Abstract
Brain deposition of Aβ in the form of amyloid plaques is a pathological hallmark of Alzheimer's disease. There are two major species of Aβ in the brain: Aβ42 and Aβ40. Although Aβ40 is several-fold more abundant than Aβ42 in soluble form, Aβ42 is the major component of amyloid plaques. Structural knowledge of Aβ42 fibrils is important both for understanding the process of Aβ aggregation and for designing fibril-targeting drugs. Here we report site-specific structural information of Aβ42 fibrils at 22 residue positions based on electron paramagnetic resonance data. In combination with structure prediction program Rosetta, we modeled Aβ42 fibril structure at atomic resolution. Our Aβ42 fibril model consists of four parallel in-register β-sheets: βN (residues ~7-13), β1 (residues ~17-20), β2 (residues ~32-36), and βC (residues 39-41). The region of β1-loop-β2 in Aβ42 fibrils adopts similar structure as that in Aβ40 fibrils. This is consistent with our cross seeding data that Aβ42 fibril seeds shortened the lag phase of Aβ40 fibrillization. On the other hand, Aβ42 fibrils contain a C-terminal β-arc-β motif with a special turn, termed “arc”, at residues 37-38, which is absent in Aβ40 fibrils. Our results can explain both the higher aggregation propensity of Aβ42 and the importance of Aβ42 to Aβ40 ratio in the pathogenesis of Alzheimer's disease.
Keywords: Alzheimer disease, amyloid, Aβ, protein aggregation, spin labeling
INTRODUCTION
Deposition of Aβ in the amyloid plaques is a pathological hallmark of Alzheimer's disease. The major component of amyloid plaques is fibrillar Aβ42, a 42-residue peptide resulting from proteolytic processing of amyloid precursor protein (Fukumoto et al., 1996; Gravina et al., 1995; Iwatsubo et al., 1995; Iwatsubo et al., 1994; Mak et al., 1994; Miller et al., 1993). Fibrils are the primary product of Aβ42 aggregation. Therefore, structural knowledge of Aβ42 fibrils is important for drug development targeting the fibril itself or the process of Aβ42 aggregation. Despite intensive studies, the structure of Aβ42 fibrils has remained elusive. In the brain, the major soluble Aβ species is Aβ40, which shares the same amino acid sequence as the first 40 residues of Aβ42. Several high resolution structural models based on solid-state NMR data have been proposed for Aβ40 fibrils (Bertini et al., 2011; Lu et al., 2013; Paravastu et al., 2008; Petkova et al., 2006). The common structural features of Aβ40 fibrils are two parallel in-register β-sheets: one consisting of approximately residues 17-21, and the other one spanning approximately residues 31-35 (e.g., PDB ID 2LMN). Although Aβ40 is several-fold more abundant than Aβ42 in the brain, it is less commonly found in the amyloid plaques.
Strong structural evidence supports a parallel in-register β-sheet structure for Aβ42 fibrils, as shown, for example, by the solid-state NMR data on inter-molecular distances at Ala21 and Leu34 (Antzutkin et al., 2002). Several earlier NMR studies on Aβ42 fibrils (Ahmed et al., 2010; Colvin et al., 2015; Masuda et al., 2005; Masuda et al., 2008; Sato et al., 2006) provided some structural constraints, but these constraints were not enough to allow high resolution modeling. Two hydrogen exchange studies (Lührs et al., 2005; Olofsson et al., 2006) have identified two highly protected regions at approximately residues 17-25 and 30-42. The protected amide protons are generally involved in hydrogen bonds, and are thus interpreted as adopting β-sheet structures in Aβ42 fibrils. Mutagenesis to introduce β-sheet breakers such as proline has also been used to identify β-strand regions in Aβ42 fibrils (Morimoto et al., 2004). Electron microscopy (Schmidt et al., 2015; Zhang et al., 2009) and computational approaches (Ma and Nussinov, 2011) have also been used to model the structure of Aβ42 fibrils. Recently, Xiao et al (2015) reported a structure of Aβ42 fibrils based on solid-state NMR constraints and molecular dynamics simulations. The primary feature of this Aβ42 fibril structure is a triple-β motif, which consists of three β-sheets spanning residues 12-18, 24-33, and 36-40. Xiao et al's structure bears little resemblance to the structure of Aβ40 fibrils, at the level of both secondary structure and tertiary packing. In terms of previous studies on Aβ42 fibrils, the structure by Xiao et al. also has notable differences. For example, the side chain contact between Phe19 and Leu34, which were identified in previous solid-state NMR studies of Aβ42 fibrils (Ahmed et al., 2010), is missing in Xiao et al's model. Furthermore, data from two previous hydrogen exchange studies (Lührs et al., 2005; Olofsson et al., 2006) can not be accounted for by Xiao et al's model. One likely explanation is that, via repeated seeding, Xiao et al obtained a polymorph that is structurally distinct from other more common polymorphs studied by other investigators. Therefore, more work needs to be done to better understand the structure of Aβ42 fibrils.
