Abstract
Cardiac fibroblasts help maintain the normal architecture of the healthy heart and are responsible for scar formation and the healing response to pathological insults. Various genetic, biomechanical, or humoral factors stimulate fibroblasts to become contractile smooth muscle-like cells called myofibroblasts that secrete large amounts of extracellular matrix. Unfortunately, unchecked myofibroblast activation in heart disease leads to pathological fibrosis, which is a major risk factor for the development of cardiac arrhythmias and heart failure. A better understanding of the molecular mechanisms that control fibroblast plasticity and myofibroblast activation is essential to develop novel strategies to specifically target pathological cardiac fibrosis without disrupting the adaptive healing response. This review highlights the major transcriptional mediators of fibroblast origin and function in development and disease. The contribution of the fetal epicardial gene program will be discussed in the context of fibroblast origin in development and following injury, primarily focusing on Tcf21 and C/EBP. We will also highlight the major transcriptional regulatory axes that control fibroblast plasticity in the adult heart, including transforming growth factor β (TGFβ)/Smad signaling, the Rho/myocardin-related transcription factor (MRTF)/serum response factor (SRF) axis, and Calcineurin/transient receptor potential channel (TRP)/nuclear factor of activated T-Cell (NFAT) signaling. Finally, we will discuss recent strategies to divert the fibroblast transcriptional program in an effort to promote cardiomyocyte regeneration. This article is a part of a Special Issue entitled “Fibrosis and Myocardial Remodeling”.
Keywords: Cardiac fibroblast, fibrosis, heart, myocardial infarction, myofibroblast, transcription
1. Introduction
The heart is composed of three main cell types: contractile cardiomyocytes (CMCs), vascular cells, and fibroblasts. Fibroblasts contribute to ~10–30% of the total cardiac cell population, providing basic structural support via secretion of extracellular matrix (ECM) into the interstitial space [1–3]. In addition to generating an ECM scaffold that other cells adhere to, cardiac fibroblasts (CFs) play many underappreciated functions, including paracrine signaling, electrical coupling, and tissue repair [4]. Thus, CFs are emerging as a malleable cell type that is coaxed down various pathways based upon regional requirements and physiological conditions.
As the major source of ECM, fibroblasts play a stereotypical role in tissue replacement and repair following injury. The wound healing process is typified by the transformation of quiescent fibroblasts into a state of high contractility and ECM production, often referred to as a myofibroblast. However, the characteristics of myofibroblasts that allow for efficient wound repair are also responsible for the development of pathological fibrosis and scar formation when left unchecked. In the heart, aberrant scar formation disrupts electrical signaling and muscle contraction and leads to heart failure, the most common cause of death in the U.S. [5]. Thus, tight control of fibroblast plasticity is essential for the maintenance of normal cardiac function. This review highlights the transcriptional control of CF phenotype in the healthy heart and following injury or disease.
2. Fibroblast sources and plasticity
2.1. Fibroblast origins
The heart is lined by a single cell layer of mesothelium called the epicardium. The epicardium is a source of cardiovascular progenitor cells that undergo epithelial-to-mesenchymal-transition (EMT) and differentiate into various cardiac lineages including coronary vascular cells and CFs [6–10]. The CF population can be roughly grouped into three categories: ventricular CFs, atrial CFs, and CFs in specific structures within the heart such as around the sinoatrial node, valves, and annulus fibrosis. Compared to ventricular fibroblasts, atrial fibroblasts display a more robust response to congestive heart failure [11, 12]. Fibroblasts within the valves and annulus fibrosis share significant resemblance to ventricular and atrial fibroblasts, but are more densely packed and retain more specialized phenotypes that are possibly determined during EMT [13, 14]. Fibroblasts within these structures secrete high levels of ECM and create an electrically inert extension of the atrioventricular valves that separates the atria and ventricles to allow asynchronous contraction [15].
In the adult, new fibroblasts are hypothesized to derive from multiple sources including preexisting CFs, fibrocytes, circulating bone marrow stem cells, the epicardium and endothelium [10, 16–21] (Figure 1). Defining the source of fibroblasts remains difficult however, largely due to their heterogenous nature and lack of a specific marker. Commonly used markers such as FSP1/S100A4, vimentin, discoidin domain receptor tyrosine kinase 2 (DDR2), periostin (Postn), and collagen 1a1 (Col1a1), and THY1/CD90 are also expressed by other cell types [22–25]. Although multiple groups have detected fibroblasts arising from circulating cells or EMT, a growing consensus is that resident fibroblasts are the primary source of myofibroblasts, at least in mouse models of pressure overload and ischemia-reperfusion (IR) induced remodeling [19–21].
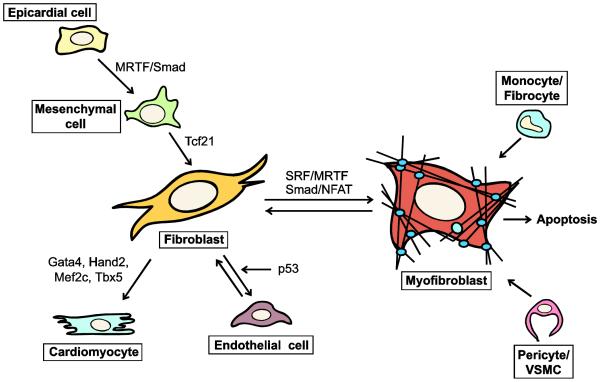
Figure 1. Fibroblast and myofibroblast origins.
Multiple cell types have been hypothesized to differentiate or transdifferentiate into myofibroblasts including pericytes/vascular smooth muscle cells (VSMC), circulating monocytes and fibrocytes, endothelial cells, epicardial and mesenchymal cells, and quiescent fibroblasts. In development, quiescent fibroblasts primarily differentiate from epicardial and mesenchymal cells in a mechanism known as epithelial-to-mesenchymal transition (EMT). EMT is largely governed by TGFβ signaling through SMAD proteins and the MRTF and TCF21 transcription factors. Disease can cause fibroblasts to differentiate into myofibroblasts and cause tissue fibrosis. Resolution of fibrosis can occur through two mechanisms: apoptosis or dedifferentiation. Myofibroblasts that dedifferentiate into quiescent fibroblasts may also further transdifferentiate into endothelial cells in a p53-dependent manner. Chronic disease often results in cardiomyocyte death, and reprogramming fibroblasts into cardiomyocytes may offer potential therapies to reduce fibrosis.
2.2. Myofibroblast activation
Fibroblasts proliferate and become myofibroblasts in response to various genetic, mechanical, and humoral cardiac insults [26–30]. The expression of a number of characteristic genes distinguishes myofibroblasts from quiescent fibroblasts, none of which is a particularly specific or defining feature in isolation. Myofibroblasts express high levels of genes encoding contractile proteins that are typically associated with smooth muscle cells (SMC), including smooth muscle α actin (Acta2, Sma) and Transgelin (Tagln, Sm22), although they generally lack smooth muscle myosin (Myh11) [31]. Indeed, ACTA2 incorporation into stress fibers is among the most accepted myofibroblast markers, albeit with the obvious limitations with regards to cell specificity. Myofibroblasts also possess mature focal adhesions consisting of vinculin, paxillin, integrin αvβ3, focal adhesion kinase, and actin [32, 33], allowing for a directed migration to the source of injury in MI. Finally, myofibroblasts express and secrete an abundance of ECM proteins, including collagen 1, collagen 3, fibronectin 1 (FN1), fibronectin splice variant ED-A, tenascin-C (TNC), POSTN, and MMPs [30, 34, 35]. This ECM provides temporary structural support for disrupted tissue. It can also act as an anchor for static myofibroblasts to adhere to, which allows for the contraction of surrounding tissue. ECM components can also trigger mechanical signals via activation of cell surface receptors such as integrins and TRPC6, inducing downstream signaling pathways that contribute to changes in fibroblast and CMC gene expression and phenotype.
Many organs, including the heart, share this stereotypical fibroblast response to injury or disease. Pathological stresses on the heart including high blood pressure, ischemic heart or coronary artery disease, and inherited cardiomyopathy mutations can lead to CMC apoptosis and replacement by CFs. Indeed, following a cardiac insult such as myocardial infarction (MI), myofibroblasts are essential for necrotic tissue replacement and prevention of cardiac wall rupture [28, 36, 37]. However, key differences distinguish the CF injury response from that of other organs. First, ECM deposition in tissues such as the skin and lung is often followed by proliferation and replacement by other specialized cell types, which ultimately leads to the repair of organ structure and function [38–40]. Unlike organisms such as zebrafish that retain the ability to regenerate the adult heart after resection [41], adult mammalian CMCs are postmitotic and do not support cardiac repair [42]. Therefore, damage resulting in CMC death is considered virtually irreparable. Cardiac fibrosis thus serves as a compensatory mechanism to prevent the disastrous loss of cardiac integrity. Second, the heart becomes more rigid upon accumulation of interstitial fibrosis during the healing process [43]. While this increased rigidity is an adaptive response to preserve tissue integrity, cardiac fibrosis reduces muscle contractility and is a risk factor for arrhythmia and heart failure. Third, in the absence of continued pathological stress, myofibroblasts are eventually lost from most tissues, either by reverting back to quiescent fibroblasts or through apoptotic cell death [44–46]. For reasons that are not fully understood, clearance of myofibroblasts from the diseased heart appears to be an inefficient process, leading to persistent fibrosis and deterioration of cardiac function. Because increased cellular tension is a major mediator of myofibroblast activation [47–49] (Figure 2), the healing heart is an ideal substrate for persistent myofibroblast activation that may lead to a pathological feed-forward loop.
