be averaged (lower inset). Panels C & E show 2.3 nm xy-slices from a cryo-electron tomogram of the ventral disc. C slices through the more dorsal arranged microribbons, according to the position marked in the xz-projection shown in D, while E slices through the lumen of the microtubule. Corresponding areas within the disc are marked with red and green lines. The barrel sketches at the lower right corners indicate the orientation of ventral disc microtubules within the images, either corresponding to a side view (A, B, C, & E; xy-slices), or an axial end-on view (D; xz-slices). All scale bars are 50 nm.
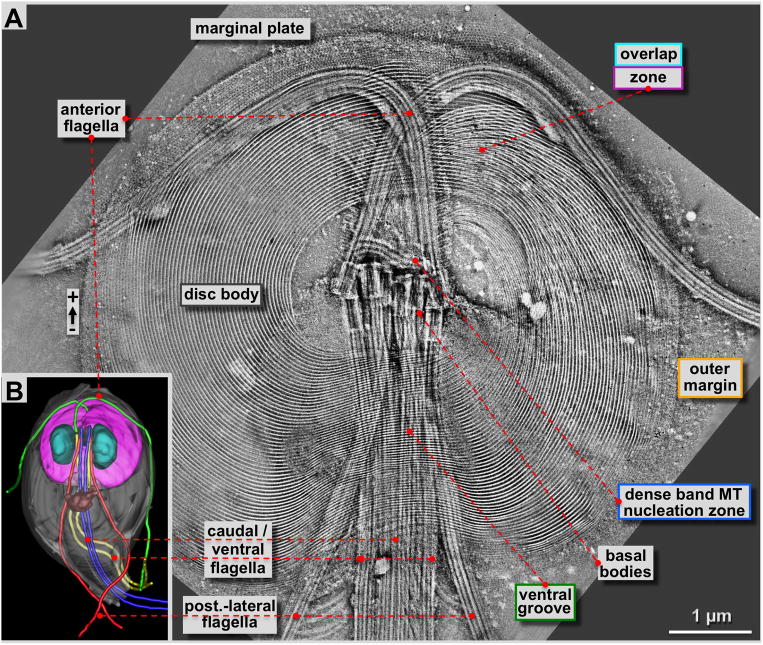
Figure 2. 4×4 tomogram montage of an entire Giardia ventral disc and some of its associated cytoskeletal structures such as flagella and the marginal plate.
A: Approximately 180 nm thick slice through a tomographic reconstruction obtained from a negatively stained, Giardia cytoskeleton preparation (see: Holberton and Ward, 1981). The most prominent features are the highly ordered array of microtubule-microribbon complexes of the ventral disc that start at the dense band MT nucleation zone or the inner disc edge, and end at the outer margin or the ventral overlap zone. Furthermore, four pairs of flagella originate from basal bodies arranged at the center of the spiral. One pair (anterior flagella) points towards the front of the cell, and then curves outwards. All other flagella leave the cell towards the posterior end. The ventral disc microtubules form a large right-handed spiral assembly with an overlap zone at the top right. Distinct regions of the ventral disc are marked with the corresponding color-coding of figure 1B and all figures that follow. B: 3-D reconstructions of Giardia lamblia from Schwartz et al. (2012) by a procedure called 3View (Denk & Horstmann, 2004). This SEM and microtomy 3-D reconstruction approach reveals the overall architecture of the Giardia cell with the ventral disc (pink) the nuclei (cyan), the median body (brown; Woessner & Dawson, 2012), and the four flagellar pairs in green (anterior), blue (caudal), yellow (ventral) and red (post-lateral; Dawson & House, 2010B).
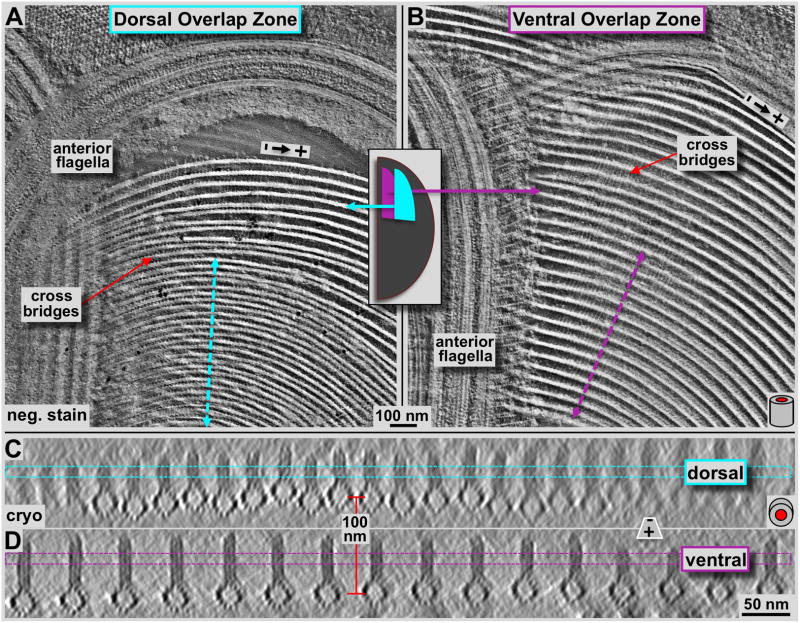
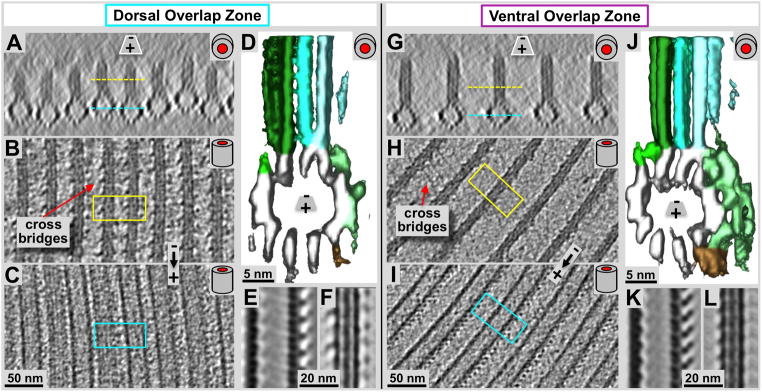
Figure 5. Tomographic 3-D analysis of the Giardia ventral disc overlap zone using negative staining tomography (A & B) and cryo-tomography (C & D).
