Summary
Cytochrome P450 (P450) 4X1 is one of the so-called “orphan” P450s without assigned biological function. Codon-optimized P450 4X1 and a number of N-terminal modified sequences were expressed in Escherichia coli. Native P450 4X1 showed a characteristic P450 spectrum but low expression in E. coli DH5α cells (<100 nmol P450/L). The highest level of expression (300-450 nmol P450/L culture) was achieved with a bicistronic P450 4X1 construct (N-terminal MAKKTSSKGKL, change of E2A, amino acids 3-44 truncated). Anandamide (arachidonoyl ethanolamide) has emerged as an important signaling molecule in the neurovascular cascade. Recombinant P450 4X1 protein, co-expressed with human NADPH-P450 reductase in E. coli, was found to convert the natural endocannabinoid anandamide to a single monooxygenated product, 14,15-epoxyeicosatrienoic (EET) ethanolamide. A stable anandamide analog (CD-25) was also converted to a monooxygenated product. Arachidonic acid was oxidized more slowly to 14,15- and 8,9-EETs but only in the presence of cytochrome b5. Other fatty acids were investigated as putative substrates but showed only little or minor oxidation. Real-time PCR analysis demonstrated extrahepatic mRNA expression, including several human brain structures (cerebellum, amygdala, and basal ganglia), in addition to expression in human heart, liver, prostate, and breast. The highest mRNA expression levels were detected in amygdala and skin. The ability of P450 4X1 to generate anandamide derivatives and the mRNA distribution pattern suggest a potential role for P450 4X1 in anandamide signaling in the brain.
Keywords: Cytochrome P450, anandamide, heterologous expression, mRNA localization
(Introduction)
Cytochrome P450 (P450, EC 1.14.14.1) monooxygenases are involved in tissue specific conversions of many naturally occurring substances; e.g., vitamins, hormones, and signaling molecules, including the diverse group of the so-called eicosanoids [2]. P450 Families 1-3 are primarily involved in the metabolism of therapeutic drugs and other xenobiotic chemicals, while Families 4-51 consist of enzymes involved in the endogenous metabolism of important biological compounds; e.g., steroids, fatty acids, vitamins, and eicosanoids [3]. P450 Subfamily 4F members are known to primarily oxidize endogenous compounds, e.g. fatty acids and arachidonic acid derivatives [4]. The primary site of P450 metabolism is the liver, and the amount of P450 found in brain is relatively low compared with liver [3], ranging from 1-10%. P450 metabolism of fatty acids may be of importance in brain, i.e. neurotransmitters and fatty acids are oxidized by P450s [4,5].
Arachidonic acid derivatives have been implicated in a large number of physiologically important processes. The arachidonic acid derivative anandamide (arachidonoyl ethanolamide) is a natural endocannabinoid found in most human tissues and acts as an important signaling mediator in both neurological and other physiological functions [6,7]. Anandamide was originally found in human brain, binding to the cannabinoid receptor CB1, and is believed to elicit cannabinoid-like pharmacological activity, i.e. nociception and hypomotility, with a 30-fold higher affinity in the brain than in the periphery [7,8]. 2-Arachidonoyl glycerol (2-AG) is another natural endogenous endocannabinoid [9]. Unlike 2-AG, the naturally occurring level of anandamide is relatively low in the central nervous system. When administrated as a drug, anandamide elicits pharmacological effects mimicking the effects of Δ9-tetrahydrocannabinol, the active component of marijuana (Cannabis sativa L.) [10]. Anandamide has recently been shown to be oxidized by P450s in mouse liver and brain microsomes [6] and human liver and kidney microsomes [11], forming a number of P450-derived hydroxyeicosatrienoic (HETE) and epoxyeicosatrienoic (EET) ethanolamides in the latter case.
At least one-fourth of the known 57 human P450 (CYP) genes (http://drnelson.utmem.edu/CytochromeP450.html) remain “orphans,” based on the terminology used for receptors and other proteins without known ligands. The largest number of orphans is found within P450 Family 4. P450 Family 4 consists of six human subfamilies: 4A, 4B, 4F, and the recently discovered 4X, 4V, and 4Z [3,12].