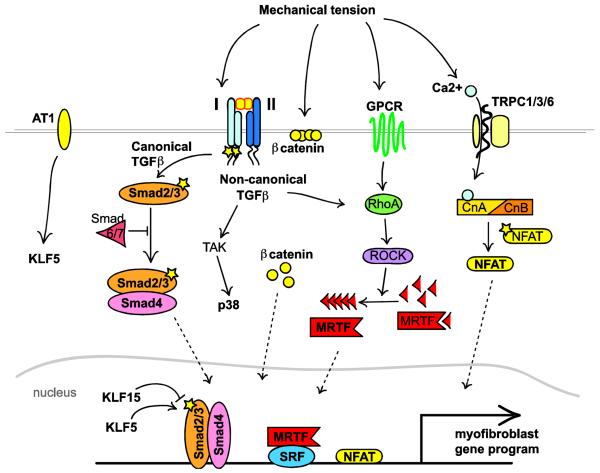
Figure 2. Major signaling pathways that promote fibroblast activation.
Multiple pathways converge on the myofibroblast phenotype. Mechanical stress or ligand mediated receptor activation can induce TGFβ signaling, GPCR activation, or calcium influx. TGFβ signaling plays a central role where the canonical arm results in nuclear localization of SMAD2/3/4. Inhibitory SMADs 6/7 or KLF15 can block SMAD2/3/4-dependent transcription whereas angiotensin/KLF5 signaling can enhance canonical TGFβ signaling. Non-canonical TGFβ signaling not only promotes MAPK/p38/JNK/ERK-dependent transcription, but can feed into the Rho/ROCK signaling pathway to promote MRTF nuclear localization. Rho/ROCK can also be activated by G-protein coupled receptors (GPCRs). Finally, mechanical tension can also disrupt β-catenin localization in the adherens junctions to prime the cell for further stimulation or cause calcium influx which activates calcineurin/NFAT signaling.
Thus, despite the short-term advantages of CF activation in adaptive remodeling, this process remains a double-edged sword that ultimately leads to pathological fibrosis, maladaptive remodeling, and heart failure. Novel therapeutic strategies that directly target the fibroblast are needed to limit fibrosis following injury and perhaps coax activated fibroblasts towards a cardiac fate. The transcriptional changes that underlie fibroblast origin and plasticity may form a scaffold for the development of such reprogramming strategies in the pursuit of cardiac regenerative medicine.
3. Transcriptional regulators of the cardiac fibroblast phenotype
The CF transcriptome is altered by various pathological signals including mechanical tension, activation of cell surface receptors, and alterations in calcium signaling (Figure 2). The culmination of transcriptional changes following a cardiac insult leads to dramatic changes in fibroblast function. The transcriptional mechanisms contributing to fibroblast phenotypic plasticity in development and disease are highlighted in the following sections.
3.1. Fetal gene program
Reactivation of the fetal cardiac gene program, which is thought to provide the basis of compensatory remodeling during heart disease, is commonly considered a CMC response. However, recent studies suggest that fibroblast biology may also be impacted by the induction of developmental programs in the damaged heart. In fact, adult CFs display heterogeneous expression of early cardiogenic and stem cell markers such as TBX20 and SCA1. SCA1-positive fibroblasts have reduced Acta2 expression, suggesting they may reflect a more stem-like population that may be resistant to activation. Furthermore, conditional knockout of Tbx20 in CFs results in an increase in BMP10 expression and myocardial hypoplasia, providing further evidence of CF – CMC crosstalk [50].
During embryonic development, epicardial cells are identified by the heterogeneous expression of various transcription factors, including Wilms Tumor 1 (Wt1), C/EBP, RALDH2/ALDH1A2, TBX18, TCF21 (also known as Capsulin or POD1), HAND2, and Myocardin-Related Transcription Factors (MRTF) which influence EMT and epicardial-derived progenitor cell (EPDC) differentiation [51–57]. This gene signature is silenced after birth, but is reactivated by disease or injury, potentially mobilizing EPDCs and leading to the generation of nascent fibroblasts. TCF21 is expressed in a population of EPDCs and appears to be essential for the formation of CFs during embryonic development at the expense of the SMC lineage [58]. Animals deficient in Tcf21 fail to produce CFs, instead accumulating cells expressing SMC markers on the surface of the heart [58, 59]. RNA-sequencing of Tcf21-deficient coronary SMC combined with Ingenuity Pathway Analysis revealed that TCF21 promotes a gene expression signature consistent with cell proliferation and migration while inhibiting SMC differentiation [60]. ChIP-sequencing identified 5' – CAGCTG – 3' as the canonical binding sequence for TCF21 and suggests shared genomic occupancy with other transcription factors, including AP-1, TEAD, C/EBP, and ATF. Currently, the direct transcriptional targets of TCF21 that mediate the differentiation of epicardial derived cells into fibroblasts are not clear. However, these findings hint at a potential cooperative regulation between TCF21 and AP-1, which was previously shown to bind to and activate the type 1 collagen promoter in fibroblasts and regulate CF migration [61, 62]. The Olson group recently defined a requisite early upstream role of C/EBP in the reactivation of Raldh and Wt1 following MI or IR injury [52]. Animals that lack C/EBP in the epicardium have improved cardiac function following ischemic injury, at least partially stemming from reduced inflammatory cell recruitment. Although this study did not test the possibility that C/EBP may directly modulate the fibroblast phenotype, it is interesting to speculate that C/EBP and TCF21 may also coordinate the generation of epicardial-derived fibroblasts in the adult heart. Taken together, these studies highlight the transcriptional regulation of CF formation in the embryo and suggest a combinatorial transcriptional code in the epicardium that may contribute to the adult injury response.
3.2. TGF-β signaling
One of the best-characterized regulators of fibroblast activation in the adult is transforming growth factor (TGF)-β [63]. Most tissues harbor high levels of biologically inactive latent TGFβ that is cleaved into an active form by proteases, thrombospondin 1, integrins, and reactive oxygen species [64–66]. The MI injury response also leads to the accumulation of additional TGFβ, which is secreted from inflammatory cells or resident fibroblasts [63, 67]. TGFβ signaling is mediated through the stimulation of a heterodimer of the TGFβRI/ALK5 and TGFβRII receptors [68]. The canonical TGFβ pathway is defined by the subsequent phosphorylation and activation of the intracellular SMAD2/3 proteins. SMAD2/3 then interacts with SMAD4 and enters the nucleus, binding to and activating SMAD-binding elements (minimally 5' – GTCT – 3') in the promoters of target genes [69–71] (Figure 2). The inhibitory SMADs 6/7 prevent SMAD2/3/4 nuclear accumulation and the activation of TGFβ-SMAD targets, which includes the core myofibroblast gene program such as Col1a, Acta2, Tagln [72–77]. CFs isolated from Smad3-deficient animals secrete less collagen and have fewer ACTA2 positive stress fibers compared to fibroblasts from control animals [78]. Furthermore, loss of Smad3 attenuates fibrotic remodeling in mouse models of MI, idiopathic pulmonary fibrosis, and diabetes mellitus [67, 79, 80]. Animals heterozygous for Smad3 appear to be protected from diabetes-induced cardiac hypertrophy suggesting a dose-dependent role of SMAD3-regulated TGFβ signaling [79]. Expression of the inhibitory SMAD7 is reduced in the infarcted rat heart, which is thought to relieve the repression of the TGFβ-Smad axis and promote fibroblast activation in vivo [81]. Indeed, overexpression of Smad7 in vivo prevented angiotensin (Ang) II-induced fibrosis and loss of contractility while overexpression in vitro prevented ROS-induced expression of MMP and collagen [82, 83].
The consensus SMAD binding element (SBE) consists of only four bases and is found in nearly every promoter. Thus, interactions between Smads and other transcriptional activators or repressors, such as AP-1, SP1, TFE3, KLF15 and P300, confer the magnitude and specificity of target gene expression. Various points of intersection mediate a coordinated response to the TGF β-Smad axis and other signal transduction pathways. For example, AngII signaling induces the expression of the Kruppel-like transcription factor KLF5, which subsequently activates the expression of TGFβ, linking these signaling axes [84, 85]. Conversely, TGFβ activation in myofibroblasts attenuates the expression of KLF15, an inhibitor of SMAD3-dependent expression of connective tissue growth factor (CTGF) (Figure 2). Consistent with this finding, Klf15-null mice exhibit increased CTGF levels and fibrosis in response to pressure overload induced cardiac remodeling [86, 87]. Finally, a unique interaction exists between SMAD3 and the basic helix-loop-helix transcription factor scleraxis, which is induced by TGFβ-Smad activation and subsequently synergizes with SMAD3 to activate Col1a expression [88].