Site-directed spin labeling in combination with electron paramagnetic resonance (EPR) spectroscopy is capable of revealing secondary structures in Aβ42 fibrils. The general strategy of site-directed spin labeling is that a residue of interest is first replaced with cysteine and then modified with a spin labeling reagent to introduce the spin label side chain. Within the parallel in-register β-sheets, identical side chains stack on top of each other. As a result of side chain stacking, spin labels, when introduced into β-strand sites, interact with each other through spin exchange interactions (Margittai and Langen, 2008). Strong spin exchange interactions lead to the collapse of the three-line EPR spectrum, which is typical of a nitroxide spin label, to a single-line spectrum, providing a signature for the parallel in-register β-sheet structure (Agopian and Guo, 2012; Chen et al., 2007; Ngo et al., 2011; Török et al., 2002). Previous work shows that quantitative analysis of the spin exchange interaction can identify the location of β-strands and turns (Ngo et al., 2012). The rationale is that the side chain stacking of introduced spin labels leading to the single-line EPR spectrum requires the residue to be located on well-ordered β-strands. In general, turns are not as ordered as β-strands, and thus spin labels located on the turn would have much weaker spin-spin interactions than a β-strand site (Ngo et al., 2012).
To this end, we introduced spin labels, one at a time, at 22 residue positions in Aβ42 fibrils and studied spin exchange interactions with EPR. Single-line spectra were identified at residues 17-20 and 32-36, corresponding to two β-strands, which we called β1 and β2. Quantitative analysis revealed that residues 37 and 38 have weak spin exchange interactions, followed by residues 39-41 with strong spin exchange interactions. These results suggest the presence of a C-terminal turn at residues 37-38 and a C-terminal β-strand (βC) at residues 39-41. Aided by protein structure prediction program Rosetta, we present an atomic-level structural model of Aβ42 fibrils.
MATERIALS AND METHODS
Preparation of Aβ peptides and spin labeling
The DNA constructs of wild-type GroES-ubiquitin-Aβ (Shahnawaz et al., 2007) and the deubiquitylating enzyme Usp2cc (Baker et al., 2005) were kindly provided by Dr. Il-Seon Park at Chosun University (South Korea) and Dr. Rohan T. Baker at Australian National University (Australia). Cysteine mutations were introduced into Aβ42 sequence using QuikChange kit (Agilent) and confirmed with DNA sequencing.
GroES-ubiquitin-Aβ and Usp2cc proteins were expressed in E. coli and purified with nickel column as previously described (Agopian and Guo, 2012; Gu et al., 2014; Ngo and Guo, 2011). Full-length Aβ was cleaved from the fusion protein with Usp2cc using a previously described protocol (Gu and Guo, 2013; Gu et al., 2013).
For spin labeling of Aβ42 cysteine mutants, dithiothreitol was added to purified protein fraction to a final concentration of 10 mM and was then incubated at room temperature for 20 min. Then the Aβ42 sample was buffer exchanged to labeling buffer (20 mM MOPS, 7 M guanidine hydrochloride, pH 6.8) using a HiTrap desalting column (GE Healthcare). The spin labeling reagent MTSSL (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate, Enzo Life Sciences) was added at 10-fold molar excess at room temperature for 1 h. The spin-labeled Aβ42 was further buffer exchanged to 30 mM ammonium acetate, pH 10.0, using a HiTrap desalting column (GE Healthcare), and lyophilized. MALDI-TOF mass spectrometry was performed to ensure that the mass of Aβ42 is correct, and the extent of labeling is >95%. Wild-type Aβ40 and Aβ42 proteins were simply buffer exchanged to 30 mM ammonium acetate, pH 10.0, and then lyophilized and stored at −80°C.
Aβ42 fibril preparation
For fibril formation, Aβ42 was suspended in 100% 1,1,1,3,3,3 hexafluoro-2-propanol (HFIP) at 1 mM and bath sonicated for 5 min. Then the sample was incubated at room temperature for 30 min. HFIP was evaporated overnight in the fume hood and then under vacuum for 1 h. Finally the Aβ sample was dissolved in CG buffer (20 mM CAPS, 7 M guanidine hydrochloride, pH 11) and then diluted 20-fold to PBS buffer (50 mM phosphate, 140 mM NaCl, pH 7.4) to a final concentration of 50 μM. Then the Aβ solution was placed on a digital vertex mixer with a speed of 600 rpm at room temperature. Fibril formation was monitored daily using thioflavin T fluorescence for all the mutants. Fibrils were collected by centrifugation at 14,000 g for 20 min after thioflavin T binding has reached plateau (~5–7 days). Soluble proteins were removed by washing the pellet with PBS buffer.
Transmission electron microscopy
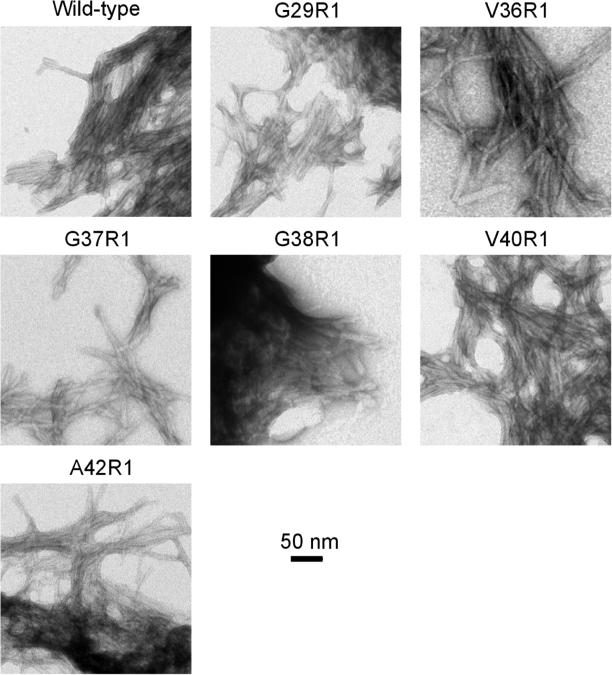
The Aβ fibril sample (5 μl) was placed on glow-discharged copper grids covered with 400 mesh formvar/carbon film (Ted Pella). The sample was negatively stained with 2% uranyl acetate. Samples were examined using a JEOL JEM-1200EX transmission electron microscope at 80 kV. Fibrils of the wild-type and 6 spin-labeled mutants were studied and the electron micrographs are shown in Figure 3.