Human P450 4X1 (NM_178033.1) is located on chromosome 1p33 (www.ncbi.nlm.nih.gov) close to P450s 4Z1, 4Z2P, 4A11, 4A22, and 4B1. The gene has 12 exons and the predicted protein includes 509 amino acids. Homologous genes have been found in several mammalian species, including rat (70% amino acid similarity), mouse (71%), and dog (75%) (www.ensembl.org). Rat P450 4X1 was originally cloned using reverse transcriptase (RT) polymerase chain reaction (PCR) and found to be specifically expressed in several brain regions (e.g. brain stem, hippocampus, cortex, and cerebellum) as well as in vascular endothelial cells [13]. The mouse ortholog, P450 4x1, has been proposed to be a major brain P450, with protein localization demonstrated primarily in brain neurons, choroidal epithelial cells, and vascular endothelial cells [14]. Human P450 4X1 mRNA has been reported in kidney, brain, heart, and liver [15,16]. Expression was detected in brain by expressed sequence tag (EST) analysis and in aorta by mRNA blotting. However, no quantitation of the mRNA expression of P450 4X1 in tissues has been reported. A major limitation of these studies has been that no heterologous expression system has been published to date, and so far no catalytic activity has been reported in order to establish a putative physiological function.
We report the expression and purification of an N-terminal modified codon-optimized version of P450 4X1 in Escherichia coli. Recombinant P450 4X1 oxidized anandamide rather specifically to the 14,15-EET ethanolamide derivative, at a slow rate. Arachidonic acid formed trace amounts of 14,15- and 8,9-EETs but only in the presence of cytochrome b5 as an auxiliary factor. The rates of oxidation of a number of other arachidonic acid derivatives, neurosteroids (e.g. dopamine and tyramine), and common drugs (e.g. loratadine and clotrimazole) were below the limits of detection. Quantitative PCR indicated highest levels of P450 4X1 mRNA in brain regions and skin. The oxidation of anandamide (and a stable analog of anandamide and Δ9-tetrahydrocannabinol), albeit slow, suggests a potential role for P450 4X1 in neurovascular function in human brain.
Results
Synthesis of codon-optimized P450 4X1 cDNA
A cDNA was prepared for heterologous expression using polymerase chain assembly (PCA) with 63 overlapping oligonucleotides (Supplementary material Table S1). The sequence was codon-optimized for heterologous E. coli expression, a protocol previously used in this laboratory for successful expression of other P450s [17,18]. A product with a perfect P450 4X1 sequence was used for further studies and expression. The P450 4X1 insert was also integrated into a bicistronic vector (containing the cDNA for human NADPH-P450 reductase, EC 1.6.2.4) [19].
Expression of N-terminal variants
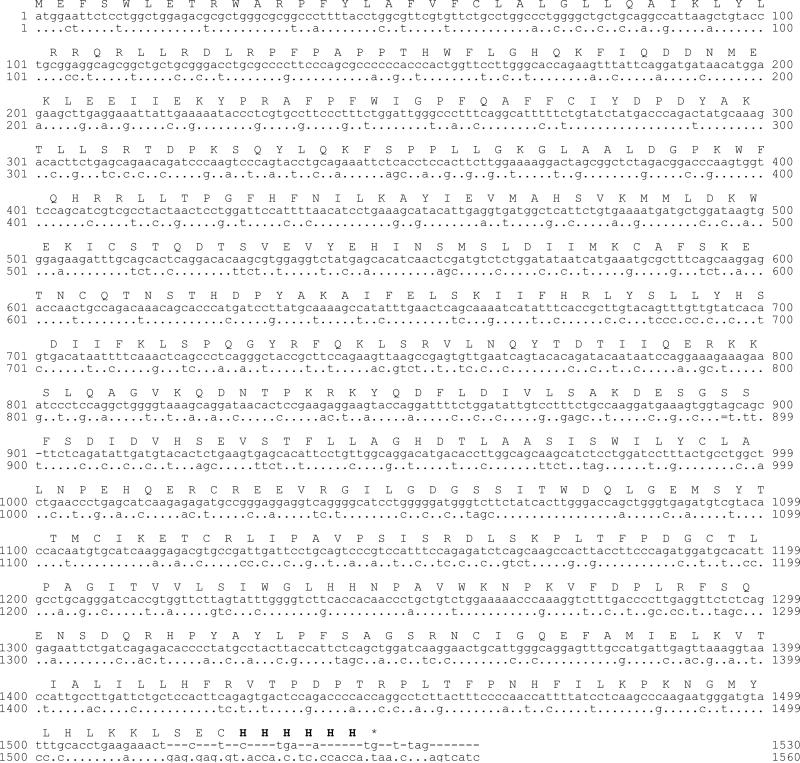
The alignment of the codon-optimized P450 4X1 sequence was compared to the native P450 4X1 sequence reported in the NCBI database (NM_178033) (Fig. 1). The modifications introduced at the N-terminus were based on alignments with close P450 family members. For P450 Family 4 enzymes most heterologous expression work to date has been performed in yeast, and a limited amount of information about E. coli expression is available. In the case of P450 4B1 [20] the best expression was achieved with a sequence adapted from bovine P450 17A1 [21] in front of the third codon (corresponding to P450 4X1 construct 2) (Fig. 2). In order to optimize expression levels, the first 45 amino acids were truncated based on predictions from the program SOPMA (Pôle Informatique Lyonnais, http://npsa-pbil.ibcp.fr/cgi-bin/secpred_sopma.pl) which indicated the presence of two α-helix structures in the N-terminal part of the protein (1-11 and 15-44). N-Terminal modified P450 4X1 constructs 3 and 4 (Fig. 2) were based on modifications previously used for rabbit P450 2C3 [22] and rat P450 2C11[23]. Both constructs have sequences truncated before the well-conserved proline rich region found at amino acid residues 44-50. P450 4X1 construct 1 used the bovine P450 17A1 sequence [21] along with a Δ2-44 truncation (Supplementary material Table S2, Figs. S1, S2).
