Abstract
A MYB transcription factor gene, RsMYB1, from radish was introduced into the chrysanthemum cultivars ‘Peach ND’, ‘Peach Red’, and ‘Vivid Scarlet’ under the control of the cauliflower mosaic virus 35S promoter. Presence of RsMYB1 in transgenic lines was confirmed using polymerase chain reaction (PCR). Results of reverse-transcription-PCR analysis revealed that the expression of RsMYB1 was stable in all transgenic lines and could enhance the expression levels of three key biosynthetic genes (F3H, DFR, and ANS) involved in anthocyanin production. Accordingly, anthocyanin content was significantly higher in transgenic lines than in control of all the cultivars, although the increasement was not visually observed in any of the transgenic lines. Therefore, these results demonstrate that RsMYB1 has potential to enhance anthocyanin content in the chrysanthemums.
Keywords: Anthocyanin, RT-PCR, Spectrophotometer, Transcription factor
Introduction
Chrysanthemum morifolium Ramat. is one of the most important floricultural crops that occupies a prominent position in flower markets worldwide. In 2012, its cultivation area was 31 % of the total cut-flower cultivation area (1724 ha) in Korea (Ministry of Agriculture, Food, and Rural Affairs, 2012), and it has been increasing on a yearly basis. In summer, the heat stress due to elevated temperatures causes degradation of anthocyanin accumulation in flower petals, leading to low flower quality that causes economic loss in chrysanthemum fields (Eun et al. 2008).
Temperature has been considered as an important factor that influences anthocyanin biosynthesis in various plants, such as apple (Ubi et al. 2006; Lin-Wang et al. 2011), grape (Yamane et al. 2006), petunia (Shvarts et al. 1997), red orange (Lo Piero et al. 2005), and rose (Dela et al. 2003). Elevated temperatures decrease the accumulation of anthocyanin. Huh et al. (2008) claimed that flower coloration was observed in red chrysanthemum cultivars when they were grown at an elevated temperature (35 °C).
Genetic engineering with MYB transcription factors has been used to enhance anthocyanin accumulation in several plant species, since MYB transcription factors up-regulated anthocyanin biosynthesis in tobacco, petunia, apple, rose, and lily (Espley et al. 2007; Lin-Wang et al. 2010; Pattanaik et al. 2010; Quattrocchio et al. 2006; Yamagishi et al. 2010). Pattanaik et al. (2010) reported that overexpression of MYB (NtAn2) in tobacco enhanced anthocyanin accumulation and expression levels of chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS) genes. In apples, red fruit color is controlled by MdMYB10 (Espley et al. 2007). The purple phenotype of sweet potato is also strongly associated with IbMYB1, which induces all anthocyanin biosynthetic genes (Mano et al. 2007). LhMYB6 and LhMYB12 increase the expression levels of anthocyanin biosynthetic genes and determine the accumulation of anthocyanin (Yamagishi et al. 2010). Radish MYB transcription factor belongs to the R2R3-MYB transcription factor family and is highly expressed in the skin and flesh of three radish cultivars (‘Seo Ho’, ‘Man Tang Hong’, and ‘Hong Feng No.1’); strong anthocyanin accumulation has been observed in the radish cultivars (Park et al. 2011). Expression of the MYB transcription factor in petunia enhanced the expression level of anthocyanin biosynthetic genes (CHI, CHS, F3H, DFR, and ANS) as well as anthocyanin accumulation in our previous studies. Since the main anthocyanin biosynthesis pathway in chrysanthemum is the cyanidin-based pathway, CHS, CHI, F3H, DFR, and ANS are considered as the five key biosynthetic genes responsible for anthocyanin accumulation in chrysanthemums. Thus, it is quite interesting to investigate the role of RsMYB1 in anthocyanin accumulation in chrysanthemum.
