Abstract
Cardiac fibroblasts produce the extracellular matrix (ECM) scaffold within which the various cellular components of the heart are organized. As well as providing structural support, it is becoming evident that the quality and quantity of ECM is a key factor for determining cardiac cell behavior during development and in pathological contexts such as heart failure involving fibrosis. Cardiac fibroblasts have long remained a poorly characterized cardiac lineage. Well characterized markers are now paving the way for a better understanding of the roles of these cells in various developmental and disease contexts. Notably, the relevance of processes including endothelial-tomesenchymal transition and the recruitment of circulating fibroblast progenitors in heart failure has been challenged. This review describes the latest findings on cardiac fibroblast markers and developmental origins, and discusses their importance in myocardial remodeling. Effective modulation of cardiac fibroblast activity would likely contribute to successful treatment of various cardiac disorders.
Keywords: fibroblast, fibrosis, hypertrophy, myocardial infarction, fibroblast markers, cardiac lineage
Introduction
Heart failure results from adverse remodeling of the myocardium following injury and is a major clinical issue[1]. Remodeling of the adult myocardium involves multiple cellular processes including myocyte hypertrophy, immune cell infiltration and fibrosis. The latter refers to the activation and accumulation of cardiac fibroblasts that deposit large amounts of ECM, a process considered largely to be maladaptive, although the latter point is context-dependent. Myocardial infarction involves the loss of an area of myocardium, most often in the left ventricle following the obstruction of a coronary artery, and commonly underlies heart failure. Hemodynamic overload endured by the remaining myocardium triggers pathologic “reactive fibrosis”, characterized by excessive deposition of ECM around myocytes and vascular cells causing increased tissue rigidity. However, the loss of viable myocardium also activates “replacement fibrosis”, characterized by the formation of a scar that ensures continual structural integrity of the chamber. Hence, although reactive fibrosis is undesirable within viable myocardium, replacement fibrosis can be vital for compensating the lack of regenerative capacity of the adult heart in the case of significant myocyte loss. Moreover, it is likely that replacement fibrosis constitutes an essential initial step for myocardial repair. Indeed, it has been reported that the neonatal mouse heart can regenerate, and that fibrosis precedes regeneration[2].
Fibroblasts have long been recognized as a major cell population of the myocardium, but the lack of specific markers has hindered efforts to gain detailed insight into their origins and functions. Recent studies, employing new markers, are revealing new aspects of cardiac fibroblast origins during development and in pathological contexts. It has been demonstrated that resident cardiac fibroblasts, rather than fibroblasts arising from circulating bone marrow cells or from resident endothelial cells, are the major cell type producing fibrotic extracellular matrix in the setting of pressure overload and myocardial infarction models[3, 4]. These efforts have re-centered attention to resident cardiac fibroblast activation in the context of fibrosis.
During heart development, fibroblasts are important for stimulating myocyte proliferation, thus contributing to normal heart development[5]. Understanding distinct pathways activated in fibroblast populations during normal heart development and in the setting of adult heart disease might allow for selective promotion of one, and inhibition of the other, to assist in replacement of functional heart tissue in disease settings.
Cardiac fibroblast markers
Much of the literature on cardiac fibroblast lineage development and adult myocardial fibrosis has stemmed from studies not relying on robust fibroblast markers. The intermediate filament Vimentin is often used as a fibroblast marker, but it is expressed in many cell types including endothelial cells[6]. CD90 (Thy1), another commonly used marker, is expressed by immune cells[7], lymphatic endothelium[8] and pericytes[9]. Of note, Fibroblast Specific Protein 1 (FSP1) has been extensively used as a fibroblast marker[10, 11]. However, several groups including ours have since shown that FSP1, or S100 calcium binding protein A4 (S100a4), is more frequently associated with immune cells than fibroblasts[3, 12, 13]. Among arguably more robust markers is Discoidin Domain Receptor 2 (DDR2), which labels fibroblasts, but not endothelium, smooth muscle or myocytes[14]. DDR2 is a collagen specific receptor tyrosine kinase and mediates signaling for cell growth and migration[15, 16]. Transcription factor 21 (TCF21), a marker of proepicardium among other mesothelial populations, has also been used to identify fibroblasts in embryonic heart[17-19].