Fig. 1.
Optimizations introduced into the P450 4X1 cDNA for E. coli expression. Upper line; predicted amino acid sequence; middle line, nucleotide sequence predicted from genomic sequence; lower line, nucleotide sequence optimized for E. coli expression.
Fig. 2.
N-Terminal modifications used for heterologous expression of P450 4X1 membranes in E. coli [18] (see also Supplementary material Fig. S2). Amino acid changes are in italics and underlined.
The levels of expression of native and N-terminally modified monocistronic P450 4X1 constructs were initially very modest in E. coli DH5α cells. For the native monocistronic P450 4X1 construct, the normal expression level was >100 nmol P450/L, with the highest level of expression ~ 200 nmol P450/L; however the apparent P450:cytochrome P420 ratio was ~1:20 and the weak P450 spectral peak was shifted (to 455 nm). We considered numerous changes to improve the ratio of P450 to cytochrome P420. A similar pattern was found for the four N-terminal modifications, with expression levels of ~25 nmol P450/L (30 °C, 48 h); at 24 h only P450 4X1 construct 2 showed expression (60 nmol P450/L). Expression trials with P450 4X1 constructs 1-4 (Fig. 2) were also carried out, using these constructs with co-expression of the molecular chaperones pGroES/EL12 in E. coli DH5α (induced by arabinose, 4 mg/mL); in this case P450 4X1 construct 1 showed an expression level of 150 nmol P450/L, and the remainder yielded <25 nmol/L (detection limit).
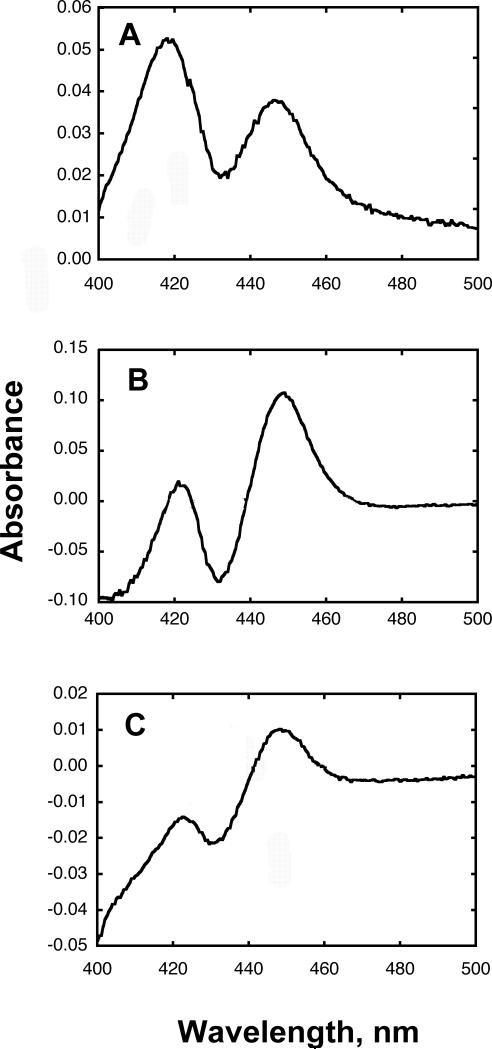
The inserts were moved into a bicistronic vector (containing human NADPH-P450 reductase). Expression trials were carried out using these constructs with and without co-expression of the molecular chaperones pGroES/EL12 in E. coli DH5α under different conditions of temperature and time. In E. coli DH5α cells none of these constructs expressed >25 nmol P450/L, while with co-expression of the molecular chaperones pGroES/EL12 in E. coli DH5α the expression levels of P450 4X1 construct 3 were considerably better. The optimal expression temperature for construct 3 was found to be 28 °C and a strong P450 peak was detected (Fig. 3A) 17-21 h following induction (150 - 450 nmol P450/L), with expression levels then decreasing with time to <70 nmol P450/L after 48 h. The OD600 at the time of induction proved to very important, because almost no expression was detected if the value was much lower or higher than 0.5.