In this study, we used three different red chrysanthemum cultivars that have been observed to show coloration in summer. RsMYB1 was introduced into the cultivars by using Agrobacterium-mediated transformation. Expression levels of anthocyanin biosynthetic genes regulated by RsMYB and associated anthocyanin accumulation were analyzed.
Materials and methods
Three red chrysanthemum cultivars (C. morifolium Ramat.), namely, ‘Peach ND’, ‘Peach Red’, and ‘Vivid scarlet’, were obtained from Gumi Research Station. The cultivars were then proliferated in vitro, according to the method described by Naing et al. (2015a). Briefly, shoot tips with about 1 cm2 in size were cultured on Murashige and Skoog medium (MS; Duchefa Biochemie, Netherlands) containing 3 % (w/v) sucrose, 0.8 % plant agar, and 0.01 % activated charcoal. Then, the cultures were maintained at 25 ± 2 °C and a 16-h photoperiod (37 μmol m−2 s−1)
Plasmid construction and transformation
In this study, Agrobacterium tumefaciens strain C58C1 carrying a binary vector pB7WG2D with RsMYB1 isolated from radish (Raphanus sativus L.) was used. RsMYB1 was placed under the control of the cauliflower mosaic virus 35S promoter. In addition, the vector contained the gene bar for resistance to phosphinothricin (PPT) in transgenic plants.
Prior to transformation, infection solution of Agrobacterium strain harboring the binary vector pB7WG2D were cultured as described by Naing et al. (2014), and transformation was performed according to protocol described by Naing et al. (2016). Briefly, 100 leaf segments (5 cm2) excised from in vitro plants of the three different cultivars were incubated in the Agrobacterium infection solution. The leaf segments were cultured on an MS co-cultivation medium with 0.5 mg L−1 of BA and 0.5 mg L−1 of NAA, 3 % sucrose, and 3 g L−1 of Gelrite (pH 5.8) and placed in the dark at 25 ± 2 °C for 3 days. Then, the leaf segments were cultured on the same medium containing 125 mg L−1 Clavamox (Zoetis, India) and placed in the dark at 25 ± 2 °C for 7 days. The leaf segments were further cultured on the same medium containing 1 mg L−1 of PPT and 125 mg L−1 of Clavamox under a 16-h photoperiod (37 μmol m−2 s−1). They were transferred to a fresh medium every 3 weeks to suppress Agrobacterium growth. After 6 weeks, green shoots resistant to PPT were collected and transferred to plant growth regulator-free MS medium with 1 mg L−1 PPT and 125 mg L−1 of Clavamox to assess plant growth. PPT-resistant plants that were 4–5 cm in size were transplanted to a tray with vermiculite soil and acclimatized in a greenhouse at 25 °C. The plantlets were then transferred to pots filled with peat soil and placed in the greenhouse.
DNA extraction and polymerase chain reaction
Genomic DNA was extracted from young leaves of 6-week-old plants selected using 1 mg L−1 PPT with the RBC HiYield™ Genomic DNA Mini Kit (Real Biotech Corporation, Taiwan), according to manufacturer’s instructions. Genomic DNA was then amplified using polymerase chain reaction (PCR) with specific primers and PCR conditions mentioned in Table 1. To detect presence of RsMYB1 and bar, amplified DNA was observed under UV (UVITEC Cambridge, UK) irradiation after electrophoresis for 30 min using 2 % agarose gel and staining with ethidium bromide.
Table 1.