Recently, more dependable markers directly associated with fibroblast function have emerged. These include a Collagen1a1-GFP reporter line[20], that labels fibroblasts during development and in pathological contexts (Figure 3)[3, 21]. Platelet-derived growth factor receptor, Alpha polypeptide (PDGFRα) is a tyrosine kinase receptor targeted by the PDGF mitogens that is extensively expressed in the mesenchymal of early embryos, subsequently becoming restricted to stromal populations [22, 23]. Collagen1a1-GFP and PDGFRα expression are tightly correlated, reinforcing the suitably of each of these markers to identify cardiac fibroblasts[3, 21].
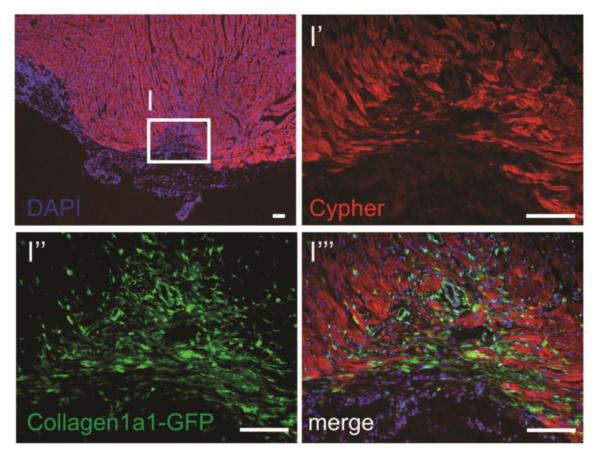
Figure 3.
Collagen type I expressing fibroblasts (green) are active in the area of regeneration, tightly associated with myocytes (Cypher+, red) following resection of neonatal mouse heart (unpublished observations, Evans S.M. et al.). Images represent the apex of a heart 2 weeks after resection, during the window of regeneration previously described [2]. Scale bars represent 100μm.
PDGRFα is strongly expressed by fibroblasts, whereas high PDGFRβ expression is associated with pericytes[3, 19]. The latter are also considered a type of mesenchymal stem cell (MSC) of the heart[24]. However, pluripotent/colony forming MSCs expressing PDGFRα and CD90, but not the pericyte marker NG2 (chondroitin sulfate proteoglycan 4), have also been identified[25]. In fact, fibroblasts express many markers previously associated with MSCs, including the PDGFRα, making the identification of putative MSCs in the heart challenging [26]. However, PDGFRαHIGH and PDGFRβHIGH expression broadly identify fibroblasts and pericytes, respectively.
In a pathological context, fibroblasts can co-express a number of markers more generally associated with progenitors such as epicardium[27]. Hence, as well as expressing the markers mentioned above, some activated fibroblasts upregulate Wilms Tumor 1 (WT1), T-Box 18 (TBX18) and TCF21[28]. Subsets, but not all, of fibroblasts also express the classic myofibroblast marker α-smooth muscle actin (αSMA)[3, 28]. Fibroblast activation protein (FAP) has also been shown to be expressed in cardiac fibroblasts in the context of fibrosis[29]. The extent and distribution of these co-markers depends on the disease model and localization of fibroblasts within the heart[28]. Notably, in the context of pressure overload, some interstitial fibroblasts express WT1, whereas subsets of perivascular fibroblasts express TBX18 and FSP1[3, 28].Finally, in pathological contexts, activated fibroblasts upregulate specific ECM proteins, including ED-A fibronectin (Fn)[30]. The latter is pro-inflammatory[31] and ED-A Fn-deficiency attenuates loss of cardiac function following myocardial infarction[32].
Origins of fibroblasts during development and heart failure
For some time the epicardium, an epithelial layer that forms over the mid-gestation heart and is maintained in adult, has been recognized as the major source of cardiac fibroblasts in various development models[19, 21, 33-35]. This observation was first made in early vital dye or retroviral tagging of proepicardial progenitors experiments in chick embryos[33]. Other lineages, including, pericytes, smooth muscle cells and, perhaps, myocytes and endothelium[33-37] also derive from epicardium. Furthermore, expression of the transcription factor TCF21 in epicardial cells is required for cardiac fibroblast development[19].
Fibroblast-like cells of the mural leaflets of the cardiac valves, known as valve interstitial cells (VICs), are produced by endothelial-to-mesenchymal transition of endocardium that occurs during development of the endocardial cushions[38, 39]. Recent studies have shown that fibroblasts produced during this process during development also invade the myocardium, resulting in the presence of a substantial number of endothelially-derived fibroblasts, mostly located within the interventricular septum, overall presenting a distribution pattern complementary to that of epicardially derived fibroblasts (Figure 2)[3].