Figure 3. Transmission electron microscopy images of wild-type and spin-labeled Aβ42 fibrils.
Similar morphology from wild-type and spin-labeled Aβ42 fibrils suggest that spin labeling does not perturb the process of fibril formation.
EPR spectroscopy
EPR measurements were performed at X-band frequency on a Bruker EMX spectrometer equipped with the ER 4102ST cavity. Approximately 20 μL of fibril sample was loaded into glass capillaries (VitroCom) sealed at one end. A modulation frequency of 100 kHz was used. Measurements were performed at 20 mW microwave power at room temperature. Conversion time was set at 10.24 ms and time constant was set at 81.92 ms for typical single-line EPR spectra. The sweep time for each scan was ~20 s. Modulation amplitude was optimized to individual spectrum (typically ~4 G). We varied the number of scans based on the signal to noise ratio of the EPR spectrum. Typically 10 to 30 scans were performed to obtain each EPR spectrum. EPR spectra in each figure panel were normalized to the same number of spins.
EPR spectral simulations
Spectral simulations were performed using the program MultiComponent of Dr. Christian Altenbach at University of California, Los Angeles, which provides a LabVIEW (National Instruments) interface of the program NLSL developed by Freed and co-workers (Budil et al., 1996; Schneider and Freed, 1989). A microscopic order macroscopic disorder model was used as previously described (Budil et al., 1996). A least-squares fit of the user-defined spectral parameters was performed using the Levenberg-Marquardt algorithm. For all fits, the values for the magnetic tensors A and g were fixed as Axx = 6.2, Ayy = 5.9, Azz = 37.0, and gxx = 2.0078, gyy = 2.0058, gzz = 2.0022, which were determined previously for spin label R1 (Columbus et al., 2001). An anisotropic model for the motion of the spin label was assumed and was found to give better fits than isotropic models. For anisotropic simulations, diffusion tilt angles were fixed to (α,β,γ) = (0,36°,0) for z-axis anisotropy as previously reported (Columbus et al., 2001). The diffusion tilt angels are the Euler angles relating the axes of the diffusion tensor and the magnetic tensor. The number of fitted parameters was kept at a minimum, which in this work includes the rotational diffusion constant (R), an order parameter (S), and Heisenberg exchange frequency (ω). We found that satisfactory fits were obtained with only these three parameters. Rotational correlation time (τ) was calculated using τ = 1/ (6R). For N-terminal sites (1, 4, 7, 10, 12, and 13), an additional spectral component without spin exchange interactions was required to obtain the best fits. This non-exchange component is fitted with an isotropic motion. All fitted parameters are reported in Table S1.
Modeling of Aβ42 fibrils with Rosetta
The structural model of Aβ42 fibril was built using the structure prediction program Rosetta (Leaver-Fay et al., 2011). According to EPR data, the four stretches of strong exchange residues are βN (residues 7-13), β1 (residues 17-20), β2 (residues 32-36) and βC (residues 39-41). First, each of the four segments of 7-13, 17-20, 32-36 and 39-41 was modeled as a β-sheet. The β-sheets of these segments were assembled by exploring all possible arrangements, and the assembled structures were then filtered by the length limitation of the connecting segment and energetic evaluation of current models. The complete structures of Aβ42 were built by modeling the conformation of loops connecting these segments at residues 1-6, 14-16, 21-31 and 37-38. Finally, the fibril structures of Aβ42 were refined by simultaneously optimizing the rigid-body degree of freedom between symmetrical copies, side chain and backbone torsions of Aβ42 subunits, guided by full-atom Rosetta energy functions (Kuhlman et al., 2003). Taking advantage of the recently developed symmetry implementation in Rosetta (Andre et al., 2007), the fibril symmetry is restrained to assure that symmetrical geometry is satisfied during the whole optimization process. The models were inspected based on Rosetta energy, additional experimental restraint of the residue pair of Asp23-Lys28, the secondary structure agreement and the packing between different β sheets.
Aβ aggregation kinetics
For aggregation experiments, HFIP-treated Aβ40 was first dissolved in CG buffer to a concentration of 1.5 mM, and then diluted 20-fold to PBS containing 50 μM thioflavin T (final concentration). Three Aβ40 samples were prepared without any fibril seeds, and three Aβ40 samples were prepared with 2% Aβ42 fibrils seeds. The final Aβ40 concentration in the aggregation reaction is 75 μM. The Aβ42 fibrils seeds were prepared by sonicating the wild-type Aβ42 fibrils using a Branson Digital Sonifier model 450 (microtip, 10% amplitude, 15 20-second pulses, with 20 seconds pause in between pulses). 50 μL of each sample was transferred to a 384-well Nonbinding Surface microplate with clear bottom (Corning 3655). The plate was then sealed with a plastic film (Corning 3095). All these steps were performed on ice. The aggregation was initiated by placing the plate in a Victor 3V plate reader (Perkin Elmer) at 37°C without agitation. The thioflavin T fluorescence was measured through the bottom of the plate at every 3 min (with excitation filter of 450 nm and emission filter of 490 nm).