Primers and PCR conditions used in this study
| Gene | Primer | Size (bp) | PCR condition |
|---|---|---|---|
| RsMYB1 | F-ATG GAG GGT TCG TCC AAA GG | 700 | 98 °C for 30 s, followed by 25 cycles including 10 min at 98 °C, 1 min at 98 °C and 1 min at 72 °C, and final elongation step at 72 °C for 1 min |
| R-GAA ACA CTA ATC AAA TTA CAC AGT CTC TCC | |||
| Bar | F-GGT CTG CAC CAT CGT | 496 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 30 s at 55 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-TCA GAT TTC GGT GAC GGG CA | |||
| CHS | F-CAA CGG TTT TCT CCA TTA GGT | 299 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 40 s at 57 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-GAG GAC CAC GGT TTC GAC | |||
| CHI | F-TGG TGC AAC CAT TGA CAA GT | 300 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 40 s at 55.6 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-AAA TTT GGT TCA GCA TCT GTA GTT | |||
| F3H | F-ACC CGG TTC GTC CGT GAT GAG G | 804 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 40 s at 64 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-TGC CTG GTG GTC CGC ATT CT | |||
| DFR | F-ATG AAA GAA GAC TCA CCA GCC A | 1048 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 40 s at 59 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-CTT CGT GAG TGT CCG CCT TT | |||
| ANS | F-ATA CAT CCG AAC ACA AGA TG | 432 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 40 s at 60 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-AAT CGC TAG GTG TCG AGG GCC | |||
| Actin | F-ACA ACG TTT TAC AAT GAG CTT CG | 196 | 95 °C for 2 min, followed by 30 cycles including 20 s at 95 °C, 40 s at 57 °C and 30 s at 72 °C, and final elongation step at 72 °C for 5 min |
| R-CCG TTC AGC AGT TGT AGT AA |
RNA extraction and reverse transcription-PCR
To evaluate the expression level of RsMYB1 and anthocyanin biosynthetic genes in the transgenic lines already confirmed using PCR, RNA of each line was extracted for reverse transcription (RT)-PCR analysis. Prior to RNA extraction, all equipment, including reagent bottles, were cleaned using RNA eraser (MP Bio, USA). Total RNA was isolated from 100 mg of leaf tissue of the transgenic and wild type chrysanthemum plants by using TRI Reagent™ Solution (Ambion, USA), according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 100 ng of total RNA by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA), according to the manufacturer’s protocol. Primers and PCR conditions for RsMYB1 remained unchanged, and those for the biosynthetic genes (CHI, CHS, F3H, DFR, and ANS) are listed in Table 1; Actin was used as the internal control. PCR products were observed under UV (UVITEC Cambridge, UK) irradiation after electrophoresis for 30 min using 2 % agarose gel and staining with ethidium bromide.
Analysis of anthocyanin content
Total anthocyanin contents of the transgenic lines and control plants were analyzed according to the protocol used by Naing et al. (2015b), with some modifications. Briefly, approximately 500 mg of leaf materials excised from the plants grown in the greenhouse was crushed, and pigments were extracted with 5 mL of distilled water. They were then incubated overnight at 4 °C by adding 5 mL of 1 % (w/v) hydrochloric acid in methanol solution. The supernatant was collected after centrifugation at 3000 rpm for 20 min and transferred to a 2 mL collection tube. Absorbance was measured at 430–630 nm by using a spectrophotometer (U-2800; Hitachi, Tokyo, Japan). Anthocyanin content of each sample was calculated according to the method reported by Sung et al. (2013).
Statistical analysis
For measurement of anthocyanin content, five samples from each plant (wild type and two independent transgenic lines) of the three chrysanthemum cultivars were detected. Data were analyzed using Duncan Multiple Range Test (DMRT) (p > 0.05).
Results
Generation of chrysanthemum transgenic plants that express RsMYB1
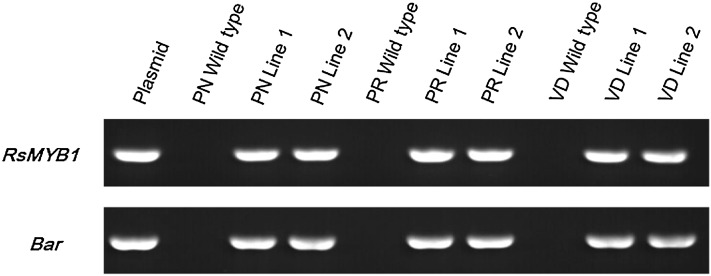
To investigate whether RsMYB1 could enhance the expression level of biosynthetic genes responsible for anthocyanin biosynthesis, we produced two independent transgenic lines from the cultivars ‘Peach ND’, ‘Peach Red’, and ‘Vivid Scarlet’, in which the presence of the selection marker and transgene were detected using PCR (Fig. 1). Interestingly, enhanced anthocyanin expression was not observed in the transgenic plants of the cultivars. Thus, it was necessary to investigate the expression levels of the transgene and anthocyanin biosynthetic genes.
