Abstract
Background
We sought to determine the location, expression, and characterization of cardiac stem cells (CSCs) in children with end-stage heart failure (ESHF). We hypothesized ESHF myocardium would contain an increased number of CSCs relative to age-matched healthy myocardium, and ESHF-derived CSCs would have diminished functional capacity as evidenced by reduced telomere length.
Methods
Tissue samples were obtained from the explanted hearts of children undergoing heart transplantation with ESHF, defined as New York Heart Association class III or IV and ejection fraction less than 0.20, and from age-matched congenital heart disease patients with normal myocardium. The expression profile of cardiac-specific stem cell markers was determined using quantitative real time polymerase chain reaction and immunofluorescence. Cardiac stem cell growth reserve was assessed with telomere length.
Results
There were 15 ESHF and 15 age-matched congenital heart disease patients. End-stage heart failure myocardium demonstrated increased expression of c-kit+ and islet-1+ CSCs by 2.0- and 2.5-fold, respectively, compared with myocardium from congenital heart disease patients. There was no difference in expression of c-kit+ CSCs with advancing age from infants to children in ESHF myocardium. The c-kit+ CSCs isolated from ESHF patients demonstrated significantly reduced telomere length, suggesting a diminished functional capability in these cells (8.1 ± 0.6 kbp versus 6.3 ± 0.3 kbp; p = 0.015).
Conclusions
End-stage heart failure myocardium demonstrated an age-independent increase in CSCs relative to healthy myocardium; however, these CSCs from ESHF patients may have diminished proliferative ability and reduced functionality as an autologous cell therapy candidate. Further investigation is necessary to determine the role of ESHF-derived CSCs within the myocardium.
The prevalence of pediatric heart failure has increased dramatically in the last three decades, resulting in frequent hospitalizations, development of significant comorbidities, and protracted heart transplantation waiting-list times [1]. Given the current limitations in medical therapy and the limited number of donors available for pediatric heart transplantation, the potential for an effective cell-based therapy for pediatric heart failure patients is an attractive option. During the last decade, the discovery and characterization of a resident pool of cardiac stem cells (CSCs) has led to a surge of preclinical research and the recent completion of two promising phase I trials in adults with ischemic cardiomyopathy [2, 3]. Specifically, c-kit+ CSCs have been shown to be multipotent, clonogenic, and self-renewing in in vitro and in vivo regenerative assays, and are characterized by a phenotype negative for the hematopoietic and endothelial markers CD45, CD34, and CD31 [2, 4]. The cellular and molecular processes underlying these beneficial effects have yet to be defined, but are thought to be driven by paracrine-mediated signaling, epigenetic modifications, and interaction with endogenous cardiac- or bone marrow–derived progenitor cells [5]. As mechanistic studies evolve, however, a number of important translational questions remain regarding the role of resident CSCs in the developing myocardium of pediatric patients.
We and others have previously shown the enhanced regenerative capacity of CSCs derived from pediatric patients compared with adults, and also the increased number of resident CSCs present in neonatal and infant myocardium when compared with that from older children, adolescents, and adults [6–8]. However, the relative role of endogenous CSCs in the setting of pediatric end-stage heart failure (ESHF) has not been extensively studied, nor has the effect of ESHF on stem cell functionality. Recently, the correlation between a more robust telomere-telomerase axis and population doubling time of the CSCs was found to be highly predictive of negative left ventricular remodeling after coronary artery bypass grafting and improved ventricular function [9]. These results imply the importance of maintaining a strong telomere-telomerase axis that will directly influence the functional abilities of the c-kit+ CSCs. Whether a similar association exists between the ESHF myocardium-derived CSCs and their telomere-telomerase axis has yet to be explored, and could provide important insights into the functionality of ESHF-derived c-kit+ CSCs as a potential cell therapy product for clinical use.