Panels A and B show tomographic 4.7 nm thick xy-slices of negatively stained dorsal (A) and ventral (B) overlap zones. The overall orientation is analogous to figure 2 with the marginal plate near the top of the image, and the slice cuts through the microribbon part of the microtubule-microribbon complexes. The cyan (A) and pink (B) lines mark the positions of the corresponding frames in C and D, respectively. The lateral packing in the dorsal overlap zone is much tighter than in the ventral overlap zone. The tomographic xz-slices in C & D are taken from a cryo-electron tomogram of the overlap zone and represent an approximately 16 nm projection along the microtubule axis. Dorsal overlap zone (C) and ventral overlap zone slices (D) are from the same tomogram but slightly rotated towards each other as indicated by the arrows in A and B, to align each projection along the microtubule axes. This was not possible over the entire width of the picture. Hence, the fuzzy microtubule-microribbon complexes, particularly in C are not precisely aligned to the projection axis, but are equally well preserved. The distance between dorsal (C) and ventral microtubule layers (D) is according to that measured in the tomogram and varies between approximately 100–120 nm.
Results
Variation in structural organization defines regions of the ventral disc
We analyzed the ventral disc of Giardia lamblia by negative staining electron tomography (ns-ET) and cryo-electron tomography (cryo-ET). This extends our previous detailed cryo-ET 3-D structure of the microtubule-microribbon complexes, including inner and outer microtubule-associated proteins (Schwartz et al., 2012). That previous work focused exclusively on the ventral disc body that comprises the largest part of the disc (grey region in Fig. 1A). Here we compare our findings from the disc body to the structural and functional variations of other important regions of the ventral disc, the so-called ventral and dorsal parts of the overlap zone (Fig. 1A: pink and cyan regions respectively), as well as the disc margin (orange region in Fig. 1A). Thereby, we find striking variations in the 3-D structure of microtubule-microribbon complexes, especially regarding the structure and mass of associated proteins, and within the cross-bridges that connect adjacent microribbons.
Our classification of distinct disc regions and the distribution of microtubule-microribbon complexes are schematically illustrated in figure 1 with the labeling color-coded according to figure 1B and maintained through all figures of this paper. The approximate regional microtubule composition of the ventral disc is illustrated in figure 1D. We followed 7 representative microtubules (Fig. 1A and C) evenly spaced across the disc from the center to the margin to show how microtubules enter and exit seven disc regions according to distinct structural and functional features, representing distinct disc ‘zones’. In total, we found the disc to be composed of approximately 95 microtubules (± 5 MTs; N=6) that, if connected head to tail would accumulate a total length of ~1.23 mm (± 0.05 mm; N=6). This corresponds to about 100 x the longest axis of the cell (approx. 12-μm). The shortest microtubule was 2.34 μm (± 1.34 μm; N=6). The longest microtubule we measured was 18.36 μm long (± 0.75 μm; N=6). Structural evidence from microtubule capped minus-ends, flared versus blunt ends, and the reproducible orientation of attached side arms on their margin-facing side (see also: Schwartz et al., 2012) strongly indicates that all microtubules are parallel with their minus-ends towards the dense bands or the disc center and the plus-ends towards the margin or ventral overlap zone (see polarity markers in all figures). The Giardia ventral disc is a right-handed spiral with most of its microtubules bending at various degrees of curvature to the right (concave) (see Fig. 1E, positive kappa values). Only one small area called the ventral groove has a convex curvature (green in Fig. 1A; negative kappa values in Fig. 1E).
Montaged negative-stain tomograms reveals the 3-D structure of a complete ventral disc
The results we obtained with ns-ET are shown in figures 2A, 3A, 4 and 5A & B. All other tomographic data shown here have been obtained by cryo-ET (Fig. 3C–E, Fig. 5C & D, Figs. 6 & 7; reviewed in Hoenger, 2014; see also Schwartz et al., 2012). Negative staining electron microscopy (ns-EM) has the advantage over cryo-electron microscopy (cryo-EM) that it is significantly less electron beam sensitive but still produces reliable molecular 3-D data to approximately 2 nm resolution. Here we employed ns-ET to gain a large-field overview of a complete Giardia ventral disc by montaging 4 × 4 tomograms into one continuous volume of which a ~180 nm thick xy-slice is shown in figure 2A. The overall orientation of the disc in figure 2A is the same as in 2B, a 3View® reconstruction (Gatan Inc. Pleasanton CA; Denk & Horstmann, 2004; see also Schwartz et al., 2012) where the ventral disc is shown in pink, with the two nuclei in cyan, the median body in brown (Woessner & Dawson, 2012), and the four pairs of flagella in green (anterior), blue (caudal), yellow (ventral), and red (post-lateral; Dawson & House, 2010B). The flagellar basal bodies are clearly visible in the tomographic reconstructions of the disc preparations (Figs. 2A & 3A). They are between the two nuclei, roughly in the center of the ventral disc, which is known as the bare area. The dense band microtubule nucleation zone (Fig. 3A & 4) forms two sets of three ribbons each (Fig. 4). This is the nucleation site of the supernumerary microtubules (Fig. 4B), and the disc microtubules that form the dorsal overlap zone (Fig. 5A), which eventually end at the disc margin (Fig. 7). The microtubules that originate from the disc center end in the ventral overlap zone (Figs. 3A & 5B).
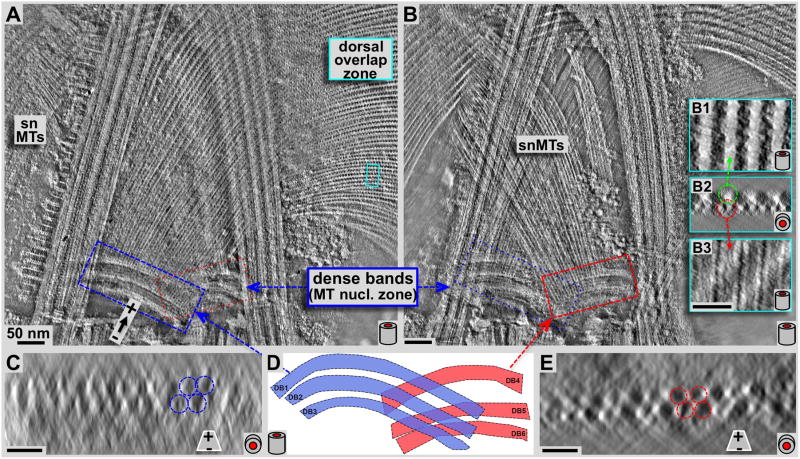
Figure 4. Negative staining tomography study on the structure and arrangement of microtubules leaving the dense band nucleation zone.