TGFβ activity is also mediated by the non-canonical pathway via TGFβ-activated kinase (TAK1) stimulation of mitogen activated protein kinases (MAPKs) including ERK1/2, c-Jun N-terminal kinase (JNK), and p38 (MAPK14) [89, 90] (Figure 2). TAK1/p38α has been specifically implicated in promoting myofibroblast activation; pharmacological inhibition of p38 blunts TGFβ-dependent Acta2 expression and the development of fibrosis in multiple organs, including the heart [91–94]. TGFβ stimulation of human dermal fibroblasts also triggers ERK phosphorylation and CTGF expression, which contributes to myofibroblast activation and cytoskeletal rearrangements [95, 96]. ERK also transduces mechanical tension through focal adhesion kinase (FAK) in fibroblasts [97, 98]. Finally, non-canonical TGFβ intersects with canonical TGFβ signaling to induce expression of TIMP-3 in human gingival fibroblasts in a synergistic manner [94].
3.3. MRTF/SRF/RhoA
Serum response factor (SRF) is an ubiquitously expressed and highly conserved transcription factor that is essential for life. SRF binds to and activates promoters harboring a DNA element called a CArG box (CC(A/T)6GG) [99, 100]. More than 8000 evolutionarily conserved CArG elements exist [101, 102] that are predicted to regulate the expression of thousands of protein coding genes [103]. SRF target gene selection and the magnitude of transcriptional activation depends upon interactions with various tissue-restricted or signal responsive co-factors.
The expression of genes encoding SMC contractile proteins, which are nearly always regulated by a CArG element, is potently stimulated by interactions between SRF and members of the myocardin family of transcriptional co-activators [104, 105]. The founding member of this family, myocardin is restricted to SMC and CMCs and constitutively induces the SMC gene program in vascular and visceral smooth muscle [106–110]. In contrast, myocardin-related transcription factor (MRTF)-A (also called MAL/MKL1/BSAC) and MRTF-B (MKL2) are broadly expressed, signal responsive transcription factors [111, 112]. Under basal conditions, MRTFs interact with monomeric (G)-actin through an N-terminal RPEL domain, masking a nuclear localization signal [113–115]. Polymerization of filamentous (F)-actin reduces the pool of G-actin, allowing MRTFs to enter the nucleus and bind to SRF, activating components of the SMC gene program such as Acta2 [116–120] (Figure 2). Conversely, inhibiting F-actin polymerization with latrunculin B or other means blocks MRTF-dependent Acta2 expression [114]. Thus, MRTFs control fibroblast phenotypic plasticity by linking changes in the actin cytoskeleton to regulation of the SMC gene program [105, 121].
Recent studies have uncovered a dominant role for SRF / MRTF-dependent transcriptional activation in regulating the myofibroblast phenotype [103, 122]. Exogenous expression of MRTF-A in fibroblasts or epithelial cells is sufficient to induce phenotypic transformation into migratory and contractile myofibroblasts [123–126]. Tomasek et al. first demonstrated in a dermal wound healing model that expression of Acta2 in granulation tissue fibroblasts requires binding sites for both SRF and Smads [74]. Induction of Acta2 in myofibroblasts was later shown to depend upon Rho/Rho kinase (ROCK1) signaling [127]. Consistent with this, myofibroblasts express higher levels of RhoA than quiescent fibroblasts [128]. Further evidence linking Rho to MRTF target genes comes from pharmacological inhibition of ROCK1 with fasudil or Y-27632, which prevents remodeling and fibrosis in vitro and in vivo [129–131]. We and others have since demonstrated overlapping functions of TGFβ and Rho-ROCK1 signaling in mediating F-actin polymerization and MRTF activation in myofibroblasts of various sources [120, 124, 130, 132–134]. Indeed, myocardin family members and SMAD3 synergistically activate SBE/CArG element-containing promoters in SMCs and during EMT or myofibroblast activation [73, 135, 136].
It is important to note the context-dependent role of Rho-ROCK1 activation in regulating MRTF activity and myofibroblast differentiation. Although the fibroblast phenotype is directly affected by substrate stiffness and growth factors, external factors such as cell density and contact inhibition are also important modulators of fibroblast activation. In cell culture, confluent monolayers of fibroblasts and epithelial cells form adherens junctions consisting of cadherins and β-catenin [137–140]. Disruption of adherens junctions leads to the release of β-catenin from the cell membrane, which is typically rapidly degraded. TGFβ stimulation is not only sufficient to prevent degradation of β-catenin, but stabilization of β-catenin in cells with reduced intercellular contacts is required for TGFβ-induced Acta2 expression [141–144]. Cytoplasmic β-catenin may indirectly promote nuclear localization of MRTFs by competing for GSK3β-mediated ubiquitination [145, 146]. Furthermore, stabilized β-catenin can function as a transcriptional activator of Wnt signaling, which promotes expression of ECM in epithelial cells and mouse embryonic fibroblasts (MEFs), pointing to a potential role of WNTs in modifying the fibroblast phenotype [147, 148] (Figure 2).
MRTF stability and activity is also influenced by post-translational modifications, including phosphorylation, sumoylation and ubiquitination [149, 150], and proteasome inhibitors lead to MRTF-A accumulation [124]. In line with these findings, the four-and-a-half LIM-only protein 2 (FHL2) protein, which is thought to inhibit MURF3-dependent ubiquitination, prevents proteosomal degradation of MRTF-A [151]. In contrast, FHL2 also competes with MRTF-A for SRF binding and inhibits expression of SRF target genes [151, 152]. Consistent with the latter data, FHL2-knockout animals treated with bleomycin had increased pulmonary fibrosis and expression of TNC [153]. It is interesting to note that SRF induces Fhl2 expression in embryonic stem cells, suggesting the possibility of a negative feedback loop that limits myofibroblast activation [152].
The importance of regulating MRTFs in fibroblast activation is becoming increasingly clear. Global knockout of Mrtfb or Srf results in embryonic lethality [154, 155]. In contrast, deletion of Mrtfa results in viable and fertile adults although female dams fail to nurse their young due to a defective mammary myoepithelial cell differentiation [156]. The contribution of MRTF-A in cardiac fibrosis was determined when MRTF-A-deficient animals were subjected to myocardial infarction; MRTF-A-null animals had reduced scar formation after MI [130]. Similarly, bleomycin-induced pulmonary fibrosis is reduced in MRTF-A-deficient animals [129, 157]. Taken together, this suggests that MRTF-A is central in promoting the myofibroblast phenotype.
Although therapeutic strategies typically target receptor-ligand interactions or intercellular kinases, manipulation of upstream signaling molecules can potentially have a wide range of off-target effects. Many of the studies mentioned previously demonstrate the efficacy and specificity of controlling MRTF expression and function in vitro and in vivo. In the adult, MRTFs are generally inactive and tethered in the cytoplasm, and can become precociously activated in response to pathological signals that lead to alterations in the actin cytoskeleton. In an attempt to harness the therapeutic potential of MRTF activity, recent studies have identified small molecules that specifically inhibit MRTFs. In an Acta2 promoter-based luciferase screen, Velasquez et al. identified N-cyclopropyl-5-(thiophen-2-yl)-isoxazole-3-carboxamide (isoxazole/ISX) as a stimulator of fibroblast activation in a CArG-box and MRTF-dependent manner. These results were confirmed in human foreskin fibroblasts and in cutaneous wound healing experiments where isoxazole promoted more rapid wound healing compared to control-treated animals [132]. A similar screen for modulators of RhoA-mediated signaling led to the identification of CCG-1432 [158]. Compounds related to CCG-1432 bind the nuclear localization signal within the RPEL domain of MRTFs, inhibiting importin-dependent nuclear translocation [159]. Subsequent studies have revealed that inhibition of MRTF activity with this class of compound blocks dermal, colonic, and lung fibrosis in vivo [160–162]. Together, these studies demonstrate the potential of targeting signal responsive transcription factors, such as MRTF-A, to regulate the fibroblast response.