Fig. 3.

Fe2+-CO vs. Fe2+ difference spectra. A, P450 4X1 construct 3 expression was done in E. coli (with pGroES/EL12). The spectrum was recorded using 1/2 dilutions of whole cell extracts and reducing with Na2S2O4. B, Solubilized P450 4X1 (1.5 μM) C, Difference spectrum of purified P450 4X1 (0.14 μM).
Purification of P450 4X1
Solubilization of the bicistronic P450 4X1 membranes was achieved in the presence of 1% sodium 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid (CHAPS) (w/v) (Fig. 3B) and purification was performed using a Ni-nitrilotriacetic acid (NTA) column (elution with imidazole, Fig. 3C) (39% yield). Purified P450 4X1 (Fig. 4) was found to aggregate (in the first trial, after removal of detergent and KCl and lowering the ionic strength to 100 mM); therefore, subsequent dialysis utilized a final storage buffer of 200 mM potassium phosphate buffer (containing 1 mM EDTA and 20% glycerol, v/v), which appeared to prevent aggregation.
Fig 4.
SDS-polyacrylamide gel electrophoresis of purified recombinant P450 4X1. Lane 1, Mr markers; lane 2, purified P450 4X1 (4 pmol).
Real-time Quantitative PCR Analysis of P450 4X1
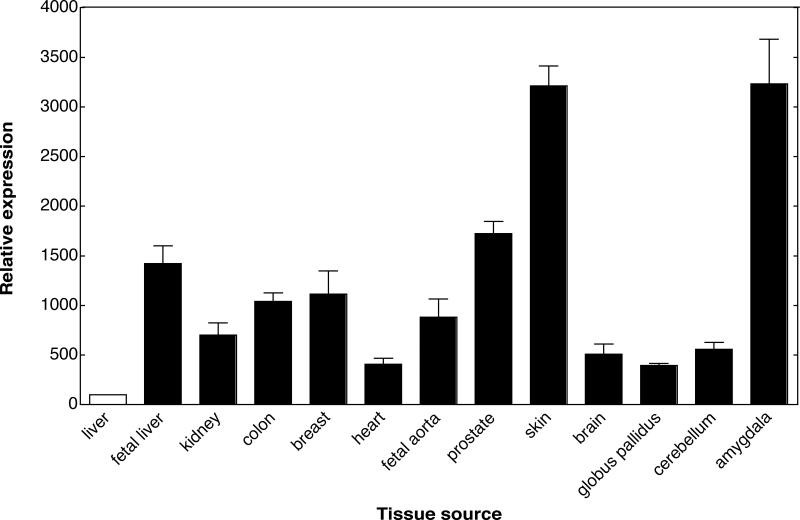
In order to investigate the quantitative tissue distribution pattern of P450 4X1 in human tissues, real-time PCR was used to compare the mRNA levels of P450 4X1 expression to an internal housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The results of the panels have, for graphic representation (Fig. 5), been normalized to human adult liver (at 100), and all the other values are compared with adult liver. The expression level in adult heart is 2- to 3-fold higher than in adult liver, and the mRNA levels in kidney, colon, breast, and fetal liver and aorta were 6- and 10-fold higher than in adult liver. The highest levels were detected in prostate, skin, and particularly amygdala. Whole brain levels were 2- to 3-fold higher than in liver, cerebellum was 3-fold higher, and amygdala was 20-fold higher (Fig. 5). However, the caveat should be added that all of the adult mRNA samples were from single donors (the fetal samples were from a pool of 5 individuals) and the issue of inter-individual variation has not been addressed. Due to the difficulty of obtaining human mRNA from multiple donors for some of these tissues, we were limited to investigating the expression levels with single donors in most cases.
Fig. 5.
Tissue distribution of P450 4X1 mRNA measured by real-time PCR. The relative levels of P450 4X1 mRNA were determined using real-time PCR in the tissues indicated below, using GAPDH as a reference standard. Different human cDNAs were used as templates, and SYBR Green was used for detection. The mRNA levels are shown as the ratio of P450 4X1 to GAPDH and represent the mean of triplicate measurements from each sample. The relative expression was calculated using the ΔCt method (Livak). For graphic presentation the graphs have standard deviations shown.