RESULTS
Quantitative analysis of spin exchange interactions to reveal secondary structures in Aβ42 fibrils
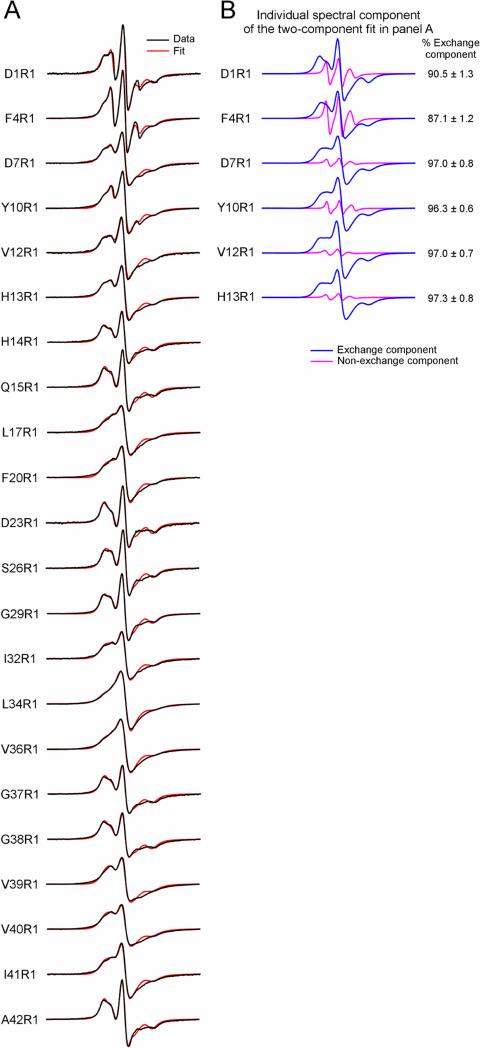
We prepared a total of 22 spin-labeled Aβ42 fibril samples, and each sample contains a spin label at a unique residue position. The spin label is termed R1 in this work. We followed the fibril formation with thioflavin T binding and collected fibrils after thioflavin T fluorescence reached a plateau, typically within 5-7 days. The EPR spectra of these spin-labeled Aβ42 fibrils are shown in Figure 1 (black traces). Most of these EPR spectra are characterized by the single-line feature, which results from strong spin exchange interactions between spin labels. The strong spin exchange interaction requires spin labels to be close, approximately <5.5 Å apart (Agopian and Guo, 2012). Overall, these single-line spectra suggest Aβ42 fibrils adopt a parallel in-register β-sheet structure.
Figure 1. EPR spectra of Aβ42 fibrils spin-labeled at indicated residue positions.
(A) Experimental spectra (black) are superimposed with best fits from spectral simulations (red). From spectral simulations we can extract spin exchange frequency as a measure of the strength of spin exchange interactions. (B) Individual spectral components of the two-component fit for indicated positions are shown in blue and magenta. The rest of the labeled positions were fitted with just the exchange component. Note that most EPR spectra are characterized by the single-line feature, resulting from strong spin exchange interactions between spin labels. The single-line feature is a signature of parallel in-register β-sheet structure in amyloid fibrils. R1 represents the spin label. Scan width is 200 G.
To reveal structural details of Aβ42 fibrils, we performed quantitative analysis with spectral simulations to obtain the strength of spin exchange interactions. The best fits to the experimental spectra are shown in Figure 1 (red traces). Most spectra were fitted with a single spectral component, except residues in the range of 1 to 13 that require a minor spectral component (~3 to 13% of total spin label population), representing a locally disordered state. From spectral simulations, we obtained spin exchange frequencies as a measure of the strength of spin exchange interactions, which are plotted as a function of residue positions in Figure 2.
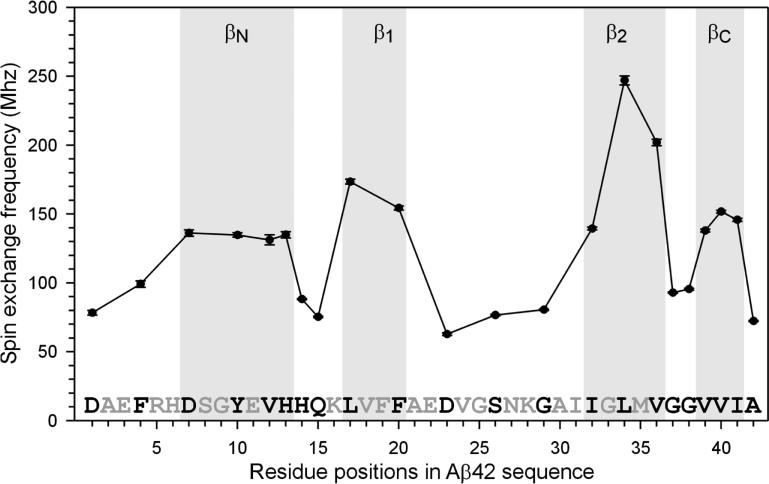
Figure 2. Plot of spin exchange frequencies reveals secondary structure in Aβ42 fibrils.