We sought to explore the differences in CSC expression between ESHF myocardium and normal myocardium from pediatric patients with congenital heart disease (CHD). Others have shown that the number of resident CSCs increase by 29- and 14-fold in adults with acute and chronic myocardial infarction, respectively [10], and by 3-fold in the right ventricle of children with pressure-overload single ventricles [11]. Accordingly, we hypothesized that in the setting of ESHF, pediatric patients would demonstrate enhanced expression of endogenous CSCs relative to age-matched CHD patients with normal myocardium. Furthermore, we predicted that c-kit+ CSCs isolated from ESHF patients would exhibit differential growth properties when compared with c-kit+ CSCs isolated from CHD patients with normal myocardium, as evidenced by differences in telomere length and population doubling time.
Patients and Methods
Study Patients
This study was approved by the University of Maryland and Lurie Children’s Hospital, Chicago Institutional Review Board, and parental consent was provided for use of all cardiac tissue biopsy specimens. Tissue specimens were obtained from the right atrial appendage (RAA) of pediatric patients with normal myocardium or ESHF during congenital cardiac surgery or at the time of heart transplantation. End-stage heart failure was defined as New York Heart Association class III or IV and ejection fraction less than 0.20. Right atrial appendage samples were also obtained from age-matched patients with normal myocardium and no clinical evidence of heart failure undergoing routine congenital cardiac surgery for CHD.
Generation of Cardiac Stem Cells
C-kit+ CSCs were isolated from RAA samples as previously described [3]. Briefly, samples were minced in Ham’s F12 medium (cat. no. 12-615F; Lonza Group Ltd, Basel, Switzerland) and digested with 1 to 2 mg/mL of collagenase type II (cat. no. 4177; Worthington Biochemical Corp, Lakewood, NJ). Samples were then incubated on a shaker for 30 to 45 minutes at 37ºC. After collagenase treatment, cells were washed twice with growth medium (Ham’s F12, 10% fetal bovine serum, 10 ng/mL recombinant human fibroblast growth factor-basic, 0.2 mmol/L L-glutathione, and 5 U/mL human erythropoietin 250) before being plated. At subconfluency, cells were trypsinized and sorted for c-kit+ with microbeads (CD117 MicroBead Kit human cat. no. 130-091-332; Miltenyi Biotec, Bergisch Gladbach, Germany) as per kit instructions. The c-kit+ cells were collected and cultured in c-kit growth media.
Characterization of c-kit+ Cardiac Stem Cells
Cardiac stem cells at P3 to P5 were evaluated by flow cytometry using a Becton-Dickinson FACS caliber (San Jose, CA) with 10,000 events collected. Cells were incubated with fluorochrome-conjugated primary antibodies against c-kit, the hematopoietic lineage surface markers (CD34 and CD45), the mesenchymal stromal lineage markers (CD90 and CD105), and the endothelial cell surface marker (CD31). Isotype controls were run for each immunologic subtype.
RNA Isolation and Real Time Polymerase Chain Reaction Analysis
Total RNA was isolated from ESHF and CHD RAA tissues using RNeasy Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. Five hundred nano-grams of RNA per reaction was reverse-transcribed using a cDNA synthesis kit according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA). Five nanograms of cDNA was used for each sample in a 20-μL polymerase chain reaction. Each reaction was performed using an ABI Fast SYBR Green reaction mix on a StepOnePlus (Applied Biosystems) polymerase chain reaction machine. The following Quantitect primer assays for each primer set were obtained from Qiagen: interleukin 8 (cat. no. QT0000032); angiopoietin 2 (cat. no. QT00100947); vascular endothelial growth factor (cat. no. QT01682072); stromal-derived factor 1 (cat. no. QT00087591); fibroblast growth factor (cat. no. QT00047579); insulin growth factor 1 (cat. no. QT00029785); oxidative stress-induced growth inhibitor (cat. no. QT00092155); hypoxia-inducible factor 1 (cat. no. QT00083664); and platelet-derived growth factor (cat. no. QT00001260). The CT values of the housekeeping gene (glyceraldehyde 3-phosphate dehydrogenase) were subtracted from the correspondent gene of interest (ΔCT). Relative RNA abundance was calculated using the following equation: 2−ΔΔCT. The final values were averaged, and results are presented as fold expression with the standard error.