Fig. 1.
PCR detection of selection marker (bar), target (RsMYB1) genes in transgenic chrysanthemum (cvs. Peach ND (PN), Peach Red (PR), and Vivid Scarlet (VD) lines 1 and 2
Expression analysis of RsMYB1 and anthocyanin biosynthetic genes
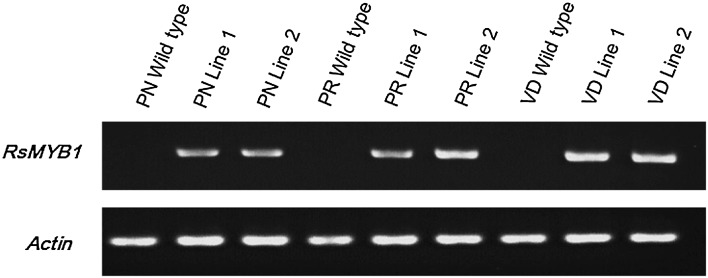
We used the two independent transgenic lines derived from each cultivar to perform an expression analysis of RsMYB1 and anthocyanin biosynthetic genes. Results of RT-PCR showed that distinct and stable expression of RsMYB1 in all individual lines of the same genotype (Fig. 2), but its expression level was different along with different genotype tested. Surprisingly, distinct and stable expression of RsMYB1 in the cultivars did not enhance anthocyanin production.
Fig. 2.
RT-PCR analysis of RsMYB1 expression in the non-transgenic lines (wild type) and the transgenic lines of the chrysanthemum cultivars, namely, Peach ND (PN), Peach Red (PR), and Vivid Scarlet (VD)
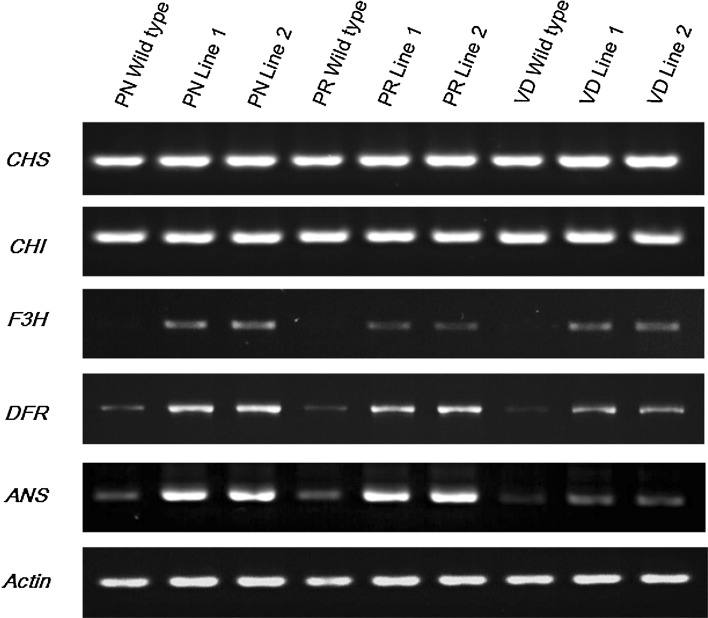
Expression analysis of biosynthetic genes by RT-PCR revealed that the key biosynthetic genes (CHI, CHS, F3H, DFR, and ANS) were expressed in all transgenic lines, including the control. However, expression of RsMYB1 in chrysanthemum could regulate the enhanced expression of the biosynthetic genes, in particular, F3H, DFR, and ANS, as compared to the expression of these genes in the control. Expression levels of other biosynthetic genes such as CHI and CHS did not differ between the controls and transgenic lines (Fig. 3). Interestingly, higher expression of RsMYB1 is not associated with that of biosynthetic genes in the transgenic lines because expression levels of biosynthetic genes in ‘Peach ND’, which contains low transcripts of RsMYB1, were slightly higher than those in ‘Peach Red’ and ‘Vivid Scarlet’, which express high transcripts of RsMYB1 (Figs. 2, 3).
