SUMMARY
Histone variant H2A.Z occupies the promoters of active and poised, bivalent genes in ESCs to regulate developmental programs, yet how it contributes to these contrasting states is poorly understood. Here, we investigate the function of H2A.Z.1 mono-ubiquitylation (H2A.Z.1ub) by mutation of the PRC1 target residues (H2A.Z.1K3R3). We show that H2A.Z.1K3R3 is properly incorporated at target promoters in murine ESCs (mESCs), however, loss of mono-ubiquitylation leads to de-repression of bivalent genes, loss of Polycomb binding, and to faulty lineage commitment. Using quantitative proteomics, we find that tandem bromodomain proteins, including the BET family member Brd2, are enriched in H2A.Z.1 chromatin. We further show that Brd2 is gained at de-repressed promoters in H2A.Z.1K3R3 mESCs whereas Brd2 inhibition restores gene silencing at these sites. Together, our study reveals an antagonistic relationship between H2A.Z.1ub and Brd2 to regulate the transcriptional balance at bivalent genes to enable proper execution of developmental programs.
Keywords: H2A.Z, Polycomb, BRD2, chromatin, embryonic stem cells
INTRODUCTION
Pluripotent cells must translate signaling cues into rapid transcriptional responses during development to specify the many cell types in the adult. The histone H2A variant H2A.Z has essential, but poorly understood roles in early metazoan development (Faast et al., 2001; van Daal and Elgin, 1992) and murine embryonic stem cell (mESC) differentiation (Creyghton et al., 2008; Hu et al., 2013). In mESCs, H2A.Z is enriched at H3K4me3-marked promoters of active genes and at silent, but poised bivalent promoters marked by both H3K4me3 and H3K27me3 (Creyghton et al., 2008; Hu et al., 2013; Ku et al., 2012; Subramanian et al., 2013). Bivalent genes, which encode the majority of developmental regulators in mESCs, are silent yet maintain the capacity to be activated during differentiation (Surface et al., 2010; Voigt et al., 2013). These data suggest that H2A.Z acts as a molecular rheostat for transcriptional output (Subramanian et al., 2015), however, we currently lack a mechanistic understanding of how H2A.Z regulates the balance between gene activation and repression.
Post-translational modification (PTM) of histones is important for regulation of gene expression, and can mark functional regions of chromatin (Zhou et al., 2011). H2A.Z is subject to similar PTMs as canonical histones including acetylation and ubiquitylation. For example, H2A.Z amino-terminal acetylation correlates with gene activation (Bruce et al., 2005; Hu et al., 2013; Ku et al., 2012). Moreover, H2A.Z carboxy-terminal lysine residues (K120, K121, and K125) can be mono-ubiquitylated by the Polycomb Repressive Complex 1 (PRC1) (Draker et al., 2011; Ku et al., 2012; Sarcinella et al., 2007), however, the role of this H2A.Z modification is not known. Prior work suggested that PRC1-mediated H2A ubiquitylation (H2Aub) is important for silencing bivalent genes in mESCs (de Napoles et al., 2004; Endoh et al., 2012), possibly through pausing of RNA Polymerase II (RNAPII) at promoters (Stock et al., 2007). More recently, non-canonical PRC1 that includes distinct subunits such as RYBP and Kdm2b/Fbxl10 was shown to catalyze H2Aub, which in turn recruits and stimulates PRC2 activity, reinforcing gene repression (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). Given that PRC1 also modifies H2A.Z, and that H2A fails to compensate for loss of H2A.Z during early development (Creyghton et al., 2008; Faast et al., 2001; Hu et al., 2013), we hypothesized that H2A.Zub also plays important roles in regulating transcriptional outcomes in mESCs.
H2A.Z facilitates interactions with specific chromatin-associated proteins to affect gene expression in a context dependent manner (Draker et al., 2012; Fujimoto et al., 2012; Li et al., 2012). Moreover, histone PTMs, including mono-ubiquitylation, often serve as binding platforms for downstream effectors (Braun and Madhani, 2012). Here, we dissect the function of H2A.Z.1 mono-ubiquitylation (H2A.Z.1ub) by generating site-specific mutations in the C-terminal PRC1 target lysine residues (K120R, K121R, K125R; denoted H2A.Z.1K3R3). Using a transgenic mESC system, we show that H2A.Z.1ub is necessary for both proper induction of developmental programs and for the maintenance of bivalent chromatin, including PRC2 recruitment. Furthermore, using a quantitative proteomics approach, we find that Brd2, a member of the BET family of transcriptional activators, is enriched in H2A.Z.1 chromatin. Brd2 enrichment is significantly increased at bivalent genes in H2A.ZK3R3 mESCs coincident with gene activation, and Brd2 inhibition restores silencing of bivalent genes. Collectively, our study suggests that H2A.Z.1ub antagonizes Brd2 to maintain the balance between gene activation and repression in response to developmental cues.
RESULTS
H2A.Z.1ub is dispensable for mESC self-renewal
H2A.Z mono-ubiquitylation (H2A.Zub) is enriched in H3K27me3 containing nucleosomes in mESCs (Ku et al., 2012), and on the inactive X chromosome in human 293T cells (Sarcinella et al., 2007), however, the function of this H2A.Z modification is not known. Using tandem mass spectrometry (MS/MS), we confirmed that Ring1b, a catalytic component of PRC1, mono-ubiquitylates three C-terminal lysines of H2A.Z (K120, K121, and K125) in mESCs (Figures S1A–H). We next investigated whether these lysine residues are subject to other modifications. By analyzing raw mass spectrometry data from mESCs (Ku et al., 2012), we confidently assigned signals to the unmodified and ubiquitylated forms of H2A.Z118–127. In contrast, we were unable to reliably detect acetylation or methylation of these sites (Figures S1G–F and Table S1). We also find that H2A.Zub levels decrease upon mESC differentiation (Figure S1I), coincident with decreased co-localization of H2A.Z and Polycomb complexes at target genes (Creyghton et al., 2008; Ku et al., 2012).
We focused further analysis on the H2A.Z.1 isoform because it is 20-fold more abundant than H2A.Z.2 in mESCs, and it is essential for early development (Faast et al., 2001; Subramanian et al., 2013). To this end, mESC lines were generated that harbor either a doxycycline-inducible H2A.Z.1 transgene with a C-terminal YFP fusion (denoted H2A.Z.1WT) or an H2A.Z.1-YFP transgene with the three lysine residues mutated to arginine (K120R, K121R, K125R; denoted H2A.Z.1K3R3) (Figure 1A and S2A). These mutations resulted in near complete loss of H2A.Z mono-ubiquitylation in mESCs as determined by immunoblot of histone extracts (Figure 1B and S2A). We next engineered our H2A.Z.1WT and H2A.Z.1K3R3 transgenic mESC lines to constitutively express a short hairpin targeting endogenous H2A.Z.1, resulting in ~90% depletion of its mRNA and protein levels with minimal effect on transgene levels (Figures S2A–C). Similar results were observed with an independent hairpin (Figure S2B). Expression of the RNAi-resistant H2A.Z.1WT-YFP transgene fully rescues H2A.Z.1 depletion (Subramanian et al., 2013), establishing a system for directly investigating the role of H2A.Z.1ub. Similar to induction of H2A.Z.1WT, we found that expression of H2A.Z.1K3R3 in H2A.Z.1-depleted mESCs did not appear to affect morphology, levels of pluripotency markers, cell cycle dynamics, or response to DNA damaging agents (Figures S2D–G). These data suggest that H2A.Z.1ub is largely dispensable for mESC self-renewal and may play critical roles in early lineage commitment.