Four regions with strong spin exchange interactions (>120 MHz) are categorized as β-strands.
The spin exchange frequency plot reveals four stretches of residues that give rise to strong spin exchange interactions (Figure 2). Previously, our quantitative analysis of spin exchange interactions in Aβ40 fibrils suggests that residues with strong spin exchange interactions (i.e., >120 MHz) correlate with β-strand residues as revealed by solid state NMR studies (Agopian and Guo, 2012). For Aβ42 fibrils, all four stretches of strong exchange residues have spin exchange frequencies of >120 MHz, suggesting that they adopt β-strand structures. The residues that correspond to the two β-strands in Aβ40 fibrils are named β1 (residues 17-20) and β2 (residues 32-36) (Figure 2). EPR data also reveal two additional β-strands, which we named βN (residues 7-13) and βC (residues 39-41). The strength of spin exchange interactions suggest that β2 is the most ordered strand while βN is the least ordered stand.
While the strong exchange interactions must originate from parallel in-register stacking of spin label side chains in the β-sheet, weak exchange interactions could result from other possibilities, which we exclude one by one below. First, spin labeling at residues such as 37 or 38 might lead to formation of amorphous aggregates, and thus would result in weak spin exchange interactions. We investigated the morphology of Aβ42 fibrils spin-labeled at 6 selected positions, including residues 37 and 38 (Figure 3). In all cases, transmission electron microscopy images show fibrillar morphology that is similar as wild-type, suggesting that spin labeling at these residue positions did not affect fibril formation. Second, low labeling efficiency would result in weaker spin exchange interactions. We monitored all our spin labeling reactions with mass spectrometry. The labeling efficiency is generally >95%. Samples with <95% labeling efficiency were discarded and not used in fibril preparation. Third, loss of spin labels during fibril formation might also lead to weaker spin exchange interactions. We checked EPR signals in the soluble fraction after fibrils were collected and did not observe significant amount of free spin labels.
Rosetta modeling of Aβ42 fibrils
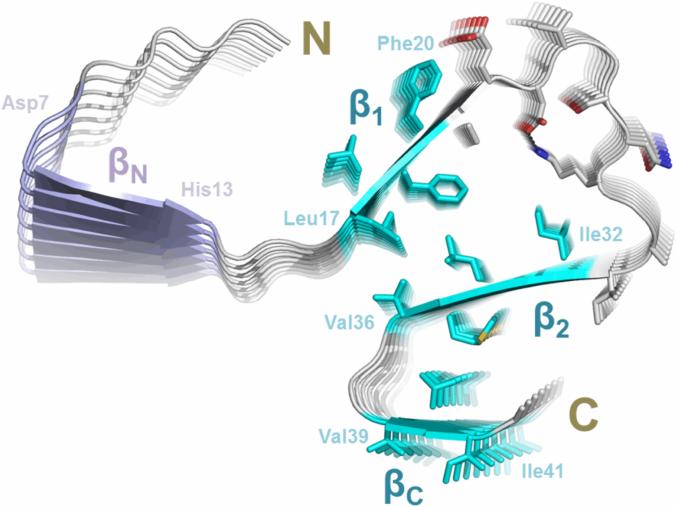
To determine the 3D structural model of Aβ42 fibrils, we incorporate the EPR-derived restraints into protein structure prediction using Rosetta (Leaver-Fay et al., 2011). The four β-strands: βN (7-13), β1 (17-20), β2 (32-36) and βC (39-41), were modeled as parallel in-register β-sheets as identified in EPR experiments. Guided by full-atom Rosetta energy functions while restraining the fibril symmetry, structural models of Aβ42 fibrils were simultaneously optimized and refined. Finally, the models were assessed by a combination of full-atom Rosetta energy and satisfaction of additional experimental constraints (i.e., salt bridge restraint between residues D23 and K28 derived from the mutagenesis studies (Lührs et al., 2005)). The final model with favorable Rosetta energy and good agreement to the experimental restraints was selected. As shown in Figure 4, our model of Aβ42 protofilament consists of four parallel in-register β-sheets. The sheet βN does not pack together with β1, β2 and βC, consistent with EPR data suggesting βN being the least ordered β-sheet. Interdigitating side chains of Leu17, Phe19, Ile32 and Leu34 constitute the hydrophobic core between the sheets β1 and β2. The salt bridge D23–K28 is located at the loop region of residues 22–30 connecting the sheets β1 and β2. The sheets β2 and βC pack side-by-side via hydrophobic contacts of residues Met35 and Val40, connected by the Gly-Gly turn of residues 37-38. The presence of the C-terminal turn at residues 37-38 distinguishes our Aβ42 fibril model from other models in the literature.
Figure 4. The structural model of Aβ42 fibrils.
A model of Aβ42 protofilament was built based on EPR restraints and Rosetta prediction. The side chains of residues 17-42 are shown in sticks, elucidating possible side chain interactions in the fibril core. The four stretches of strong exchange residues are colored based on how well they are structurally ordered. The three ordered regions, β1 (residues 17-20), β2 (residues 32-36) and βC (residues 39-41), are colored in cyan, while the less ordered region βN (residues 7-13) is in purple. The salt bridges between Asp23 and Lys28 are shown as black dashed lines.