Search for Catalytic Properties of P450 4X1
A number of putative substrates were investigated, based on both the P450 4X1 mRNA tissue distribution and other well-known P450 Family 4 substrates (e.g. fatty acids and prostaglandins). In all but two cases, no oxidation to possible mono- or dioxygenated products was detected under our conditions (Supplementary material Table S3). Anandamide, considered the endogenous ligand of endocannabinoid receptors, exhibits cannabinoid-like pharmacological activity [6] and is known to be oxidized to prostaglandin-like products by cyclooxygenases [24].
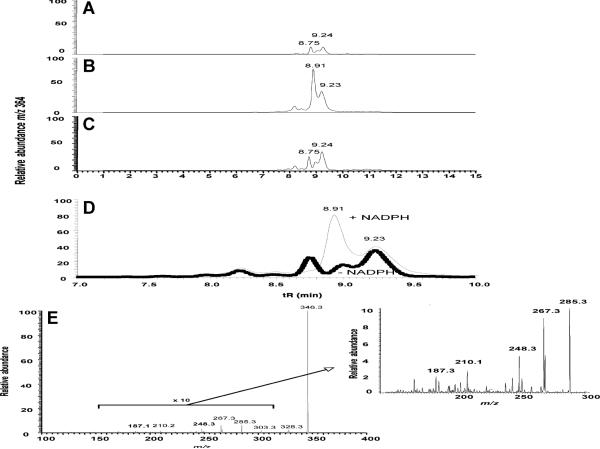
P450 4X1 did not form 20-HETE ethanolamide; however, one of the four potential epoxide (EET) products was found to increase in the presence of NADPH (Fig. 6A-6E). The MS/MS spectrum of the product was very similar to those previously described for EET ethanolamides [11] and to a 14,15-EET ethanolamide standard, with major fragments at m/z 346 (M-18, −H2O), 328 (M-36, −2 × H2O), 303 (M-61, loss of the ethanolamide group), 285 (loss of 18 (H2O) from m/z 303), and 267 (loss of 18 (H2O) from m/z 303). The characteristic fragment m/z 248 was readily detectable, and a minor m/z 187 peak was also found (Fig. 6E) [11]. We conclude that the peak at tR 8.91 is 14,15-EET ethanolamide. A Km of 65 (± 19) μM and kcat of 65 (± 9) pmol product formed/min/nmol P450 were measured, using bicistronic membranes (Supplementary material Figs. S3, S4). None of the other EET ethanolamides was formed by P450 4X1. An experiment with a second preparation of bicistronic membranes yielded a rate of 130 pmol 14,15-EET formed/min/nmol P450.
Fig 6.
LC-MS analysis of the oxidized product formed from anandamide. The chromatogram show selective ion monitoring of m/z 364 (MH+ of ananamide + 16). A, Control reaction (no protein); B, P450 4X1, NADPH-P450 reductase, and NADPH; C, P450 4X1 (and NADPH-P450 reductase) in the absence of NADPH; D, Overlay of the product formation chromatograms (from parts B and C). Top (—): P450 4X1 in the presence of NADPH, bottom (--------): P450 4X1 in the absence of NADPH; E, MS/MS spectra of 14,15-EET ethanolamide formed by P450 4X1, with the insert showing the ×10 expansion of the indicated section of the spectrum.
Formation of the epoxide was inhibited by pre-incubation (10 min) of P450 4X1 with 1-aminobenzotriazole (ABT) (and in the presence of NADPH) [25], providing further evidence for P450-dependent formation of 14,15-EET ethanolamide from anandamide. One of two stable anandamide analogs [26] also yielded a monooxygenated product. CB-25 (N-cyclopropyl-11-(3-hydroxy-5-pentylphenoxy)-undecanamide), a stable analog of both anandamide and THC, was converted to both a mono- and a dioxygenated product, though the position of the oxygen group has not been determined due to the lack of available standards (see Supplementary material Fig. S5). Another anadamide analog, CB-52 (N-cyclopropyl-11-(2-hydroxy-5-pentylphenoxy)-undecanamide), did not form any products under these conditions.
