Abstract
Traditionally a pursuit of large pharmaceutical companies, high-throughput screening assays are becoming increasingly common within academic and government laboratories. This shift has been instrumental in enabling projects that have not been commercially viable, such as chemical probe discovery and screening against high risk targets. Once an assay has been prepared and validated, it must be fed with screening compounds. Crafting a successful collection of small molecules for screening poses a significant challenge. An optimized collection will minimize false positives whilst maximizing hit rates of compounds that are amenable to lead generation and optimization. Without due consideration of the relevant protein targets and the downstream screening assays, compound filtering and selection can fail to explore the great extent of chemical diversity and eschew valuable novelty. Herein, we discuss the different factors to be considered and methods that may be employed when assembling a structurally diverse compound screening collection. Rational methods for selecting diverse chemical libraries are essential for their effective use in high-throughput screens.
Keywords: Drug-like Molecule: A molecule with molecular properties that overlap with the majority of existing drugs., High-throughput Screening: A screening process that utilises robotics and rapid data processing to perform millions of assays in a short space of time., Molecular Similarity: A measure of the relatedness of two molecules. This would ideally quantify the similarity in biological effect but in practice tends to quantify the similarity in structure., Molecular Diversity: A measure of how well a subset of molecules represents a larger set of molecules. A more diverse subset will tend to have a lower molecular similarity between molecules., Frequent Hitter: A molecule or molecular substructure that hits numerous screening assays on different drug targets with a mode of action that is assumed to be non-specific., Substructure Filter: A computational filter used to remove molecules containing molecular substructures that are considered to give rise to non-specific binding or deleterious pharmacodynamic properties.
Introduction
The earliest efforts in drug discovery focused on crude extracts from natural sources and success relied mainly on trial and error. Work in the middle of last century established the concept of a molecular disease(1), moving drug discovery in a more rational direction and toward screening compounds against a molecular target. Natural products provided the majority of early drugs and still remain as an invaluable source of chemicals for screening, along with semi-synthetic derivatives(2). In more recent times, the advent of combinatorial chemistry provided a radical increase in the number of available screening compounds and this was coupled with high-throughput screening (HTS) of large chemical libraries(3). Despite many failures amongst the successes, HTS remains a widely used method for initiating the process of drug and chemical probe discovery(4-9). The concept of a drug-like molecule has existed for many years(10) and includes optimized parameters for physicochemical properties as well as functional groups to be avoided. This concept has been extended to consider lead-like instead of drug-like molecules(11), and this progresses naturally to the identification of hit-like molecules, which are geared to provide true positive results in HTS assays and yield a basis for lead generation(12). The vastness of chemical space means that there are currently tens of millions of molecules available for purchase and screening. Even using harsh filters to remove unwanted compounds, there are in the order of a million hit-like molecules available commercially(13-14). However, identifying a representative subset of these molecules to screen is a complex task, with multiple scientific, financial and logistical considerations. Whilst this review article is unable to comprehensively cover the multifold aspects of library design, its aim is to highlight the key issues that must be taken into account. This is now important in academic groups and government labs as well as in industry(15). Here we review current methods for crafting screening compound collections and outline the traps and pitfalls. This will be done in three sections: compound sourcing, compound filtering and compound selection. Finally, we highlight key challenges to the field and outline future directions.
Compound Sourcing
There are many suppliers of screening compounds, ranging from small chemical suppliers with hundreds of compounds to large ones with over a million compounds. Many collections of small molecules have been analyzed for drug-like and lead-like properties (13, 16-20) and chemical supplier libraries are being increasingly tailored toward these parameters. Details of the main screening libraries from six chemical suppliers with varied collections of over 300,000 screening compounds are reported in Table 1. At present, all have a high pass rate for commonly employed drug-like and lead-like filters. However, compound collections turn over rapidly and should be analysed in this way prior to selecting suppliers. Compound prices per milligram vary widely dependent on the number of compounds purchased and the sample weight per compound required, with significantly lower prices per compound if thousands or tens of thousands are purchased. Theoretically, searching the entirety of currently available chemical space encompasses the maximum commercially available molecular diversity. In practice, a great expanse of available diversity can be sampled by selecting large numbers of compounds from a few chemical suppliers with diverse collections. Many chemical suppliers also sell pre-selected diverse libraries at reduced cost. These are generally selected by rational means, but the compound filters employed may have been too harsh or too lenient, dependent on the nature of the screening assay and the target. Furthemore, although the compounds tend to be relatively diverse, they are also much more likely to have been tested by other laboratories, as they are for sale off-the-shelf. Including novelty in HTS is a vital aspect of drug discovery and many firms offer unlisted libraries at higher costs, promising an easier path to intellectual property rights.
Table 1.
Details of the screening libraries for six chemical suppliers, the ZINC database of purchasable molecules and the Drugbank database of experimental drugs. All physicochemical properties were generated with Qikprop and filtering was performed with Canvas.The compound collection refers to the subset of molecules that was analyzed from each source.
| Compound Source | Compound Collection | URL | Number Of Compounds | % Lipinski Passes | % REOS Passes |
|---|---|---|---|---|---|
| Asinex | Gold and Platinum Collections | http://www.asinex.com | 364407 | 79.6 | 73.0 |
| Chembridge | Express Pick Library | http://www.chembridge.com | 442051 | 84.0 | 66.6 |
| ChemDiv | Discovery Chemistry | http://www.chemdiv.com | 789603 | 73.8 | 72.1 |
| Enamine | HTS Collection | http://www.enamine.net | 1116406 | 90.7 | 79.6 |
| Life Chemicals | Stock | http://www.lifechemicals.com | 327211 | 84.9 | 76.6 |
| Vitas M Labs | HTS Stock | http://www.vitasmlab.com | 476184 | 75.1 | 65.8 |
| Drugbank | All Drugs | http://www.drugbank.ca | 4886 | 71.4 | 51.7 |
| Zinc | Purchasable Compounds | http://zinc.docking.org | 18671085 | 87.2 | 73.1 |
Compound Databases
In addition to compound libraries direct from chemical suppliers, there are a number of preassembled online data repositories including ZINC(21) (http://zinc.docking.org/), emolecules (http://www.emolecules.com/) and Chemspider (http://www.chemspider.com/). The ZINC repository currently has the largest number of compounds, including the complete compound libraries of the majority of chemical suppliers. The number of molecules in the ZINC set of purchasable compounds currently stands at just under 18.7 million. However, chemical suppliers commonly update their libraries every few months, which may not be reflected in data repositories such as ZINC. Despite the huge number of commercially available compounds, existing chemistry efforts have only probed a small proportion of chemical space. The number of synthetically feasible, drug-like molecules is estimated to be in excess of 1060 (22) and only a small subset of this has been explored. For example, data compiled in the Generated Database of Molecules (http://www.dcb-server.unibe.ch/groups/reymond/gdb/start.html) demonstrates that less than 0.5% of the synthetically feasible compounds comprised of up to 11 atoms of C, N, O and F are recorded in public databases as having been synthesised(23). Recent studies have also highlighted a large number of novel ring systems that are not currently represented in available chemical space(24). Many sources of diversity are excluded from existing compound collections and this greatly restricts the coverage of chemical space. In particular, the bias against chirality skews commercially available compounds toward flat compounds with many aromatic rings(25). This in turn may negatively impact on the properties related to absorption, distribution, metabolism, elimination and toxicity (ADMET) and increase the risk of attrition during development(26). Shelat and Guy have questioned whether libraries of synthetic molecules are suitable for addressing novel drug targets and suggest the use of natural products in HTS, particularly for phenotypic and high-content screens.