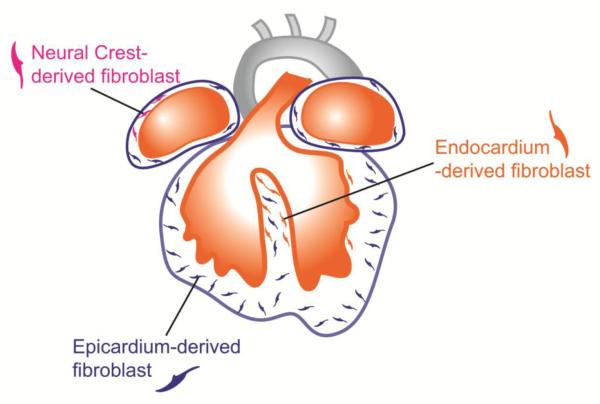
Figure 2.
Fibroblasts from cardiac lineages populate the heart during embryonic development. Most fibroblasts derived from the epicardium (blue), a protective epithelial layer that entirely covers the four cardiac chambers. Smaller populations are derived from the endocardium (orange) and cardiac neural crest, and are mostly found in the interventricular septum and right atrium, respectively.
Finally, a small subset of fibroblasts within myocardium, mostly found in the right atrium, appear to be derived from the neural crest[4]. Neural crest cells represents a very heterogeneous population derived from the dorsal aspect of the neural tube and giving rise to various lineages including neurons, melanocytes and mesenchymal cells[40]. A specific population of neural crest cells contributes to valves within the outflow region of the heart, giving rise to mesenchyme of the semilunar valves, as well as melanocytes[41-43]. In the valves, neural crest derived mesenchyme is eventually replaced by endocardium derived mesenchyme, leaving melanocytes as the only neural crest contribution to adult valves[44]. However, fibroblasts identified in atrial myocardium seem to persist, at least to some extent, into adulthood[4].
The origin of fibroblasts constituting fibrotic lesions in failing hearts has been controversial. Indeed, some reports have shown that fibrotic fibroblasts are derived solely from developmentally-derived resident fibroblast lineages, whereas others have suggested that other cell types transdifferentiate into fibroblasts and significantly contribute to fibrotic matrix deposition. The latter point of view has been put forward in studies reporting that endothelial-to-mesenchymal transition and recruitment of circulating fibroblast progenitors contributed significantly to cardiac fibrosis[45-47]. These studies include transaortic banding[45], chronic rejection[45], myocardial infarction [46-48] diabetes [49], and angiotensin-induced cardiac fibrosis models[50], suggesting that EndoMT and blood progenitor recruitment were general mechanisms of cardiac fibrosis. Importantly, these processes represent potential therapeutic targets. Notably, inhibiting EndoMT would promote preservation of the vasculature whilst reducing fibroblast accumulation. However, these studies overlooked developmental heterogeneity of cardiac fibroblasts and relied on markers including FSP1 and αSMA that, as mentioned above, present limitations. For example, as FSP1 labels immune cells, it should not be used as a marker of hematopoietic-derived fibroblasts. Also, FSP1 and αSMA do not label fibroblasts in control conditions, underlining the importance of using more robust markers for making an initial assessment of fibroblast presence and diversity[3]. More recent studies have revealed that heterogeneity in fibroblast lineages observed in a pathological context was the result of the activation of distinct developmentally derived lineages. Indeed, these studies show that expansion of the two main developmentally derived fibroblast lineages, of epicardial and endocardial/endothelial origin, accounted for the vast majority of fibroblasts prior to and during fibrosis following pressure overload[3, 4]. Notably, bone marrow transplants[4] and adult endothelial-lineage tracing[3], in combination with markers such as PDGFRα, provided strong evidence against contributions from blood and endothelium to fibrosis.