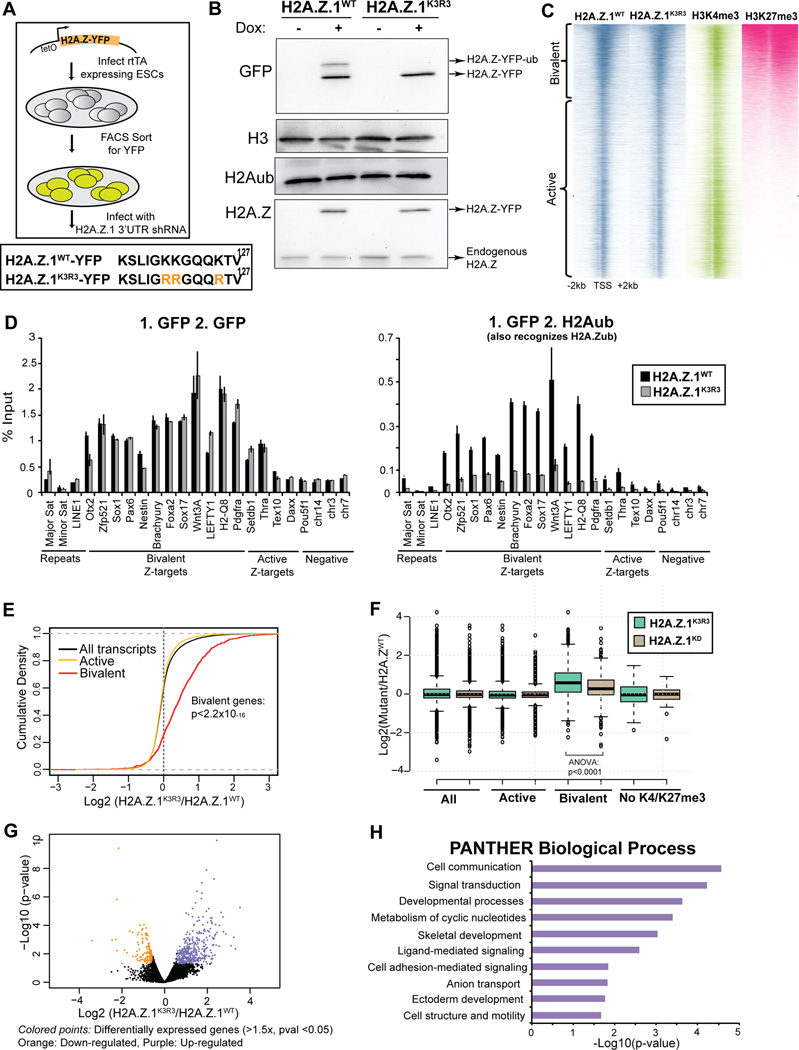
Figure 1. H2A.Z.1ub regulates bivalent genes in mESCs.
A. mESCs expressing rtTA were infected with lentiviral particles with H2A.Z.1K3R3 and H2A.Z.1WT transgenes harboring a C-terminal YFP, under the control of a tetOn promoter. Lower panel shows the modified C-terminal lysine H2A.Z.1 residues. Upon addition of doxycycline, YFP+ cells were sorted and infected with a hairpin against the 3’UTR of endogenous H2A.Z.1. B. Immunoblots of H2A.Z.1WT and H2A.Z.1K3R3 mESC histone extracts were probed with GFP, H3, H2Aub and H2A.Z antibodies. C. Heatmap of ChIP-seq in H2A.Z.1WT and H2A.Z.1K3R3 mESCs displays similar incorporation at active and bivalent target genes. H3K4me3 and H3K27me3 (Wamstad et al., 2012) are included for comparison. Heatmaps are centered on transcriptional start sites (TSSs) of all genes ranked by H2K27me3 and extend +/− 2kb. Bivalent gene classification is from Subramanian et al., 2013. D. Sequential ChIP using GFP followed by GFP or H2Aub (also recognizes H2A.Zub) antibodies. % Input is calculated using 2(Cp(Input)-Cp(ChIP))))*(%of total input). Error bars represent standard deviation of triplicate reactions. E. Distribution of the log2 fold change in expression in H2A.Z.1K3R3 mESCs was plotted using cumulative density function of log2(H2A.Z.1K3R3/H2A.Z.1WT). Black trace represents all genes with RPKM of at least 1 in any sample and 5 unique reads in each sample. Active (yellow trace) and bivalent (red trace) gene classes that pass threshold were plotted independently. F. Box plots represent the log2 fold change in expression of either H2A.Z.1K3R3 or H2A.Z.1KD mESCs relative to H2A.Z.1WT mESCs at all (13857 genes), active (11010), bivalent (1611) or K4me3−/K27me3− (61) genes. Classification of bivalent and active genes are from Subramanian (2013). Center line represents median value; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. G. Volcano plot of all genes that passed detection threshold were plotted by p-value vs. change in expression. H. Top 10 enriched categories using PANTHER Biological Process database are displayed for up-regulated genes compared to all genes. See also Figure S1–2.
H2A.Z.1ub is enriched at bivalent promoters
H2A.Z is enriched at discrete genomic sites in mESCs including the majority of H3K4me3-marked TSSs as well as a small fraction of distal enhancers (Hu et al., 2013; Ku et al., 2012; Subramanian et al., 2013). Our prior work showed that H2A.Z.1-YFP incorporation is highly similar to endogenous H2A.Z as determined by ChIP-seq (Subramanian et al., 2013). Thus, we asked whether loss of H2A.Z.1 mono-ubiquitylation affected its chromatin incorporation. We found that H2A.Z.1WT and H2A.Z.1K3R3 are similarly enriched in chromatin by fractionation of mESCs lysates (Figure S2H). We also show that H2A.Z.1WT and H2A.Z.1K3R3 nucleosomes contain overall similar global histone modification patterns (Figure S2I). In addition, we found that H2A.Z.1K3R3 displays similar dynamics to H2A.Z.1WT in mESCs by Fluorescence Recovery after Photobleaching (FRAP) (Figure S2J), unlike mutation of the divergent acidic patch which leads to an increase in H2A.Z.1 dynamics (Subramanian et al., 2013). We further demonstrate that conditional ablation of Ring1b did not alter H2A.Z incorporation at target genes (Figure S2K).
We next performed ChIP-seq in H2A.Z.1WT and H2A.Z.1K3R3 mESCs using GFP antibodies. Both wild type and mutant transgenes are similarly enriched at a large set of active (H3K4me3+) and bivalent (H3K4me3+, H3K27me3+) genes (Spearman correlation=0.972; Figure 1C) as well as at a small number of H2A.Z-enriched enhancers (Figure S2L). In total, 13,684 and 13,000 genes show H2A.Z.1 enrichment within 2kb of a TSS in H2A.Z.1WT and H2A.Z.1K3R3 mESCs, respectively. As examples, H2A.Z.1K3R3 occupies both active promoters including Thra as well as bivalent promoters including Nestin and genes associated with the HoxA cluster, in a pattern similar to H2A.Z.1WT (Figure S2M).
Our ChIP-seq data shows that H2A.Z.1K3R3 occupies promoters similar to endogenous H2A.Z.1WT, however, the localization pattern of H2A.Z.1ub is not known due to the lack of specific antibodies that distinguish H2Aub and H2A.Zub (Figure S2I). To address this limitation, we performed sequential ChIP by first enriching for H2A.Z.1 nucleosomes in H2A.Z.1WT and H2A.Z.1K3R3 mESCs using a GFP antibody followed by re-ChIP with an antibody that recognizes H2A/Zub. H2A.Z.1K3R3 mESCs that lack H2A.Z.1ub were used as a negative control. Sequential ChIP demonstrates that H2A.Z.1ub is largely enriched at bivalent promoters and low at active promoters (Figure 1D). As a control, re-ChIP with GFP antibodies shows that both H2A.Z.1WT and H2A.Z.1K3R3 are incorporated similarly (Figure 1D). Collectively, these data suggest loss of H2A.Z.1 mono-ubiquitylation does not affect global H2A.Z.1 incorporation or dynamics in mESCs.
H2A.Z.1ub regulates developmental gene expression programs
Given that the majority of enriched regions map to TSSs, we asked whether loss of H2A.Z.1 mono-ubiquitylation affects gene expression. We profiled the transcriptome of H2A.Z.1K3R3 mESCs by RNA-seq and found that bivalent genes were expressed at higher levels compared to H2A.Z.1WT controls (p<2.2×10−16, Kolmogorov-Smirnov test, two-sided) (Figure 1E). In contrast, levels of active genes were largely unaffected in H2A.Z.1K3R3 mESCs, similar to H2A.Z.1 depletion (Figures 1E and 1F) (Hu et al., 2013; Subramanian et al., 2013). Because bivalent genes are lowly expressed (Mikkelsen et al., 2007), we applied stringent threshold criteria to reduce false positives in our analysis (expression of ≥ 1 RPKM in at least one sample, and at least 5 reads in every sample). A total of 9667 active genes and 816 bivalent genes passed these criteria. Specifically, 370 and 107 of these genes are up- and down-regulated, respectively, in H2A.Z.1K3R3 mESCs using a cutoff of 1.5-fold change and p-value≤0.05 (Figure 1G). The set of 370 up-regulated genes significantly overlap the 816 bivalent genes (p<2.65×10−87, hypergeometric test).