When purified P450 4X1 was incubated with anandamide, 14,15-EET ethanolamide was also detected (Fig. 6). The measured rate was 200 pmol product formed/min/nmol P450. The addition of cytochrome b5 did not significantly change the amount of product formed (180 pmol 14,15-EET ethanolamide formed/min/nmol P450). However, when arachidonic acid was used as the substrate, 14,15- and 8,9-EETs were formed (rates of 18 and 9 pmol/min/nmol P450, respectively) but only in the presence of cytochrome b5 (molar ratio of 1:1) (Supplementary material Fig. S6). When another naturally occurring endocannabinoid, 2-AG, was incubated with purified P450 4X1 (and NADPH-P450 reductase), no product formation was detected (<5 pmol/min/nmol P450). The coupling efficiency was low. In the absence of substrate, P450 4X1 oxidized 27 (± 5) nmol NADPH/min/nmol P450 (22 ± 6 with the addition of cytochrome b5). With the substrate anandamide present, the NADPH oxidation rate was 70 (± 7) nmol/min/nmol P450 (88 ± 10 with cytochrome b5 added). When arachidonic acid was added as the substrate, the NADPH oxidation rate was 36 (± 5) nmol/min/nmol P450 (29 ± 2 with cytochrome b5 added).
Discussion
P450 4X1 was heterologously expressed in E. coli and found to selectively oxidize the endocannabinoid anandamide to 14,15-EET ethanolamide (Fig. 6). In addition, a stable analog of both anandamide and the cannabinoid Δ9-tetrahydrocannabinol, CB-25, was oxidized to both mono- and dioxygenated products. P450 4X1 formed two arachidonic acid epoxides but only in the presence of cytochrome b5 and at much lower rates (Supplementary material Figs. S5, S6).
Anandamide is an arachidonic acid derivative found in most tissues and an important signaling mediator in neurological, immune, and cardiovascular functions [27]. It binds to the CB1 cannabinoid receptor and has been proposed to be an endogenous cannabinoid receptor ligand [7,8]. Recent reports also indicate that anandamide, at concentrations higher than those needed to activate the CB1 cannabinoid receptors, is a full agonist of vanilloid receptor (VR)-1 mediated functional response, i.e., vasodilatation of small arteries (not dependent on the endothelium). VR1 may be involved in the transduction of acute and inflammatory pain signals [28,29]. In brain and liver, anandamide is rapidly converted to arachidonic acid and ethanolamine by a fatty acid amide hydrolase. P450 oxidations of anandamide are also kinown. Studies of mouse liver microsomes incubated with NADPH showed the generation of ≥20 anandamide products determined by HPLC-UV [6]. Human liver and kidney microsomes produced a single monoohydroxy product, 20-HETE ethanolamide, in addition to four epoxides, 5,6-, 8,9-, 11-12, and 14,15-EET ethanolamides [11].
In this work, P450 4X1 oxidized anandamide to 14,15-EET ethanolamide as judged by comparison with commercial standards and previously reported MS spectra (Fig. 6E), and no other products were detected (Fig. 6). Another member of P450 Family 4, P450 4F2 (expressed in liver and kidney), has been reported to form a single monooxygenated product from anandamide (20-HETE-arachinodoyl ethanolamide), and P450 3A4 (in the liver and small intestine) has been reported to form all four epoxides (EETs) of anandamide [11]. Administration of anandamide to rats increased the levels of P450 in the 2C and 3A subfamilies in rat liver and brain [30]. The in vivo formation and biological relevance of the P450-derived HETE and EET ethanolamides remains to be determined, but they may be important signaling molecules in human brain. The high level of P450 4X1 (mRNA) in skin (Fig. 5) may be relevant to a function there. Anandamide concentrations have been measured in rat and mouse skin [31-33] but apparently not human skin, to our knowledge and analysis of database searches. We are currently working to procure skin samples for analysis of anandamide and the 14,15-EET product.
In our initial experiments, P450 4X1 was found not to oxidize either arachidonic acid or a number of other long chain fatty acids. However, when cytochrome b5 was added, P450 4X1 formed both 14,15- and 8,9-EETs from arachidonic acid, albeit at very low rates. A number of P450s, primarily from Subfamilies 2C, 2J, 4A, and 4F, are known to oxidize arachidonic acid to EETs and HETEs, which have been implicated as important signaling mediators with relevance to blood pressure regulation and other physiological processes, i.e. mitogenesis, vasodilatation, modulation of cellular Ca2+, Na+, and K+ fluxes, and activation of Ca2+-dependent K+ channels [2]. Most P450 Family 4 members are recognized for their fatty acid hydroxylation activity but some drugs are also oxidized, e.g. P450 4F3 oxidizes erythromycin and imipramine [34].