Natural Products
The vast majority of commercially available small molecules are obtained from synthetic chemistry. Nonetheless, nature is an important source of biologically active compounds and natural products have played a key role in drug discovery efforts. It has been estimated that as many as 50% of marketed small molecule drugs have been derived from natural products(27). However, of the compounds currently approved for marketing each year, natural products represent a much lower percentage. Many chemical suppliers sell natural products for HTS and some chemical suppliers specialize in natural product chemistry. The natural product collections are usually separated from synthetic compounds and can be significantly more expensive. However, they can provide unique chemical structures, and may show more drug-like ADMET properties(28). Natural products have proven particularly powerful as anti-cancer and anti-infective agents(2) and tend to be well suited to phenotypic screening. Recent analysis shows that there are many ring systems present in natural products that are not found in screening libraries and many have suggested that screening compounds should be further biased toward biogenic scaffolds(29-30). However, the advantages of natural products must be balanced against their often greater structural complexity that may lead to difficulties in synthesis and purification of analogues during lead generation and optimization. There is still great controversy over the relative merits of screening natural products or natural product derivatives versus screening libraries from combinatorial chemistry or diversity oriented synthesis(31). Both have advantages and disadvantages and thus HTS library commonly combine both sources, though typically with more synthetic small molecules. Recently, it has been suggested that compounds balancing the properties of natural products and synthetic molecules may be optimal(32).
In summary, there are multiple sources of potential screening compounds and successful libraries typically strike a balance between synthetic compounds and natural products. However, whilst the growth in commercially available chemical space should always be capitalized upon, many compounds are unsuitable for screening in HTS assays and should be filtered out of any quality screening collection.
Compound Filtering
In order to obtain commercially available hit-like compounds, computational filters are commonly used to remove compounds with undesirable properties. Ideal drug-like and lead-like molecules have differing properties and these differ again from hit-like molecules. In general, the physicochemical properties of a lead-like molecule can be improved during lead optimization toward a drug-like molecule by tailoring the lipophilicity. Similarly, the binding affinity of a hit-like molecule can be improved during the process of hit explosion to yield a lead-like molecule. However, hit-like molecules must be large and lipophilic enough to gain sufficient binding affinity that they can be identified in a screening assay, but not so large that they have a very small probability of binding. Larger and more complex molecules have a lower probability of exhibiting perfect shape and electrostatic complementarity with any given target and this suggests that smaller and less complex molecules will more commonly provide starting points for drug development(33). An ideal hit molecule should also be amenable to chemical elaboration, show reasonable levels of cell permeability and have a range of commercially available analogues, some of which have also been tested in the same assay.
Computational Filters
There are numerous computational filters used to mark compounds that may have problems due to assay interference or downstream ADMET properties. The most commonly used of these are physicochemical property filters that specifically attempt to remove compounds that may lead to low levels of drug absorption and distribution. An exception that is ignored by these filters is compounds that are substrates for drug transporters, which recent works suggests may be a significant proportion of molecules(34). In addition to Lipinski’s well known rule of five(35), Ghose filters(36) and Veber filters(37) are commonly employed to filter compounds. Noteworthy analysis has also been performed by Walters(38), Oprea(39), Egan(40), Lee(41), Baurin(13) and Martin(42). The key properties that determine drug absorption and distribution for an oral drug are the lipophilicity measures of the octanol/water partition coefficient (logP) and surface area of the polar atoms in the molecule (PSA)(43-45). Analysis of trends in launched drugs has highlighted a significant increase in molecular weight in the last fifty years, but a negligible increase in logP values(46). This is not surprising, as drugs with increased logP tend to be more promiscuous binders and can thus be expected to have a higher attrition rate in later development(47). However, studying the most recent trends in molecules being synthesized in leading drug discovery companies suggests an increase in both molecular weight and logP(46). This has been attributed to the fact that more lipophilic drugs have the potential to be more efficacious, as they tend to have increased binding affinity. It has been suggested that this may adversely affect drug attrition rates in the future due to an increased likelihood of toxicity(48). However, as discussed, larger and more complex molecules have a lower probability of exhibiting perfect shape and electrostatic complementarity with any given target and they are thus expected to show greater specificity(33). This predicted increase in promiscuity due to increased lipophilicity may thus be ameliorated by increased complexity. Despite the noted increase in molecular weight, there is great pressure during the development process to lower the molecular weight, likely because larger molecules show reduced passive absorption across cell membranes, increased number of toxic pharmacophores or rapidly metabolized moieties(49). One caveat when filtering on lipophilicity or solubility is to note whether you are using experimental values or predicted values. Solubility predictions based on clogP values or PSA can be accurate in some circumstances, but are inaccurate in others and tend to perform particularly badly for charged compounds(50). Charged compounds may be better represented by the octanol/water distribution coefficient logD, which takes into account the different protonation states. It is vital to carefully consider whether compounds should be excluded based on predicted insolubility, when such predictions can be inaccurate.
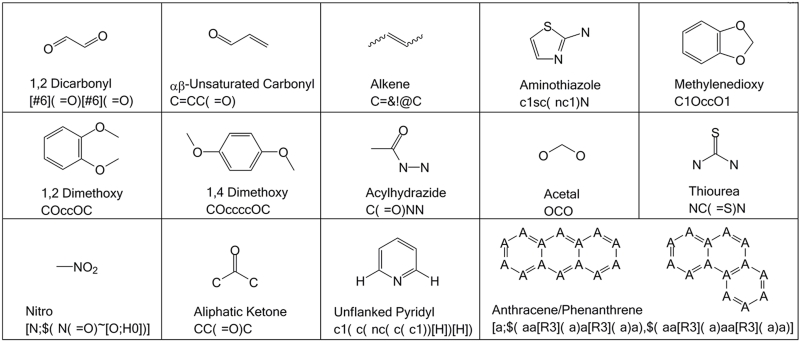
One other significant method for marking ADMET risks are the Rapid Elimination of Swill(51) (REOS) filters. As well as physicochemical properties, REOS filters remove molecules containing certain functional groups, as described by SMILES or SMARTS patterns(52). Some of these are shown in Figure 1. REOS filters flag compounds containing functional groups that may lead to false positives due to reactivity or assay interference, which have long been noted as a problem in HTS efforts(53). They also remove compounds containing functional groups known to be risks for ADMET. However, it is important to note that many known drug molecules fail the common physicochemical and substructure filters. The Drugbank(54) (http://www.drugbank.ca/) contains structural data for over 1,350 FDA approved small molecule drugs and nearly 5000 experimental drug entries. Analysis of the Drugbank experimental drugs is shown in Table 1 and reveals that only 71.4% pass all of the Lipinski filters and only 51.7% pass all of the REOS substructure filters. This data highlights that compound filtering is used to reduce risk, but will also eliminate useful molecules from further consideration. More recently, a Herculean analysis of compounds hitting multiple orthogonal HTS assays has lead to the identification of pan assay interference compounds (PAINS)(55). As increasing amounts of assay data from different HTS efforts around the world is becoming publically available, a clearer picture of compounds and functional groups that tend to yield false positives is developing(56). This development is vital, as frequent hitters are likely to be over represented in compounds from chemical vendors due to an increased likelihood that they will be ordered as analogues of apparent hits. Research has also specifically highlighted substructures that alert when a compound may be a DNA-reactive genotoxin(57). Whilst this may be acceptable in a screening hit, it would almost certainly have to be removed in the hit to lead process.
Figure 1. Chemical structures used in compound filtering.