Panels A and B show 7.5 nm tomographic slices through the planes of the disc microtubules (A) and the supernumerary microtubules (B) respectively. Panels C and E show ~31 nm microtubule end-on projections very close to the origin of disc microtubules (C) and supernumerary microtubules (E) respectively. These views suffer from the inherent non-isotropic resolution along the z-axis in tomograms due to the limited tilt range of ±60 degrees only, and also due to some drying and flattening effects that are typical for negative staining. Two sets of triple-layered dense bands are clearly visible. One set nucleates a portion of the ventral disc microtubules (A: blue frame, C: end-on projection, blue layers in D), the other one triggers growth of the supernumerary microtubules (B, red frame, E: end-on projection, red in D). Insets B1-3 show 3.1 nm xy- (B1 & B3) and 15.5 nm xz- (B2) projections from the marked region in the dorsal overlap zone (A: cyan frame). Microtubules originate at the dense bands without attached microribbons (see C & E), and only show up once the dorsal overlap zone is reached (center, top insets in B). All scale bars are 50 nm.
Figure 6. Detailed cryo-electron tomography 3-D analysis of the microtubule-microribbon complexes within the Giardia ventral disc overlap zones.
Panels A and G are microtubule end-on views (~20 nm tomographic xz-slices) of the dorsal (A) and ventral (G) overlap zones. B & C, and H & I are 2.8 nm slices through the microribbons (B & H) and through the microtubules (C & I) respectively. Yellow and cyan lines in A & G mark the equivalent zones in B & C, as well as H & I. Volume averaging produced in the surface rendered 3-D maps are shown in D & J. Panels E & F (dorsal), and K & L (ventral) show 0.77 nm slices through averaged microtubule (E & K) and microribbon portions (F & L). The most obvious differences between ventral (D–F) and dorsal (J–L) overlap zones are the much larger amounts of associated protein densities in the case of the ventral microtubule-microribbon complexes (D & E) that revealed a similar pattern of associated proteins as previously found in the disc body (pale green (sidearms) and brown (paddles) rendered densities; see also Fig. 6C and Schwartz et al., 2012). The dorsal overlap zone microtubules, however, shows much less associated density (E & F), which could be a consequence of the denser lateral packing of the microtubule-microribbon complexes (compare panel A with F).
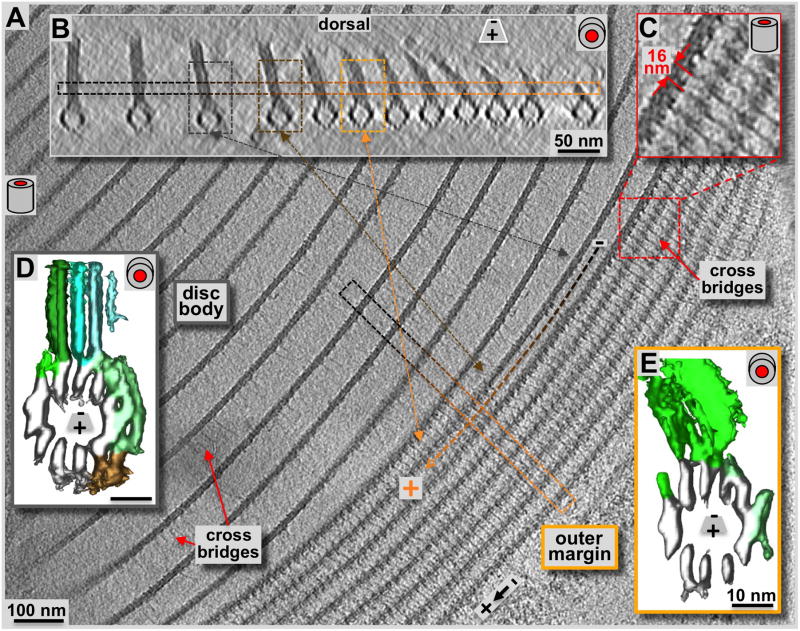
Figure 7. Cryo-electron tomography of the ventral disc outer disc margin.
Compared to the disc body region (A & B: loosely packed zone to the left), the margin (A: tomographic, 2.3 nm xy-slice, tightly packed 7–8 parallel structures to the right; B: 20 nm xz-slice) shows a strikingly different microtubule-packing pattern. The cyan arrow marks the orientation of the boxed region of B within panel A. Black to orange lines in A, and the corresponding frames in B mark the transition of a microtubule-microribbon complex from the disc body to a margin configuration. C: The marginal microtubule-microribbon complexes are laterally connected by regular tethers or cross-bridges. They form a 16 nm axial repeat that corresponds to the length of two sequential tubulin dimers along the microtubules. The lateral tethers are also present in the disc body forming the same repeat (see also figure 4), but with much less density, possibly a consequence of their extended length. Comparing volume-averages along microtubule-microribbon complexes of the disc body (D; see also Schwartz et al., 2012) with them from the margin (E) reveals much less microtubule-associated densities in the margin than found on the disc body complexes (green (side-arms) and brown (paddles) rendered densities in D and E). The low amount of microtubule-associated proteins resembles the situation of the dorsal overlap zone (see figure 5D). Due to the outermost positioning the sheets of the marginal microtubule-microribbon complexes are bending toward the disc center. The bending ribbons show a higher structural variation that produces much noisier averages, especially in the microribbon region (E).
Figure 3A represents an approximately 50 nm thick xy-slice through a single ns-ET volume at a slightly more ventral (lower) position than in figure 2A. Throughout figures 2–7 the schematic tube symbol (e.g. lower right corner in Fig. 3A–E) indicate the projection plane of the tomographic slices according to the orientation of microtubules within. Hence, xy-slices are marked with a tube symbol from the side, while xz-slices are marked with an end on tube symbol. The more ventral position of the xy-slice in figure 3A now cuts through the microtubule-microribbons of the ventral overlap zone with the dorsal zone towards the observer, above the volume presented. This tomogram reveals details of the anterior flagellar microtubules, such as the doublets, the radial spokes attached to the central pair, and associated striated fibers, which form a regular brush-like border along the flagella (Elmendorf et al., 2003). Figure 3B–E shows 2.3 nm thick cryo-ET slices (xy: Fig. 3B, C & E; xz: Fig. 3D). Cryo-ET reveals finer molecular detail than ns-ET, showing planes through the microtubule lumen (Fig. 3E) and through the microribbons (Fig. 3C). Figure 3D shows a microtubule-microribbon complex in an end-on projection with the microtubule at the bottom. The relative positions of the views in figure 3C–E are linked to the corresponding regions in figure 3A. However, figure 3A is ns-ET while figure 3B–E are cryo-ET data and therefore from a different tomogram. The disc in figure 3A is slightly tilted with respect to the xy-slice plane. Therefore, at the left side of the ventral overlap zone the xy-slice cuts the microtubule-microribbon complexes through the microribbons (green frames and line connecting Fig. 3A to C and D), but then reaches the level of microtubules (red line connecting to Fig. 3A to E; lower part in D).