A molecular model for P450 4X1 has been built on the basis of bacterial P450 102A1 (BM3) (26% sequence identity) and has a substrate pocket that is L-shaped with the heme located in an angle, with substrates being either short-chain or longer chain fatty acids, not oxidized at the ω-ends but rather within the hydrocarbon chain [14]. The model [14] may be consistent with the observed selective oxidation, although the model is based on low sequence similarity and provides not explanation for the preference for oxidation of fatty acid amides over fatty acids. We found that P450 4X1 did not catalyze the oxidation of any other fatty acids investigated, nor the neurotransmitters. It is conceivable that some function has been lost due to the N-terminal modification and truncation introduced into our P450 4X1, and we cannot unambiguously rule out the possibility that a native P450 4X1 construct expressed in a different system might oxidize these fatty acids.
In the mouse studies of Bornheim et al. [6], liver microsomes produced 20 different anandamide oxidation products at rates of 8 to 386 pmol/min/mg protein. Mouse brain microsomes produced only two products, distinct from the liver products, at rates of 7 and 17 pmol/min/mg protein. None of the products were identified. In the study of Snider et al. [11], the rates of production of anandamide oxidation products by human kidney microsomes were 44-480 pmol/min/mg protein (Vmax). Exactly how the mouse results relate to the human is unclear, in that none of the (unidentified) anandamide products matched in brain and liver microsomes in mice [6] but 14,15-EET ethanolamide, the only anandamide product formed by the brain-selective P450 4X1 (Fig. 6), is also reported to be formed by the liver enzyme P450 3A4 [11]. Another outstanding issue is that the catalytic efficiency (kcat/Km) of recombinant human P450 4X1 is relatively low because of the Km of 65 μM (Supplementary material Fig. S3), i.e. ~ 3 × 103 M−1 min−1, compared to 1.5 × 106 M−1 min−1 for 20-hydroxylation by P450 4F2 [11]. Steady-state kinetic parameters for P450 3A4 were not reported but the values measured with liver microsomes indicated that the four epoxidations (by P450 3A4) [11] are more efficient than the P450 4X1-catalyzed 14,15-epoxidation we characterized. However, it is possible that the selective formation of 14, 15-EET ethanolamide in brain has some particular significance. It should also be noted that the administration of anandamide to rats increased the levels of Subfamily 2C and 3A P450s in rat liver and brain [30]. We tried to examine the binding of potential substrates to P450 4X1 using the heme spectral perturbation method [35] but neither anandamide nor arachidonic acid induced a spectral change in three separate attempts (at concentrations up to 35 μM). However, the lack of induction of a spectral change has been noted before with some bona fide substrates [36].
P450 4X1 is located on chromosome 1 close to another orphan P450, P450 4Z1, and P450s 4A11, 4A22, and 4B1 (www.ncbi.nih.gov/). The Subfamily 4F P450s are clustered on chromosome 19p13.1. P450 4X1 is also well-conserved across species, sharing 84, 80, 81, and 99.6% nucleotide sequence identity with the dog, rat, mouse, and chimpanzee orthologs, respectively. Kidney, breast, and aorta all expressed P450 4X1 mRNA at 5-and 10-fold higher levels than adult liver, and in prostate the expression was found to be >10-fold higher than in liver (Fig. 5). Whole brain mRNA expression was 5-fold higher than liver, while individual brain structures exhibited both lower (e.g., globus pallidus) and considerably higher (e.g. amygdala) levels. The highest mRNA expression was found in amygdala and skin. Conventional PCR analysis detected transcripts in kidney, skeletal muscle, breast, ovary, and uterus and higher expression in trachea and aorta [15,16]. Our real-time PCR analyses confirm and extend these results (Fig. 5), in general, and are consistent with the expression profiles suggested by EST sequences reported to the National Center for Biotechnology Information (NCBI). A relatively large number of P450 4X1 single nucleotide polymorphisms have been reported (www.hapmap.org) and we cannot exclude the possibility that the inter-individual mRNA levels of P450 4X1 may vary, because these results are not based on pooled populations (except for fetal liver and aorta, pool of five). Rat brain regions showing high P450 4X1 mRNA expression using Northern blot and in situ hybridization were hippocampus, cerebellum, and cortex. P450 4X1 mRNA has also been detected in rat cerebral vessels in in situ hybridization analysis [13]. In mouse brain, the orthologous protein was estimated to be present at a level of 10 ng per mg microsomal protein, suggesting that this may be one of the major P450s in mouse brain [14]. Mouse P450 4x1 protein was found not to be induced by phenobarbital, dioxin, dexamethasone, or the peroxisome proliferators activated receptor (PPAR) α agonist ciprofibrate in brain, liver, or kidney [14]. Some of the P450 Family 4 enzymes are known to be induced by PPARα agonists [37], and the PPARα agonist Wyeth 14,643 induced human P450 4X1 in a human hepatoma cell line over-expressing PPARα [15].