Telomere Length
Telomere length was determined using flow-fluorescent in situ hybridization using a fluorescein-conjugated peptide nucleic acid probe (Dako, Carpinteria, CA) according to kit instructions (peptide nucleic acid is a synthetic analog of DNA with highly sensitive and specific binding to human DNA). Telomere length was calculated as the ratio between the telomere signal of each sample relative to a control cell line (1301) with a known telomere length (30 kbp).
Population Doubling Time
End-stage heart failure– and CHD-derived c-kit+ CSCs were seeded at 5.0 × 104 cells per well in Costar 6-well plates (Sigma, St. Louis, MO). Cells were trypsinized and manually counted at 24, 36, 48, and 72 hours using a hemocytometer. The number of cells present during the exponential phase of the growth curve was transformed to a logarithmic scale to generate a linear relationship between cell number and hours in culture. Population doubling time was then computed by linear regression of the log2 values of cell number.
Immunohistochemistry
Heart biopsy specimens were fixed in 4% para-formaldehyde and then cryopreserved using 30% sucrose and embedded in OCT (Sakura Finetek, Zoeterwoude, The Netherlands). Sections were cut to 7 μm, using a commercial cryostat, and stained for sarcomeric-α-actin (Santa Cruz Biotechnology, Santa Cruz, CA), transcription factor GATA-4 (Santa Cruz Biotechnology), or c-kit+ (BD Biosciences, Franklin Lakes, NJ) primary antibodies. For islet-1 (ISL-1) staining, tissue sections were permeabilized, incubated overnight with ISL-1 antibody (R&D Systems, Minneapolis, MN), and incubated with fluorescein isothiocyanate–labeled secondary antibody. Tissue sections were counterstained with nuclear 4,′6-diamidino-2-phenylindole stain (Sigma).
Statistical Analysis
Data were analyzed using GraphPad Prism 5 software (La Jolla, CA). Comparisons between two groups were made using an unpaired, two-tailed Student’s t test or Mann-Whitney U test, when appropriate. Probability values of less than 0.05 were considered significant. Data are presented as fold change between groups or mean and standard error of the mean.
Results
Reversal to Fetal Gene Program in Pediatric End-Stage Heart Failure Patients
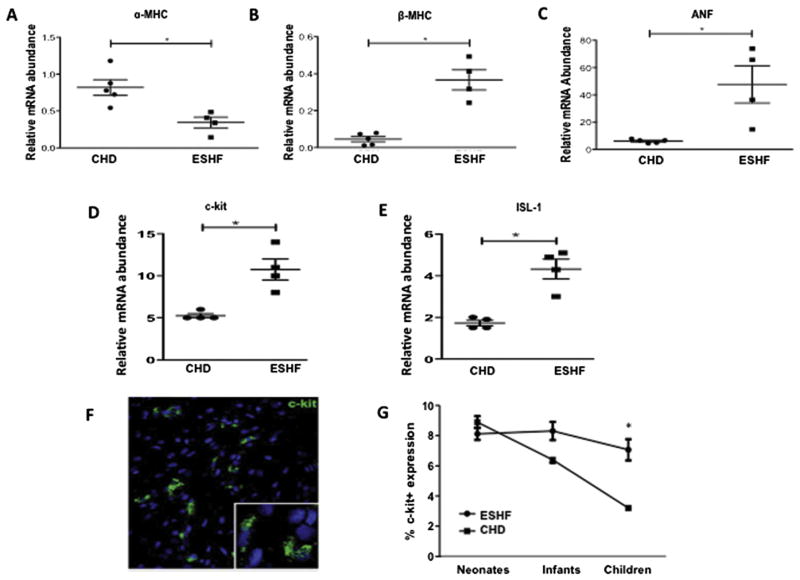
All ESHF samples were obtained from pediatric patients with New York Heart Association class III or IV and ventricular ejection fraction less than 0.20. The origins of the ESHF in pediatric patients were mostly idiopathic or dilated cardiomyopathy, whereas in CHD patients, all had a diagnosis of ventricular septal defect and no clinical evidence of heart failure (New York Heart Association class III or IV and ejection fraction <0.20; Table 1). Because previous studies demonstrated ESHF myocardium reverses to a fetal expression pattern of contractile and metabolic genes, we verified that at the biochemical level the ESHF myocardium had converted to the fetal gene expression [12]. When compared with age-matched control patients, ESHF myocardium had a reduction of α-myosin heavy chain mRNA expression by 2.4-fold (p = 0.015) and enhanced expression of β-myosin heavy chain by 7.5-fold (p = 0.015). Additionally, atrial natriuretic factor expression was upregulated by 8-fold in ESHF myocardium (p = 0.015; Figs 1A–1C). These results confirmed that all the study patients truly had ESHF, both at a clinical and biochemical level, and the CHD patients had no biochemical evidence of heart failure.