Another structurally very interesting region of the Giardia ventral disc is the marginal plate (Fig. 3A (ns-ET) and Fig. 3B (cryo-ET)). The marginal plate locates to the front of the cell (with regard to the swimming direction: top in Fig. 2B). Most of its part constitutes of a hexagonal 2-D crystalline pattern (see diffraction in Fig. 3B). However, despite strong efforts to identify as many Giardia disc associated proteins as possible (e.g. see Marshall & Holberton, 1993; Hagen et al., 2011), most of the components that form the marginal plate are unknown. Cryo-ET of the marginal disc (Fig. 3B) reveals further molecular detail. Figure 3B shows a 0.77 nm thick tomographic slice from a single-axis cryo-tomogram of the marginal plate at the same orientation of figure 3A. The insets in figure 3B are an optical diffraction pattern of that region, and a sub-volume average of the hexagonally repeating units in the marginal plate.
The dense bands microtubule nucleation zone
Our previous work established that microtubules nucleate with their minus ends, and expose their plus-ends at the margin or ventral overlap zone (Schwartz et al., 2012). Microtubules of nucleation zones 4–7 originate from the dense bands in the center of the disc (Figs. 2–4). These microtubules account for about 59% of all microtubules in the disc. 39% originate from the disc inner edge, and another 2% appear to have minus ends in the main body of the ventral disc. The dense bands are divided into two separate areas; one of them (blue frame in Fig. 4A) nucleates disc microtubules, which continue into the dorsal overlap zone (Figs. 4A, 5A) and the disc body. Most of these microtubules eventually end in the outer margin (Fig. 1A). The other dense band area (red frame in Fig. 4B) nucleates the so-called supernumerary microtubules (Figs. 3A, 4B, marked snMTs), a bundle of approximately 20 short microtubules that curve slightly opposite to the main disc spiral. They end shortly after crossing over the left anterior flagellum. Both dense band regions are composed of three distinct bands. The disc microtubule nucleate from bands DB1–3, while the snMTs nucleate from bands DB4-6 (Fig. 4D). Figures 4C and 4E show microtubules end-on (xz) projections over ~31 nm length along the microtubule axis, and very close to the dense bands. Accordingly, microtubules are nucleated in 2–3 layers (arrows and circles in Fig. 3C and E), which renders the nucleation zone a very dense area filled with tightly packed microtubules that eventually fan out into a nearly perfect single plane of microtubules. For both, early disc microtubules (Fig. 4C blue zone) and supernumerary microtubules (Fig. 4E, red zone), microribbons are not present. Microribbons eventually appear once the microtubules reach the dorsal overlap zone (Fig. 4B, insets B1-3). The insets 4B1-3 are magnifications from the cyan frame in Fig. 4A. Figure 4B1 and 4B3 are 3.2 nm xy-slices through the microribbons (B1) and further ventral through the microtubule lumen (B3). Figure 4B2 shows a microtubule end-on (xz) projection over about 15.5 nm.
Structural comparisons between dorsal and ventral overlap zones
Using a combination of ns-ET (Fig. 5A & B), cryo-ET (Figs. 5C & D, 6), and tomographic subvolume averaging (Fig. 6D & J) we investigated the 3-D structure of the ventral disc overlap zone. The overlap zone is the region where the dense band nucleated microtubules of nucleation zones 4–7 form the dorsal overlap zone (Fig. 1: pink) and overlay with the microtubules from nucleation zones 1–3 that originated from the inner disc edge and end within the ventral overlap zone (see Fig. 1A and C; cyan). These two regions are structurally very distinct and feature different patterns of microtubule-associated proteins. There is about a 100 nm vertical distance between the microtubules of the two layers (red line between Fig. 5C & D), which, in an intact cell could be even much wider. The planes of figure 5C and 5D are slightly rotated towards each other and there position is marked by the dotted lines in figure 5A and 5B. Because of the curved microtubules in the dorsal overlap zone (Fig. 1D), only few of them are properly projected along their microtubule axis. The microribbons in both dorsal and ventral overlap zones show the three-layered structure (Figs. 5C & D, 6) that has been found in the disc body as well (Fig. 7D, Schwartz et al., 2012).
The dorsal overlap zone
The dorsal overlap zone (Figs. 5A, 5C, 6A–F) follows after the dense band microtubule nucleation zone (Fig. 4A) where microtubules are extremely tightly packed. Microribbons are absent in the nucleation zone, but appear at the dorsal overlap zone region (Fig. 4). Nevertheless, they are much shorter (approximately 30–40 nm; Figs. 5C & 6A) than the ones found in the disc body (>100 nm, see also Holberton & Ward, 1981) or the ventral overlap zone (~50–60 nm). The lateral spacing between adjacent microtubule-microribbon complexes is much tighter (~25–30 nm, center-to-center of microtubules; Figs. 5A & C, 6A–C) than in the ventral overlap zone (~60 nm; Figs. 5B & D & 6G–L), or in the disc body region (~70–80 nm Fig. 7A & B). The lateral spacing of the dorsal overlap zone is similar to the disc margin (Fig. 7A & B), but the microribbons do not bend sideways. Due to this tight lateral spacing, the overall amount of microtubule-associated proteins (e.g. the side-arms or paddles) is much reduced (Fig. 6D & E) compared to the ventral overlap zone (Fig. 6J & K), or in the disc body (Fig. 7D). Nevertheless, volume averages reveal the reduced microtubule-associated side-arms with a clearly visible 8 nm repeat (Fig. 5E), corresponding to the length of an αβ-tubulin dimer. The microribbon domains show strong lateral connections or cross-bridges (see also Holberton & Ward, 1981) that link adjacent microribbons and repeat along the microtubule axis with a 16 nm repeat pattern (Fig. 6B). The 16 nm repeat corresponds to two consecutive αβ-tubulin dimers along microtubule protofilaments (see also Fig. 7C: cross-bridges within the disc margin).
The ventral overlap zone
The ventral overlap zone (Figs. 5B & D, 6G–L) is between the dorsal overlap zone and the ventral cell membrane, the part of the cell that makes contact with the microvilli of epithelial cells (see Holberton, 1973). It represents the plus-end termination region of microtubule-microribbon complexes that originate from nucleation zones 1–3 at the inner disc edge (Fig. 1C). The microtubule-microribbon complexes in the ventral overlap zone exhibit a lateral spacing of approximately 60 nm (Figs. 5D, 6G), similar to most parts of the disc body (Schwartz et al., 2012). The microtubule portions carry a large, asymmetric load of outer (marginal) associated protein mass that was previously named the side-arms (green: first described in Holberton, 1981) and paddles (brown: Schwartz et al., 2012). The overall shape of the microtubule-associated density is again very similar to that of the disc body microtubules (Fig. 7D; Schwartz et al., 2012; For a video see DOI: 10.1371/journal.pone.0043783, Video S7). Microtubule end-on views in cryo-ET data (Figs. 5D, 6G & J) reveal the 13 protofilaments and their lateral spacing. The tomographic sub-volume averaging procedure (Fig. 6J) visualizes the distribution, axial frequency, and shape of regularly associated proteins. The microribbons are about 55 nm in height and show three distinct layers (Fig. 5D, 6G & L) as seen elsewhere in the disc. The microribbons show faint cross-bridge densities that seem to control the microtubule-microribbon lateral distance. These tether-like structures seem partially absent, but where visible, they form very faint connections that in closest sequence repeat every 16 nm along the microtubule-microribbon axis (Fig. 6H). However, due to partial absence, averaged data only reveals just barely detectable hints of cross-bridges (see Figs. 4 and 5 in Schwartz et al., 2012) is in (Fig. 6L). In contrast, the microtubule-associated side-arms (Fig. 6J, green) and paddles (Fig. 6J, brown) form strong densities every 8 nm, according to the length of one αβ-tubulin dimer along a protofilament (Fig. 6I & K).