Fig. 3.
RT-PCR analysis of the expression patterns of biosynthetic genes in non-transgenic lines (wild type) and two individual transgenic lines of the chrysanthemum cultivars, namely, Peach ND (PN), Peach Red (PR), and Vivid Scarlet (VD) containing RsMYB1
Analysis of anthocyanin content
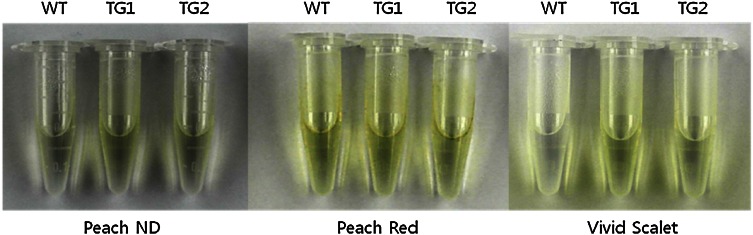
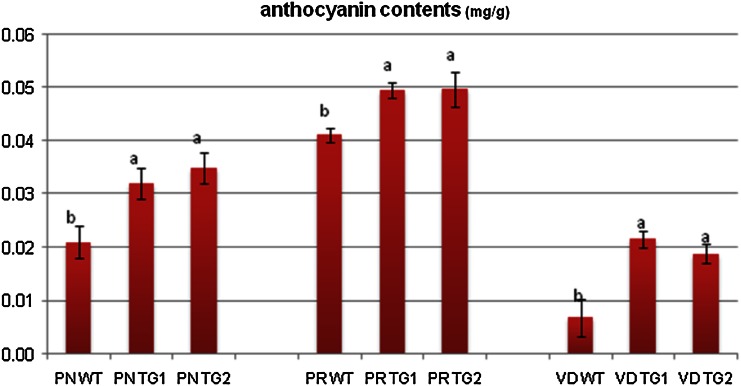
Although variations in biosynthetic gene expression were observed between the controls and transgenic lines (Fig. 3), enhanced anthocyanin accumulation was not visually observed in the transgenic lines. Hence, we assumed that anthocyanin might have accumulated inside the leaves without being expressed phenotypically. However, when anthocyanin was extracted from the leaves of the transgenic lines and compared to the extracts from the control plants, only slight significant color difference was observed in ‘Peach Red’ and ‘Peach ND’ while a distinct variation was seen in ‘Vivid Scarlet’ (Fig. 4). This was further confirmed using spectrophotometry results, which were not consistent with the results for anthocyanin color in (Fig. 5) in ‘Peach Red’, ‘Peach ND’ because anthocyanin contents were observed to be significantly different in the cultivars (Fig. 5), where the contents in transgenic lines were approximately 0.01–0.015 mg g−1 higher than those in the controls, regardless of genotypes. Thus, higher expression levels of the three biosynthetic genes were associated with anthocyanin accumulation in the cultivars.
Fig. 4.
Comparative analysis of anthocyanin content between the non-transgenic lines (wild type) and transgenic lines of the chrysanthemum cultivars, namely, Peach ND (PN), Peach Red (PR), and Vivid Scarlet (VD) containing RsMYB1
Fig. 5.