Table 1.
Patient Characteristics
| Patient | Age | ESHF Diagnosis | CHD Diagnosis |
|---|---|---|---|
| 1 | 3 mo | Congenital cardiomyopathy (HLHS) | VSD |
| 2 | 5 mo | Congenital cardiomyopathy (DORV, UAVSD) | VSD |
| 3 | 8 mo | Congenital cardiomyopathy (HLHS) | VSD |
| 4 | 12.5 mo | Dilated cardiomyopathy | VSD |
| 5 | 1 y | Congenital cardiomyopathy (HLHS) | VSD |
| 6 | 13 mo | Dilated cardiomyopathy | VSD |
| 7 | 2 y | Dilated cardiomyopathy | VSD |
| 8 | 7 y | Myocarditisa | VSD |
| 9 | 9 y | Dilated cardiomyopathy | VSD |
| 10 | 10 y | Dilated cardiomyopathy | VSD |
| 11 | 11 y | Dilated cardiomyopathy | VSD |
| 12 | 12 y | Idiopathic cardiomyopathy | VSD |
| 13 | 12 y | Dilated cardiomyopathy | VSD |
| 14 | 13 y | Dilated cardiomyopathy | VSD |
| 15 | 14 y | Dilated cardiomyopathy | VSD |
Decompensated heart failure ensued after recovery from acute myocarditis.
CHD = congenital heart disease; DORV = double-outlet right ventricle; ESHF = end-stage heart failure; HLHS = hypoplastic left heart syndrome; UAVSD = unbalanced atrioventricular septal defect; VSD = ventricular septal defect.
Fig 1.
End-stage heart failure (ESHF) myocardium switches back to a fetal gene program and increases expression of endogenous c-kit+ cells. Quantitative real-time polymerase chain reaction from ESHF myocardium demonstrated decreased expression of (A) α-myosin heavy chain (α-MHC), increased expression of (B) β-myosin heavy chain (β-MHC), and (C) atrial natriuretic factor (ANF) compared with CHD myocardium. Quantitative real-time polymerase chain reaction for (D) c-kit suggests a 2-fold increase in the right atrium of ESHF as compared with CHD, and (E) a 2.5-fold increase of islet-1 (ISL-1) mRNA expression. (F) Confocal microscopy image showing c-kit+ cells (white arrow) from a patient with normal myocardium (magnification, ×40). (G) C-kit expression in myocardium from ESHF patients is age-independent and shows high expression with increasing age, in contrast to normal myocardium from CHD patients. *p < 0.05.
Age-Independent Number of Cardiac Stem Cells in End-Stage Heart Failure Myocardium
We studied the expression of two well-established markers of CSCs: the tyrosine kinase receptor, c-kit, and ISL-1, a transcription factor that marks undifferentiated cells in the secondary heart field [13]. Compared with normal myocardium, the RAA of ESHF hearts demonstrated a 2-fold increase in c-kit (p = 0.03; Fig 1D) and a 2.5-fold increase in ISL-1 (p = 0.03; Fig 1E). A comparison of ESHF versus CHD myocardium among age-matched samples from infancy through childhood demonstrated c-kit+ cells were upregulated in ESHF patients when compared with cells from CHD patients despite advancing age (6.9% ± 1.0% versus 3.1% ± 0.8% for children; p < 0.02; Fig 1G).