Cardiac fibroblast origins and implications for remodeling
Fibroblasts in different tissues and locations have been shown to have distinct transcriptional profiles[51]. Notably, compared to fibroblasts from other tissues, cardiac fibroblasts express high levels of the cardiogenic transcription factors T-Box 20 (TBX20) and GATA binding protein 4 (GATA4)[52], a result of particular relevance to studies reporting “direct cardiac reprogramming” of cardiac fibroblasts into myocytes in vivo [53, 54]. As the origins of cardiac fibroblasts during development and in pathological contexts become clearer, the extent to which the origin of a fibroblast determines its functional properties remains uncertain. In adult hearts, fibroblasts with endothelial and epicardial signatures presented similar transcriptional signatures and responded similarly, notably in terms of proliferative response, in the pressure overload model[3, 4]. Independently of their origin, the location of fibroblasts within the myocardium with respect to the vasculature is associated with the expression of specific markers in fibrotic conditions [3, 28]. Hence, the plasticity of fibroblasts means their phenotype may depend more on their environment rather than their developmental origin.
The above-mentioned studies focus on models that ultimately lead to heart failure, but further insight into how fibroblast origin might determine their biological properties could come from complementary studies in regeneration models. Indeed, the hearts of some vertebrates including zebrafish, and even neonatal mouse have been shown to have regenerative potential[2, 55]. On one hand, regeneration depends on re-entry of myocytes into the cell cycle, following sarcomere disassembly[56]. However, new studies are also providing evidence that specific properties of other cell populations are required. Notably, macrophages play an active role in mediating regeneration by promoting angiogenesis[57, 58], It is likely that the activation of resident fibroblast populations, known to support cardiomyocyte growth and maturation through the secretion of specific ECM components[5], also plays a key role and has yet to be investigated. Indeed, regeneration of zebrafish heart as well as neonatal mouse heart is associated with the formation of cardiac fibroblast-rich scar tissue[2, 55, 59, 60] (also, see Figure 3, Evans S.M. unpublished observations). Whether these fibroblasts are generated by resident fibroblast proliferation, epicardial activation or hematopoietic progenitors remains to be determined. Furthermore, determining specific properties of these fibroblasts may be of interest for promoting regeneration in adult heart.
Concluding remarks
The use of more extensively characterized fibroblast markers has allowed recent progress in defining the origins of these cells during development and in fibrotic conditions. During development, the majority of these cells derive from epicardium, with subsets also deriving from the endothelium/endocardium and neural crest. Lineage tracing studies in murine models of pressure overload and myocardial infarction show that fibrosis results from intense proliferation of resident fibroblast populations and/or re-activation of epicardial EMT rather that endothelial-to-mesenchymal transition or circulating progenitor recruitment. Whether the origin of a fibroblast determines its function during fibrosis to any extent is currently unclear. Fibrosis is pathogenic when occurring in viable myocardium. However, examples such as the replacement fibrosis occurring following MI and the fibrosis preceding regeneration in recently described neonatal resection/infarction models demonstrate that fibrosis is an important component for cardiac repair. Modulating fibroblast activity could help to provide a suitable environment for tissue repair to occur in a clinical context, by allowing the development of vasculature and integration of myocytes from various sources.
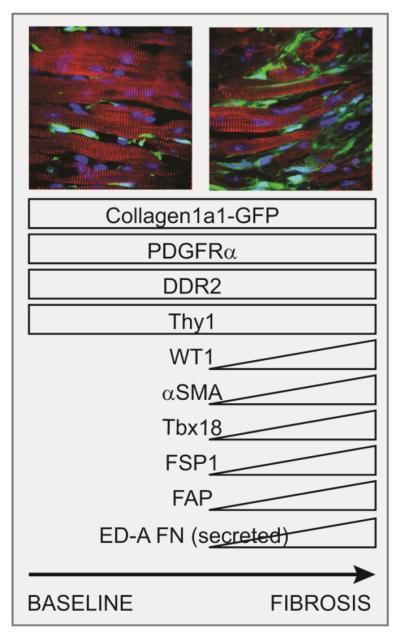
Figure 1.
Markers expressed by fibroblasts prior to and during fibrosis. Collagen1a1-GFP and PDGFRα are constitutively expressed by fibroblasts in normal and pathological contexts. Subsets of fibroblasts up-regulate a number of additional markers in fibrotic conditions, including αSMA and WT1, whose expression depends on the disease context and the location, notably with respect to large coronary vessels. Images: Cypher (myocyte markers, red), Collagen1a1-GFP(green), DAPI (blue).
Highlights.