Chemical structures of functional groups commonly used to remove compounds from consideration in HTS assays. The functional group name and SMILES/SMARTS string used in the filter are reported.
Physicochemical Property Filters
The majority of physicochemical property filters are simple to understand. Eight drug-like filters and one lead-like filter are described in Table 2. There is general agreement, although the exact properties vary slightly. Any of these rules can be used, alone or in conjunction, to filter a set of compounds and it is worth noting that many of the properties are highly correlated, such as logP and PSA. However, due consideration must be given to the details of the screening assay and the nature of the target as this affects the desired physicochemical properties of the screening compounds. For example, a fragment with a molecular weight of 200 may be too small to show measurable binding in typical HTS assays or compete with high-affinity ligands. However, if the assay is tailored to identify smaller molecules, fragment based methods have been shown to be very useful, with higher ligand efficiencies(58) and a greater potential for chemical elaboration and linking(59). Compound filters for fragments are completely different to filters for traditional small molecules. Phenotypic screens also place a different pressure on the screening library, with considerably more emphasis on cell permeability at the initial stage. As well as the importance of the assay format, the composition of an ideal screening library also varies with the protein target. Many existing screening libraries and are tailored toward screening against a narrow range of targets such as kinases and GPCRs(60). A screening library tailored toward screening against protein-protein interactions would have a very different profile. Recent analysis collected in the TIMBAL database(24) suggests that inhibitors of protein-protein interactions have higher molecular weights and lipophilicity than inhibitors of buried binding sites, as well as a greater number of hydrogen bond donors, hydrogen bond acceptors and rotatable bonds. Whilst the general applicability of this approach to generating approved drugs remains to be seen, it is an important consideration. As well as traditional physicochemical property filters, there are now a number of flags for more complex properties(61). Increasing evidence shows that small molecules may cause non-specific protein aggregation(62) and thus lead to false positives in some assays. Experimental work has shown that a significant number of compounds may act in this way and potential risks can be identified and removed from consideration(63). There are also experimental methods to identify compound that are reactive, such as ALARM NMR(64), and also for compounds containing fluorophores(65). However, whilst the latter is of great importance for fluorometric assays, it is of little or no importance in other assays. Experimental studies such as PAINS have identified molecular scaffolds that form the basis for promiscuous inhibitors and thus yield false positives in many screening assays(55, 66). Defining the mechanism underlying the promiscuous inhibition of these PAINS compounds will no doubt provide significant but interesting challenges in the next decade. In addition there are now methods for predicting compounds that disrupt particular screening assays(67), but these methods are approximate and should be used with this understanding.
Table 2. Details of physicochemical property filters to mark drug-like and lead-like compounds for screening libraries. LTE stands for less than or equal to.
| MW | PSA (A2) | HBA | HBD | logP | Rotatable Bonds | # Atoms | Charge | |
|---|---|---|---|---|---|---|---|---|
| Lipinski (1997) | LTE 500 | 0 to 10 | 0 to 5 | LTE 5.0 | ||||
| Ghose (1999) | 160 to 480 | −0.4 to +5.6 | 20 to 70 | |||||
| Oprea Drug-Like (2000) | 2 to 9 | 0 to 2 | 2 to 8 | |||||
| Egan (2000) | LTE 130 | −1.0 to +5.8 | ||||||
| Walters (2000) | 200 to 500 | LTE 120 | 0 to 10 | 0 to 5 | 0 to 8 | 20 to 70 | −2 to +2 | |
| Oprea Lead-Like (2001) | LTE 450 | 0 to 8 | 0 to 5 | −3.5 to +4.5 | ||||
| Veber (2002) | LTE 140 | 0 to 10 | ||||||
| REOS (2002) | 200 to 500 | 0 to 10 | 0 to 5 | −5.0 to +5.0 | 0 to 8 | −2 to +2 | ||
| Martin (2005) | LTE 150 |
Substructure Filters
Many filters simply remove compounds with specific functional groups that are known to interfere with HTS assays or cause problems later in drug development. The importance of removing these functional groups has been discussed in numerous papers(38, 53). The majority of screening libraries contain very few if any of the most troublesome compounds such as aldehydes, epoxides or α-halo ketones. The prevalence of these three groups in the six supplier databases is on average 0.3%, 0.01% and 0.04% respectively. However, many still contain potential risks such as isolated alkenes (12.3%), αβ-unsaturated carbonyls (8.5%) or nitro groups (7.6%). The prevalence of the more common functional groups can be seen in Table 3. Each of these substructures is a potential liability for the reasons described in Box 1.
Table 3.
Percentage of compounds failing common drug-like filters for unfavourable physiochemical properties and unwanted substructures for the six combined chemical supplier libraries, the ZINC database of purchasable molecules and the Drugbank database of experimental drugs. All physicochemical properties were generated with Qikprop and filtering was performed with Canvas.
| Combined Suppliers | Drugbank | ZINC | |
|---|---|---|---|
| clogP > 5 | 15.8 | 7.0 | 10.7 |
| HBA > 10 | 3.8 | 23.0 | 6.7 |
| HBD > 5 | 0.0 | 13.1 | 0.1 |
| MW > 500 | 4.9 | 13.3 | 1.7 |
| PSA > 150 | 1.8 | 22.0 | 3.3 |
| Rotatable Bonds > 10 | 1.5 | 20.3 | 2.5 |
| Isolated Alkene | 9.1 | 12.3 | 8.7 |
| αβ-Unsaturated Carbonyl | 8.5 | 8.5 | 6.9 |
| 1,2-Dimethoxy | 7.6 | 6.0 | 7.6 |
| Nitro | 7.4 | 6.6 | 6.5 |
| Acylhydrazide | 4.0 | 4.6 | 4.1 |
| Aminothiazole | 4.0 | 4.8 | 3.1 |
| Thiourea | 3.3 | 4.3 | 1.6 |
| Anthracene/Phenanthrene-like | 3.1 | 5.9 | 1.2 |
| Unflanked Pyridyl | 3.1 | 5.9 | 2.5 |
| Acetal | 2.7 | 13.0 | 2.0 |
| Methylene-Dioxy | 2.3 | 4.6 | 1.5 |
| Aliphatic Ketone | 2.1 | 10.6 | 2.0 |
| 1,2 dicarbonyl | 1.6 | 5.6 | 1.0 |
| 1,4-dimethoxy | 1.5 | 4.5 | 1.6 |
Box 1. Screening Liabilities.
1,2 dicarbonyls – Metabolically unstable/Potential toxicity due to mutagenicity.
1,2 dimethoxys – Prone to oxidation yielding reactive quinones.
1,4 dimethoxys – Very prone to oxidation yielding reactive quinones.
αβ-Unsaturated Carbonyls – Prone to reactivity by acting as a Michael acceptor.
Acetals – Metabolically unstable due to acetal hydrolysis.
Acylhydrazides – Metabolically unstable due to acyl hydrolysis.
Aliphatic Ketones – Metabolically unstable due to nucleophilic attack.
Alkenes – Metabolically unstable due to epoxidation.
Aminothiazoles – Potential toxicity.
Anthracene/Phenanthrene-likes – Known DNA intercalation.
Nitro Groups – Prone to reduction yielding reactive species/Potential hepatocarcinogens.
Methylenedioxys – Metabolically unstable due to acetal hydrolysis/Prone to oxidation yielding reactive quinones.
Thioureas – Metabolically unstable due to flavin oxidation/Potential non-specific protein binding.
Unflanked Pyridyls – Potential interference with cytochrome P450s due to metal ion coordination.