3.5. TRPC/Calcineurin/NFAT
Calcium influx into the CMC is critical for maintaining cardiac function in part by regulating nuclear factor of activated T-cells (NFAT)-dependent target genes implicated in cardiac hypertrophy. High intracellular levels of Ca2+ permits binding of a calcineurin (Cn) A/B heterodimer to calmodulin to induce a conformational change. This conformational change exposes the active site of CnA, leading to NFAT dephosphorylation and nuclear translocation, where it induces gene expression [163]. A number of factors, including mechanical tension, increase intracellular calcium levels and thus activate NFAT in fibroblasts. Activated Cn/NFAT signaling can then trigger the expression of the hypertrophic gene response in CMCs, Col3 and Mrtfa in fibroblasts, or Acta2 in SMCs [164, 165] (Figure 2). Not only is CnA overexpression sufficient to induce myofibroblast differentiation both in vivo and in vitro in an NFAT-dependent manner, but this activity can be blocked by Cn inhibitors [163, 166–168].
Recent work has focused on the transient receptor potential (TRP) family of proteins as mediators of myofibroblast differentiation [169]. TRP channels form heterotrimeric channels in vivo and control Ca2+ influx levels in response to various stimuli including mechanical signals and oxidative stress. Formation of the TRPC channel depends on the expression of TRPC1, which is strongly expressed in rat CMCs and transcriptionally upregulated in CFs in response to TGFβ stimulation [166, 170]. Several TRPC channels are upregulated in models of heart failure such as TRPC1, 3, 5, and 6 [171–173]. TRPC3 is sufficient to drive myofibroblast differentiation in atrial and renal fibroblasts in an NFAT- and ERK1/2-dependent manner, respectively, and may play a more important role in atrial function or fibrosis of other tissues [174, 175]. The most notable of the TRPC proteins in fibroblast plasticity is TRPC6, was identified using an in vitro overexpression screen in MEFs. Trpc6 expression is induced by non-canonical TGFβ signaling and SRF; SRF overexpression was sufficient to increase Trpc6 transcription, but this increase was blocked with a p38-specific inhibitor [166]. Overexpression of TRPC6 is specifically required to promote TGFβ- or AngII-dependent myofibroblast differentiation in cell culture and is required to prevent cardiac wall rupture after myocardial infarction [166].
4. Fibroblast resolution and reprogramming
Resolution of fibrosis typically culminates in fibroblast apoptosis, however, a subset of CFs are resistant to apoptosis and remain within the scar [176–178]. A recent study revealed that P53+COL1A2+ cells express the endothelial cell (EC) marker VE-cadherin 3 days after IR injury [21]. Loss of p53 in CFs is correlated with decreased cardiac function due to reduced mesenchymal – endothelial transformation and capillary density. This not only suggests that there are inherent transcriptional differences between the types of CFs that become activated in disease, but that CFs that escape P53-dependent gene regulation may transdifferentiate into ECs [21] (Figure 1). CF-EC transdifferentiation in disease may improve cardiovascular function by both promoting neovascularization within a fibrotic infarct and reducing the number of activated fibroblasts. These studies may have further implications as a recent study demonstrated that ECs comprise up to 63% of cardiac cells and suggested that the role of ECs in cardiac physiology may be underappreciated [3]. Further support of this hypothesis demonstrates that some CFs can spontaneously adopt a proliferative myofibroblast phenotype in vitro whereas others adopt a non-proliferative TGFβ-induced myofibroblast phenotype [177]. Cells that retain a proliferative phenotype regress to a more quiescent fibroblast transcriptional profile after removing the TGFβ stimulus, including a decrease in Mrtfa transcription and susceptibility to apoptosis. Conversely, non-proliferative myofibroblasts remain activated after removal of TGFβ stimulation [177]. Further support of this comes from recent data from D'Souza et al, who demonstrate reduced activation in CFs isolated from rats treated with ACE inhibitors. One proposed mechanism is apoptosis of activation-prone CFs and survival of more quiescent CFs [179, 180]. These data suggest that a subpopulation of CFs may retain the ability to revert back into a quiescent state or undergo apoptosis. Taken together, these studies challenge the previously held notion that CFs are terminally differentiated and further emphasize the need to identify novel methods of manipulating transcriptional regulators of the myofibroblast state.
In line with the intrinsic plasticity of CFs, reprogramming strategies might be utilized as a means to repopulate lost myocardial tissue with functioning CMCs. Recent studies have defined transcription factor cocktails that can coax CFs into a beating CMC-like cell. Albeit a small percentage of cells, mouse and human CFs can be transformed into CMCs with viral overexpression of the core set of transcription factors: Gata4, Mef2c, and Tbx5 (called GMT) [181]. Other early cardiac transcription factors, such as Nkx2.5, Mesp1, and myocardin are less critical, or even inhibit reprogramming whereas Hand2 can increase the percentage of cells transformed into atrial, ventricular, and pacemaker CMCs [182–184]. Similar reports have used small molecules in combination with the pluripotency factor OCT4 to produce CMC-like cells from fibroblasts [185, 186]. Importantly, cellular reprogramming strategies have proven efficacious in blunting cardiac dysfunction and remodeling in rodent models of MI [182, 187, 188]. It is interesting to speculate that forced expression of reprogramming factors may lead to improved cardiac performance by diverting CFs away from the pro-fibrotic phenotype in addition to stimulating CMC production [189]. Of note, suppression of pro-fibrotic signaling with ROCK or TGFβ inhibitors dramatically improves CF transdifferentiation into CMCs in vitro, implying a potential mutual antagonism between reprogramming factors and pro-fibrotic signaling [184]. This study also suggests an intriguing similarity between embryonic stem cell-derived CMCs and CF reprogramming strategies, which are both inhibited by TGFβ signaling, and adds further support to the concept of fibroblast multipotency. Indeed, fibroblasts seem uniquely capable of re-programming, given current reports that ECs or other non-myocyte cell types do not efficiently transdifferentiate into CMCs [188]. Defining the genomic occupancy of GMT during fibroblast reprogramming or in response to inhibitors of reprogramming such as pro-fibrotic signals or Nkx2-5 might provide clues as to the combinatorial interactions that control fibroblast plasticity. Altogether, these studies provide a foundation for developing potential therapeutic strategies to promote cardiac repair.
5. Conclusion
Fibroblasts are no longer relegated to merely structural and supportive roles and are now appreciated as a highly plastic cell type that contributes to maintaining tissue homeostasis and wound repair. Interactions between CFs and CMCs, inflammatory cells, and other cell types promote a balanced environment that can quickly respond to the changing needs of a healthy heart. However, it has become increasingly clear that CFs also play a central role in the progression of heart failure. Resolution of activated myofibroblasts at the culmination of the cardiac injury response is an inefficient process that often leads unchecked fibrosis. Myofibroblast activation is rapidly induced by a growing number of signaling pathways that converge on a limited number of transcription factors. Targeting TGFβ/Smad/scleraxis or Rho-ROCK/MRTF/SRF pathways has already proven efficacious in blocking the progression of fibrosis in animal models of disease. Recently, fibroblasts were coaxed into beating CMC-like cells with exogenous expression of select transcription factors, both in vitro and in vivo. Additional therapeutic strategies that harness fibroblast phenotypic plasticity may stem from studies that better define CF origin and heterogeneity. While major hurdles include the development of better markers of fibroblast identity and improved tools that specifically target the fibroblast, manipulating the CF phenotype in disease is certainly a challenging yet attainable goal.
Highlights.
Cardiac fibroblasts are a uniquely plastic cell type.
The myofibroblast is a primary source of extracellular matrix during cardiac repair.
Excessive stimulation of the myofibroblast phenotype leads to cardiac fibrosis.
Transcriptional regulators of fibroblast plasticity are reviewed.
Diverting the fibrotic gene program may contribute to the benefits of reprogramming.
Acknowledgements
We thank M.A. Trembley for assistance with graphics. JKL received funding from a T32 training grant from the National Institutes of Health (T32 HL066988-13) and the American Heart Association (15POST25550114). E.M.S. was supported in part by the National Institutes of Health (R01HL120919) and an University of Rochester/Aab CVRI Pilot Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures No conflicts of interest exist.