The disc margin
The outermost region of the ventral disc is the margin, shown here in a top view (Fig. 7A & C: 2.3 nm xy-slice), and microtubule-end on view (Fig. 7B: 16 nm xz-slice). The margin is the plus-end termination point for all the microtubules that nucleate at the dense band microtubule nucleation zone (zones 4 – 7: Fig. 1C). The area of ventral disc margin shown in figure 7A & B consists of the outer 7–8 microtubule-microribbon complexes; all complexes further in show a configuration typical of the disc body. There is a clear transition where the same microtubule-microribbon complex has disc body morphology and then changes to disc margin morphology (see Fig. 7). Overall we found that the margin may consist of anywhere between 4 and 9 microtubule-microribbon complexes. The microribbons of the marginal complexes are getting shorter as they approach the plus-end and the very margin of the disc (Fig. 7B). They appear bent towards the disc center, which coincides with a pronounced appearance of shortened, lateral cross-bridges (see also: Holberton, 1973). As elsewhere in the disc, lateral cross-bridges repeat axially every 16 nm (Fig. 7C: magnification of the red framed region in A). Cross-bridge densities are much fainter within the disc body region, but that may be a consequence of the much longer distance to span, indicating that cross-bridges may have flexible and stretchable properties.
Volume averaging of microtubule-microribbon complexes revealed the rendered densities for the disc body in figure 7D and for the margin in figure 7E. The marginal complexes showed much higher structural variability that is reflected in a higher noise level of the margin map (data not shown). However, here we do not focus on fine structural detail, but on the large-scale differences between regions. Accordingly, the microtubule associated protein density and distribution in the margin was similar to the dorsal overlap zone (Fig. 6D), but much lower than in the disc body or the ventral overlap zone. The inward bend configuration of the microribbons does not lend itself well to averaging procedures, and neither do the cross-bridges. Despite being very apparent in the tomogram before averaging (Fig. 7C), both elements are not well resolved in averages due to structural flexibility.
Discussion
The Giardia lamblia ventral disc has been extensively imaged in the 1970s and 1980s by Holberton and colleagues (e.g. see: Holberton, 1973, 1974, 1981; Holberton and Ward, 1981). These marvelous data have been mostly collected from thin-sections of plastic-embedded specimens and originate from a time before cryo-electron microscopy (Dubochet et al., 1988) became popular. Likewise, only recently the computational possibilities reached a level that allowed reconstructions of large tomographic volumes (Patwardhan et al., 2012) of plastic-embedded or frozen-hydrated cellular specimens (reviewed in Hoenger & McIntosh, 2009; Hoenger, 2014; Asano et al., 2015). In our work we optimize specimen for preparation for cryo-electron tomography, and therefore we worked with isolated Giardia cytoskeletons (Holberton & Ward, 1981), rather than intact or sectioned cells. These cytoskeleton preparations (see Fig. 2; Holberton & Ward, 1981) are close to in vivo conditions and include the ventral disc and other associated organelles such as all flagella including basal bodies, some parts of the nuclei and the marginal plate (Figs. 2 & 3). The procedure places the disc in a reproducible orientation on the grid, and spares us the tedious, and potentially disruptive process of cutting vitreous sections (Bouchet-Marquis & Hoenger, 2011).
The work presented here is a follow-up attempt to our work published previously in Schwartz et al. (2012) that has focused extensively on a detailed 3-D reconstruction from the ventral disc body region, the best-conserved and reproducible part of the disc. Here we combine negative staining (ns-ET), and cryo-electron tomography (cryo-ET) and focus in more detail on the structural variations of the different regions within the ventral disc, especially the dorsal and ventral overlap zones (Figs. 5, & 6), the dense band microtubule nucleation zone (Fig. 4), and the disc margin (Fig. 7). It is obvious from our data that, at least for a large, complex specimen like the intact Giardia ventral disc, cryo-ET, where applicable (Figs. 3B–E; 5C & D, 6, and 7) reveals significantly more structural detail than ns-ET (Figs. 2, 3A, 4, 5A, B). However, ns-ET proved to be a great method for large overviews (Figs. 2, 3A), and to reach locations where cryo-ET was not a good option due to increased ice thickness. Microtubules in the dense bands microtubule nucleation zones (Fig. 4) were too tightly packed for an average-based cryo-ET image reconstruction, but ns-ET delivered enough detail to see that microtubules in that region do not yet have any associated microribbons (Fig. 4C & E). Some material within the dense bands indeed seem to actively nucleate microtubules, but it is unclear if that is resembling the nucleation process we know from basal bodies or that of microtubule organizing centers in centrosomes. γ-tubulin is present at the basal bodies during interphase (Nohynková et al., 2000), although with the resolution of light microscopy, it is unclear if it is present at the dense bands. As far as we can say from the negative staining tomograms in figure 4C and E, the microtubules exit the dense bands from two layers of densely packed microtubule ends. Unlike microtubule doublets that originate from basal bodies, both ventral disc and supernumerary microtubules are all single tubes. Hence, the nucleation process may be closer to a centrosome-based process than that of basal bodies.
Although ns-ET did not give us fine molecular detail or data that is amenable to sub-volume averaging, it allowed us to gather important information about the overall structure of the disc and the spatial relations between different disc zones. We were able to map 6 distinct regions of the disc based on location and morphology (Fig. 1A). Because the disc was in three dimensions, we were able to model individual microtubules and obtain quantitative data on numbers of microtubules present and microtubule lengths. We established that individual microtubules pass through multiple regions within the disc (Fig. 1C). These regions have distinct morphological characteristics (i.e. short microribbons in dorsal overlap zone, or reduced side-arms at disc margin). Therefore, each microtubule must transition through these regions (Fig. 7) and change the protein complement at each region as needed.