Using the gene annotation tool PANTHER (Huang et al., 2009a; 2009b), we found that the up-regulated genes function in cell communication, signaling, and development (Figure 1H). In contrast, we did not find significant overlap with bivalent genes or enriched gene ontology (GO) terms among the down-regulated genes. Expression changes were validated using a second independent H2A.Z.1 hairpin (Figure S2N). Notably, although H2A.Z.1 depletion also leads to de-repression of bivalent genes (Hu et al., 2013; Subramanian et al., 2013), we observed overall higher expression of these genes in H2A.Z.1K3R3 mESCs (Figure 1F, ANOVA p<0.0001), suggesting that H2A.Z incorporation is critical for gene activation in response to developmental signals.
H2A.Z.1ub is required for proper mESC differentiation
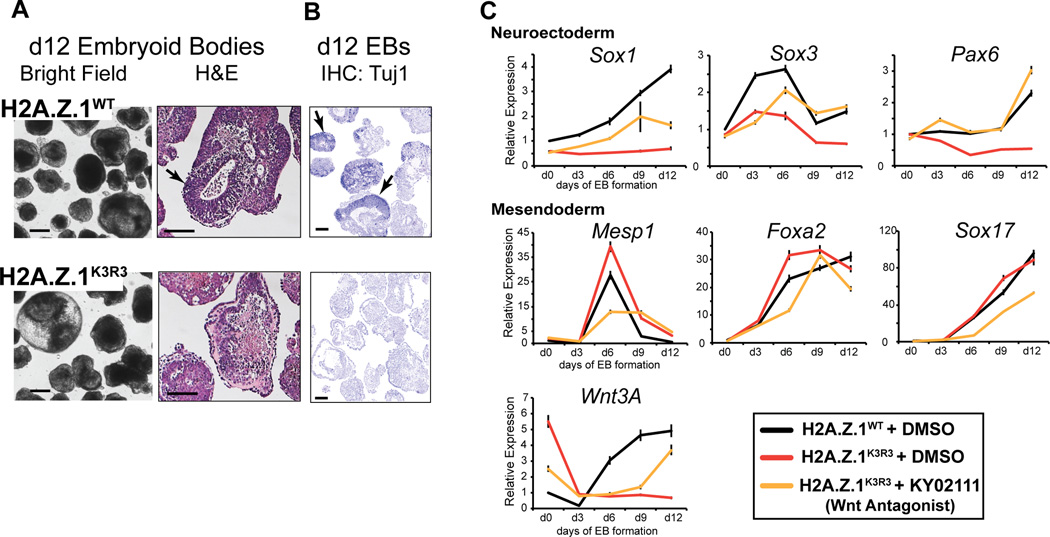
Because the precise regulation of bivalent genes is key for proper lineage commitment (Subramanian et al., 2015), we next examined the differentiation capacity of H2A.Z.1K3R3 mESCs by allowing cells to aggregate into embryoid bodies (EBs), a process that leads to multi-lineage differentiation similar to the gastrulating embryo (Berge et al., 2008). Induction of the H2A.Z.1WT transgene restores proper mESC differentiation as evidenced by appropriate expression of germ layer markers (Figures 2A–C). In contrast, EBs generated from H2A.Z.1K3R3 mESCs failed to undergo multi-lineage differentiation, as evidenced by H&E stained sections displaying distinct differences in tissue representation relative to H2A.Z.1WT (Figure 2A). In particular, H2A.Z.1K3R3 EBs lack neuroepithelial structures and failed to activate the neural marker Tuj1 compared to H2A.Z.1WT EBs (Figure 2B). Additionally, we found that genes involved in neuroectoderm lineages (e.g. Sox1, Sox3, Pax6) are not properly induced in H2A.Z.1K3R3 EBs, whereas mesendodermal genes are highly expressed (Figure 2C and S3A). Hyper-activation of WNT signaling during EB formation promotes cardiac mesoderm and inhibits neuroectodermal differentiation (Berge et al., 2008). We found that WNT signaling genes including Wnt3A and Axin2 were up-regulated in H2A.Z.1K3R3 mESCs (Figures S3B and S3C). H2A.Z.1K3R3 EBs also display increased nuclear β-catenin levels, reduced binding of TCF3 at WNT target genes, and increased activity of a WNT signaling reporter compared to H2A.Z.1WT controls (Figures S3D–F). Furthermore, treatment with the WNT antagonist KY02111 partially rescued their differentiation defects as shown by activation of the neuroectodermal genes Sox3 and Pax6 (Figure 2C). These results suggest that H2A.Z.1ub is necessary for coordinating specific transcriptional outputs in response to developmental cues.
Figure 2. H2A.Z.1ub is necessary for proper lineage commitment.
A. Bright field and H&E staining of day 12 embryoid bodies from H2A.Z.1WT and H2A.Z.1K3R3 mESCs. Arrows indicate examples of neuroectoderm present in H2A.Z.1WT EB sections. B. IHC staining of d12 H2A.Z.1WT and H2A.Z.1K3R3 EB sections with Tuj1, a marker of neural differentiation. Arrows indicate examples of Tuj1 positive regions. Scale bar for all =100um. C. Expression of lineage markers during EB time course indicates lack of neuroectoderm differentiation in H2A.Z.1K3R3 mESCs (red v. black lines). Addition of WNT inhibitor KY02111 to H2A.Z.1K3R3 mESCs 48 hours prior to and during EB formation restores multi-lineage differentiation (yellow lines). Expression is analyzed by qRT-PCR with values normalized to H2A.Z.1WT+DMSO using Tubb5. Error bars represent SD of triplicate reactions. See also Figure S3.
H2A.Z.1ub is necessary for maintenance of bivalent chromatin
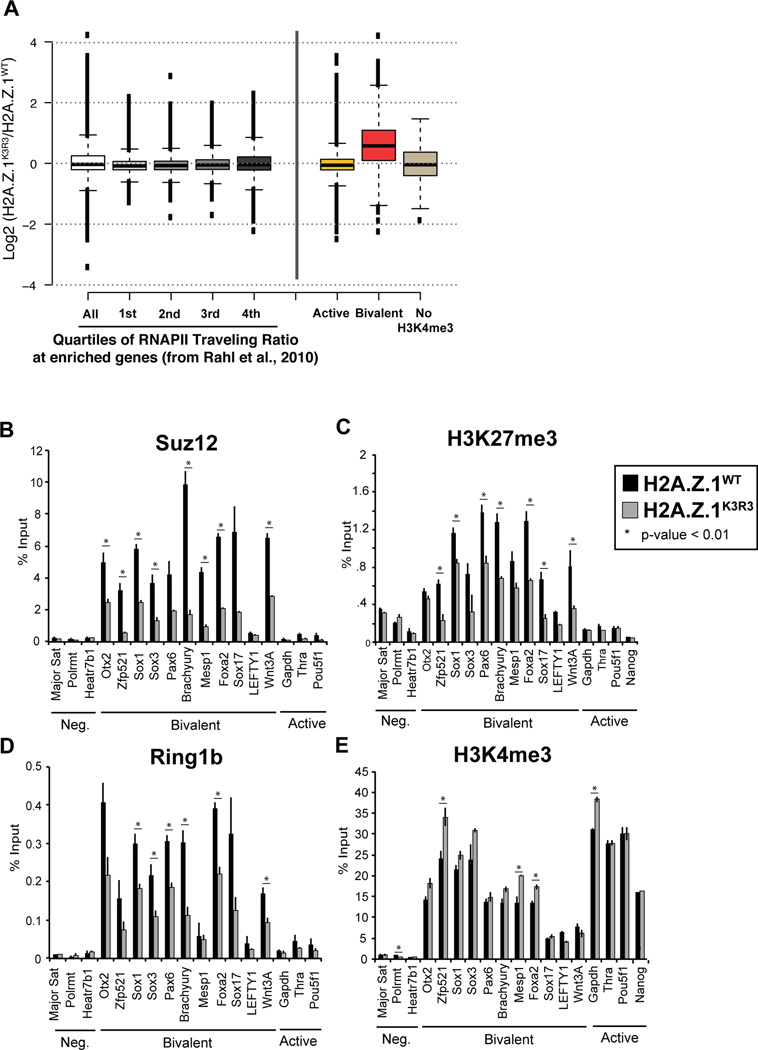
The activity of PRC1 regulates bivalent gene expression potentially through RNAPII pausing (Stock et al., 2007), and/or recruitment of PRC2 (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). However, we found that expression of highly paused genes in mESCs (Rahl et al., 2010) is not significantly affected in H2A.Z.1K3R3 mESCs (Figure 3A), consistent with the markedly lower levels of RNAPII at bivalent promoters compared to active genes (Figure S4A) (Williams et al., 2015). Thus, we next asked whether H2A.Z.1ub, like H2Aub, contributes to maintenance of PRC2 at bivalent genes in mESCs. We performed ChIP-qPCR in H2A.Z.1K3R3 mESCs and observed a significant decrease in the enrichment of the PRC2 component Suz12 at bivalent promoters, as well as its corresponding mark H3K27me3 (Figures 3B and 3C). In addition, PRC1 components including Ring1b as well as Kdm2b, a key factor responsible for recruitment of non-canonical PRC1 to CpG islands (Blackledge et al., 2014; He et al., 2013; Wu et al., 2013) were reduced at bivalent promoters in H2A.Z.1K3R3 mESCs (Figures 3D and S4B). Conversely, loss of mono-ubiquitylation did not affect H3K27ac, H4ac or H2A.Zac levels, and led to a slightly increase in levels of H3K4me3, modifications associated with transcriptional activity (Figures 3E and S4C–E). Together, these results suggest that H2A.Z.1ub plays a key role in maintaining bivalent chromatin and gene silencing in mESCs.