References
- [1].Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293(3):H1883–91. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- [2].Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res. 2009;105(12):1164–76. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D'Antoni M, Debuque RJ, et al. Revisiting Cardiac Cellular Composition. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Evans SM. The Multifaceted Roles of Cardiac fibroblasts. Journal of Molecular and Cellular Cardiology. 2015 JMCC9734. [Google Scholar]
- [5].Kochanek KD, Murphy SL, Xu J. Deaths: Final Data for 2011. Natl Vital Stat Rep. 2015;63(3):1–120. [PubMed] [Google Scholar]
- [6].Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108(12):e15–26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dettman RW, Denetclaw W, Jr., Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193(2):169–81. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- [8].Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46(8):1005–13. [PubMed] [Google Scholar]
- [9].Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174(2):221–32. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- [10].Fang M, Xiang FL, Braitsch CM, Yutzey KE. Epicardium-derived fibroblasts in heart development and disease. Journal of Molecular and Cellular Cardiology. 2015 doi: 10.1016/j.yjmcc.2015.12.019. JMCC9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Snider P, Standley KN, Wang J, Azhar M, Doetschman T, Conway SJ. Origin of cardiac fibroblasts and the role of periostin. Circ Res. 2009;105(10):934–47. doi: 10.1161/CIRCRESAHA.109.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Burstein B, Libby E, Calderone A, Nattel S. Differential behaviors of atrial versus ventricular fibroblasts: a potential role for platelet-derived growth factor in atrial-ventricular remodeling differences. Circulation. 2008;117(13):1630–41. doi: 10.1161/CIRCULATIONAHA.107.748053. [DOI] [PubMed] [Google Scholar]
- [13].Liu AC, Joag VR, Gotlieb AI. The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. Am J Pathol. 2007;171(5):1407–18. doi: 10.2353/ajpath.2007.070251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hinton RB, Yutzey KE. Heart valve structure and function in development and disease. Annu Rev Physiol. 2011;73:29–46. doi: 10.1146/annurev-physiol-012110-142145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhou B, von Gise A, Ma Q, Hu YW, Pu WT. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 2010;338(2):251–61. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13(8):952–61. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- [17].Haudek SB, Xia Y, Huebener P, Lee JM, Carlson S, Crawford JR, et al. Bone marrow-derived fibroblast precursors mediate ischemic cardiomyopathy in mice. Proc Natl Acad Sci U S A. 2006;103(48):18284–9. doi: 10.1073/pnas.0608799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res. 2012;110(12):1628–45. doi: 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest. 2014;124(7):2921–34. doi: 10.1172/JCI74783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res. 2014;115(7):625–35. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- [21].Ubil E, Duan J, Pillai IC, Rosa-Garrido M, Wu Y, Bargiacchi F, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514(7524):585–90. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kong P, Christia P, Saxena A, Su Y, Frangogiannis NG. Lack of specificity of fibroblast-specific protein 1 in cardiac remodeling and fibrosis. Am J Physiol Heart Circ Physiol. 2013;305(9):H1363–72. doi: 10.1152/ajpheart.00395.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sempowski GD, Borrello MA, Blieden TM, Barth RK, Phipps RP. Fibroblast heterogeneity in the healing wound. Wound Repair Regen. 1995;3(2):120–31. doi: 10.1046/j.1524-475X.1995.30204.x. [DOI] [PubMed] [Google Scholar]
- [24].Krenning G, Zeisberg EM, Kalluri R. The origin of fibroblasts and mechanism of cardiac fibrosis. J Cell Physiol. 2010;225(3):631–7. doi: 10.1002/jcp.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Moore-Morris T, Guimaraes-Camboa N, Yutzey KE, Puceat M, Evans SM. Cardiac fibroblasts: from development to heart failure. J Mol Med (Berl) 2015;93(8):823–30. doi: 10.1007/s00109-015-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127(3):526–37. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- [27].Turner NA, Porter KE. Function and fate of myofibroblasts after myocardial infarction. Fibrogenesis Tissue Repair. 2013;6(1):5. doi: 10.1186/1755-1536-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J. Myocardial remodeling after infarction: the role of myofibroblasts. Nat Rev Cardiol. 2010;7(1):30–7. doi: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- [29].Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, et al. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest. 2010;120(10):3520–9. doi: 10.1172/JCI42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Serini G, Bochaton-Piallat ML, Ropraz P, Geinoz A, Borsi L, Zardi L, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol. 1998;142(3):873–81. doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mayer DC, Leinwand LA. Sarcomeric gene expression and contractility in myofibroblasts. J Cell Biol. 1997;139(6):1477–84. doi: 10.1083/jcb.139.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hinz B, Gabbiani G. Cell-matrix and cell-cell contacts of myofibroblasts: role in connective tissue remodeling. Thromb Haemost. 2003;90(6):993–1002. doi: 10.1160/TH03-05-0328. [DOI] [PubMed] [Google Scholar]
- [33].Manso AM, Kang SM, Plotnikov SV, Thievessen I, Oh J, Beggs HE, et al. Cardiac fibroblasts require focal adhesion kinase for normal proliferation and migration. Am J Physiol Heart Circ Physiol. 2009;296(3):H627–38. doi: 10.1152/ajpheart.00444.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101(3):313–21. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ignotz RA, Massague J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem. 1986;261(9):4337–45. [PubMed] [Google Scholar]
- [36].Weber KT, Sun Y, Bhattacharya SK, Ahokas RA, Gerling IC. Myofibroblast-mediated mechanisms of pathological remodelling of the heart. Nat Rev Cardiol. 2013;10(1):15–26. doi: 10.1038/nrcardio.2012.158. [DOI] [PubMed] [Google Scholar]
- [37].Benjamin IJ, Jalil JE, Tan LB, Cho K, Weber KT, Clark WA. Isoproterenol-induced myocardial fibrosis in relation to myocyte necrosis. Circ Res. 1989;65(3):657–70. doi: 10.1161/01.res.65.3.657. [DOI] [PubMed] [Google Scholar]
- [38].Katoonizadeh A, Nevens F, Verslype C, Pirenne J, Roskams T. Liver regeneration in acute severe liver impairment: a clinicopathological correlation study. Liver Int. 2006;26(10):1225–33. doi: 10.1111/j.1478-3231.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- [39].Sun M, Kisseleva T. Reversibility of liver fibrosis. Clin Res Hepatol Gastroenterol. 2015 doi: 10.1016/j.clinre.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–38. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- [42].Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Doering CW, Jalil JE, Janicki JS, Pick R, Aghili S, Abrahams C, et al. Collagen network remodelling and diastolic stiffness of the rat left ventricle with pressure overload hypertrophy. Cardiovasc Res. 1988;22(10):686–95. doi: 10.1093/cvr/22.10.686. [DOI] [PubMed] [Google Scholar]
- [44].Elsharkawy AM, Oakley F, Mann DA. The role and regulation of hepatic stellate cell apoptosis in reversal of liver fibrosis. Apoptosis. 2005;10(5):927–39. doi: 10.1007/s10495-005-1055-4. [DOI] [PubMed] [Google Scholar]
- [45].Lee YA, Wallace MC, Friedman SL. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64(5):830–41. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Camelo A, Dunmore R, Sleeman MA, Clarke DL. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol. 2014;4:173. doi: 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B. Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. J Cell Biol. 2006;172(2):259–68. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3(5):349–63. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- [49].van Putten S, Shafieyan Y, Hinz B. Mechanical control of cardiac myofibroblasts. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.025. JMCC9700. [DOI] [PubMed] [Google Scholar]
- [50].Furtado MB, Costa MW, Pranoto EA, Salimova E, Pinto AR, Lam NT, et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res. 2014;114(9):1422–34. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous EE. Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech Dev. 1998;73(1):33–43. doi: 10.1016/s0925-4773(98)00031-8. [DOI] [PubMed] [Google Scholar]