However, many of these functional groups do appear in certified drug molecules(68), as shown in Table 3, and many show no activity in HTS assays(69). When eliminating functional groups due to any ADMET risk, the nature of the functional group should be considered. It may be easier to replace a potentially risky side-group at the hit-to-lead stage than a potentially risky core group. For example, a nitroaromatic side-group can be replaced with another similar side-group such as a trifluoromethanesulfonyl side-group to retain or increase binding affinity without disrupting the structure of the molecule(70). The same is not true for a 2-aminothiazole core group, as its shape and hydrogen bonding characteristics are more difficult to mimic without disrupting the structure of the molecule. Despite this, scaffold hopping can be achieved and is increasingly common(71). When eliminating functional groups due to the risk of cytotoxicity, it is important to consider the target, as some therapies (for cancer in particular) are damaging to cells. For example, 2-aminothiazoles may lead to cytotoxicity but they form the basis of a number of potent CDK inhibitors for cancer therapy(72). Functional groups implicated in organ toxicity may also be acceptable in chemical probe discovery.
Filtering Tools
There are a number of software packages used to predict chemical properties and/or filter screening compounds. This includes Accelrys’ Pipeline Pilot(73), MOE’s sdfilter(74), Schrodinger’s qikprop(75) and Openeye’s filter(76), which is freely available to academics. Once the filtering process is complete, it is important to inspect a subset of the resulting structures. No matter how sophisticated the filtering criteria and algorithms, a scientist should always ensure that the remaining compounds meet their requirements. Despite the importance of filtering compounds to prevent screening potentially problematic compounds, it is common to screen a small proportion of “wildcards” that do not pass all of the filters. As seen in Tables 1 and 2, many drug molecules do not pass the drug-like or lead-like filters and contain significant proportions of functional groups that are commonly removed by HTS filters. For example, the REOS rule to exclude compounds with more than four joined rings, removes all steroids and nearly 10% of the Drugbank experimental drugs. It is important to realise that the process of compound filtering is about minimising risk and downstream expenditure rather than maximising hit-rate. For example, reactive groups may present the risk of false positives, but work has shown that this is not always the case(69). In some cases, reactive groups can act as covalent inhibitors, inactivating the target by binding irreversibly, and thus provide an advantage over non-covalent inhibitors. However, this activity may be difficult to extract from HTS data as it can be hard to discriminate from unwanted reactivity. Potentially reactive compounds should remain, at most, a small percentage of any screening library, unless there is a clear plan to extract useful data on covalent inhibition from the screening assay.
In summary, it may be necessary to rethink the process of designing libraries for screening against the more diverse range of targets now being considered. Research at Harvard(77), the NIH(6, 78), and the DDU in Dundee(9) amongst others has shown that HTS is feasible in a non-industrial center and can be vital in developing treatments for neglected diseases. Whilst such drug development projects must also select screening compounds with care, many of the functional group and physicochemical property filters are unsuitable for screening efforts aimed at development of chemical probes. Compounds causing assay interference or low solubility should be avoided, but compounds causing liver toxicity or poor oral absorption may be acceptable. Recent analysis suggests that the nature of screening hits is shifting to larger and more lipophilic molecules as a result of the increased use of in vitro assays over in vivo assays(79). This is expected to shift or widen the nature of screening libraries. However, the exact nature of the assay and the target must be considered when selecting compound exclusions as, for a diversity library aiming to span multiple assays and targets, it may not be appropriate to remove all potential risks. A balance must be reached between filtering out all compounds that are a risk in any drug development program and only filtering compounds that are a risk in all programs. There is now a critical mass of published data highlighting risks for compound interference and this can easily be applied to hits post screening, along with experimental methods to detect false positives such as dose-response plotting. This should ensure that screening libraries take advantage of the enormous diversity in chemical space, whilst assessing risk appropriately. With respect to chemical diversity, chemical suppliers will only provide chiral compounds if there is a market for them and thus filtering out chiral compounds from screening libraries will drive the purchasable chemical space further in this direction and away from biogenic chemical space.
Compound Selection
Aggressive filtering may remove up to 50% of compounds from consideration, but huge numbers of commercially available compounds still remain. The main aim of compound selection is to pick a subset of these compounds for testing. In general, it is wasteful to test many compounds with similar structures in frontline assays, at the expense of more diverse compounds. Analysis has shown that if a compound is biologically active, a molecule with very high similarity will have a similar biologically activity and thus testing the second molecule in the frontline assay is unlikely to be worthwhile(80-81). It is thus common to select a structurally diverse subset of compounds that represents the chemical space being considered. However, chemical space grows very rapidly with molecular size and, in 200 years of chemical synthesis, we have covered only a tiny fraction of chemical space up to a molecular weight of 500. The biggest screening libraries, which are of the order of tens of millions of molecules, can never hope to cover this space. Approaching compound selection in a sensible manner is thus very important(82).
Measuring Chemical Diversity
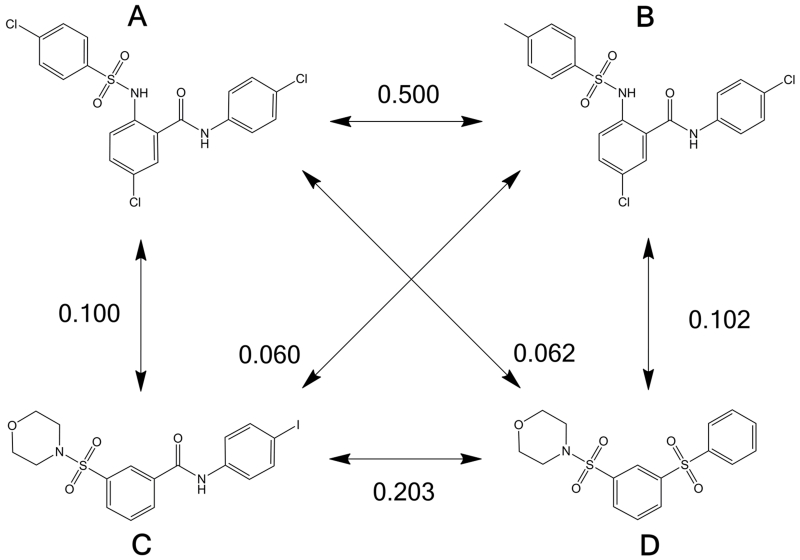
Molecular similarity is a key prerequisite in assessing molecular diversity(83). There are many different techniques to measure whether two compounds are similar(81, 84) but none of them are entirely satisfactory. From a pharmaceutical perspective, the ideal metric would predict that two compounds are similar if they elicit the same biological effect by hitting the same biological target and binding in the same pose. Unfortunately such a metric does not exist. Currently used metrics predict that two compounds are similar if they have similar chemical connectivity or similar shape and electrostatic form. One important issue in assessing chemical similarity is that a compound can be very different in its various conformations, tautomers and protonation states. Two compounds that are calculated to be similar in specific tautomeric states may be calculated to be different in other states. However, there are numerous computational methods for the enumeration of protonation and tautomeric states. This includes Schrodinger’s Ligprep(85), the Openeye toolkit(76), CCG’s MOE(74), Tripos Sybyl(86) and Accelrys’ Discovery Studio(73). Three of the most common methods for predicting similarity are fingerprint(87), shape-based(88) and pharmacophore(71) methods. These methods are commonly used in virtual screening when a known active compound has been identified. Fingerprint methods are relatively simple and usually two-dimensional. Each molecule is assessed for a number of atom and bond connectivities. Each of these connected units is termed a bit/key and the combination of bits/keys that are present in a given molecule is its fingerprint. Two molecules with similar fingerprints have similar atoms in similar bonding environments and are likely to bind in similar ways to a protein target. There are a number of fingerprinting techniques as well as a number of atom-typing schemes and close reading of the current literature is recommended before selecting a method, as this is still a rapidly developing field(89). Recent analysis has shown that atom-type based radial fingerprints perform well(90) but other work suggests that fingerprints based on physicochemical properties or pharmacophores may perform better(91). Different fingerprinting methods can yield very different similarities and thus an exact comparison with literature is not always appropriate. There are also a number of similarity/difference metrics(92) and, whilst the Tanimoto metric is most commonly used, close reading of the current literature is again recommended. The molecules in Figure 2 were analyzed using radial fingerprints based on daylight atom types using Schrodinger’s Canvas software and Tanimoto similarity scores were then generated. As can be seen, molecule with a high similarity such as A and B are very similar and would likely give similar assay results, whereas molecule A and D are significantly different and should ideally both be tested in a frontline assay. Shape based methods compare molecules by analyzing whether they have the same shape and electronic form. This is implemented in Openeye’s ROCS and EON software(76), which is widely used and is freely available to non-commercial groups working toward public disclosure(93). Pharmacophore methods have the obvious advantage of including the three dimensional geometry of the molecules. As noted, chemical similarity is a very important concept in assessing chemical diversity. Whilst three dimensional methods have the potential to provide a much more accurate model of molecular similarity, there is great difficulty in applying them when the bioactive conformation is unknown, as is the case in diversity analysis. Thus, two dimensional methods such as fingerprinting remain the tool of choice at present.