Spectrophotometeric analysis of anthocyanin content between the non-transgenic (wild type) and transgenic lines of the chrysanthemum cultivars, namely, Peach ND (PN), Peach Red (PR), and Vivid Scarlet (VD) containing RsMYB1. Bar stands for standard error. Means marked with same letter within the same cultivar are not significantly different
Discussion
Transcription factors can regulate genes involved in the anthocyanin biosynthetic pathway in various plants, including model plants and floricultural or food crops (Nishihara and Nakatsuka 2011). R2R3 MYB transcription factors, which regulate anthocyanin biosynthesis, cloned from petunia (Quattrocchio et al. 2006), apple, strawberry, plum and rose (Lin-Wang et al. 2010), lily (Yamagishi et al. 2010), snapdragon (Jackson et al. 1991), morning glory (Morita et al. 2006), gentian (Nakatsuka et al. 2008) and tobacco (Pattanaik et al. 2010) have been shown to regulate sets of genes in the anthocyanin biosynthetic pathway. Therefore, we assumed that it is possible to use the radish RsMYB1 transcription factor, which belongs to the R2R3-MYB transcription factor family and was found to be highly expressed in the skin and flesh of three radish cultivars and responsible for anthocyanin accumulation (Park et al. 2011), to enhance anthocyanin biosynthesis in the chrysanthemum cultivars ‘Peach ND’, ‘Peach Red’, and ‘Vivid Scarlet’, which showed anthocyanin degradation under elevated temperatures.
Here, we produced two individual transgenic lines from each cultivar. However, not all shoots that showed positive results for the introduced RsMYB1 exhibited anthocyanin-enhancing phenotypes. Butaye et al. (2005) claimed that this type of variation among individual transgenic plants/genotypes is often observed in plant transformation experiments and can be explained by variation in the expression of the introduced RsMYB1. Analysis of RsMYB1 expression in the transgenic lines was shown to be stable using RT-PCR; this was not consistent with the claim of Butaye et al. (2005).
In addition, many researchers have documented that anthocyanin accumulation in chrysanthemum correlated with the expression pattern of biosynthetic genes (Chen et al. 2012; He et al. 2013; Sung et al. 2013). Since the main anthocyanin biosynthesis pathway in chrysanthemum is the cyanidin-based pathway (He et al. 2013), CHS, CHI, F3H, DFR, and ANS are considered as five key biosynthetic genes responsible for anthocyanin accumulation in chrysanthemum. Our results show that the expression of RsMYB1 in chrysanthemum could enhance the expression level of key biosynthetic genes (F3H, DFR, and ANS), although anthocyanin accumulation is not visible in the cultivars. However, analysis of anthocyanin content using a spectrophotometer revealed that anthocyanin contents in transgenic lines of the cultivars were significantly higher than those in the controls. Thus, anthocyanin accumulation in the cultivars was correlated with the expression pattern of biosynthetic genes as reported by the researchers (Chen et al. 2012; He et al. 2013; Sung et al. 2013), but anthocyanin content produced by RsMYB1 seemed to be genotype-dependent because enhancement of anthocyanin contents in cultivars ‘Peach ND’ and ‘Vivid Scarlet’ seemed to be higher than that in cultivar ‘Peach Red’ (Fig. 5). Quattrocchio et al. (1999) claimed that although MYB transcription factors could enhance anthocyanin production, different pigmentation control activities were observed in different species of Antirrhinum and Petunia. Moreover, different pigmentation effects of MYB transcription factors on berry skin color in grape (Kobayashi et al. 2004, 2005) and tuber color in potato (De Jong et al. 2004) have also been reported. There seems to be some variation in the ability of transcription factors to control anthocyanin production in different genotypes.