- [52].Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338(6114):1599–603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lu J, Richardson JA, Olson EN. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Dev. 1998;73(1):23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- [54].Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Drager UC, et al. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199(1):55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- [55].Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126(9):1845–57. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- [56].Barnes RM, Firulli BA, VanDusen NJ, Morikawa Y, Conway SJ, Cserjesi P, et al. Hand2 loss-of-function in Hand1-expressing cells reveals distinct roles in epicardial and coronary vessel development. Circ Res. 2011;108(8):940–9. doi: 10.1161/CIRCRESAHA.110.233171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Trembley MA, Velasquez LS, de Mesy Bentley KL, Small EM. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development. 2015;142(1):21–30. doi: 10.1242/dev.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Braitsch CM, Combs MD, Quaggin SE, Yutzey KE. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol. 2012;368(2):345–57. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139(12):2139–49. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sazonova O, Zhao Y, Nurnberg S, Miller C, Pjanic M, Castano VG, et al. Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci. PLoS Genet. 2015;11(5):e1005202. doi: 10.1371/journal.pgen.1005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ponticos M, Harvey C, Ikeda T, Abraham D, Bou-Gharios G. JunB mediates enhancer/promoter activity of COL1A2 following TGF-beta induction. Nucleic Acids Res. 2009;37(16):5378–89. doi: 10.1093/nar/gkp544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ponticos M, Papaioannou I, Xu S, Holmes AM, Khan K, Denton CP, et al. Failed degradation of JunB contributes to overproduction of type I collagen and development of dermal fibrosis in patients with systemic sclerosis. Arthritis Rheumatol. 2015;67(1):243–53. doi: 10.1002/art.38897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bujak M, Frangogiannis NG. The role of TGF-beta signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res. 2007;74(2):184–95. doi: 10.1016/j.cardiores.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Schultz-Cherry S, Lawler J, Murphy-Ullrich JE. The type 1 repeats of thrombospondin 1 activate latent transforming growth factor-beta. J Biol Chem. 1994;269(43):26783–8. [PubMed] [Google Scholar]
- [65].Sarrazy V, Koehler A, Chow ML, Zimina E, Li CX, Kato H, et al. Integrins alphavbeta5 and alphavbeta3 promote latent TGF-beta1 activation by human cardiac fibroblast contraction. Cardiovasc Res. 2014;102(3):407–17. doi: 10.1093/cvr/cvu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(Pt 2):217–24. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- [67].Bujak M, Ren G, Kweon HJ, Dobaczewski M, Reddy A, Taffet G, et al. Essential role of Smad3 in infarct healing and in the pathogenesis of cardiac remodeling. Circulation. 2007;116(19):2127–38. doi: 10.1161/CIRCULATIONAHA.107.704197. [DOI] [PubMed] [Google Scholar]
- [68].Frangogiannis NG. The inflammatory response in myocardial injury, repair, and remodelling. Nat Rev Cardiol. 2014;11(5):255–65. doi: 10.1038/nrcardio.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Derynck R. SMAD proteins and mammalian anatomy. Nature. 1998;393(6687):737–9. doi: 10.1038/31593. [DOI] [PubMed] [Google Scholar]
- [70].Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- [71].Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1(4):611–7. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- [72].Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993;122(1):103–11. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Qiu P, Ritchie RP, Fu Z, Cao D, Cumming J, Miano JM, et al. Myocardin enhances Smad3-mediated transforming growth factor-beta1 signaling in a CArG box-independent manner: Smad-binding element is an important cis element for SM22alpha transcription in vivo. Circ Res. 2005;97(10):983–91. doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- [74].Tomasek JJ, McRae J, Owens GK, Haaksma CJ. Regulation of alpha-smooth muscle actin expression in granulation tissue myofibroblasts is dependent on the intronic CArG element and the transforming growth factor-beta1 control element. Am J Pathol. 2005;166(5):1343–51. doi: 10.1016/s0002-9440(10)62353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Gan Q, Yoshida T, Li J, Owens GK. Smooth muscle cells and myofibroblasts use distinct transcriptional mechanisms for smooth muscle alpha-actin expression. Circ Res. 2007;101((9):883–92. doi: 10.1161/CIRCRESAHA.107.154831. [DOI] [PubMed] [Google Scholar]
- [76].Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389(6651):622–6. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- [77].Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- [78].Dobaczewski M, Bujak M, Li N, Gonzalez-Quesada C, Mendoza LH, Wang XF, et al. Smad3 signaling critically regulates fibroblast phenotype and function in healing myocardial infarction. Circ Res. 2010;107(3):418–28. doi: 10.1161/CIRCRESAHA.109.216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Biernacka A, Cavalera M, Wang J, Russo I, Shinde A, Kong P, et al. Smad3 Signaling Promotes Fibrosis While Preserving Cardiac and Aortic Geometry in Obese Diabetic Mice. Circ Heart Fail. 2015;8(4):788–98. doi: 10.1161/CIRCHEARTFAILURE.114.001963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhao J, Shi W, Wang YL, Chen H, Bringas P, Jr., Datto MB, et al. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol. 2002;282(3):L585–93. doi: 10.1152/ajplung.00151.2001. [DOI] [PubMed] [Google Scholar]
- [81].Wang B, Hao J, Jones SC, Yee MS, Roth JC, Dixon IM. Decreased Smad 7 expression contributes to cardiac fibrosis in the infarcted rat heart. Am J Physiol Heart Circ Physiol. 2002;282(5):H1685–96. doi: 10.1152/ajpheart.00266.2001. [DOI] [PubMed] [Google Scholar]
- [82].Yu H, Huang J, Wang S, Zhao G, Jiao X, Zhu L. Overexpression of Smad7 suppressed ROS/MMP9-dependent collagen synthesis through regulation of heme oxygenase-1. Mol Biol Rep. 2013;40(9):5307–14. doi: 10.1007/s11033-013-2631-2. [DOI] [PubMed] [Google Scholar]
- [83].Wei LH, Huang XR, Zhang Y, Li YQ, Chen HY, Yan BP, et al. Smad7 inhibits angiotensin II-induced hypertensive cardiac remodelling. Cardiovasc Res. 2013;99(4):665–73. doi: 10.1093/cvr/cvt151. [DOI] [PubMed] [Google Scholar]
- [84].Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8(8):856–63. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- [85].Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, et al. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120(1):254–65. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wang B, Haldar SM, Lu Y, Ibrahim OA, Fisch S, Gray S, et al. The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. J Mol Cell Cardiol. 2008;45(2):193–7. doi: 10.1016/j.yjmcc.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Leenders JJ, Wijnen WJ, Hiller M, van der Made I, Lentink V, van Leeuwen RE, et al. Regulation of cardiac gene expression by KLF15, a repressor of myocardin activity. J Biol Chem. 2010;285(35)):27449–56. doi: 10.1074/jbc.M110.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bagchi RA, Czubryt MP. Synergistic roles of scleraxis and Smads in the regulation of collagen 1alpha2 gene expression. Biochim Biophys Acta. 2012;1823(10):1936–44. doi: 10.1016/j.bbamcr.2012.07.002. [DOI] [PubMed] [Google Scholar]
- [89].Choi ME, Ding Y, Kim SI. TGF-beta signaling via TAK1 pathway: role in kidney fibrosis. Semin Nephrol. 2012;32(3):244–52. doi: 10.1016/j.semnephrol.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115(Pt 15):3193–206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- [91].Kompa AR, See F, Lewis DA, Adrahtas A, Cantwell DM, Wang BH, et al. Long-term but not short-term p38 mitogen-activated protein kinase inhibition improves cardiac function and reduces cardiac remodeling post-myocardial infarction. J Pharmacol Exp Ther. 2008;325(3):741–50. doi: 10.1124/jpet.107.133546. [DOI] [PubMed] [Google Scholar]
- [92].Matsuoka H, Arai T, Mori M, Goya S, Kida H, Morishita H, et al. A p38 MAPK inhibitor, FR-167653, ameliorates murine bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2002;283(1)):L103–12. doi: 10.1152/ajplung.00187.2001. [DOI] [PubMed] [Google Scholar]
- [93].See F, Thomas W, Way K, Tzanidis A, Kompa A, Lewis D, et al. p38 mitogen-activated protein kinase inhibition improves cardiac function and attenuates left ventricular remodeling following myocardial infarction in the rat. J Am Coll Cardiol. 2004;44(8):1679–89. doi: 10.1016/j.jacc.2004.07.038. [DOI] [PubMed] [Google Scholar]
- [94].Leivonen SK, Lazaridis K, Decock J, Chantry A, Edwards DR, Kahari VM. TGF-beta-elicited induction of tissue inhibitor of metalloproteinases (TIMP)-3 expression in fibroblasts involves complex interplay between Smad3, p38alpha, and ERK1/2. PLoS One. 2013;8((2):e57474. doi: 10.1371/journal.pone.0057474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Stratton R, Rajkumar V, Ponticos M, Nichols B, Shiwen X, Black CM, et al. Prostacyclin derivatives prevent the fibrotic response to TGF-beta by inhibiting the Ras/MEK/ERK pathway. FASEB J. 2002;16(14):1949–51. doi: 10.1096/fj.02-0204fje. [DOI] [PubMed] [Google Scholar]
- [96].Chichger H, Vang A, O'Connell KA, Zhang P, Mende U, Harrington EO, et al. PKC delta and betaII regulate angiotensin II-mediated fibrosis through p38: a mechanism of RV fibrosis in pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2015;308(8):L827–36. doi: 10.1152/ajplung.00184.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rustad KC, Wong VW, Gurtner GC. The role of focal adhesion complexes in fibroblast mechanotransduction during scar formation. Differentiation. 2013;86(3):87–91. doi: 10.1016/j.diff.2013.02.003. [DOI] [PubMed] [Google Scholar]