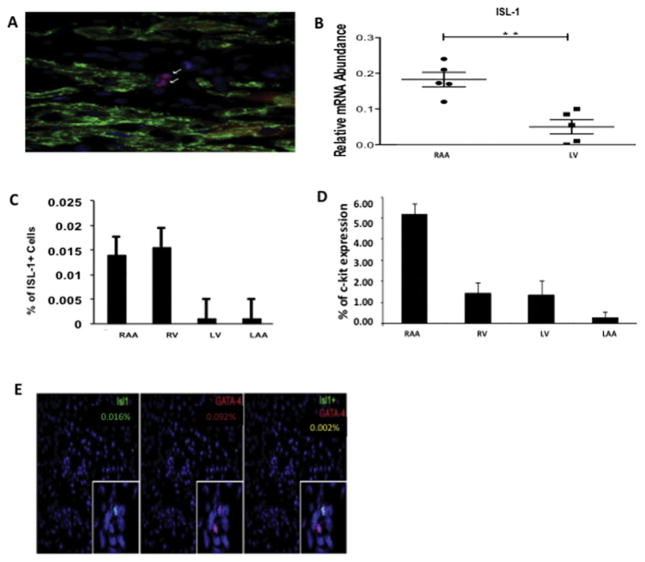
Explanted ESHF hearts revealed increased expression of ISL-1 in right-side cardiac structures relative to left-side structures, including a fourfold increase in ISL-1 mRNA levels in the RAA compared with the left ventricle as determined by quantitative real time–polymerase chain reaction (p = 0.008; Figs 2B, 2C). This expression pattern is consistent with previously reported differential levels of ISL-1 in the secondary heart field, responsible for development of right-side cardiac structures [13, 14]. This trend was also observed in the percentage of cells expressing c-kit, which was highest in the RAA, followed by the right ventricle, the left ventricle, and the left atrial appendage (Fig 2D). The proportion of ISL-1+/GATA-4+ cells was low, present in only 0.20% of ISL-1+ cells (Fig 2E).
Fig 2.
Expression profile of cardiac progenitor markers from explanted end-stage heart failure (ESHF) hearts. (A) Confocal microscopy image of islet-1 (ISL-1) cells within ESHF myocardium (magnification, × 40). Analysis of cardiac chambers from explanted ESHF revealed (B) a fourfold increase in ISL-1 mRNA levels in the right atrial appendage (RAA) compared with the left ventricle (LV) as determined by quantitative real-time polymerase chain reaction, and (C) increased expression of ISL-1 in right-side heart chambers relative to left. (D) Increased expression of c-kit in the right atrial appendage relative to other structures in ESHF hearts. (E) Representative images of RV myocardium from ESHF hearts showing coexpression of ISL-1 with the cardiac lineage marker GATA-4 was only present in 0.002% of ISL-1+ cells. *p< 0.05. (LAA = left atrial appendage.)
Characterization of c-kit+ Cardiac Stem Cells
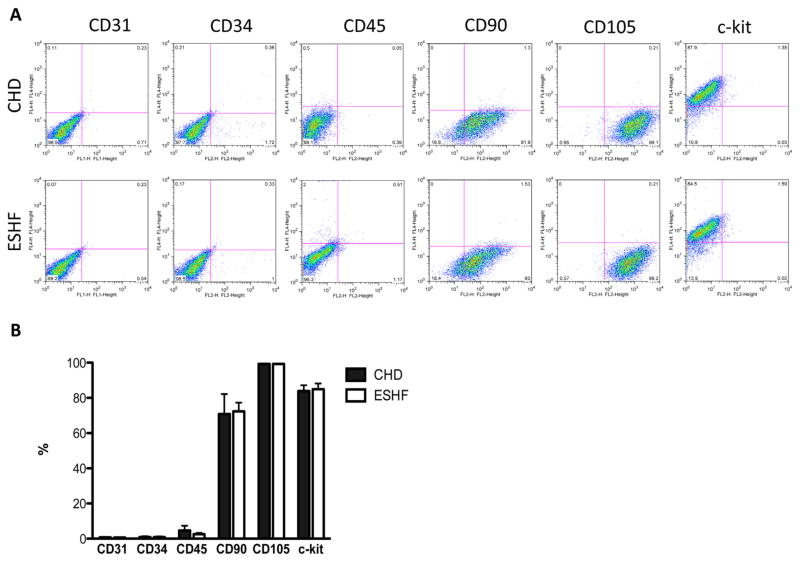
The two different types of c-kit+ CSCs derived from CHD or ESHF myocardium had the same cellular morphology and isolation characteristics (Fig 3A). These results indicated that the generation and expansion of the c-kit+ CSCs was independent of patient diagnosis. Next, c-kit+ CSCs were examined for their expression of stemness markers and cardiac lineage commitment markers to determine whether the expanded c-kit+ CSCs remained in an uncommitted state or a differentiated state. Characterization by fluorescence-activated cell sorting revealed 84.5% ± 2.1% of isolated cells was positive for c-kit+ (Fig 3B). A low percentage of cells expressed the hematopoietic markers, CD34 and CD45, and no isolated cells were positive for the endothelial surface marker, CD31. In addition to c-kit+ expression, isolated cells were found to express the mesenchymal stromal cell markers, CD90 and CD105. A comparison of c-kit+ CSCs from CHD versus ESHF patients revealed no difference in the expression profile of these surface markers.