Aβ42 fibrils seed Aβ40 fibrillization
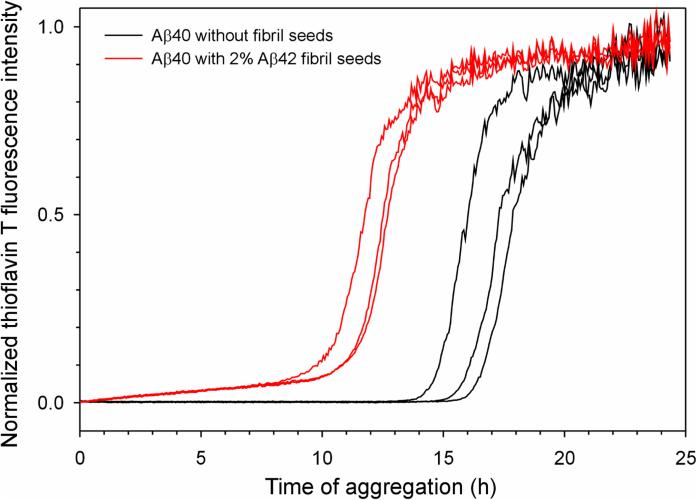
Our Aβ42 fibril model share similar structural features at the region of residues 17 to 35 with other Aβ40 fibril models based on solid-state NMR data (Bertini et al., 2011; Paravastu et al., 2008; Petkova et al., 2006). This may be the structural basis for the observation that Aβ42 and Aβ40 form interlaced fibrils (Gu and Guo, 2013). The structural similarity between Aβ42 and Aβ40 fibrils also suggest that Aβ42 fibrils should be able to seed the fibril formation of Aβ40. To investigate this, we studied the aggregation kinetics of Aβ40 in the absence and presence of Aβ42 fibril seeds. Clearly, the lag time of Aβ40 fibrillization was shortened by the presence of Aβ42 seeds (Figure 5), supporting the common structural feature between Aβ42 and Aβ40. It should also be noted that some studies found that Aβ42 fibril seeds were incapable of promoting the aggregation of Aβ40 (Lu et al., 2013; Xiao et al., 2015), in contrast to the results in this work (Figure 5) and elsewhere (Jan et al., 2008; Pauwels et al., 2012). The causes of this discrepancy are not yet defined. Polymorphic nature of the fibrils, aggregation conditions, and how Aβ is prepared could all contribute to these contradicting observations.
Figure 5. Aβ42 fibrils seed the aggregation of Aβ40.
Aggregation of Aβ40 was followed with thioflavin T fluorescence. Three repeats of Aβ40 in the absence of fibril seeds (black traces) and in the presence of 2% Aβ42 fibril seeds (red traces) are shown. Note that the lag time is shortened by the presence of seeds, suggesting that Aβ42 fibril seeds promote the aggregation of Aβ40.
DISCUSSION
Aβ42 deposition in amyloid plaques precedes Aβ40 in the development of Alzheimer's disease (Gravina et al., 1995; Iwatsubo et al., 1995; Iwatsubo et al., 1994; Mak et al., 1994; Miller et al., 1993) and in normal aging (Fukumoto et al., 1996). Yet the structure of Aβ42 fibrils, unlike its Aβ40 isoform, still remains elusive. Based on EPR data and Rosetta predictions, we report an atomic model of Aβ42 fibrils. Our structural model consists of four β-sheets at residues 7-13 (βN), 17-20 (β1), 32-36 (β2), and 39-41 (βC). The sheets of β1, and β2 have a structure that is very similar to the same region in Aβ40 fibrils. The C-terminal sheet βC distinguishes the structure of Aβ42 from Aβ40 fibrils.
Our structural model of Aβ42 fibrils is consistent with a large body of the existing biochemical and biophysical data in the literature. Lührs et al. (2005) and Olofsson et al.(2006) reported two sets of hydrogen exchange data, which revealed two stretches of protected backbone amide protons. The first stretch of protected residues starts from residue 17 in Lührs et al. (2005) or residue 11 in Olofsson et al. (2006) and ends at residue 25. The second stretch of protected residues covers from residue 30 to C-terminus. Our structural model is largely consistent with both sets of hydrogen exchange data. Residues 37 and 38 in our model adopt a special turn conformation, called “arc” (Hennetin et al., 2006; Kajava et al., 2010), and backbone hydrogen bonds are maintained in the arc. Our model is also consistent with the close contact between residues 19 and 34 identified in solid-state NMR studies (Ahmed et al., 2010). Moreover, the C-terminal β-arc-β motif in our model can also explain the tolerance to proline mutagenesis in the arc region as identified previously (Morimoto et al., 2004). A recent solid-state NMR study of Aβ42 fibrils also suggests a loop or turn structure at residues 37 and 38 (Colvin et al., 2015). Additionally, our EPR data also revealed an N-terminal β-strand region, βN. This is in agreement with the hydrogen exchange data by Lührs et al. (2005), which showed that approximately half of the amide population for the N-terminal region (residues 3-16) is well protected. At the same time, subtle differences exist between our structural model and models from investigations, and among other models themselves. These differences include the existence of βN, the exact length of each β-strand, and the presence or absence of certain inter-residue distances. This suggests that, just like Aβ40, Aβ42 fibrils may also be highly polymorphic. Our understanding of Aβ42 fibril structures will not be complete without the knowledge of their structural diversity.