Figure 2. Example of similarity between compounds.
Four compounds and the Tanimoto similarity between them. The compounds were assigned radial fingerprints using Schrodinger’s Canvas software at 64-bit precision using daylight invariant atom types.
Rational Selection
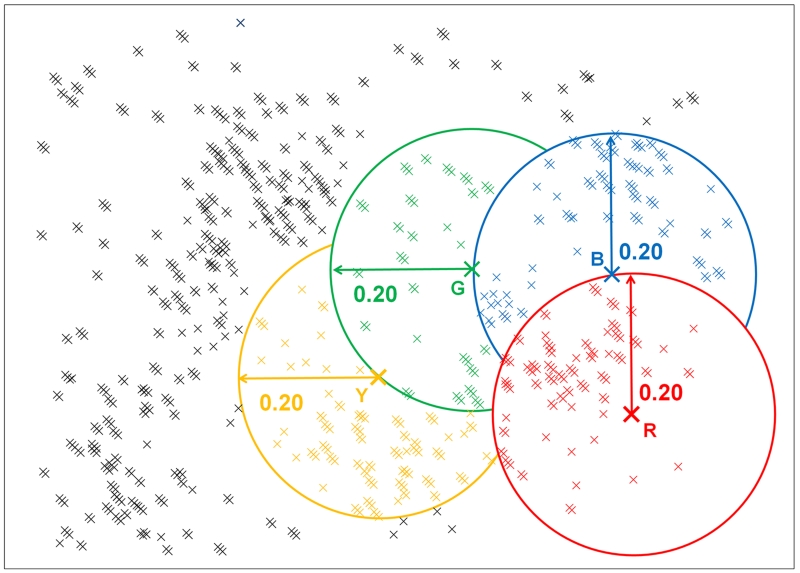
Once a set of compounds has been analyzed on the basis of similarity it is possible to select a diverse set of compounds. In some cases it is possible to consider the average similarity between compounds and optimise this as an objective function. However, this requires generation of an N by N similarity matrix, which may become prohibitively large as N increases(94). Heuristic clustering methods are thus more commonly used(94). Such methods include k-means clustering(95), sphere exclusion(96), directed sphere exclusion(97) and maxmin(98). The aim of such methods is that, for each selected molecule, no similar molecules are then selected. This is illustrated using a two dimensional representation for a simple sphere exclusion method in Figure 3. The centroid molecules R, B, G and Y represent all the molecules within a similarity of greater than 0.2. Iterative selection in this chemical space will finally encompass all molecules. A secondary aim of compound selection is to pick clusters of two or more structurally similar compounds in each cluster, such that the initial assay results immediately provide some QSAR data to inform decision-making. In many cases the aim of compound selection is to augment an existing compound collection. In this case, the existing compound structures can be used as an input to the diverse selection algorithm. This can be used to select new compounds that “fill the gaps” in chemical space. Despite this usefulness of diversity selection methods, the use of virtual screening methods should always be considered in a resource constrained environment, with sufficient knowledge of the protein target and its structure. Both molecular docking(99) and pharmacophore analysis(100) can improve hit rates in HTS assays and are commonly used.
Figure 3. Clustering of compounds in chemical space.
A two dimensional representation of chemical space being partitioned into clusters of similar compounds using a simple sphere exclusion method.
In summary, the process of selecting a representative subset of compounds from a large collection relies heavily on the ill-defined concept of molecular similarity. However, the concept is vital as it allows lead molecules to be identified at reduced cost and effort through hit identification and explosion.
Conclusions and Discussion
Shrewd selection of screening compounds is one of the most vital enabling steps in the drug development process. There are no strict rules, only rules of thumb. No compound filters are globally applicable and no diversity metrics or selection methods can be proven as optimal. However, misapplication of filtering can reduce chemical diversity within a project and preclude many novel discoveries. Conversely, careful filtering reduces the risk of false positives and downstream ADMET failures, whilst sensible compound selection can yield libraries that cover larger regions of chemical space and increase true positive hit rates. ADMET concerns may not be as important for chemical probes developed in academic groups, but solubility, cell permeability and potential chemical reactivity are all still important considerations and chemical diversity is still highly desirable. There are numerous sources of compound interference, which plague HTS assays. However, recent large-scale analyses have identified molecular scaffolds that appear as frequent hitters in numerous assays. The resultant data is very useful and should be incorporated either into library filtering or triaging of assay data. However, if every group used the same filters then every group would test similar compounds and many useful molecules could be missed. Large screening libraries in industry include a substantial fraction of commercially available compounds. Thus, if an academic group sources from commercial vendors and uses traditional industry filters then they will develop smaller relatives of the big industrial libraries with little or no chemical novelty. It may thus be advisable for academics to consider synthesizing or purchasing molecules in untapped regions of chemical space, particularly embodying multiple stereogenic centers, to maximise chemical diversity and increase the number of unique chemical entities tested. Diversity should also be maximised by considering natural products and biogenic scaffolds, which may show improved ADMET properties. At present, commercially available chemical space is heavily skewed toward flat compounds with many aromatic rings. Whilst this makes synthesis more tractable, it excludes many sources of chemical diversity and shifts screening libraries away from biogenic scaffolds and toward pharmacological risks. These risks have been recently quantified and the results are compelling(26). This problem will only be remedied by customers changing their practices to incentivise chemical suppliers.
A screening library must have the correct balance of molecular weight and logP, tailored to the constraints of the assay. Once a true positive hit has been identified, increasing size and complexity in tandem with lipophilicity is expected to increase both affinity and specificity. It is important to note that the ideal range of chemical and physicochemical properties of an HTS library differs when considering different assay platforms or protein targets. An optimal screening library for a fragment-based screen or targetting a protein-protein interaction will thus be different from a traditional kinase set and should be carefully designed. Due to the economies of scale with respect to purchasing a screening library, cost sharing between academic and government labs can increase the scope of screening efforts. Some companies may be willing to share portions of their screening libraries, in return for IP rights, on projects focused on commercially viable, validated targets. With respect to compound selection, there are numerous existing methods for measuring chemical similarity and selecting diverse sets of compounds, but no ideal metric can exist. Whilst current work has highlighted the best applications of fingerprinting, shape-based and pharmacophore methods, these are all evolving fields and no technique can be proven superior in all cases. However, compound selection through analysis of molecular similarity reduces the size and cost of screening libraries whilst retaining diversity.