Color pigmentation in petunia due to RsMYB1 was visually shown in our preliminary experiment (data not shown). In addition, enhancement of anthocyanin contents in the cultivars by RsMYB1 was observed in this study. However, the reason why increased anthocyanin content was not visually observed in the cultivars is unclear. One possible explanation is that anthocyanin contents induced by RsMYB1 was not dramatic in these cultivars. Another possible explanation is that the expression levels of F3H, DFR, and ANS were not high enough to enhance high anthocyanin content. Otherwise, it might be that expression of MYB transcription factor alone is not able to enhance strong anthocyanin accumulation in chrysanthemum, because the expression of MYB transcription factor alone failed to express anthocyanin in Arabidopsis and tobacco (Lloyd et al. 1992), and grape (Hichri et al. 2011). Alternatively, it also might be due to lack of basic helix–loop–helix (bHLH) expression in the chrysanthemums because, in several other cases, co-expression of MYB transcription factor and a bHLH partner are required to strongly express anthocyanin (Butelli et al. 2008; Lloyd et al. 1992; Quattrocchio et al. 1998). Therefore, we will conduct further experiments to reveal the reason why increased anthocyanin was not visually observed in the transgenic chrysanthemums expressing RsMYB1.
Conclusions
We produced transgenic chrysanthemums that expressed RsMYB1 and demonstrated the effects of its ectopic expression on the expression levels of anthocyanin biosynthetic genes. This study revealed that RsMYB1 could enhance the expression of the biosynthetic genes (F3H, DFR, and ANS) in transgenic lines of the cultivars Peach ND, Peach Red, and Vivid Scarlet and increase anthocyanin contents in the cultivars. The reason for the lack of visual enhancement in anthocyanin contents is still unknown. Accordingly, further studies on promoter functions and environmental conditions that affect the anthocyanin production pathway are necessary to understand the regulatory mechanism underlying anthocyanin biosynthesis. Overall, our study provides important information regarding the role of RsMYB1 in the regulation of key anthocyanin biosynthetic genes in chrysanthemums.
Acknowledgments
This study was supported by the Bio-industry Technology Development Program, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea, and Kyungpook National University Research Fund, 2012.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Contributor Information
Aung Htay Naing, Email: aunghtaynaing2005@gmail.com.
Chang Kil Kim, Email: ckkim@knu.ac.kr.
References
- Butaye K, Cammue B, Delauré S, De Bolle M. Approaches to minimize variation of transgenic expression in plants. Mol Breed. 2005;16:79–91. doi: 10.1007/s11032-005-4929-9. [DOI] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- Chen SM, Li CH, Zhu XR, Deng YM, Sun W, Wang LS, Chen FD, Zhang Z. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol Plant. 2012;56:458–464. doi: 10.1007/s10535-012-0069-3. [DOI] [Google Scholar]
- De Jong WS, Eannetta NT, De Jong DM, Bodis M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet. 2004;108:423–432. doi: 10.1007/s00122-003-1455-1. [DOI] [PubMed] [Google Scholar]
- Dela G, Or E, Ovadia R, Nissim-Levi A, Weiss D, Oren-Shamir M. Changes in anthocyanin concentration and composition in ‘Jaguar’ rose flowers due to transient high-temperature conditions. Plant Sci. 2003;164:333–340. doi: 10.1016/S0168-9452(02)00417-X. [DOI] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun JH, Hak KS, Seong YC, Oh GK, Young RL. Thermosusceptible developmental stage in anthocyanin accumulation and color response to high temperature in red chrysanthemum cultivars. Korea J Hort Sci Technol. 2008;26:357–361. [Google Scholar]