- [98].Wong VW, Rustad KC, Akaishi S, Sorkin M, Glotzbach JP, Januszyk M, et al. Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat Med. 2012;18(1):148–52. doi: 10.1038/nm.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- [100].Minty A, Kedes L. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol Cell Biol. 1986;6(6):2125–36. doi: 10.1128/mcb.6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sun Q, Chen G, Streb JW, Long X, Yang Y, Stoeckert CJ, Jr., et al. Defining the mammalian CArGome. Genome Res. 2006;16(2):197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Benson CC, Zhou Q, Long X, Miano JM. Identifying functional single nucleotide polymorphisms in the human CArGome. Physiol Genomics. 2011;43(18):1038–48. doi: 10.1152/physiolgenomics.00098.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28(9):943–58. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35(6):577–93. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- [105].Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11(5):353–65. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23(7):2425–37. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Huang J, Cheng L, Li J, Chen M, Zhou D, Lu MM, et al. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J Clin Invest. 2008;118(2):515–25. doi: 10.1172/JCI33304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100(16):9366–70. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132(5):987–97. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- [110].Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105(7):851–62. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- [111].Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20(12):1545–56. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- [112].Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci U S A. 2002;99(23):14855–60. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol Cell Biol. 2008;28(2):732–42. doi: 10.1128/MCB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113(3):329–42. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- [115].Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal. 2011;4(177):ra40. doi: 10.1126/scisignal.2001750. [DOI] [PubMed] [Google Scholar]
- [116].Cen B, Selvaraj A, Burgess RC, Hitzler JK, Ma Z, Morris SW, et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol. 2003;23(18):6597–608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292(2):H1170–80. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- [118].Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol. 2004;5:13. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25(8):3173–81. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sandbo N, Kregel S, Taurin S, Bhorade S, Dulin NO. Critical role of serum response factor in pulmonary myofibroblast differentiation induced by TGF-beta. Am J Respir Cell Mol Biol. 2009;41(3):332–8. doi: 10.1165/rcmb.2008-0288OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16(11):588–96. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- [122].Small EM. The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J Cardiovasc Transl Res. 2012;5(6):794–804. doi: 10.1007/s12265-012-9397-0. [DOI] [PubMed] [Google Scholar]
- [123].Busche S, Descot A, Julien S, Genth H, Posern G. Epithelial cell-cell contacts regulate SRF-mediated transcription via Rac-actin-MAL signalling. J Cell Sci. 2008;121(Pt 7):1025–35. doi: 10.1242/jcs.014456. [DOI] [PubMed] [Google Scholar]
- [124].Elberg G, Chen L, Elberg D, Chan MD, Logan CJ, Turman MA. MKL1 mediates TGF-beta1-induced alpha-smooth muscle actin expression in human renal epithelial cells. Am J Physiol Renal Physiol. 2008;294(5):F1116–28. doi: 10.1152/ajprenal.00142.2007. [DOI] [PubMed] [Google Scholar]
- [125].Leitner L, Shaposhnikov D, Mengel A, Descot A, Julien S, Hoffmann R, et al. MAL/MRTF-A controls migration of non-invasive cells by upregulation of cytoskeleton-associated proteins. J Cell Sci. 2011;124(Pt 24):4318–31. doi: 10.1242/jcs.092791. [DOI] [PubMed] [Google Scholar]
- [126].O'Connor JW, Gomez EW. Cell adhesion and shape regulate TGF-beta1-induced epithelial-myofibroblast transition via MRTF-A signaling. PLoS One. 2013;8(12):e83188. doi: 10.1371/journal.pone.0083188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tomasek JJ, Vaughan MB, Kropp BP, Gabbiani G, Martin MD, Haaksma CJ, et al. Contraction of myofibroblasts in granulation tissue is dependent on Rho/Rho kinase/myosin light chain phosphatase activity. Wound Repair Regen. 2006;14(3):313–20. doi: 10.1111/j.1743-6109.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- [128].Copaja M, Venegas D, Aranguiz P, Canales J, Vivar R, Catalan M, et al. Simvastatin induces apoptosis by a Rho-dependent mechanism in cultured cardiac fibroblasts and myofibroblasts. Toxicol Appl Pharmacol. 2011;255(1):57–64. doi: 10.1016/j.taap.2011.05.016. [DOI] [PubMed] [Google Scholar]
- [129].Zhou Y, Huang X, Hecker L, Kurundkar D, Kurundkar A, Liu H, et al. Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J Clin Invest. 2013;123(3):1096–108. doi: 10.1172/JCI66700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Small EM, Thatcher JE, Sutherland LB, Kinoshita H, Gerard RD, Richardson JA, et al. Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ Res. 2010;107(2):294–304. doi: 10.1161/CIRCRESAHA.110.223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kagiyama S, Matsumura K, Goto K, Otsubo T, Iida M. Role of Rho kinase and oxidative stress in cardiac fibrosis induced by aldosterone and salt in angiotensin type 1a receptor knockout mice. Regul Pept. 2010;160(1–3):133–9. doi: 10.1016/j.regpep.2009.12.002. [DOI] [PubMed] [Google Scholar]
- [132].Velasquez LS, Sutherland LB, Liu Z, Grinnell F, Kamm KE, Schneider JW, et al. Activation of MRTF-A-dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proc Natl Acad Sci U S A. 2013;110(42):16850–5. doi: 10.1073/pnas.1316764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Crider BJ, Risinger GM, Jr., Haaksma CJ, Howard EW, Tomasek JJ. Myocardin-related transcription factors A and B are key regulators of TGF-beta1-induced fibroblast to myofibroblast differentiation. J Invest Dermatol. 2011;131(12):2378–85. doi: 10.1038/jid.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Luchsinger LL, Patenaude CA, Smith BD, Layne MD. Myocardin-related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. J Biol Chem. 2011;286(51):44116–25. doi: 10.1074/jbc.M111.276931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Qiu P, Feng XH, Li L. Interaction of Smad3 and SRF-associated complex mediates TGF-beta1 signals to regulate SM22 transcription during myofibroblast differentiation. J Mol Cell Cardiol. 2003;35(12):1407–20. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- [136].Morita T, Mayanagi T, Sobue K. Reorganization of the actin cytoskeleton via transcriptional regulation of cytoskeletal/focal adhesion genes by myocardin-related transcription factors (MRTFs/MAL/MKLs) Exp Cell Res. 2007;313(16):3432–45. doi: 10.1016/j.yexcr.2007.07.008. [DOI] [PubMed] [Google Scholar]
- [137].Kam Y, Quaranta V. Cadherin-bound beta-catenin feeds into the Wnt pathway upon adherens junctions dissociation: evidence for an intersection between beta-catenin pools. PLoS One. 2009;4(2):e4580. doi: 10.1371/journal.pone.0004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].El Sayegh TY, Arora PD, Laschinger CA, Lee W, Morrison C, Overall CM, et al. Cortactin associates with N-cadherin adhesions and mediates intercellular adhesion strengthening in fibroblasts. J Cell Sci. 2004;117(Pt 21):5117–31. doi: 10.1242/jcs.01385. [DOI] [PubMed] [Google Scholar]
- [139].Ho AT, Voura EB, Soloway PD, Watson KL, Khokha R. MMP inhibitors augment fibroblast adhesion through stabilization of focal adhesion contacts and up-regulation of cadherin function. J Biol Chem. 2001;276(43):40215–24. doi: 10.1074/jbc.M101647200. [DOI] [PubMed] [Google Scholar]
- [140].Banerjee I, Yekkala K, Borg TK, Baudino TA. Dynamic interactions between myocytes, fibroblasts, and extracellular matrix. Ann N Y Acad Sci. 2006;1080:76–84. doi: 10.1196/annals.1380.007. [DOI] [PubMed] [Google Scholar]
- [141].Masszi A, Fan L, Rosivall L, McCulloch CA, Rotstein OD, Mucsi I, et al. Integrity of cell-cell contacts is a critical regulator of TGF-beta 1-induced epithelial-to-myofibroblast transition: role for beta-catenin. Am J Pathol. 2004;165(6):1955–67. doi: 10.1016/s0002-9440(10)63247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Masszi A, Di Ciano C, Sirokmany G, Arthur WT, Rotstein OD, Wang J, et al. Central role for Rho in TGF-beta1-induced alpha-smooth muscle actin expression during epithelial-mesenchymal transition. Am J Physiol Renal Physiol. 2003;284(5):F911–24. doi: 10.1152/ajprenal.00183.2002. [DOI] [PubMed] [Google Scholar]
- [143].Fan L, Sebe A, Peterfi Z, Masszi A, Thirone AC, Rotstein OD, et al. Cell contact-dependent regulation of epithelial-myofibroblast transition via the rho-rho kinase-phospho-myosin pathway. Mol Biol Cell. 2007;18(3):1083–97. doi: 10.1091/mbc.E06-07-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Speight P, Nakano H, Kelley TJ, Hinz B, Kapus A. Differential topical susceptibility to TGFbeta in intact and injured regions of the epithelium: key role in myofibroblast transition. Mol Biol Cell. 2013;24(21):3326–36. doi: 10.1091/mbc.E13-04-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Charbonney E, Speight P, Masszi A, Nakano H, Kapus A. beta-catenin and Smad3 regulate the activity and stability of myocardin-related transcription factor during epithelial-myofibroblast transition. Mol Biol Cell. 2011;22(23):4472–85. doi: 10.1091/mbc.E11-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Baurand A, Zelarayan L, Betney R, Gehrke C, Dunger S, Noack C, et al. Beta-catenin downregulation is required for adaptive cardiac remodeling. Circ Res. 2007;100(9):1353–62. doi: 10.1161/01.RES.0000266605.63681.5a. [DOI] [PubMed] [Google Scholar]