Fig 3.
Surface marker expression of end-stage heart failure (ESHF) and congenital heart disease (CHD) -derived cardiac stem cells. (A) Representative dot plot of fluorescence-activated cell sorting analysis of antigenic phenotype of ESHF- and CHD-derived cardiac stem cells. (B) Expression of the stem cell marker c-kit expression was not found to be different in either population. Both kinds of cardiac stem cells were found to have similar expression of endothelial (CD31), hematopoietic (CD34, CD45), and mesenchymal (CD90, CD105) markers.
Growth Properties of c-kit+ Cardiac Stem Cells
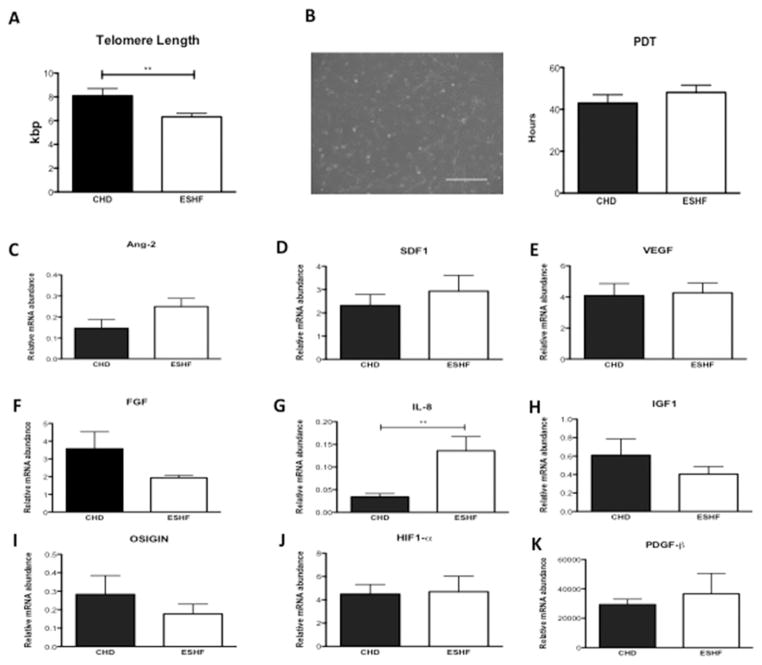
We observed a significantly reduced telomere length in c-kit+ CSCs isolated from ESHF myocardium compared with those from CHD myocardium (p = 0.015; Fig 4A). The growth reserve of c-kit+ CSCs from each of the groups was assessed by analyzing population doubling times. There was a slight increase in the population doubling time of ESHF-derived CSCs compared with CHD-derived CSCs; however, this difference did not reach statistical significance (Fig 4B).
Fig 4.