Figure 3. H2A.Z.1ub is necessary for maintenance of bivalent chromatin.
A. Box plots represent the log2 fold change in expression of either H2A.Z.1K3R3 relative to H2A.Z.1WT mESCs at RNAPII-bound genes, split into quartiles of RNAPII traveling ratios (~2459 genes/quartile) from (Rahl et al., 2010), as well as at active (11010 genes) and bivalent (1611) genes. Center lines represent the median value; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. B–E. ChIPs for the PRC2 component, Suz12 (B), H3K27me3 (C), PRC1 component Ring1b (D), H3K4me3 (E), were assayed by qPCR. Error bars represent the SD of triplicate reactions. See also Figure S4.
H2A.Z.1ub interacts with specific proteins in mESCs
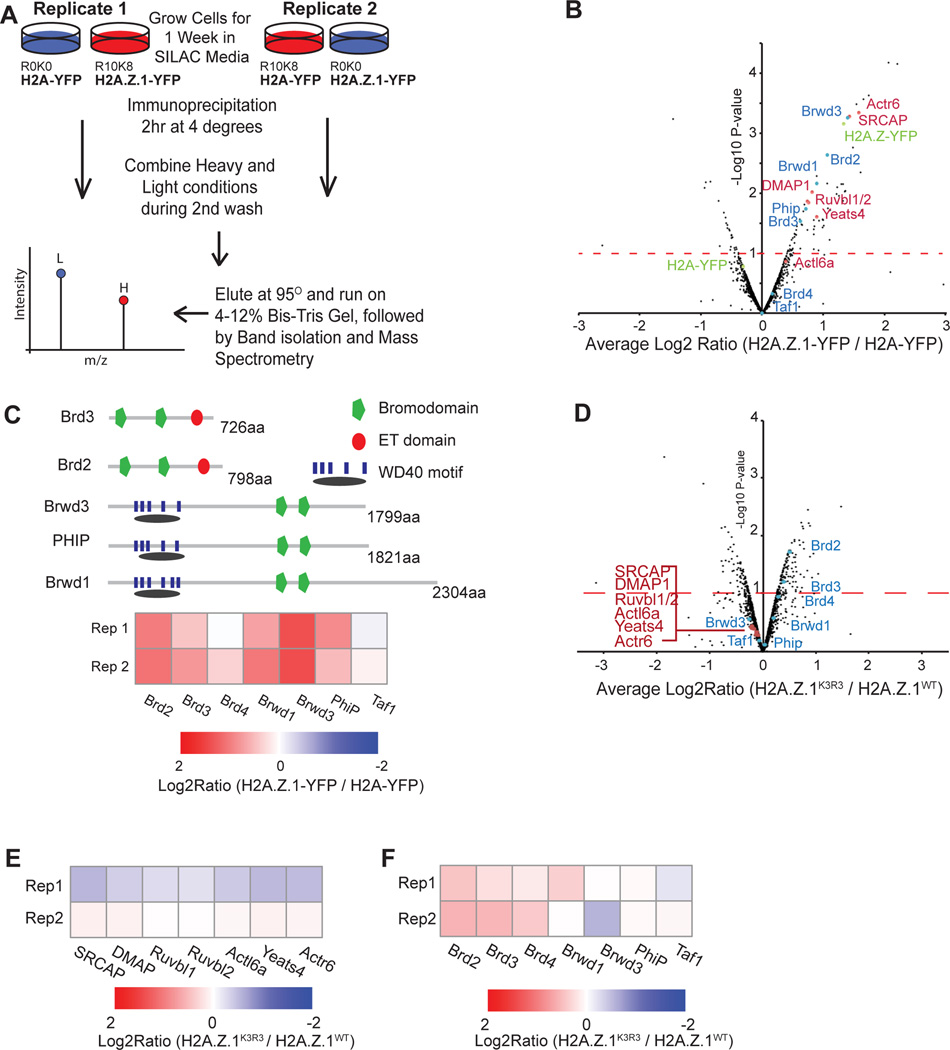
Histone post-translational modifications often function to recruit downstream regulators (Braun and Madhani, 2012) and specifically ubiquitin moieties often mediate downstream interactions with regulatory factors. Given that H2A.Z interacts with a distinct set of binding partners compared to H2A (Draker et al., 2012; Fujimoto et al., 2012), we reasoned that H2A.Z.1ub might mediate interactions with specific chromatin-associated proteins. To test this idea, we developed an approach that combines SILAC (Stable Isotope Labeling of Amino acids in Cell culture) (Ong et al., 2002) with immunoprecipitation (IP) and mass spectrometry to quantitatively assess changes in chromatin associated protein interactions (SILAC-IP) (Figure 4A). We first compared enriched proteins in mESC lines containing either a H2A.Z.1-YFP or H2A-YFP transgene (Subramanian et al., 2013) (Figures S5A, S5B and Table S2). Proteins were labeled by culturing cells in isotope containing media prior to IP, resulting in greater than 93% labeling efficiency as measured by the incorporation of the heavy isotopes. SILAC-IP was performed on the soluble fraction of micrococcal nuclease treated nuclei. Our approach allows for direct comparison between target proteins within one experiment and internally controls for nonspecific as well as YFP-associated interactions. To confidently identify H2A.Z.1-specific interactions, we required representation of multiple peptides in replicate samples from paired isotope swaps (e.g. H2A.Z.1-YFP heavy and H2A-YFP light) (Figure S5C). Differential enrichment was ranked by the fold change of ratio counts between paired samples and by a modified t-statistic for significance of reproducibility between replicates (Mertins et al., 2013; Sancak et al., 2013) (Figure 4B and Table S2). Validating our approach, we identified nearly all members of the H2A.Z specific ATP-dependent deposition complex SRCAP among the H2A.Z.1 enriched proteins (Ruhl et al., 2006) (Figures 4B and S5D).
Figure 4. H2A.Z.1ub interacts with chromatin-associated proteins in mESCs.