The main component of the Giardia ventral disc are hyper-stable microtubules (Sagolla et al., 2006), complexed microribbons (composed of 3 sheets), multiple microtubule associated proteins, and cross-bridges connecting laterally between adjacent microribbons. Hence, a lateral sliding of adjacent microtubules over more than a few nanometers seems unlikely, as it would snap the cross-bridges and other lateral connections. The most plausible mechanism may be that cross-bridges may contract or expand between adjacent microribbons with a contracted configuration seen at the margin or the dorsal overlap zone, and an expanded configuration seen in the disc body and the ventral overlap zone. It is difficult to conclude such a mechanism from static EM pictures alone. However, two observations support this idea: As demonstrated in figure 7, the very same microtubules may change from a disc-body to a margin configuration, and cross-bridges seem expandable. Previous models propose that the Giardia shape, together with a constant flow of liquid propelled by flagella between cell and microvilli may create a reversed wing-like effect (hydrodynamic model: Holberton, 1974). However, House et al., (2011) proposed a suction cup model by demonstrating that the flow underneath the cell is not required for attachment. Either way, providing space underneath the cell body, or clamping like a suction cup may be mediated by expanding or contracting cross-bridges (Figs. 5 and 7; see also images by Holberton, 1973; Erlandsen and Feely, 1984).
After sub-volume averaging, it became clear that the most reproducible components of the disc in all regions are the microtubules, whereas contributions from microribbons, associated protein densities, and cross-bridges vary substantially between different disc regions. The microtubules are also one of the few structures we can unambiguously identify. By using the microtubule portions for a normalization of surface-rendered, volume-averaged density maps from the various disc regions, we got a reasonably quantitative view of the variations in MAPs and MIPs densities between dorsal (Fig. 6D) and ventral overlap zone (Fig. 6J), the nucleation zone, the disc body (Fig. 7D) and the outer disc margin (Fig. 7E). The total mass and distribution of microtubule-associated proteins within the ventral overlap zone (Fig. 6J) resembles that of the disc body (Fig. 7D; Schwartz et al., 2012), while al other disc regions show significantly less associated protein mass. The largest visible chunks of these densities have been named side-arms (Figs. 6J & 7D: pale green in all renderings; Holberton, 1981) and paddles (brown; Schwartz et al., 2012).
In contrast to the disc body and the ventral overlap zone, microtubule-associated protein densities in the dorsal overlap zone (Fig. 6D) and the margin (Fig. 7E) are sparse. This reduction of microtubule-associated proteins at both locations coincides with a much narrower lateral spacing between adjacent microtubule-microribbon complexes (~30nm vs. 60nm: Figs. 5, 6, & 7), leaving little room for the extended side-arms we found in the disc body or the ventral overlap zone. Nevertheless, the microribbons at the margin and dorsal overlap zones show strong, and highly regular cross-bridges (Figs. 6B, 7C). The cross-bridges are much fainter at the wider spaced disc body and ventral overlap zones and may be caused by stretching these bridges to span the much larger gap (~60 nm). Interestingly, ns-ET seems to enhance the contrast of cross bridges, even at wide gaps (see Fig. 5B) over cryo-ET (Fig. 6H & 7A). The cross-bridges always disappear from the volume averages, suggesting a flexible structure that links adjacent microribbons at various angles. We do not know the nature of these bridges, but they appear to be formed by coiled-coils or other extendable elements (see also: Feely et al., 1982; Marshall & Holberton, 1993; Hagen et al., 2011). Even at the disc margin where the cross-bridges appear very strong, they do not show up well in averages, indicating structural flexibility. In the particular case of the outer disc margin, its regularity seems highly affected by the inward bending of microribbons (see Fig. 7B). The microribbon bending may be an effect of adsorbing an isolated cytoskeletal preparation onto a flat carbon film that may deform its natural geometry. Plastic-embedding and sectioning shows that the disc is naturally curved at the margin as if it would form a clamping system to hold on to the microvilli of the small intestines (see Holberton 1973, 1981). The obvious contraction of cross-bridges at the margin (Fig. 7) further support the hypothesis of a mechanism that maintains adhesion to the intestinal tract (Holberton, 1974) during that part of the Giardia life cycle.
Our tomograms and 3-D reconstructions were made from isolated cytoskeletons as explained in materials & methods. Hence, we lost the plasma membrane with that procedure and therefore have no data on connective structures between disc and membrane. However, our work carried out with plastic-embedded whole cells show that the gap between microtubules and the underlying plasma membrane is relatively narrow (see Suppl. Fig. 1; Holberton, 1974; Erlandsen & Feely, 1984), suggesting that the microtubules themselves, or some of the ventral associated proteins such as the paddle or other MAPs on that side maintain contact with the plasma membrane. Giardia has no cell wall but there are layers of proteins such as the variant protein TS-antigen-417 (Svärd et al., 1998) that create a densely stained outer layer (see Suppl. Fig. 1B). Our own data in supplementary figure 1 shows a gap of approximately 25 – 50 nm without any strong connective densities, suggesting a rather loose connection between disc and membrane. However, the gap may be variable. Possibly the most likeliest connections to the plasma membrane are provided by the paddles.
Having the ventral disc and other associated organelles available within a very close to in situ situation enhances it accessibility for cryo-ET imaging and omits the complications that are typically associated with keeping cells viable during specimen preparation steps. Here we present the Giardia ventral disc as a most complete reconstruction that places all relevant elements in a 3-D context. Furthermore, this cell-free system bears a lot of potential for detailed studies about the protein composition of the ventral disc, which is currently an active field of research. Our previous structural studies identified the body of the ventral disc as a source of various MIPs and MAPs (Hagen et al., 2011; Schwartz et al., 2012) of which 30-plus components have been identified so far. However, a precise spatial localization to particular regions of microtubules or microribbons is still lacking. Nevertheless, all of these disc-associated components will be ideal candidates for future structural and functional studies and potential drug targets. Here, we have shown that protein distribution of the MIPs and MAPs varies among the disc regions and in fact, along the length of every individual microtubule within the disc as they pass through those regions. It is clear that the ventral disc is a very complex organelle and given its importance in the pathology of giardiasis, more detailed 3-D investigations will be needed to identify these MIPs and MAPs in a system showing as much detail as presented here.
Materials and Methods
Cell culture 558TB182691A003609
WBC6-M giardia trophozoite cells were cultured in non-commercial modified TY-S-33 media in anaerobic conditions at 37°C (Keister, 1983). Cells were cultured in 15 ml culture tubes containing 12 ml of media. Large cultures were grown in 50 ml conical tubes, containing 50 ml of media. The lids of both types of tubes were screwed on tightly to prevent air exchange and protect the cells from excessive oxygen exposure. After growing a culture from a frozen cell stock, cells were allowed 1 hour at 37°C to recover and attach to the tube surface. After 1 hour, the DMSO-containing frozen storage media was removed and exchanged for 12 ml of modified TY-S-33 media. Cells were then allowed to grow to near 100% confluence before splitting the culture. In order to split the culture, the culture tubes were placed on ice (30 minutes for 15 ml tubes and 45 minutes for 50 ml tubes). As the media cooled, the cells detached from the sides of the tube and formed a suspension in the media. This cell suspension was then diluted to the desired concentration with fresh media. Culture tubes were then returned to 37°C to allow cells to reattach to the tube.