Growth properties of end-stage heart failure (ESHF) versus congenital heart disease (CHD)-derived c-kit+ cardiac stem cells and in vivo paracrine factor expression. (A) Comparison of telomere length in c-kit+ cardiac stem cells isolated from CHD versus ESHF myocardium. (B) Representative phase-contrast microscopy image of c-kit+ cardiac stem cell morphology with quantification of population doubling time. (C–K) In vivo comparison of growth factors (CHD, n = 5 versus ESHF, n = 5 for each): (C) angiopoietin 2 (Ang-2), (D) stromal-derived factor 1 (SDF-1), (E) vascular endothelial growth factor (VEGF), (F) fibroblast growth factor (FGF), (G) interleukin 8 (IL-8), (H) insulin growth factor 1 (IGF-1), (I) oxidative stress-induced growth inhibitor 1 (OSIGIN), (J) hypoxia-inducible factor 1 (HIF-1α), and (K) platelet-derived growth factor (PDGF-β). **p < 0.01.
Myocardial Expression of Growth Factors
The relative mRNA level of the interleukin 8 was significantly more abundant in ESHF myocardium (p = 0.008), and there was a trend toward significance for the angiogenic cytokine, angiopoietin 2 (p = 0.09). However, the expression of vascular endothelial growth factor, stromal-derived factor 1, fibroblast growth factor, insulin growth factor 1, oxidative stress-induced growth inhibitor, hypoxia-inducible factor 1, and platelet-derived growth factor were comparable between both groups of myocardium (Figs 4C–4K).
Comment
Although CSC-based therapies have made rapid progress toward clinical use, a number of key translational questions remain undetermined. The role of resident CSCs within the developing myocardium, as well as a clear understanding of CSC characteristics that correlate with optimal cell function, remains an active area of investigation. We show that the ESHF myocardium not only reverses to a fetal gene program but also to a developmental program by augmenting the number of CSCs. Further, we demonstrate that in contrast to our previous work with healthy pediatric myocardium, the number of CSCs present in the ESHF myocardium does not exhibit an age-dependent decline, but rather remains increased relative to healthy age-matched controls throughout childhood. We also provide the first evidence to our knowledge that CSCs isolated from ESHF myocardium have a reduced telomere length compared with CHD-derived CSCs, suggesting these cells may have a diminished functional capacity.
It has previously been shown that chronologic age is associated with shortened telomere length and diminished telomerase activity in CSCs [15]. Furthermore, it was recently shown that a weakened growth reserve in CSCs isolated from patients undergoing coronary artery bypass grafting was highly predictive of negative left ventricular remodeling at 12 months after the operation, despite equivalent baseline and postoperative patient characteristics [9]. In the present study, we exploited the telomere-telomerase axis of c-kit+ CSCs to explore the replicative history of CSCs derived from ESHF patients versus those from CHD patients and to learn more about their functional potential as a cell therapy candidate [16]. Indeed, the reduced telomere length in ESHF-derived CSCs suggests these cells have undergone an enhanced number of cell divisions, a finding consistent with the increased number of CSCs we observed in failing myocardium. However, although these findings imply that ESHF-derived CSCs may have impaired functionality as a cell therapy product intended to undergo ex vivo expansion in culture, future studies are needed to validate the functional capacity of ESHF-derived CSCs in other regenerative assays, including the myocardial infarction rodent model. Furthermore, these studies suggest that an autologous-based c-kit+ CSC trial in ESHF patients may not be ideal, but alternatively, an allogeneic cell preparation of screened, highly functional CSCs may be the ideal candidate as an off-the-shelf therapy available to heart failure patients.
In this work, we compared the number of resident CSCs identified in right atrial samples from ESHF and CHD patients. Lack of a similar comparison for other chambers was a limitation of this work, and the right atrium was chosen because RAA biopsy specimens were uniformly available in CHD patients undergoing routine congenital cardiac surgical procedures, whereas obtaining biopsy tissue from other chambers was not warranted in healthy patients. Thus, the difference between the two groups could be an underestimate as the actual number of cardiac progenitor cells may be even more increased in the failing ventricle. In a population of patients with pressure-overloaded single right ventricles, for example, Rupp and colleagues [11] identified a threefold increase in the number of resident c-kit+/mast cell tryptase−/CD45− resident CSCs compared with the number of CSCs in the right ventricle of pediatric patients with dilated cardiomyopathy. Future studies are aimed to examine the signaling mechanisms by which endogenous CSCs are activated and recruited to areas of myocardial injury.