The C-terminal β-arc-β motif distinguishes our Aβ42 fibril model from Aβ40 fibril models. Except for the C-terminal arc at residues 37-38 and a short β-strand at residues 39-41, the rest of Aβ42 sequence adopts structures similar as in previously reported Aβ40 fibril structures (Bertini et al., 2011; Tycko, 2006). With its two additional residues at the C-terminus, Aβ42 plays a pathological role in the development of Alzheimer's, while Aβ40 is considered by many investigators to be non-pathogenic or even protective (Jan et al., 2008; Kuperstein et al., 2010; McGowan et al., 2005; Yan and Wang, 2007). In our new Aβ42 fibril model with β1, β2, and βC forming the hydrophobic core, there is more hydrophobic contact in the core of Aβ42 fibrils. This may explain the higher aggregation propensity of Aβ42 than Aβ40. At the same time, the structural similarity for residues 17-35 between Aβ42 and Aβ40 fibrils provides a structural explanation for the in vitro aggregation data showing that Aβ42 and Aβ40 can form mixed fibrils (Gu and Guo, 2013) and the cross-seeding effect that Aβ42 fibrils showed on Aβ40 fibrillization (Figure 5). This structural similarity between Aβ42 and Aβ40 may be the basis for the critical role of Aβ42/Aβ40 ratio (Nutu et al., 2013), not just Aβ42 level, in Alzheimer's disease.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Christian Altenbach and Dr. Wayne Hubbell for providing EPR analysis programs, Hongsu Wang and Frederick Hsu for assistance in sample preparation and data collection.
FUNDING
This work was supported by the National Institute of Health (Grant R01GM110448) and by a Turken Research Award to Z.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agopian A, Guo Z. Structural origin of polymorphism for Alzheimer's amyloid-β fibrils. Biochem. J. 2012;447:43–50. doi: 10.1042/BJ20120034. [DOI] [PubMed] [Google Scholar]
- Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, et al. Structural conversion of neurotoxic amyloid-β(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. USA. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- Baker RT, Catanzariti AM, Karunasekara Y, Soboleva TA, Sharwood R, et al. Using deubiquitylating enzymes as research tools. Methods Enzymol. 2005;398:540–554. doi: 10.1016/S0076-6879(05)98044-0. [DOI] [PubMed] [Google Scholar]
- Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J. Am. Chem. Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J. Magn. Reson., Ser A. 1996;120:155–189. [Google Scholar]
- Chen M, Margittai M, Chen J, Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- Columbus L, Kálai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in α-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- Colvin MT, Silvers R, Frohm B, Su Y, Linse S, et al. High resolution structural characterization of Aβ42 amyloid fibrils by magic angle spinning NMR. J. Am. Chem. Soc. 2015;137:7509–7518. doi: 10.1021/jacs.5b03997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto H, Asami-Odaka A, Suzuki N, Shimada H, Ihara Y, et al. Amyloid β protein deposition in normal aging has the same characteristics as that in Alzheimer's disease. Predominance of Aβ42(43) and association of Aβ40 with cored plaques. Am. J. Pathol. 1996;148:259–265. [PMC free article] [PubMed] [Google Scholar]
- Gravina SA, Ho LB, Eckman CB, Long KE, Otvos L, et al. Amyloid β protein (Aβ) in Alzheimer's disease brain: Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Aβ40 or Aβ42(43). J. Biol. Chem. 1995;270:7013–7016. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- Gu L, Guo Z. Alzheimer's Aβ42 and Aβ40 peptides form interlaced amyloid fibrils. J. Neurochem. 2013;126:305–311. doi: 10.1111/jnc.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Liu C, Guo Z. Structural insights into Aβ42 oligomers using site-directed spin labeling. J. Biol. Chem. 2013;28:18673–18683. doi: 10.1074/jbc.M113.457739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Liu C, Stroud JC, Ngo S, Jiang L, et al. Antiparallel triple-strand architecture for prefibrillar Aβ42 ligomers. J. Biol. Chem. 2014;289:27300–27313. doi: 10.1074/jbc.M114.569004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennetin J, Jullian B, Steven AC, Kajava AV. Standard conformations of β-arches in β-solenoid proteins. J. Mol. Biol. 2006;358:1094–1105. doi: 10.1016/j.jmb.2006.02.039. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Mann DMA, Odaka A, Suzuki N, Ihara Y. Amyloid β protein (Aβ) deposition: Aβ42(43) precedes Aβ40 in Down syndrome. Ann. Neurol. 1995;37:294–299. doi: 10.1002/ana.410370305. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, et al. Visualization of Aβ42(43) and Aβ40 in senile plaques with end-specific Aβ monoclonals: Evidence that an initially deposited species is Aβ42(43). Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Jan A, Gokce O, Luthi-Carter R, Lashuel HA. The ratio of monomeric to aggregated forms of Aβ40 and Aβ42 is an important determinant of amyloid-β aggregation, fibrillogenesis, and toxicity. J. Biol. Chem. 2008;283:28176–28189. doi: 10.1074/jbc.M803159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Baxa U, Steven AC. β arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. FASEB J. 2010;24:1311–1319. doi: 10.1096/fj.09-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Kuperstein I, Broersen K, Benilova I, Rozenski J, Jonckheere W, et al. Neurotoxicity of Alzheimer's disease Aβ peptides is induced by small changes in the Aβ42 to Aβ40 ratio. EMBO J. 2010;29:3408–3420. doi: 10.1038/emboj.