One question of great importance that has not been addressed in great detail is how many compounds need to be tested to ensure a sufficient coverage of chemical space(101). This question can be answered by considering the number of lead series desired, the false positive rate, the number of molecules assayed per cluster and the hit rate of the primary screen. Such an analysis predicts that on average one lead series can be developed from testing approximately 350,000 diverse compounds in a typical HTS screen(102). This number applies only to leads successfully developed into marketed drugs and is thus not appropriate when considering chemical probe discovery. However, it is commonly accepted that some targets are more druggable than others such that this value can vary greatly and that some screens will yield no successful lead series. Due to the importance of HTS in the development of new drugs and chemical probes, high-quality screening libraries are a key asset of any research group and there are many factors to be weighed. However, each library will be unique and should be suited to the particular needs of the screening group. With the rapid increase in the number of purchasable molecules, the almost limitless volume of chemical space and the proliferation of HTS groups, rational selection of diverse hit-like compounds seems likely to continue as a lynchpin of drug development.
Acknowledgements
The authors would like to thank Andreas Bender, Bob Boyle, Mike Cherry, Jasveen Chugh, Warren Galloway, Simon Osborne, Mike Payne, William Ross Pitt and Herman Verheij for helpful discussions. We are grateful for financial support from the MRC, Wellcome Trust, CRUK, EPSRC, BBSRC and Newman Trust.
Abbreviations
- ADMET
Absorption, distribution, metabolism, elimination and toxicity
- HTS
High-throughput screening
- logP
Octanol/water partition coefficient
- PAINS
Pan assay interference compounds
- PSA
Polar surface area.REOS: Rapid elimination of swill
References
- 1.Pauling L, Itano H, Singer S, Wells I. Sickle Cell Anemia, a Molecular Disease. Science. 1949;110:543. doi: 10.1126/science.110.2865.543. [DOI] [PubMed] [Google Scholar]
- 2.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy JP, Williams L, Bridges TM, Daniels RN, Weaver D, Lindsley CW. Application of combinatorial chemistry science on modern drug discovery. J Comb Chem. 2008;10:345–354. doi: 10.1021/cc700187t. [DOI] [PubMed] [Google Scholar]
- 4.Spring DR. Chemical genetics to chemical genomics: small molecules offer big insights. Chem Soc Rev. 2005;34:472–482. doi: 10.1039/b312875j. [DOI] [PubMed] [Google Scholar]
- 5.Kodadek T. Rethinking screening. Nature Chemical Biology. 2010;6:162–165. doi: 10.1038/nchembio.303. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy A. The NIH Molecular Libraries Program: Identifying Chemical Probes for New Medicines. Chemistry & Biology. 2010;17:549–550. doi: 10.1016/j.chembiol.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Workman P, Collins I. Probing the Probes: Fitness Factors For Small Molecule Tools. Chemistry & Biology. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inglese J, Johnson RL, Simeonov A, Xia MH, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nature Chemical Biology. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 9.Brenk R, Schipani A, James D, Krasowski A, Gilbert IH, Frearson J, Wyatt PG. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. Chemmedchem. 2008;3:435–444. doi: 10.1002/cmdc.200700139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajay, Walters WP, Murcko MA. Can we learn to distinguish between “drug-like” and “nondrug-like” molecules? Journal of Medicinal Chemistry. 1998;41:3314–3324. doi: 10.1021/jm970666c. [DOI] [PubMed] [Google Scholar]
- 11.Teague SJ, Davis AM, Leeson PD, Oprea T. The design of leadlike combinatorial libraries. Angew Chem Int Edit. 1999;38:3743–3748. doi: 10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd DG, Golfis G, Knox AJ, Fayne D, Meegan MJ, Oprea TI. Oncology exploration: charting cancer medicinal chemistry space. Drug Discov Today. 2006;11:149–159. doi: 10.1016/S1359-6446(05)03688-3. [DOI] [PubMed] [Google Scholar]
- 13.Baurin N, Baker R, Richardson C, Chen I, Foloppe N, Potter A, Jordan A, Roughley S, Parratt M, Greaney P, Morley D, Hubbard RE. Drug-like annotation and duplicate analysis of a 23-supplier chemical database totalling 2.7 million compounds. Journal of Chemical Information and Computer Sciences. 2004;44:643–651. doi: 10.1021/ci034260m. [DOI] [PubMed] [Google Scholar]
- 14.Chuprina A, Lukin O, Demoiseaux R, Buzko A, Shivanyuk A. Drug- and Lead-likeness, Target Class, and Molecular Diversity Analysis of 7.9 Million Commercially Available Organic Compounds Provided by 29 Suppliers. Journal of Chemical Information and Modeling. 2010;50:470–479. doi: 10.1021/ci900464s. [DOI] [PubMed] [Google Scholar]
- 15.Editorial The academic pursuit of screening. Nature Chemical Biology. 2007;3:433–433. doi: 10.1038/nchembio0807-433. [DOI] [PubMed] [Google Scholar]
- 16.Monge A, Arrault A, Marot C, Morin-Allory L. Managing, profiling and analyzing a library of 2.6 million compounds gathered from 32 chemical providers. Mol Divers. 2006;10:389–403. doi: 10.1007/s11030-006-9033-5. [DOI] [PubMed] [Google Scholar]
- 17.Sirois S, Hatzakis G, Wei DQ, Du QS, Chou KC. Assessment of chemical libraries for their druggability. Computational Biology and Chemistry. 2005;29:55–67. doi: 10.1016/j.compbiolchem.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Verheij HJ. Leadlikeness and structural diversity of synthetic screening libraries. Molecular Diversity. 2006;10:377–388. doi: 10.1007/s11030-006-9040-6. [DOI] [PubMed] [Google Scholar]
- 19.Chuprina A, Lukin O, Demoiseaux R, Buzko A, Shivanyuk A. Drug- and Lead-likeness, Target Class, and Molecular Diversity Analysis of 7.9 Million Commercially Available Organic Compounds Provided by 29 Suppliers. J Chem Inf Model. 2010 doi: 10.1021/ci900464s. [DOI] [PubMed] [Google Scholar]
- 20.Voigt JH, Bienfait B, Wang SM, Nicklaus MC. Comparison of the NCI open database with seven large chemical structural databases. Journal of Chemical Information and Computer Sciences. 2001;41:702–712. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- 21.Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohacek RS, McMartin C, Guida WC. The art and practice of structure-based drug design: A molecular modeling perspective. Med Res Rev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Fink T, Reymond JL. Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. J Chem Inf Model. 2007;47:342–353. doi: 10.1021/ci600423u. [DOI] [PubMed] [Google Scholar]