Although the amplified number of CSCs is clearly not adequate to reverse ventricular dysfunction and prevent heart failure, an expanded population of CSCs may contribute to repair mechanisms that prevent more advanced deterioration. In this work, we explored the possibility that the elevated presence of CSCs within ESHF myocardium may correlate with increased levels of secreted paracrine factors. Previously, we demonstrated with in vitro assays of conditioned cell media that neonatal CSCs are capable of potent secretion of angiogenic cytokines [8]. However, in vivo concentrations of these factors have not been investigated in the context of a relationship to the presence of CSCs. Our results yielded an equivalent expression of cytokine mRNA between normal and ESHF myocardium, except for the angiogenic cytokine, angiopoietin 2, which trended toward significantly elevated levels of mRNA in ESHF hearts, and the proinflammatory cytokine, interleukin 8. It is important to note that although we observed a greater expression of the stem cell markers c-kit and ISL-1 in ESHF myocardium, regenerative assays are necessary to verify whether these cells indeed have regenerative capabilities.
In summary, we present evidence for the increased presence of CSCs in pediatric patients with ESHF. We provide initial evidence for the growth properties of CSCs derived from pediatric ESHF patients, which suggests that decompensated heart failure may contain CSCs with poor functional capabilities. Future studies will aim to further characterize the functionality of resident CSCs derived from pediatric patients and to correlate these findings with clinically significant changes in ventricular performance.
Acknowledgments
This work was funded by a grant from the National Institutes of Health (NHLBI R01 HL118491).
Footnotes
Presented at the Basic Science Forum of the Sixty-first Annual Meeting of the Southern Thoracic Surgical Association, Tucson, AZ, Nov 5–8, 2014.
References
- 1.Rossano JW, Shaddy RE. Heart failure in children: etiology and treatment. J Pediatr. 2014;165:228–33. doi: 10.1016/j.jpeds.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 2.Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanganalmath SK, Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ Res. 2013;113:810–34. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dey D, Han L, Bauer M, et al. Dissecting the molecular relationship among various cardiogenic progenitor cells. Circ Res. 2013;112:1253–62. doi: 10.1161/CIRCRESAHA.112.300779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnecchi M, Zhang Z, Ni A, Dzau VJ. Paracrine mechanisms in adult stem cell signaling and therapy. Circ Res. 2008;103:1204–19. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amir G, Ma X, Reddy VM, et al. Dynamics of human myocardial progenitor cell populations in the neonatal period. Ann Thorac Surg. 2008;86:1311–9. doi: 10.1016/j.athoracsur.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 7.Mishra R, Vijayan K, Colletti EJ, et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011;123:364–73. doi: 10.1161/CIRCULATIONAHA.110.971622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson DL, Mishra R, Sharma S, Goh SK, Deshmukh S, Kaushal S. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 2012;126(11 Suppl 1):S46–53. doi: 10.1161/CIRCULATIONAHA.111.084699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Amario D, Leone AM, Iaconelli A, et al. Growth properties of cardiac stem cells are a novel biomarker of patients’ outcome after coronary bypass surgery. Circulation. 2014;129:157–72. doi: 10.1161/CIRCULATIONAHA.113.006591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Urbanek K, Torella D, Sheikh F, et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci USA. 2005;102:8692–7. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupp S, Bauer J, von Gerlach S, et al. Pressure overload leads to an increase of cardiac resident stem cells. Basic Res Cardiol. 2012;107:252. doi: 10.1007/s00395-012-0252-x. [DOI] [PubMed] [Google Scholar]
- 12.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–31. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 13.Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104:933–42. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- 14.Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;138:2389–98. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cesselli D, Beltrami AP, D’Aurizio F, et al. Effects of age and heart failure on human cardiac stem cell function. Am J Pathol. 2011;179:349–66. doi: 10.1016/j.ajpath.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiyama E, Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96:1020–4. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]