- [147].Duan J, Gherghe C, Liu D, Hamlett E, Srikantha L, Rodgers L, et al. Wnt1/betacatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31(2):429–42. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Laeremans H, Rensen SS, Ottenheijm HC, Smits JF, Blankesteijn WM. Wnt/frizzled signalling modulates the migration and differentiation of immortalized cardiac fibroblasts. Cardiovasc Res. 2010;87(3):514–23. doi: 10.1093/cvr/cvq067. [DOI] [PubMed] [Google Scholar]
- [149].Muehlich S, Wang R, Lee SM, Lewis TC, Dai C, Prywes R. Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol Cell Biol. 2008;28(20):6302–13. doi: 10.1128/MCB.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Nakagawa K, Kuzumaki N. Transcriptional activity of megakaryoblastic leukemia 1 (MKL1) is repressed by SUMO modification. Genes Cells. 2005;10(8):835–50. doi: 10.1111/j.1365-2443.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- [151].Hinson JS, Medlin MD, Taylor JM, Mack CP. Regulation of myocardin factor protein stability by the LIM-only protein FHL2. Am J Physiol Heart Circ Physiol. 2008;295(3):H1067–H75. doi: 10.1152/ajpheart.91421.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Philippar U, Schratt G, Dieterich C, Muller JM, Galgoczy P, Engel FB, et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol Cell. 2004;16(6):867–80. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- [153].Alnajar A, Nordhoff C, Schied T, Chiquet-Ehrismann R, Loser K, Vogl T, et al. The LIM-only protein FHL2 attenuates lung inflammation during bleomycin-induced fibrosis. PLoS One. 2013;8(11):e81356. doi: 10.1371/journal.pone.0081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17(21):6289–99. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, et al. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci U S A. 2005;102(25):8916–21. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26(15):5797–808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bernau K, Ngam C, Torr EE, Acton B, Kach J, Dulin NO, et al. Megakaryoblastic leukemia-1 is required for the development of bleomycin-induced pulmonary fibrosis. Respir Res. 2015;16:45. doi: 10.1186/s12931-015-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Evelyn CR, Wade SM, Wang Q, Wu M, Iniguez-Lluhi JA, Merajver SD, et al. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6(8):2249–60. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- [159].Hayashi K, Watanabe B, Nakagawa Y, Minami S, Morita T. RPEL proteins are the molecular targets for CCG-1423, an inhibitor of Rho signaling. PLoS One. 2014;9(2):e89016. doi: 10.1371/journal.pone.0089016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Haak AJ, Tsou PS, Amin MA, Ruth JH, Campbell P, Fox DA, et al. Targeting the myofibroblast genetic switch: inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J Pharmacol Exp Ther. 2014;349(3):480–6. doi: 10.1124/jpet.114.213520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Johnson LA, Rodansky ES, Haak AJ, Larsen SD, Neubig RR, Higgins PD. Novel Rho/MRTF/SRF inhibitors block matrix-stiffness and TGF-beta-induced fibrogenesis in human colonic myofibroblasts. Inflamm Bowel Dis. 2014;20(1):154–65. doi: 10.1097/01.MIB.0000437615.98881.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Evelyn CR, Bell JL, Ryu JG, Wade SM, Kocab A, Harzdorf NL, et al. Design, synthesis and prostate cancer cell-based studies of analogs of the Rho/MKL1 transcriptional pathway inhibitor, CCG-1423. Bioorg Med Chem Lett. 2010;20(2):665–72. doi: 10.1016/j.bmcl.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem Biophys Res Commun. 2004;322(4):1178–91. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- [164].Herum KM, Lunde IG, Skrbic B, Florholmen G, Behmen D, Sjaastad I, et al. Syndecan-4 signaling via NFAT regulates extracellular matrix production and cardiac myofibroblast differentiation in response to mechanical stress. J Mol Cell Cardiol. 2013;54:73–81. doi: 10.1016/j.yjmcc.2012.11.006. [DOI] [PubMed] [Google Scholar]
- [165].Gonzalez Bosc LV, Layne JJ, Nelson MT, Hill-Eubanks DC. Nuclear factor of activated T cells and serum response factor cooperatively regulate the activity of an alpha-actin intronic enhancer. J Biol Chem. 2005;280(28):26113–20. doi: 10.1074/jbc.M411972200. [DOI] [PubMed] [Google Scholar]
- [166].Davis J, Burr AR, Davis GF, Birnbaumer L, Molkentin JD. A TRPC6-dependent pathway for myofibroblast transdifferentiation and wound healing in vivo. Dev Cell. 2012;23(4):705–15. doi: 10.1016/j.devcel.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93(2):215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Berry JM, Le V, Rotter D, Battiprolu PK, Grinsfelder B, Tannous P, et al. Reversibility of adverse, calcineurin-dependent cardiac remodeling. Circ Res. 2011;109(4):407–17. doi: 10.1161/CIRCRESAHA.110.228452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Yue Z, Xie J, Yu AS, Stock J, Du J, Yue L. Role of TRP channels in the cardiovascular system. Am J Physiol Heart Circ Physiol. 2015;308(3):H157–82. doi: 10.1152/ajpheart.00457.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Huang H, Wang W, Liu P, Jiang Y, Zhao Y, Wei H, et al. TRPC1 expression and distribution in rat hearts. Eur J Histochem. 2009;53(4):e26. doi: 10.4081/ejh.2009.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Kuwahara K, Wang Y, McAnally J, Richardson JA, Bassel-Duby R, Hill JA, et al. TRPC6 fulfills a calcineurin signaling circuit during pathologic cardiac remodeling. J Clin Invest. 2006;116(12):3114–26. doi: 10.1172/JCI27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Nakayama H, Wilkin BJ, Bodi I, Molkentin JD. Calcineurin-dependent cardiomyopathy is activated by TRPC in the adult mouse heart. FASEB J. 2006;20(10):1660–70. doi: 10.1096/fj.05-5560com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Makarewich CA, Zhang H, Davis J, Correll RN, Trappanese DM, Hoffman NE, et al. Transient receptor potential channels contribute to pathological structural and functional remodeling after myocardial infarction. Circ Res. 2014;115(6):567–80. doi: 10.1161/CIRCRESAHA.115.303831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Harada M, Luo X, Qi XY, Tadevosyan A, Maguy A, Ordog B, et al. Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation. 2012;126(17):2051–64. doi: 10.1161/CIRCULATIONAHA.112.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Saliba Y, Karam R, Smayra V, Aftimos G, Abramowitz J, Birnbaumer L, et al. Evidence of a Role for Fibroblast Transient Receptor Potential Canonical 3 Ca2+ Channel in Renal Fibrosis. J Am Soc Nephrol. 2015;26(8):1855–76. doi: 10.1681/ASN.2014010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Willems IE, Havenith MG, De Mey JG, Daemen MJ. The alpha-smooth muscle actin-positive cells in healing human myocardial scars. Am J Pathol. 1994;145(4):868–75. [PMC free article] [PubMed] [Google Scholar]
- [177].Driesen RB, Nagaraju CK, Abi-Char J, Coenen T, Lijnen PJ, Fagard RH, et al. Reversible and irreversible differentiation of cardiac fibroblasts. Cardiovasc Res. 2014;101(3):411–22. doi: 10.1093/cvr/cvt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Vivar R, Humeres C, Ayala P, Olmedo I, Catalan M, Garcia L, et al. TGF-beta1 prevents simulated ischemia/reperfusion-induced cardiac fibroblast apoptosis by activation of both canonical and non-canonical signaling pathways. Biochim Biophys Acta. 2013;1832(6):754–62. doi: 10.1016/j.bbadis.2013.02.004. [DOI] [PubMed] [Google Scholar]
- [179].Hale TM. Persistent Phenotypic Shift in Cardiac Fibroblasts: Impact of Transient Renin Angiotensin System Inhibition. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.027. JMCC9599. [DOI] [PubMed] [Google Scholar]
- [180].D'Souza KM, Biwer LA, Madhavpeddi L, Ramaiah P, Shahid W, Hale TM. Persistent change in cardiac fibroblast physiology after transient ACE inhibition. Am J Physiol Heart Circ Physiol. 2015;309(8):H1346–53. doi: 10.1152/ajpheart.00615.2015. [DOI] [PubMed] [Google Scholar]
- [181].Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Nam YJ, Lubczyk C, Bhakta M, Zang T, Fernandez-Perez A, McAnally J, et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141(22):4267–78. doi: 10.1242/dev.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O'Rourke R, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Fu Y, Huang C, Xu X, Gu H, Ye Y, Jiang C, et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015;25(9):1013–24. doi: 10.1038/cr.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, et al. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014;6(5):951–60. doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Mathison M, Singh VP, Gersch RP, Ramirez MO, Cooney A, Kaminsky SM, et al. “Triplet” polycistronic vectors encoding Gata4, Mef2c, and Tbx5 enhances postinfarct ventricular functional improvement compared with singlet vectors. The Journal of thoracic and cardiovascular surgery. 2014;148(4):1656–64. e2. doi: 10.1016/j.jtcvs.2014.03.033. [DOI] [PubMed] [Google Scholar]
- [188].Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–8. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Nagalingam RS, Safi HA, Czubryt MP. Gaining myocytes or losing fibroblasts: Challenges in cardiac fibroblast reprogramming for infarct repair. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.029. JMCC9596. [DOI] [PubMed] [Google Scholar]