A. Schematic of SILAC-IP approach. B. Volcano plot of average ratio of enrichment of SILAC intensity for H2A.Z.1-YFP over H2A-YFP against p-value for significance of reproducibility. Bait proteins are indicated in green. Proteins in red are members of the SRCAP complex. Proteins in blue are tandem bromodomain proteins. C. Heatmap of SILAC Log2 ratio of H2A.Z.1-YFP IP over H2A-YFP IP for all tandem bromodomain proteins expressed mESCs. D. Volcano plot of average ratio of enrichment of SILAC intensity for H2A.Z.1WT-YFP over H2A.Z.1K3R3-YFP against p-value for significance of reproducibility. Proteins labeled as above. E. Heatmap of Log2 ratio of H2A.Z.1K3R3-YFP vs. H2A.Z.1WT-YFP intensity for SRCAP complex proteins from SILAC-IP. F. Heatmap of Log2 ratio of H2A.Z.1K3R3-YFP vs. H2A.Z.1WT-YFP intensity for tandem bromodomain proteins from SILAC-IP. See also Figure S5.
Given that many protein interactions are facilitated by specific domains, we asked whether particular motifs were enriched among the set of H2A.Z.1 interacting proteins (Franceschini et al., 2013). Our analysis identified the bromodomain as the most highly enriched motif (p-value < 2e−3). Over 40 bromodomain-containing proteins exist in the mouse genome, whereas only 8 harbor a tandem bromodomain motif (Filippakopoulos et al., 2012). Five of these 8 proteins are both expressed in mESCs and enriched in H2A.Z.1 chromatin, including two BET proteins (Brd2 and Brd3) and three proteins that harbor an additional WD40 motif (Brwd2, Brwd3, and Phip) (Figures 4B and 4C). Because Brd2 is the most highly expressed of the tandem bromodomain members in mESCs and it is necessary for murine development (Sheng et al., 2009) (Figure S5E), we focused on validating its interaction with H2A.Z.1 using two independent approaches. First, we found that Brd2 is significantly enriched upon induction of H2A.Z.1-YFP using SILAC-IP compared to un-induced condition conditions (p-value <0.01) (Table S3). Second, Brd2 shows significantly greater association with H2A.Z.1 in chromatin compared to H2A (p-value <0.1, t-test) by bioluminescence resonance energy transfer (BRET) (Deplus et al., 2013) (Figure S5F and S5G). In contrast, the BET protein Brd4 exhibited neither differential enrichment by SILAC-IP nor differential interaction by BRET (Figure S5G).
We then asked if H2A.ZK3R3 alters the association with H2A.Z-enriched proteins using our unbiased SILAC-IP approach. We found 212 differentially enriched proteins between H2A.Z.1K3R3 and H2A.Z.1WT SILAC-IPs (Figure 4D and Table S4). Of these, 123 proteins including Brd2 are significantly enriched in H2A.Z.1K3R3 chromatin compared to H2A.Z.1WT SILAC-IPs. In contrast, other tandem bromodomain proteins as well as SRCAP components showed little difference in enrichment (Figures 4D–F). Brd2 transcript and protein levels are similar between wild-type and mutant cell lines indicating that differential enrichment is not due to higher levels of Brd2 in H2A.Z.1K3R3 mESCs (Figures S5H and S5I). Together, our quantitative proteomic analysis demonstrates that H2A.Z.1ub influences specific protein interactions including with Brd2.
H2A.Z.1ub inhibits Brd2 recruitment to bivalent genes
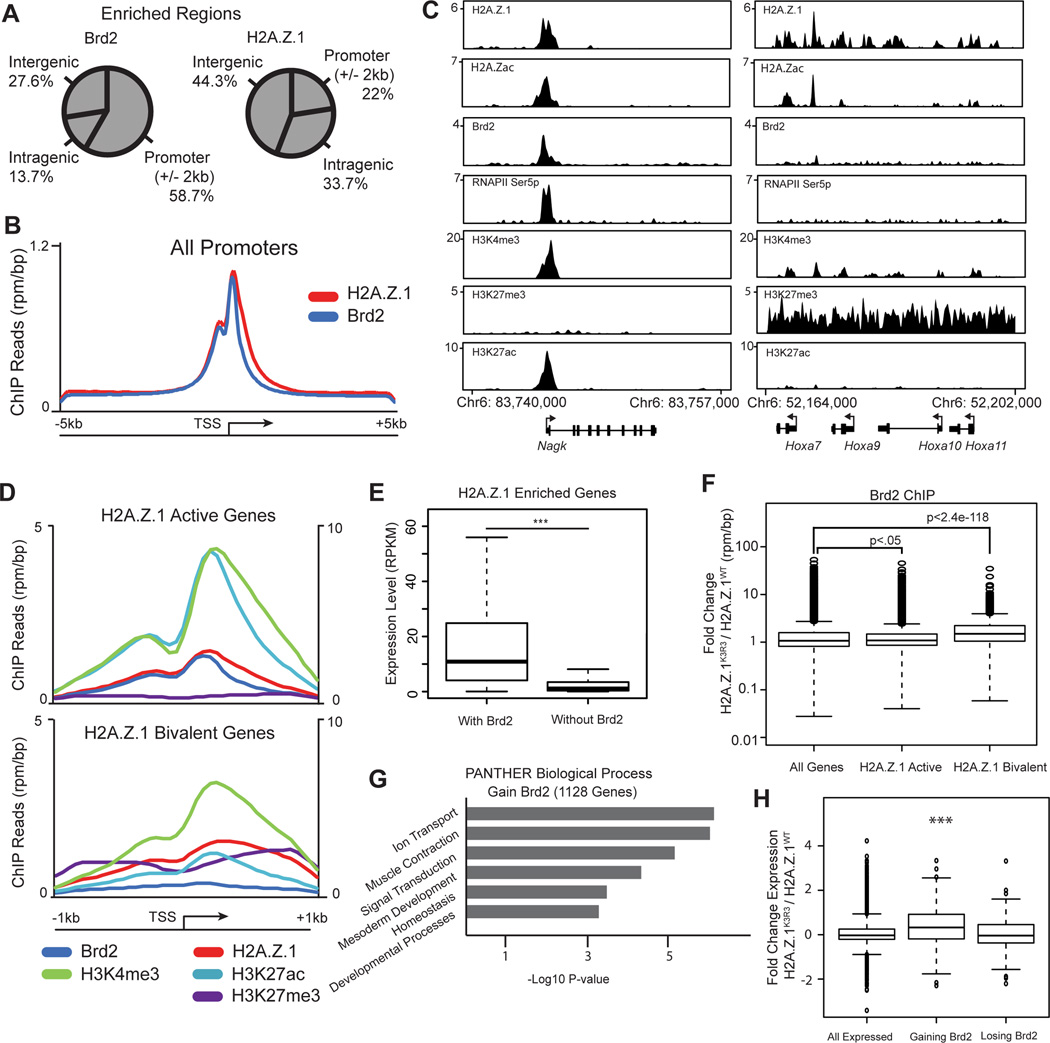
We next performed ChIP-Seq to analyze the global enrichment patterns of Brd2 in mESCs. Brd2 is enriched at 11,536 genomic regions in wild-type mESCs. Of these regions, 58.7% fall within 2kb of a known transcriptional start site (TSS), leading us to focus on the role of Brd2 at promoters (Figures 5A and 5B). In total, Brd2 localizes to the promoters of 6,856 genes of which 84% also harbor H2A.Z.1 peaks. Moreover, Brd2 enrichment at promoters coincides with active chromatin marks including H2A.Zac, H3K27ac and H3K4me3 as well as RNAPII (Figures 5C and S6A), consistent with the role of bromodomain-containing proteins in gene activation (Draker et al., 2012; Leroy et al., 2008). In contrast, Brd2 is largely absent from bivalent genes e.g. the HoxA locus (Figures 5C and 5D). Indeed, genes co-enriched with both H2A.Z.1 and Brd2 are more highly expressed than H2A.Z-enriched genes that lack Brd2 (p-value < 1e−142, t-test) (Figure 5E). These results suggest a role for Brd2 with H2A.Z.1 at active gene promoters.
Figure 5. H2A.Z.1ub prevents Brd2 recruitment to bivalent genes.