Cytoskeletal preparation
Cytoskeletons were isolated using a procedure modified from Holberton & Ward (1981) and described in Schwartz et al. (2012). Cells grown in 16 ml screw-cap culture tubes were rinsed with 37°C PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2) to remove unattached and dead cells. 2 ml of fresh PHEM buffer with 2% triton-X-100 was added to the cells and transferred to an Eppendorf tube. Cytoskeletons were vortexed on low for 30 minutes; water bath sonicated for 1 min and then checked in the light microscope (see also: Dawson and House, 2010A). The vortex/sonication procedure was repeated until the cytoskeletons were adequately extracted. They were then rinsed 3X with PHEM buffer. A 4 μl drop of cytoskeletal prep was adsorbed onto a holey carbon grid (Quantifoil, Jena, Germany) and 1 μl of 10 nm colloidal gold was pipetted into the drop. The grids were hand-blotted and plunge-frozen into liquid ethane. Grids were maintained at LN2 temperatures until visualized by cryo-electron microscopy (see: Schwartz et al., 2012).
Electron tomography data collection
All EM data was collected on an FEI Tecnai F30 FEG transmission EM (FEI-Company, Eindhoven, The Netherlands) operating at 300 kV. Tilt-series were automatically recorded using SerialEM (Mastronarde, 2005) with binning by 2 on a 4K × 4K Gatan Ultracam CCD camera (Gatan, Inc, Pleasanton, CA) after passing through a Gatan Tridium GIF energy filter (Gatan, Inc, Pleasanton, CA), set to a 20 eV slit. For ns-ET, a 4 × 4 montage tilt-series as the example shown in figure 2 was collected at a magnification of 18,000x (pixel size 1.55 nm) with a 1.5° increment from +/−60° and a target intensity of 5000 to 6000 e/Å2 for an entire tilt series. For cryo-ET, tilt-series were collected with a 2° interval from +/− 60° at nominal magnification of 27,500x (pixel size 0.77 nm) and a target dose of 80 e/Å2 for the entire tilt-series. Defocus was either −6 μm or −4 μm. All tomograms were calculated with R-weighted back projection using IMOD (Kremer et al., 1996). Cryo-ET data were corrected for the contrast transfer function (CTF) (Xiong et al., 2009) and the density contributions of the fiducial gold markers were erased.
3-D volume averaging
We used our own program suite PEET (Particle Estimation for Electron Tomography; first application: Nicastro et al., 2006; refined versions: Cope et al., 2010; Schwartz et al., 2012) to average 16 nm intervals along the axial length of the ventral disc microtubules. Sub-volumes were selected by placing a model point every 16 nm at the microtubule/microribbon interface. PEET removes duplicate particles and applies missing wedge compensation. Since we acquired multiple tomograms for each region, first we averaged within each tomogram and then combined those tomograms into a final grand average for each region. To test for reference bias, we selected multiple single sub-volumes within each tomogram and ran independent averages to see if there were differences, of which we found none. Then, for the grand average, we chose each tomogram average as reference on multiple, independent grand averaging runs, and again, found no differences between them. For each average of a different disc area we chose a single sub-volume from a tomogram as an initial reference and then averaged all of the tomograms of the same area together. After each round of alignment, a new reference was constructed to minimize reference bias. In all cases, the grand averages were virtually identical. For analysis, we calculated FSC curves to estimate resolution using the 50% criterion and used IMOD (Kremer et al., 1996) to visualize the averages and 3-D models.
Supplementary Material
The plasma membrane that attaches to the intestine microvilli leaves a gap to the ventral disc that is around 25–50 nm with no obvious densities that may act as connectors between the two organelles. The most likely candidates are microtubule-attached densities such as the paddle and other MAPs at the most ventral protofilaments 1, 2 and 13 (A; numbering according to: Schwartz et al., 2012). The 3-D reconstruction in A has been calculated from isolated cytoskeletal preparations that loose the plasma membrane (see Mat. & Meth.). Therefore, larger connective structures between plasma membrane and disc microtubules might have been lost or dislocated so that they cannot be seen on the averaged 3-D map, but may be crucial for disc function and attachment. Panel B shows a plastic-embedded preparation of a whole cell including the ventral plasma membrane.
Acknowledgments
The authors would like to thank David Mastronarde for valuable discussions on sub-volume averaging, and Christopher Nosala (UCD) for carefully reading the manuscript. This work was funded in part by National Institutes of Health-National Center for Research Resources grant (NCRR: P41RR000592 and since 05/02/12: NIGMS: P41GM103431-43, and P41GM103431-43S1) to AH. JB received support from an NSF IGERT-COSY training grant (award ID: 0801680, PI: Rafael Piestun, Engineering Univ. of Colorado, Boulder; Co-PI: AH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam RD. Biology of Giardia lamblia. Clinical microbiology reviews. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S, Engel BD, Baumeister W. In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.09.030. S0022-2836 00553-7. [DOI] [PubMed] [Google Scholar]
- Boreham PF, Upcroft JA, Upcroft P. Changing approaches to the study of Giardia epidemiology: 1681–2000. Int J Parasitol. 1990;20:479–87. doi: 10.1016/0020-7519(90)90195-s. [DOI] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Hoenger A. Cryo-electron tomography on vitrified sections: a critical analysis of benefits and limitations for structural cell biology. Micron. 2011;42:152–62. doi: 10.1016/j.micron.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Campanati L, Holloschi A, Troster H, Spring H, de Souza W, Monteiro-Leal LH. Video-microscopy observations of fast dynamic processes in the protozoon Giardia lamblia. Cell Motil Cytoskeleton. 2002;51:213–24. doi: 10.1002/cm.10026. [DOI] [PubMed] [Google Scholar]
- Cope J, Gilbert S, Rayment I, Mastronarde D, Hoenger A. Cryo-electron tomography of microtubule-kinesin motor complexes. J Struct Biol. 2010;170:257–265. doi: 10.1016/j.jsb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC, House SA. Imaging and analysis of the microtubule cytoskeleton in giardia. Methods Cell Biol. 2010A;97:307–39. doi: 10.1016/S0091-679X(10)97017-9. [DOI] [PubMed] [Google Scholar]