- 24.Higueruelo AP, Schreyer A, Bickerton GRJ, Pitt WR, Groom CR, Blundell TL. Atomic Interactions and Profile of Small Molecules Disrupting Protein-Protein Interfaces: the TIMBAL Database. Chemical Biology & Drug Design. 2009;74:457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 25.Lovering F, Bikker J, Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie TJ, Macdonald SJ. The impact of aromatic ring count on compound developability--are too many aromatic rings a liability in drug design? Drug Discov Today. 2009;14:1011–1020. doi: 10.1016/j.drudis.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 28.Singh N, Guha R, Giulianotti MA, Pinilla C, Houghten RA, Medina-Franco JL. Chemoinformatic Analysis of Combinatorial Libraries, Drugs, Natural Products, and Molecular Libraries Small Molecule Repository. Journal of Chemical Information and Modeling. 2009;49:1010–1024. doi: 10.1021/ci800426u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feher M, Schmidt JM. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. Journal of Chemical Information and Computer Sciences. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 30.Hert J, Irwin JJ, Laggner C, Keiser MJ, Shoichet BK. Quantifying biogenic bias in screening libraries. Nat Chem Biol. 2009;5:479–483. doi: 10.1038/nchembio.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spandl RJ, Bender A, Spring DR. Diversity-oriented synthesis; a spectrum of approaches and results. Org Biomol Chem. 2008;6:1149–1158. doi: 10.1039/b719372f. [DOI] [PubMed] [Google Scholar]
- 32.Clemons PA, Bodycombe NE, Carrinski HA, Wilson JA, Shamji AF, Wagner BK, Koehler AN, Schreiber SL. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. P Natl Acad Sci USA. 2010;107:18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. Journal of Chemical Information and Computer Sciences. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 34.Dobson PD, Kell DB. Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat Rev Drug Discov. 2008;7:205–220. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 35.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliver Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 36.Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 37.Veber DF. Molecular properties that influence the oral bioavailability of drug candidates. Abstracts of Papers of the American Chemical Society. 2003;225:U208–U208. [Google Scholar]
- 38.Walters WP, Murcko MA. Library Filtering Systems and Prediction of Drug-Like Properties. Methods and Principles in Medicinal Chemistry. 2000;10:15–30. [Google Scholar]
- 39.Oprea TI. Property distribution of drug-related chemical databases. Journal of Computer-Aided Molecular Design. 2000;14:251–264. doi: 10.1023/a:1008130001697. [DOI] [PubMed] [Google Scholar]
- 40.Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. Journal of Medicinal Chemistry. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 41.Lee ML, Schneider G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: Application in the design of natural product-based combinatorial libraries. J Comb Chem. 2001;3:284–289. doi: 10.1021/cc000097l. [DOI] [PubMed] [Google Scholar]
- 42.Martin YC. A bioavailability score. Journal of Medicinal Chemistry. 2005;48:3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
- 43.Palm K, Stenberg P, Luthman K, Artursson P. Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharmaceut Res. 1997;14:568–571. doi: 10.1023/a:1012188625088. [DOI] [PubMed] [Google Scholar]
- 44.Subramanian G, Kitchen DB. Computational approaches for modeling human intestinal absorption and permeability. J Mol Model. 2006;12:577–589. doi: 10.1007/s00894-005-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson TW, Dress KR, Edwards M. Using the Golden Triangle to optimize clearance and oral absorption. Bioorganic & Medicinal Chemistry Letters. 2009;19:5560–5564. doi: 10.1016/j.bmcl.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 46.Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 47.Hughes JD, Blagg J, Price DA, Bailey S, DeCrescenzo GA, Devraj RV, Ellsworth E, Fobian YM, Gibbs ME, Gilles RW, Greene N, Huang E, Krieger-Burke T, Loesel J, Wager T, Whiteley L, Zhang Y. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorganic & Medicinal Chemistry Letters. 2008;18:4872–4875. doi: 10.1016/j.bmcl.2008.07.071. [DOI] [PubMed] [Google Scholar]
- 48.Cronin D, Mark T. The role of hydrophobicity in toxicity prediction. Current Computer-Aided Drug Design. 2006;2:405–413. [Google Scholar]
- 49.Wenlock MC, Austin RP, Barton P, Davis AM, Leeson PD. A comparison of physiochemical property profiles of development and marketed oral drugs. Journal of Medicinal Chemistry. 2003;46:1250–1256. doi: 10.1021/jm021053p. [DOI] [PubMed] [Google Scholar]
- 50.Delaney JS. Predicting aqueous solubility from structure. Drug Discov Today. 2005;10:289–295. doi: 10.1016/S1359-6446(04)03365-3. [DOI] [PubMed] [Google Scholar]
- 51.Walters WP, Stahl MT, Murcko MA. Virtual screening - an overview. Drug Discov Today. 1998;3:160–178. [Google Scholar]
- 52.Weininger D. Smiles, a Chemical Language and Information-System .1. Introduction to Methodology and Encoding Rules. Journal of Chemical Information and Computer Sciences. 1988;28:31–36. [Google Scholar]
- 53.Rishton GM. Reactive compounds and in vitro false positives in HTS. Drug Discov Today. 1997;2:382–384. [Google Scholar]
- 54.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baell JB, Holloway GA. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J Med Chem. 2010 doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 56.Editorial Screening we can believe. Nature Chemical Biology. 2009;5:127–127. doi: 10.1038/nchembio0309-127. [DOI] [PubMed] [Google Scholar]
- 57.Snodin DJ. Genotoxic Impurities: From Structural Alerts to Qualification. Org Process Res Dev. 2010;14:960–976. [Google Scholar]
- 58.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 59.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 60.Miller JL. Recent developments in focused library design: Targeting gene-families. Current Topics in Medicinal Chemistry. 2006;6:19–29. doi: 10.2174/156802606775193347. [DOI] [PubMed] [Google Scholar]
- 61.Thorne N, Auld DS, Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. Journal of Medicinal Chemistry. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 63.Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 64.Huth JR, Mendoza R, Olejniczak ET, Johnson RW, Cothron DA, Liu YY, Lerner CG, Chen J, Hajduk PJ. ALARM NMR: A rapid and robust experimental method to detect reactive false positives in biochemical screens. Journal of the American Chemical Society. 2005;127:217–224. doi: 10.1021/ja0455547. [DOI] [PubMed] [Google Scholar]
- 65.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 66.Pearce BC, Sofia MJ, Good AC, Drexler DM, Stock DA. An empirical process for the design of high-throughput screening deck filters. J Chem Inf Model. 2006;46:1060–1068. doi: 10.1021/ci050504m. [DOI] [PubMed] [Google Scholar]
- 67.Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J Med Chem. 2009;53:37–51. doi: 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Axerio-Cilies P, Castaneda IP, Mirza A, Reynisson J. Investigation of the incidence of “undesirable” molecular moieties for high-throughput screening compound libraries in marketed drug compounds. Eur J Med Chem. 2009;44:1128–1134. doi: 10.1016/j.ejmech.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 69.Babaoglu K, Simeonov A, Lrwin JJ, Nelson ME, Feng B, Thomas CJ, Cancian L, Costi MP, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase. Journal of Medicinal Chemistry. 2008;51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park CM, Bruncko M, Adickes J, Bauch J, Ding H, Kunzer A, Marsh KC, Nimmer P, Shoemaker AR, Song X, Tahir SK, Tse C, Wang XL, Wendt MD, Yang XF, Zhang HC, Fesik SW, Rosenberg SH, Elmore SW. Discovery of an Orally Bioavailable Small Molecule Inhibitor of Prosurvival B-Cell Lymphoma 2 Proteins. Journal of Medicinal Chemistry. 2008;51:6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Q, Muegge I. Scaffold hopping through virtual screening using 2D and 3D similarity descriptors: Ranking, voting, and consensus scoring. Journal of Medicinal Chemistry. 2006;49:1536–1548. doi: 10.1021/jm050468i. [DOI] [PubMed] [Google Scholar]