A. Genomic distribution pattern of Brd2 and H2A.Z.1 using CEAS (Shin et al., 2009). B. Average signal of ChIP-seq reads across the TSS of all genes (+/− 5kb) for Brd2 and H2AZ.1. C. Genome tracks of representative active and bivalent H2A.Z.1 target genes. Reads are normalized to reads per million per base pair. D. Average ChIP-seq reads for Brd2, H2A.Z.1, H3K4me3, H3K27ac and H3K27me3 across the TSS of active and bivalent H2A.Z.1 enriched genes (+/− 1kb of TSS) (Subramanian et al., 2013). H3K4me3 is plotted on secondary axis to the right. E. mRNA levels as measured by RNA-seq of H2A.Z.1 enriched genes with or without Brd2 (p-value < 1e−142, two-sided t-test). RPKM=reads per kilobase per million. Boxplot indicates median and 25th and 75th percentile mRNA level, whiskers extend 1.5 times the interquartile range. F. Box and whisker plot of median fold change in Brd2 ChIP-seq reads across the promoter regions (TSS +/− 300bp) of all genes as well as H2A.Z.1 enriched active and bivalent genes. Box and whiskers defined as in E. G. GO analysis of genes gaining Brd2 at their promoter as determined by DAVID (Huang et al., 2009a; 2009b). H. Box and whisker plot of the median fold change in expression of genes gaining Brd2 >2-fold at promoters (p-value 6.3e−11, un-paired t test) and losing Brd2 (not significant). Box and whiskers defined as in E. See also Figure S6.
Our proteomics data suggests that H2A.Z.1ub inhibits Brd2 recruitment. To test this idea, we performed ChIP-seq and observed a significant increase in Brd2 enrichment at bivalent promoters in H2A.Z.1K3R3 compared to H2A.Z.1WT mESCs (Figure 5F, median fold change 1.50, Wilcox test p<2.4e−118, compared to all genes), whereas active genes showed only a slight gain (Figure 5F, median fold change 1.09, Wilcox test p<0.05, compared to all genes). Moreover, Brd2 levels were not significantly changed at either enhancers or super enhancers (two-sided t-test, median fold change enhancers 0.97, super enhancers 0.95) (Figure S6B). For example, Brd2 shows increased enrichment at the bivalent gene Mesp1 as well as across the HoxA gene cluster in H2A.Z.1K3R3 mESCs (Figure S6C), whereas active genes including Hira and Cul1 show no apparent difference in Brd2 levels (Figure S6D). These results were independently validated by ChIP-qPCR (Figure S6E). In total, we find that 1,128 genes display a 2-fold or greater change in Brd2 enrichment in H2A.Z.1K3R3 mESCs across biological replicates (Table S5). GO analysis revealed that genes gaining Brd2 have roles in Signal Transduction and Developmental Processes (Figure 5G) and showed a significant increase in expression compared to all genes (median change 1.3-fold, Wilcox test p<9.2e−17). In contrast, genes that displayed >2-fold depletion of promoter Brd2 were largely unaffected in H2A.Z.1K3R3 mESCs (Figure 5H).
Brd4, another BET family member, plays a key role in regulating gene expression and cell identity in ESCs and in many cancers (Asangani et al., 2014; Di Micco et al., 2014; Liu et al., 2014; Wu et al., 2015). In contrast to Brd2, Brd4 enrichment was more modestly affected at bivalent promoters in H2A.ZK3R3 mESCs (median fold change 1.29, Wilcox test p<1.9e−17 compared to all genes) (Figures S6H and S6I). Moreover, Brd4 was not differentially enriched in our SILAC-IP or BRET analysis (Figures 4B, 4D, 4F, S5G, Tables S2 and S4). Notably, Brd2 and H2A.Z.1 both display a highly similar bimodal distribution at promoters ChIP-seq whereas Brd4 shows a distinct single peak (Figure S6F). Furthermore, Brd4 is more highly enriched at enhancers, and particularly at super enhancer clusters (Figure S6G) (Whyte et al., 2013), whereas Brd2 and H2A.Z.1 are most highly enriched at promoters (Figures S6F and S6G). These data suggest H2A.Z.1 coordinates with Brd2 at promoters and that BET family members function independently to regulate gene expression programs in mESCs.
H2A.Z.1ub inhibits Brd2 recruitment to bivalent promoters
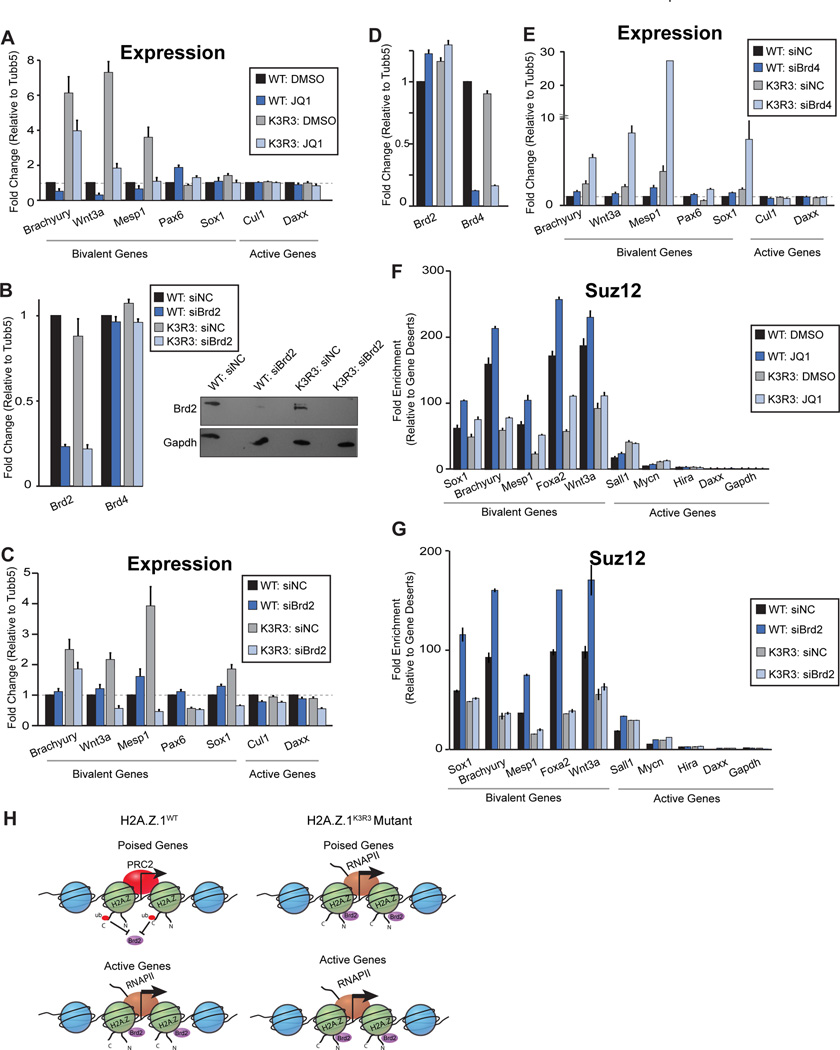
To test whether H2A.Z.1ub prevents Brd2 recruitment at bivalent promoters thereby inhibiting gene activation, we first treated mESCs with the pan-BET inhibitor JQ1 (Liu et al., 2014; Wu et al., 2015). After 24hr of treatment with 100nM JQ1, the increased expression of bivalent genes in H2A.Z.1K3R3 mESCs was restored to basal levels (Figure 6A). H2A.ZK3R3 mESCs exhibit de-repression of early mesoendoderm genes including Brachyury and Mesp1, whereas neuronal genes such as Pax6 and Sox1 are largely unaffected (Figure 2C). Consistent with these observations, Pax6 and Sox1 were largely unaffected by addition of JQ1. We next investigated whether loss of gene silencing in H2A.Z.1K3R3 mESCs can be rescued by direct Brd2 inhibition. Transfection with specific siRNAs resulted in significant Brd2 depletion after 48hrs (Figure 6B). Upon Brd2 depletion, bivalent genes showed a marked decrease in expression in H2A.Z.1K3R3 mESCs whereas genes unaffected by JQ1 treatment remained unchanged (Figure 6C). Similar results were observed with an independent Brd2 siRNA (data not shown). In contrast, Brd4 depletion led to a slight increase in bivalent gene expression in H2A.Z.1WT mESCs and to a dramatic increase in expression in H2A.Z.1K3R3 mESCs (Figures 6D and 6E). These results are consistent with a direct role for Brd2 in regulating gene promoters whereas Brd4 plays a pivotal role in regulating super enhancer associated pluripotency genes such as Oct4 (Di Micco et al., 2014; Whyte et al., 2013).