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, et al. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, et al. 3D structure of Alzheimer's amyloid-β(1-42) fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Nussinov R. Polymorphic triple β-sheet structures contribute to amide hydrogen/deuterium (H/D) exchange protection in the Alzheimer amyloid β42 peptide. J. Biol. Chem. 2011;286:34244–34253. doi: 10.1074/jbc.M111.241141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak K, Yang FS, Vinters HV, Frautschy SA, Cole GM. Polyclonals to β-amyloid(1-42) identify most plaque and vascular deposits in Alzheimer cortex, but not striatum. Brain Res. 1994;667:138–142. doi: 10.1016/0006-8993(94)91725-6. [DOI] [PubMed] [Google Scholar]
- Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q. Rev. Biophys. 2008;41:265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Irie K, Murakami K, Ohigashi H, Ohashi R, et al. Verification of the turn at positions 22 and 23 of the β-amyloid fibrils with Italian mutation using solid-state NMR. Bioorg Med Chem. 2005;13:6803–6809. doi: 10.1016/j.bmc.2005.07.071. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Uemura S, Nakanishi A, Ohashi R, Takegoshi K, et al. Verification of the C-terminal intramolecular β-sheet in Aβ42 aggregates using solid-state NMR: implications for potent neurotoxicity through the formation of radicals. Bioorg Med Chem Lett. 2008;18:3206–3210. doi: 10.1016/j.bmcl.2008.04.060. [DOI] [PubMed] [Google Scholar]
- McGowan E, Pickford F, Kim J, Onstead L, Eriksen J, et al. Aβ42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, et al. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer's disease. Arch. Biochem. Biophys. 1993;301:41–52. doi: 10.1006/abbi.1993.1112. [DOI] [PubMed] [Google Scholar]
- Morimoto A, Irie K, Murakami K, Masuda Y, Ohigashi H, et al. Analysis of the secondary structure of β-amyloid (Aβ42) fibrils by systematic proline replacement. J. Biol. Chem. 2004;279:52781–52788. doi: 10.1074/jbc.M406262200. [DOI] [PubMed] [Google Scholar]
- Ngo S, Guo Z. Key residues for the oligomerization of Aβ42 protein in Alzheimer's disease. Biochem. Biophys. Res. Commun. 2011;414:512–516. doi: 10.1016/j.bbrc.2011.09.097. [DOI] [PubMed] [Google Scholar]
- Ngo S, Gu L, Guo Z. Hierarchical organization in the amyloid core of yeast prion protein Ure2. J. Biol. Chem. 2011;286:29691–29699. doi: 10.1074/jbc.M111.269092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo S, Chiang V, Guo Z. Quantitative analysis of spin exchange interactions to identify strand and turn regions in Ure2 prion domain fibrils with site-directed spin labeling. J. Struct. Biol. 2012;180:374–381. doi: 10.1016/j.jsb.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Nutu M, Zetterberg H, Londos E, Minthon L, Nagga K, et al. Evaluation of the cerebrospinal fluid amyloid-β1-42/amyloid-β1-40 ratio measured by alpha-LISA to distinguish Alzheimer's disease from other dementia disorders. Dement Geriatr Cogn Disord. 2013;36:99–110. doi: 10.1159/000353442. [DOI] [PubMed] [Google Scholar]
- Olofsson A, Sauer-Eriksson AE, Ohman A. The solvent protection of Alzheimer amyloid-β-(1-42) fibrils as determined by solution NMR spectroscopy. J. Biol. Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels K, Williams TL, Morris KL, Jonckheere W, Vandersteen A, et al. The structural basis for increased toxicity of pathological Aβ42:Aβ40 ratios in Alzheimer's disease. J. Biol. Chem. 2012;287:5650–5660. doi: 10.1074/jbc.M111.264473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kienlen-Campard P, Ahmed M, Liu W, Li HL, et al. Inhibitors of amyloid toxicity based on β-sheet packing of Aβ40 and Aβ42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Rohou A, Lasker K, Yadav JK, Schiene-Fischer C, et al. Peptide dimer structure in an Aβ(1-42) fibril visualized with cryo-EM. Proc. Natl. Acad. Sci. USA. 2015 doi: 10.1073/pnas.1503455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DJ, Freed JH. Calculating slow motional magnetic resonance specta. In: Berliner LJ, Reuben J, editors. Spin Labeling: Theory and Applications. Plenum Press; New York: 1989. pp. 1–76. [Google Scholar]
- Shahnawaz M, Thapa A, Park IS. Stable activity of a deubiquitylating enzyme (Usp2-cc) in the presence of high concentrations of urea and its application to purify aggregation-prone peptides. Biochem. Biophys. Res. Commun. 2007;359:801–805. doi: 10.1016/j.bbrc.2007.05.186. [DOI] [PubMed] [Google Scholar]
- Török M, Milton S, Kayed R, Wu P, McIntire T, et al. Structural and dynamic features of Alzheimer's Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J. Biol. Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, et al. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease. Nat. Struct. Mol. Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Wang C. Aβ40 protects non-toxic Aβ42 monomer from aggregation. J. Mol. Biol. 2007;369:909–916. doi: 10.1016/j.jmb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Zhang R, Hu X, Khant H, Ludtke SJ, Chiu W, et al. Interprotofilament interactions between Alzheimer's Aβ1-42 peptides in amyloid fibrils revealed by cryoEM. Proc. Natl. Acad. Sci. USA. 2009;106:4653–4658. doi: 10.1073/pnas.0901085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.