Recently described robust cardiac fibroblast markers can now be used to identify fibroblasts in healthy and fibrotic heart
Cardiac fibroblasts have diverse developmental origins
Fibrosis results from the expansion of resident fibroblast lineages
Acknowledgments
Financial support:
T.M.M. was supported by AHA postdoctoral fellowship 11POST7310066. S.M.E is funded by grants from the National Heart, Lung, and Blood Institute (HL-117649 and HL119967). P.C. was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework (PIOF-623739). MP and TMM acknowledge the generosity of the Leducq Foundation (SHAPEHEART).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
None.
References
- [1].Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science (New York, NY) 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. The Journal of clinical investigation. 2014;124:2921–34. doi: 10.1172/JCI74783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, et al. Developmental Heterogeneity of Cardiac Fibroblasts Does Not Predict Pathological Proliferation and Activation. Circulation research. 2014 doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- [5].Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Developmental cell. 2009;16:233–44. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lane EB, Hogan BL, Kurkinen M, Garrels JI. Co-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryo. Nature. 1983;303:701–4. doi: 10.1038/303701a0. [DOI] [PubMed] [Google Scholar]
- [7].Raff MC. Surface antigenic markers for distinguishing T and B lymphocytes in mice. Transplantation reviews. 1971;6:52–80. doi: 10.1111/j.1600-065x.1971.tb00459.x. [DOI] [PubMed] [Google Scholar]
- [8].Jurisic G, Iolyeva M, Proulx ST, Halin C, Detmar M. Thymus cell antigen 1 (Thy1, CD90) is expressed by lymphatic vessels and mediates cell adhesion to lymphatic endothelium. Experimental cell research. 2010;316:2982–92. doi: 10.1016/j.yexcr.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell stem cell. 2008;3:301–13. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- [10].Strutz F, Okada H, Lo CW, Danoff T, Carone RL, Tomaszewski JE, et al. Identification and characterization of a fibroblast marker: FSP1. The Journal of cell biology. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zeisberg EM, Kalluri R. Origins of cardiac fibroblasts. Circulation research. 2010;107:1304–12. doi: 10.1161/CIRCRESAHA.110.231910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Osterreicher CH, Penz-Osterreicher M, Grivennikov SI, Guma M, Koltsova EK, Datz C, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:308–13. doi: 10.1073/pnas.1017547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kong P, Christia P, Saxena A, Su Y, Frangogiannis NG. Lack of specificity of Fibroblast Specific Protein (FSP)1 in cardiac remodeling and fibrosis. American journal of physiology Heart and circulatory physiology. 2013 doi: 10.1152/ajpheart.00395.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goldsmith EC, Hoffman A, Morales MO, Potts JD, Price RL, McFadden A, et al. Organization of fibroblasts in the heart. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;230:787–94. doi: 10.1002/dvdy.20095. [DOI] [PubMed] [Google Scholar]
- [15].Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Molecular cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- [16].Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Molecular cell. 1997;1:25–34. doi: 10.1016/s1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- [17].Robb L, Mifsud L, Hartley L, Biben C, Copeland NG, Gilbert DJ, et al. epicardin: A novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Developmental dynamics : an official publication of the American Association of Anatomists. 1998;213:105–13. doi: 10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- [18].Lu J, Richardson JA, Olson EN. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mechanisms of development. 1998;73:23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- [19].Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development (Cambridge, England) 2012;139:2139–49. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, et al. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology (Baltimore, Md) 2003;37:267–76. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- [21].Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circulation research. 2011;108:e15–26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ek B, Heldin CH. Characterization of a tyrosine-specific kinase activity in human fibroblast membranes stimulated by platelet-derived growth factor. The Journal of biological chemistry. 1982;257:10486–92. [PubMed] [Google Scholar]
- [23].Orr-Urtreger A, Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development (Cambridge, England) 1992;115:1045–58. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- [24].Chen WC, Baily JE, Corselli M, Diaz ME, Sun B, Xiang G, et al. Human myocardial pericytes: multipotent mesodermal precursors exhibiting cardiac specificity. Stem cells (Dayton, Ohio) 2015;33:557–73. doi: 10.1002/stem.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, et al. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell stem cell. 2011;9:527–40. doi: 10.1016/j.stem.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Furtado MB, Nim HT, Gould JA, Costa MW, Rosenthal NA, Boyd SE. Microarray profiling to analyse adult cardiac fibroblast identity. Genomics data. 2014;2:345–50. doi: 10.1016/j.gdata.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].K. Y. JMCC. 2016.