- Dawson SC, House SA. B) Life with eight flagella: flagellar assembly and division in Giardia. Current opinion in microbiology. 2010;13:480–490. doi: 10.1016/j.mib.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS biology. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf HG, Dawson SC, McCaffery JM. The cytoskeleton of Giardia lamblia. Int J Parasitol. 2003;33:3–28. doi: 10.1016/s0020-7519(02)00228-x. [DOI] [PubMed] [Google Scholar]
- Erlandsen SL, Feely DE. Trophozoite motility and the mechanism of attachment. In: Erlandsen SL, Meyer EA, editors. Giardia and Giardiasis. Plenum Press; New York: 1984. pp. 33–66. [Google Scholar]
- Feely DE, Schollmeyer JV, Erlandsen SL. Giardia spp.: distribution of contractile proteins in the attachment organelle. Exp Parasitol. 1982;53:145–54. doi: 10.1016/0014-4894(82)90100-x. [DOI] [PubMed] [Google Scholar]
- Gillin FD, Reiner DS, McCaffery JM. Cell biology of the primitive eukaryote Giardia lamblia. Annu Rev Microbiol. 1996;50:679–705. doi: 10.1146/annurev.micro.50.1.679. [DOI] [PubMed] [Google Scholar]
- Hagen K, Hirakawa M, House S, Schwartz CL, Pham JK, et al. Novel structural components of the ventral disc and lateral crest in Giardia intestinalis. PLoS Neglected Tropical Diseases. 2011;5:e1442. doi: 10.1371/journal.pntd.0001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A. High-resolution cryo-electron microscopy on macromolecular complexes and cell organelles. Protoplasma. 2014;251:417–427. doi: 10.1007/s00709-013-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, McIntosh JR. Probing the macromolecular organization of cells by electron tomography. Curr Opin Cell Biol. 2009;21:89–96. doi: 10.1016/j.ceb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberton DV. Fine structure of the ventral disk apparatus and the mechanism of attachment in the flagellate Giardia muris. J Cell Sci. 1973;13:11–41. doi: 10.1242/jcs.13.1.11. [DOI] [PubMed] [Google Scholar]
- Holberton DV. Attachment of Giardia-a hydrodynamic model based on flagellar activity. J Exp Biol. 1974;60:207–21. doi: 10.1242/jeb.60.1.207. [DOI] [PubMed] [Google Scholar]
- Holberton DV. Arrangement of subunits in microribbons from Giardia. Journal of cell science. 1981;47:167–185. doi: 10.1242/jcs.47.1.167. [DOI] [PubMed] [Google Scholar]
- Holberton DV, Ward AP. Isolation of the cytoskeleton from Giardia. Tubulin and a low-molecular-weight protein associated with microribbon structures. Journal of cell science. 1981;47:139–166. doi: 10.1242/jcs.47.1.139. [DOI] [PubMed] [Google Scholar]
- Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Marshall J, Holberton DV. Sequence and structure of a new coiled coil protein from a microtubule bundle in Giardia. J Mol Biol. 1993;231:521–30. doi: 10.1006/jmbi.1993.1303. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Nohynková E, Tumová P, Kulda J. Cell division of Giardia intestinalis: flagellar developmental cycle involves transformation and exchange of flagella between mastigonts of a diplomonad cell. Eukaryot Cell. 2006;5:753–61. doi: 10.1128/EC.5.4.753-761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Carazo JM, Carragher B, Henderson R, Heymann JB, Hill E, Jensen GJ, Lagerstedt I, Lawson CL, Ludtke SJ, Mastronarde D, Moore WJ, Roseman A, Rosenthal P, Sorzano CO, Sanz-García E, Scheres SH, Subramaniam S, Westbrook J, Winn M, Swedlow JR, Kleywegt GJ. Data management challenges in three-dimensional EM. Nat Struct Mol Biol. 2012;19:1203–7. doi: 10.1038/nsmb.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxström-Lindquist K, Palm D, Reiner D, Ringqvist E, Svärd SG. Giardia immunity--an update. Trends Parasitol. 2006;22:26–31. doi: 10.1016/j.pt.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Sagolla MS, Dawson SC, Mancuso JJ, Cande WZ. Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J Cell Sci. 2006;119:4889–4900. doi: 10.1242/jcs.03276. [DOI] [PubMed] [Google Scholar]
- Savioli L, Smith H, Thompson A. Giardia and Cryptosporidium join the “Neglected Diseases Initiative”. Trends in parasitology. 2006;22:203–208. doi: 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Heumann JM, Dawson SC, Hoenger A. A detailed, hierarchical study of Giardia lamblia’s ventral disc reveals novel microtubule-associated protein complexes. PLoS One. 2012;7:e43783. doi: 10.1371/journal.pone.0043783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd SG, Meng TC, Hetsko ML, McCaffery JM, Gillin FD. Differentiation-associated surface antigen variation in the ancient eukaryote Giardia lamblia. Mol Microbiol. 1998;30:979–89. doi: 10.1046/j.1365-2958.1998.01125.x. [DOI] [PubMed] [Google Scholar]
- Thompson RC. Giardiasis as a re-emerging infectious disease and its zoonotic potential. International journal for parasitology. 2000;30:1259–1267. doi: 10.1016/s0020-7519(00)00127-2. [DOI] [PubMed] [Google Scholar]
- Troeger H, Epple HJ, Schneider T, Wahnschaffe U, Ullrich R, Burchard GD, Jelinek T, Zeitz M, Fromm M, Schulzke JD. Effect of chronic Giardia lamblia infection on epithelial transport and barrier function in human duodenum. Gut. 2007;56:328–35. doi: 10.1136/gut.2006.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tůmová P, Kulda J, Nohýnková E. Cell division of Giardia intestinalis: assembly and disassembly of the adhesive disc, and the cytokinesis. Cell Motil Cytoskeleton. 2007;64:288–98. doi: 10.1002/cm.20183. [DOI] [PubMed] [Google Scholar]
- Woessner DJ, Dawson SC. The Giardia median body protein is a ventral disc protein that is critical for maintaining a domed disc conformation during attachment. Eukaryotic cell. 2012;11:292–301. doi: 10.1128/EC.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Morphew MK, Schwartz CL, Hoenger A, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. J Struct Biol. 2009;168:378–87. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The plasma membrane that attaches to the intestine microvilli leaves a gap to the ventral disc that is around 25–50 nm with no obvious densities that may act as connectors between the two organelles. The most likely candidates are microtubule-attached densities such as the paddle and other MAPs at the most ventral protofilaments 1, 2 and 13 (A; numbering according to: Schwartz et al., 2012). The 3-D reconstruction in A has been calculated from isolated cytoskeletal preparations that loose the plasma membrane (see Mat. & Meth.). Therefore, larger connective structures between plasma membrane and disc microtubules might have been lost or dislocated so that they cannot be seen on the averaged 3-D map, but may be crucial for disc function and attachment. Panel B shows a plastic-embedded preparation of a whole cell including the ventral plasma membrane.