Figure 6. Brd2 is necessary for gene activation and antagonizes PRC2 recruitment at bivalent genes.
A. mRNA expression upon treatment of H2A.Z.1WT and H2A.Z.1K3R3 mESCs with BET inhibitor JQ1 at 100nM or DMSO control for 24hrs. Expression is normalized to H2A.Z.1WT+DMSO using Tubb5. Error bars represent SE of a triplicate set of experiments. B. mRNA and protein levels after 48hrs of Brd2 RNAi mediated knock-down in both H2A.Z.1WT and H2A.Z.1K3R3 mESCs. Expression is normalized to a non-targeting siRNA using Tubb5. Error bars represent SE of a triplicate set of experiments. Immunoblots for Brd2 and Gapdh performed on whole cell lysates following 48 hrs siRNA treatment. C. mRNA expression for H2A.Z.1 bivalent and active target genes after Brd2 depletion. Normalized as above. D. mRNA levels after 48 hrs of Brd4 siRNA mediated depletion in both H2A.Z.1WT and H2A.Z.1K3R3 mESCs. Expression is normalized to a non-targeting siRNA using Tubb5. Error bars represent SE of a triplicate set of experiments. E. mRNA expression for H2A.Z.1 bivalent and active target genes after treatment with either a negative control siRNA or Brd4 siRNA. Normalized as above. F. Suz12 ChIP-qPCR after treatment of H2A.Z.1WT or H2A.Z.1K3R3 mESCs with either 100nM JQ1 or DMSO. Enrichment values are normalized to two gene desert regions. Error bars represent SE of a triplicate set of experiments. G. Suz12 ChIP-qPCR after treatment of H2A.Z.1WT or H2A.Z.1K3R3 mESCs with either Brd2 siRNA or negative control siRNA. Enrichment values calculated as above. H. Model for Brd2 recruitment to bivalent genes. Brd2 is localized to active promoters that lack H2A.Z.1ub (indicated by red circles) whereas Brd2 is largely absent from bivalent genes enriched for mono-ubiquitylated H2A.Z.1. H2A.Z.1K3R3 incorporation leads to Brd2 recruitment and gene activation, which further antagonizes PRC2 at promoters of developmental genes.
H2A.Z.1ub is critical for gene silencing and for maintenance of PRC2 at bivalent genes, leading us to ask whether Brd2 antagonizes PRC2 to reinforce gene activation. To test this idea, we analyzed Suz12 occupancy at a subset of target genes upon JQ1 treatment or Brd2 depletion in H2A.Z.1WT and H2A.Z.1K3R3 mESCs. BET inhibition with 100nM JQ1 for 24 hrs led to an increase in Suz12 enrichment at bivalent genes in H2A.Z.1WT with only a slight increase in H2A.Z.1K3R3 mESCs, whereas active genes were largely unaffected (Figure 6F). Similarly, direct Brd2 inhibition resulted in increased Suz12 occupancy at bivalent genes in H2A.ZWT but not in H2A.Z.1K3R3 mESCs whereas Suz12 levels at active genes were largely unaffected (Figure 6G). Taken together, these results suggest H2A.Z.1ub is required for Suz12 binding, and functions to maintain the balance between gene silencing and activation of developmental programs in mESCs (Figure 6H).
DISCUSSION
H2A.Z is incorporated at both active and bivalent promoters in mESCs, and appears to regulate contrasting gene expression states. While H2A.Z acetylation is widely associated with gene activation from yeast to mammals (Bruce et al., 2005; Hu et al., 2013; Ku et al., 2012; Millar et al., 2006), how this variant contributes to gene silencing is poorly understood. Using a defined system to directly test the function of the three C-terminal PRC1 mono-ubiquitylated lysine residues, we show that H2A.Z.1ub is necessary for both the maintenance of bivalent chromatin in mESCs and for the appropriate induction of developmental programs.
Non-canonical PRC1 containing Kdm2b catalyzes H2A ubiquitylation, and appears necessary for PRC2 recruitment and transcriptional repression both in vitro and in vivo, however a direct role for H2Aub in this process was not established in these studies (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). We show that loss of H2A.Z.1ub leads to disruption of poised, bivalent genes and to faulty mESC differentiation suggesting that H2A.Z.1ub contributes to transcriptional repression of lineage programs in mESCs. The observation that mutation of the catalytic activity of PRC1 components Ring1a/b does not affect chromatin compaction at target regions despite the partial loss of gene repression and H3K27me3 enrichment (Endoh et al., 2012; Eskeland et al., 2010; Illingworth et al., 2015) suggests neither H2Aub nor H2A.Zub contribute directly to chromatin compaction. Notably, embryos lacking Ring1b catalytic activity progress further in development compared to complete Ring1b knockout embryos are reminiscent of the partial defects in lineage commitment we observe upon differentiation of H2A.ZK3R3 mESCs, suggesting that Ring1b-mediated mono-ubiquitylation is also an important PRC1 activity. In contrast, recent work showed that the ubiquitylation activity of the Ring1a/b homolog, Sce, is not required for gene repression in Drosophila (Pengelly et al., 2015). Thus, dissecting how histone variant incorporation and PRC1 contribute to gene repression and chromatin compaction will lead to a better understanding of how these pathways coordinate developmental programs and may explain the different observations in Drosophila and mammalian systems.
Loss of H3K27me3 alone is not sufficient to confer transcriptional activation (Marks et al., 2012), suggesting that activation is mediated through a distinct mechanism. Our data suggest that modulation of H2A.Z.1 by PTMs is critical for its interaction with downstream activators such as Brd2. In support of this model, studies show that Brd2 co-localizes with H2A.Z.2 in melanoma cell lines to activate cell cycle genes (Vardabasso et al., 2015), and that H2A.Z is necessary to recruit Brd2 to activate target genes in response to hormone stimulation in LNCaP cells (Draker et al., 2012). Thus, it is possible that the H2A.Z is mono-ubiquitylated at the poised hormone-responsive genes prior to stimulation.
How Brd2 drives transcription remains unclear, as it lacks the p-TEFb interacting motif of Brd4 (Jang et al., 2005). In vitro, Brd2 directly promotes transcription on a chromatinized template by acting as a chromatin remodeler (Leroy et al., 2008). Additionally, Brd2 can bind both TBP and Mediator, both of which are normally absent from bivalent promoters (Peng et al., 2006; Pinz et al., 2015). Thus, future studies will be directed toward understanding how H2A.Z.1ub inhibits gene activation through antagonism of Brd2. More broadly, H2A.Z homologs in both yeast and C. elegans coordinate with BET proteins to regulate gene expression states (Shibata et al., 2014; Zhang et al., 2005), suggesting that coordination between H2A.Z and Brd2 is an evolutionarily conserved mechanism for controlling transcriptional output at inducible genes. Together, our work establishes a previously unknown role for H2A.Z.1ub in maintaining the transcriptional balance of developmental programs during early lineage commitment.
EXPERIMENTAL PROCEDURES
Growth of murine embryonic stem cells (mESCs)
V6.5 (129SvJae and C57BL/6; male) mESCs were plated with irradiated murine embryonic fibroblasts (MEFs) and cultured using standard conditions on gelatinized tissue culture plates as described (Subramanian et al., 2013). For WNT signaling inhibition, mESCS were treated with DMSO or WNT signaling inhibitor KY02111 (10µM) for 48 hours and then aggregated to form EBs in the absence of LIF and in the presence of the inhibitor. Cells were treated with JQ1 at 100nM or DMSO as a control. mESCs were transfected with specific siRNAs to Brd2 and Brd4 (Origene) using 10ul DharmaFect 1 (GE Healthcare). Expression was assayed 48hrs after the start of siRNA transfection. Detailed protocols are included in Supplementary Methods.
Transgenic mESC lines
H2A.Z.1WT- and H2A.Z.1K3R3-GFP constructs (which contain the H2A.Z.1 isoform) from (Sarcinella et al., 2007) were modified by replacement with YFP as described (Subramanian et al., 2013). Detailed protocols are provided in Supplementary Methods.
Embryoid body differentiation
mESCs were aggregated to form embryoid bodies (EBs) by plating them in Corning Ultra-Low Attachment Tissue Culture Plates (Corning) at a density of 100,000 cells/ml in mESC media lacking LIF. Media was changed every other day and EBs were cultured for the indicated number of days.