- 72.Misra RN, Xiao HY, Kim KS, Lu SF, Han WC, Barbosa SA, Hunt JT, Rawlins DB, Shan WF, Ahmed SZ, Qian LG, Chen BC, Zhao RL, Bednarz MS, Kellar KA, Mulheron JG, Batorsky R, Roongta U, Kamath A, Marathe P, Ranadive SA, Sack JS, Tokarski JS, Pavletich NP, Lee FYF, Webster KR, Kimball SD. N-(Cycloalkylamino)acyl-2-aminothiazole inhibitors of cyclin-dependent kinase - 2. N-[5-[[[5-(1,1-dimethylethyl)-2-oxazolyl]methyl]thio]-2-thiazolyl]-4-piperidinecarboxamide (BMS-387032), a highly efficacious and selective antitumor agent. Journal of Medicinal Chemistry. 2004;47:1719–1728. doi: 10.1021/jm0305568. [DOI] [PubMed] [Google Scholar]
- 73.Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- 74.Syed N, Smith P, Sullivan A, Spender LC, Dyer M, Karran L, O’Nions J, Allday M, Hoffmann I, Crawford D, Griffin B, Farrell PJ, Crook T. Transcriptional silencing of Polo-like kinase 2 (SNK/PLK2) is a frequent event in B-cell malignancies. Blood. 2006;107:250–256. doi: 10.1182/blood-2005-03-1194. [DOI] [PubMed] [Google Scholar]
- 75.Inglis KJ, Chereau D, Brigham EF, Chiou SS, Schobel S, Frigon NL, Yu M, Caccavello RJ, Nelson S, Motter R, Wright S, Chian D, Santiago P, Soriano F, Ramos C, Powell K, Goldstein JM, Babcock M, Yednock T, Bard F, Basi GS, Sham H, Chilcote TJ, McConlogue L, Griswold-Prenner I, Anderson JP. Polo-like kinase 2 (PLK2) phosphorylates alpha-synuclein at serine 129 in central nervous system. J Biol Chem. 2009;284:2598–2602. doi: 10.1074/jbc.C800206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Carcer G, Perez de Castro I, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem. 2007;14:969–985. doi: 10.2174/092986707780362925. [DOI] [PubMed] [Google Scholar]
- 77.Stein RL. High-throughput screening in academia: The Harvard experience. J Biomol Screen. 2003;8:615–619. doi: 10.1177/1087057103260741. [DOI] [PubMed] [Google Scholar]
- 78.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 79.Keseru GM, Makara GM. The influence of lead discovery strategies on the properties of drug candidates. Nat Rev Drug Discov. 2009;8:203–212. doi: 10.1038/nrd2796. [DOI] [PubMed] [Google Scholar]
- 80.Matter H. Selecting optimally diverse compounds from structure databases: A validation study of two-dimensional and three-dimensional molecular descriptors. Journal of Medicinal Chemistry. 1997;40:1219–1229. doi: 10.1021/jm960352+. [DOI] [PubMed] [Google Scholar]
- 81.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? Journal of Medicinal Chemistry. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 82.Rishton GM. Molecular diversity in the context of leadlikeness: compound properties that enable effective biochemical screening. Curr Opin Chem Biol. 2008;12:340–351. doi: 10.1016/j.cbpa.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 83.Bender A, Glen RC. Molecular similarity: a key technique in molecular informatics. Org Biomol Chem. 2004;2:3204–3218. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]
- 84.Willett P, Barnard JM, Downs GM. Chemical similarity searching. Journal of Chemical Information and Computer Sciences. 1998;38:983–996. [Google Scholar]
- 85.Malumbres M, Barbacid M. Cell cycle kinases in cancer. Curr Opin Genet Dev. 2007;17:60–65. doi: 10.1016/j.gde.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 86.Sybyl. Tripos; St. Louis, MO: [Google Scholar]
- 87.Hert J, Willett P, Wilton DJ. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. Journal of Chemical Information and Computer Sciences. 2004;44:1177–1185. doi: 10.1021/ci034231b. [DOI] [PubMed] [Google Scholar]
- 88.Naylor E, Arredouani A, Vasudevan SR, Lewis AM, Parkesh R, Mizote A, Rosen D, Thomas JM, Izumi M, Ganesan A, Galione A, Churchill GC. Identification of a chemical probe for NAADP by virtual screening. Nature Chemical Biology. 2009;5:220–226. doi: 10.1038/nchembio.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duan JX, Dixon SL, Lowrie JF, Sherman W. Analysis and comparison of 2D fingerprints: Insights into database screening performance using eight fingerprint methods. Journal of Molecular Graphics & Modelling. 2010;29:157–170. doi: 10.1016/j.jmgm.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 90.Bender A, Jenkins JL, Scheiber J, Sukuru SCK, Glick M, Davies JW. How Similar Are Similarity Searching Methods? A Principal Component Analysis of Molecular Descriptor Space. Journal of Chemical Information and Modeling. 2009;49:108–119. doi: 10.1021/ci800249s. [DOI] [PubMed] [Google Scholar]
- 91.Steffen A, Kogej T, Tyrchan C, Engkvist O. Comparison of Molecular Fingerprint Methods on the Basis of Biological Profile Data. Journal of Chemical Information and Modeling. 2009;49:338–347. doi: 10.1021/ci800326z. [DOI] [PubMed] [Google Scholar]
- 92.Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 93.Hawkins PCD, Skillman AG, Nicholls A. Comparison of shape-matching and docking as virtual screening tools. Journal of Medicinal Chemistry. 2007;50:74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- 94.Willett P. Dissimilarity-based algorithms for selecting structurally diverse sets of compounds. J Comput Biol. 1999;6:447–457. doi: 10.1089/106652799318382. [DOI] [PubMed] [Google Scholar]
- 95.Higgs RE, Bemis KG, Watson IA, Wikel JH. Experimental designs for selecting molecules from large chemical databases. Journal of Chemical Information and Computer Sciences. 1997;37:861–870. [Google Scholar]
- 96.Hudson BD, Hyde RM, Rahr E, Wood J. Parameter based methods for compound selection from chemical databases. Quant Struct-Act Rel. 1996;15:285–289. [Google Scholar]
- 97.Gobbi A, Lee ML. DISE: Directed Sphere Exclusion. Journal of Chemical Information and Computer Sciences. 2003;43:317–323. doi: 10.1021/ci025554v. [DOI] [PubMed] [Google Scholar]
- 98.Schmuker M, Givehchi A, Schneider G. Impact of different software implementations on the performance of the Maxmin method for diverse subset selection. Mol Divers. 2004;8:421–425. doi: 10.1023/b:modi.0000047503.82461.e8. [DOI] [PubMed] [Google Scholar]
- 99.Shoichet BK, McGovern SL, Wei BQ, Irwin JJ. Lead discovery using molecular docking. Curr Opin Chem Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 100.Wolber G, Langer T. LigandScout: 3-d pharmacophores derived from protein-bound Ligands and their use as virtual screening filters. Journal of Chemical Information and Modeling. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 101.Lipkin MJ, Stevens AP, Livingstone DJ, Harris CJ. How large does a compound screening collection need to be? Comb Chem High T Scr. 2008;11:482–493. doi: 10.2174/138620708784911492. [DOI] [PubMed] [Google Scholar]
- 102.Harper G, Pickett SD, Green DV. Design of a compound screening collection for use in high throughput screening. Comb Chem High Throughput Screen. 2004;7:63–70. doi: 10.2174/138620704772884832. [DOI] [PubMed] [Google Scholar]