- He H, Ke H, Keting H, Qiaoyan X, Silan D. Flower colour modification of chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLoS One. 2013;8:e74395. doi: 10.1371/journal.pone.0074395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- Jackson D, Culianez-Macia F, Prescott AG, Roberts K, Martin C. Expression patterns of myb genes from Antirrhinum flowers. Plant Cell. 1991;3:115–125. doi: 10.1105/tpc.3.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin-color mutants. J Jpn Soc Hort Sci. 2005;74:196–203. doi: 10.2503/jjshs.74.196. [DOI] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Micheletti D, Palmer J, Volz R, Lozano L, Espley R, Hellens RP, Chagne D, Rowan DD, Troggio M, Iglesias I, Allan AC. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011;34:1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Walbot V, Davis RW. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science. 1992;258:1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- Lo Piero AR, Puglisi I, Rapisarda P, Petrone G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem. 2005;53:9083–9088. doi: 10.1021/jf051609s. [DOI] [PubMed] [Google Scholar]
- Mano H, Ogasawara F, Sato K, Higo H, Minobe Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol. 2007;143:1252–1268. doi: 10.1104/pp.106.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006;47:457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- Naing AH, Park KI, Lim SH, Kim CK. Appropriate choice of antibiotics for plant regeneration and optimization of selective agents to be used in genetic transformation of chrysanthemum. Plant Omics. 2014;7:237–243. [Google Scholar]
- Naing AH, Park KI, Chung MY, Lim KB, Kim CK. Optimization of factors affecting efficient shoot regeneration in chrysanthemum cv. Shinma. Braz J Bot. 2015 [Google Scholar]
- Naing AH, Lim KB, Kim CK. The usage of snapdragon Delila (Del) gene as a visible selection marker for the antibiotic-free transformation system. J Plant Biol. 2015;58:110–116. doi: 10.1007/s12374-014-0423-4. [DOI] [Google Scholar]
- Naing AH, Ai TN, Jeon SM, Park KI, Lim SH, Lim KB, Kim CK (2016) Novel antibiotics enhance regeneration and genetic transformation with RsMYB1 gene of recalcitrant chrysanthemum cv. Shinma. Plant Biosystems. doi:10.1080/11263504.2015.1103800
- Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, Shimada N, Yamamura S, Nishihara M. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008;49:1818–1829. doi: 10.1093/pcp/pcn163. [DOI] [PubMed] [Google Scholar]
- Nishihara M, Nakatsuka T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol Lett. 2011;33:433–441. doi: 10.1007/s10529-010-0461-z. [DOI] [PubMed] [Google Scholar]
- Park NI, Xu H, Li X, Jang IH, Park S, Ahn GH, Lim YP, Kim SJ, Park SU. Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus) J Agric Food Chem. 2011;59:6034–6039. doi: 10.1021/jf200824c. [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta. 2010;231:1061–1076. doi: 10.1007/s00425-010-1108-y. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, NM Mol J, Koes R. Analysis of bHLH and MYB domain proteins: speciesspecific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998;13:475–488. doi: 10.1046/j.1365-313X.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with Basic-Helix-Loop-Helix transcription factors of the anthocyanin pathway. Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts M, Borochov A, Weiss D. Low temperature enhances petunia flower pigmentation and induces chalcone synthase gene expression. Physiol Plant. 1997;99:67–72. doi: 10.1111/j.1399-3054.1997.tb03432.x. [DOI] [Google Scholar]
- Sung SY, Kim SH, Velusamy V, Lee YM, Ha BK, Kim JB, Kang SY, Kim HG, Kim DS. Comparative gene expression analysis in a highly anthocyanin pigmented mutant of colorless chrysanthemum. Mol Biol Rep. 2013;40:5177–5189. doi: 10.1007/s11033-013-2620-5. [DOI] [PubMed] [Google Scholar]
- Ubi BE, Honda C, Bessho H, Kondo S, Wada M, Kobayashi S, Moriguchi T. Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci. 2006;170:571–578. doi: 10.1016/j.plantsci.2005.10.009. [DOI] [Google Scholar]
- Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K. Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 2010;51:463–474. doi: 10.1093/pcp/pcq011. [DOI] [PubMed] [Google Scholar]
- Yamane T, Jeong ST, Goto-Yamamoto N, Koshita Y, Kobayashi S. Effects of temperature on anthocyanin biosynthesis in grape berry skins. Am J Enol Vitic. 2006;57:54–59. [Google Scholar]