Histone Extracts and Immunoblot
Histone extracts were prepared as described (Subramanian et al., 2013). For immunoblot analysis, samples were resolved on SDS-PAGE gels, proteins were transferred to a PVDF membrane, blocked with 5% milk in PBST (0.1% Tween-20 in phosphate-buffered saline, pH 7.4), blotted overnight with primary antibody in PBST. The presence of the antigen was detected by HRP-conjugated secondary antibody. Antibodies listed in Supplementary Methods.
RNA isolation, quantitative real-time PCR, and RNA-seq
Total RNA was extracted using TRIzol (Invitrogen) and reverse transcribed using SuperScript III (Invitrogen) or M-MLV reverse transcriptase (Invitrogen) and random hexamers according to manufacturer protocols. qPCR reactions were performed with SYBR Green (KAPA Biosystems) and primers listed in Table S6. Relative mRNA levels were determined in triplicate for each transcript using the manufacturer’s software (Advanced Relative Quantification with Roche Lightcycler 480 Software Version 1.5) using Tubb5 transcript levels for normalization. RNA-seq libraries were prepared as described (Subramanian et al., 2013). Detailed protocols are described in Supplementary Methods.
Chromatin Immunoprecipitation
ChIP was performed with 10–25 million cells as described (Wamstad et al., 2012). Detailed protocols including site-specific qPCR and high-throughput sequencing as well as antibodies are described in Supplementary Methods.
SILAC Based Immunoprecipitation and mass spectrometry
Cells were grown for one week in SILAC medium containing either R0K0 or R10K8. Immunoprecipitation was performed using 109 cells for each condition. Native ChIP was performed on a modified version as described in (Umlauf et al., 2004). Eluted proteins were separated by SDS-gel. In-gel digestion and mass spectrometry was conducted essentially as described (Sancak et al., 2013). MS data was analyzed as in (Cox and Mann, 2008; Cox et al., 2011). Detailed explanation can be found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
H2A.Z.1WT- and H2A.Z.1K3R3-GFP vectors were a kind gift from Peter Cheung, Ontario Cancer Institute. Ring1bfl/−; CreERT2 mESCs were a kind gift from Maarten van Lohuizen. GFP antibody was kindly provided by Iain Cheeseman. JQ1 was a kind gift from Jay Bradner. We thank Charles Lin and members of the MIT BioMicro Center especially Fugen Li, Vincent Butty, Jie Wu, and Stuart Levine for assistance with data analysis. We thank Danette Daniels and Jacqui Mendez from Promega for technical advice and reagents for the BRET assay. Raw LCMS data from Ku et al. (2012) were used with permission of Bradley Bernstein. We are also grateful to members of the Boyer lab for helpful and stimulating discussions. This work was supported in part by the NIH Pre-Doctoral Training Grant T32GM007287 (L.E.S. and P.A.F), the MIT School of Science Fellowship in Cancer Research (P.A.F.), NHLBI Bench to Bassinet Program (U01HL098179) and by the Massachusetts Life Sciences Center to L.A.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
All sequencing data reported in this article have been deposited under the GEO accession ID GSE53208 and GSE67944.
AUTHOR CONTRIBUTIONS
Conceptualization, L.E.S., P.A.F., L.A.B.; Methodology L.E.S., P.A.F., V.S., N.U., J.J., L.A.B.; Investigation, L.E.S., P.A.F., V.S., R.B., N.U., S.E.P., J.D.J.; Writing – Original Draft, L.E.S., P.A.F., L.A.B.; Writing – Review & Editing, L.E.S., P.A.F., V.S., L.A.B.; Funding Acquisition, L.E.S., P.A.F., L.A.B.; Supervision, L.A.B.
REFERENCES
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge, ten D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LLP, Ito S, Cooper S, Kondo K, Koseki Y, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Madhani HD. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012;13:619–630. doi: 10.1038/embor.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Research. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. Targeting Polycomb to Pericentric Heterochromatin in Embryonic Stem Cells Reveals a Role for H2AK119u1 in PRC2 Recruitment. Cell Reports. 2014 doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. Embo J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fontanals-Cirera B, Low V, Ntziachristos P, Yuen SK, Lovell CD, Dolgalev I, Yonekubo Y, Zhang G, Rusinova E, et al. Control of Embryonic Stem Cell Identity by BRD4-Dependent Transcriptional Elongation of Super-Enhancer-Associated Pluripotency Genes. Cell Rep. 2014;9:234–247. doi: 10.1016/j.celrep.2014.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Sarcinella E, Cheung P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Research. 2011;39:3529–3542. doi: 10.1093/nar/gkq1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Isono K-I, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. Ring1B Compacts Chromatin Structure and Represses Gene Expression Independent of Histone Ubiquitination. Molecular Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Mering, von C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Seebart C, Guastafierro T, Prenni J, Caiafa P, Zlatanova J. Proteome analysis of protein partners to nucleosomes containing canonical H2A or the variant histones H2A.Z or H2A.X. Biological Chemistry. 2012;393:47–61. doi: 10.1515/BC-2011-216. [DOI] [PubMed] [Google Scholar]
- Hardy S, Jacques P-É, Gévry N, Forest A, Fortin M-È, Laflamme L, Gaudreau L, Robert F. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS Genet. 2009;5:e1000687. doi: 10.1371/journal.pgen.1000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nature Cell Biology. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes & Development. 2015;29:1897–1902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PWTC, Müller CW, Vermeulen M, Müller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nature Structural & Molecular Biology. 2014 doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biology. 2012;13:R85. doi: 10.1186/gb-2012-13-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z Mediate Nucleosome Depletion during Embryonic Stem Cell Differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Stein P, Cheng X, Yang W, Shao N-Y, Morrisey EE, Schultz RM, You J. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 2014;21:1950–1960. doi: 10.1038/cdd.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes & Development. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Peng J, Dong W, Chen L, Zou T, Qi Y, Liu Y. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol. Cell. Biochem. 2006;294:45–54. doi: 10.1007/s11010-006-9223-6. [DOI] [PubMed] [Google Scholar]
- Pinz S, Unser S, Buob D, Fischer P, Jobst B, Rascle A. Deacetylase inhibitors repress STAT5-mediated transcription by interfering with bromodomain and extra-terminal (BET) protein function. Nucleic Acids Res. 2015:gkv188. doi: 10.1093/nar/gkv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovács-Bogdán E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarcinella E, Zuzarte PC, Lau PNI, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Molecular and Cellular Biology. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Sawa H, Nishiwaki K. HTZ-1/H2A.z and MYS-1/MYST HAT act redundantly to maintain cell fates in somatic gonadal cells through repression of ceh-22 in C. elegans. Development. 2014;141:209–218. doi: 10.1242/dev.090746. [DOI] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature Cell Biology. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Fields PA, Boyer LA. H2A.Z: a molecular rheostat for transcriptional control. F1000Prime Rep. 2015;7:01. doi: 10.12703/P7-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Mazumder A, Surface LE, Butty VL, Fields PA, Alwan A, Torrey L, Thai KK, Levine SS, Bathe M, Boyer LA. H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation. PLoS Genet. 2013;9:e1003725. doi: 10.1371/journal.pgen.1003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol. Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- van Daal A, Elgin SC. A histone variant, H2AvD, is essential in Drosophila melanogaster. Molecular Biology of the Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardabasso C, Gaspar-Maia A, Hasson D, Pünzeler S, Valle-Garcia D, Straub T, Keilhauer EC, Strub T, Dong J, Panda T, et al. Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Mol. Cell. 2015 doi: 10.1016/j.molcel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee W-W, Reinberg D. A double take on bivalent promoters. Genes & Development. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. Pausing of RNA Polymerase II Regulates Mammalian Developmental Potential through Control of Signaling Networks. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Molecular Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Wu T, Pinto HB, Kamikawa YF, Donohoe ME. The BET Family Member BRD4 Interacts with OCT4 and Regulates Pluripotency Gene Expression. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-Wide Dynamics of Htz1, a Histone H2A Variant that Poises Repressed/Basal Promoters for Activation through Histone Loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Brickner JR, Majid MC, Mosammaparast N. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends in Cell Biology. 2014;24:426–434. doi: 10.1016/j.tcb.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.