- [28].Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, Yutzey KE. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. Journal of molecular and cellular cardiology. 2013;65:108–19. doi: 10.1016/j.yjmcc.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tillmanns J, Hoffmann D, Habbaba Y, Schmitto JD, Sedding D, Fraccarollo D, et al. Fibroblast activation protein alpha expression identifies activated fibroblasts after myocardial infarction. Journal of molecular and cellular cardiology. 2015;87:194–203. doi: 10.1016/j.yjmcc.2015.08.016. [DOI] [PubMed] [Google Scholar]
- [30].Ffrench-Constant C, Van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. The Journal of cell biology. 1989;109:903–14. doi: 10.1083/jcb.109.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schoneveld AH, Hoefer I, Sluijter JP, Laman JD, de Kleijn DP, Pasterkamp G. Atherosclerotic lesion development and Toll like receptor 2 and 4 responsiveness. Atherosclerosis. 2008;197:95–104. doi: 10.1016/j.atherosclerosis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- [32].Arslan F, Smeets MB, Riem Vis PW, Karper JC, Quax PH, Bongartz LG, et al. Lack of fibronectin-EDA promotes survival and prevents adverse remodeling and heart function deterioration after myocardial infarction. Circulation research. 2011;108:582–92. doi: 10.1161/CIRCRESAHA.110.224428. [DOI] [PubMed] [Google Scholar]
- [33].Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Developmental biology. 1996;174:221–32. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- [34].Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circulation research. 1998;82:1043–52. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- [35].Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–8. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9504–8. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circulation research. 2009;105:408–21. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Developmental biology. 2012;366:111–24. doi: 10.1016/j.ydbio.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Le Douarin NM, Dupin E. The neural crest in vertebrate evolution. Current opinion in genetics & development. 2012;22:381–9. doi: 10.1016/j.gde.2012.06.001. [DOI] [PubMed] [Google Scholar]
- [41].Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science (New York, NY) 1983;220:1059–61. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- [42].Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Developmental biology. 1998;196:129–44. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- [43].Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development (Cambridge, England) 2000;127:1607–16. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- [44].Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circulation research. 2006;98:1547–54. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- [45].Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nature medicine. 2007;13:952–61. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- [46].Mollmann H, Nef HM, Kostin S, von Kalle C, Pilz I, Weber M, et al. Bone marrow-derived cells contribute to infarct remodelling. Cardiovascular research. 2006;71:661–71. doi: 10.1016/j.cardiores.2006.06.013. [DOI] [PubMed] [Google Scholar]
- [47].van Amerongen MJ, Bou-Gharios G, Popa E, van Ark J, Petersen AH, van Dam GM, et al. Bone marrow-derived myofibroblasts contribute functionally to scar formation after myocardial infarction. The Journal of pathology. 2008;214:377–86. doi: 10.1002/path.2281. [DOI] [PubMed] [Google Scholar]
- [48].Aisagbonhi O, Rai M, Ryzhov S, Atria N, Feoktistov I, Hatzopoulos AK. Experimental myocardial infarction triggers canonical Wnt signaling and endothelial-to-mesenchymal transition. Disease models & mechanisms. 2011;4:469–83. doi: 10.1242/dmm.006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Widyantoro B, Emoto N, Nakayama K, Anggrahini DW, Adiarto S, Iwasa N, et al. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-tomesenchymal transition. Circulation. 2010;121:2407–18. doi: 10.1161/CIRCULATIONAHA.110.938217. [DOI] [PubMed] [Google Scholar]
- [50].Xu J, Lin SC, Chen J, Miao Y, Taffet GE, Entman ML, et al. CCR2 mediates the uptake of bone marrow-derived fibroblast precursors in angiotensin II-induced cardiac fibrosis. American journal of physiology Heart and circulatory physiology. 2011;301:H538–47. doi: 10.1152/ajpheart.01114.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS genetics. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Furtado MB, Costa MW, Pranoto EA, Salimova E, Pinto AR, Lam NT, et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circulation research. 2014;114:1422–34. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–8. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nagalingam RSSH, Czubryt MP. JMCC. 2016.
- [55].Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science (New York, NY) 2002;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- [56].Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, et al. Macrophages are required for neonatal heart regeneration. The Journal of clinical investigation. 2014;124:1382–92. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].DL. HJaM. JMCC. 2016.
- [59].Chablais F, Veit J, Rainer G, Jazwinska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC developmental biology. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development (Cambridge, England) 2011;138:1663–74. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]