Although the function of this orphan P450 enzyme must still be considered largely unknown, the expression pattern and ability to selectively convert anandamide to the epoxide 14,15-EET ethanolamide suggest a potential role in neurovascular function, and further studies may reveal other catalytic functions and an overall pharmacological role in physiological function.
Experimental procedures
Optimization of P450 4X1 and vector preparation
Automated codon optimization and oligonucleotide design for PCR-based gene synthesis were performed in silico, using DNAWorks 3.1 from the National Institutes of Health (http://helixweb.nih.gov/dnaworks) [17] (Fig. 1, Supplementary material Table S1). The amino acid sequence and the native cDNA sequence information for human P450 4X1 were obtained from NCBI GenBank sequences (see Supplementary material Table S2), and codon optimization was performed in order to match the codon preference biases of E. coli. Four different N-terminal constructs were prepared, along with the native codon-optimized sequence construct (with the change E2A) (Supplementary material Table S1). In brief, a number of overlapping oligomers were designed to span the cDNA sequence and used for primary PCA followed by a one-step PCR reaction (94 °C, 5 min; 94 °C, 30 s; 58 °C, 30 s; 72 °C, 2 min (30 cycles); 72 °C, 10 min). The sequence was prepared in one synthon containing an NdeI restriction site (spanning the start codon at the 5’-end) and an XbaI restriction site (at the 3’-flanking end of the sequence). The insert of the correct size was ligated into the pCW vector, in both the monocistronic and bicistronic versions (the latter containing an NADPH-P450 reductase gene downstream of the P450 4X1 cDNA insert, between the NdeI and XbaI sites) [19]. Positive selected clones were sequenced using an Applied Biosystems Big Dye system in the Vanderbilt facility. In order to facilitate purification using Ni-NTA chromatography, a (His)6 tag was added to the C-terminal end of the native protein.
Four different N-terminal modifications (based on previous literature, see Fig. 2, Supplemental Table S2), were introduced into the native construct (pCWmc_P450 4X1native) by PCR-based mutagenesis. Advantage™ DNA polymerase (Stratagene, La Jolla, CA, USA) was used for the PCR amplification, at an annealing temperature of 60 °C. All PCR products were purified using preparative electrophoresis on 1.5 to 2 % (w/v) agarose gels prior to restriction digestion using NdeI and XbaI. The digested insert was ligated into the monocistronic pCW vector and transformed, and positive clones were selected. All modifications were confirmed by nucleotide sequencing analysis. All modified and native 4X1 insert cDNAs were ligated into a bicistronic pCW vector containing an NADPH-P450 reductase vector [19].
Heterologous Expression of P450 4X1
Expression of P450 4X1 native and modified constructs was performed in both E. coli DH5α cells and the same cells co-expressing the chaperones pGroEL/ES12. Plasmids pGroES/EL12 and each of the constructs were transformed and selected on Luria Bertani (LB) plates (containing 50 μg/ml ampicillin or 50 μg/mL ampicillin plus 20 μg/mL kanamycin, respectively). Single colonies were grown overnight in LB media (100 μg/mL ampicillin alone or with 50 μg/mL kanamycin, in the case of pGroES/EL12) at 37 °C, with 225 rpm gyrorotary shaking, and used to inoculate 1-l cultures (1:100 dilution). Large-scale expression for P450 4X1 bicistronic construct 3 was performed in 2.8-L Fernbach flasks containing 1 L Terrific broth (TB) (with 100 μg/mL ampicillin, plus 50 μg/mL kanamycin in the case of pGroES/EL12) containing 0.025% (v/v) of a mixture of trace elements [38] in an Innova 4300 shaker (New Brunswick Scientific, Edison, NJ, USA) with gyrorotary shaking at 225 rpm until the OD600 reached 0.5. D-Isopropyl-β-galactoside (1.0 mM) and 5-aminolevulinic acid (0.5 mM) were added to start induction, along with arabinose (4 mg/mL) to initiate pGroEL/ES12 transcription, when included. Incubation continued at 28 °C with gyrorotary shaking at 190 rpm for another 17-21 h. Expression levels were monitored over 48 h.
Purification of Recombinant P450 4X1
E. coli membranes were prepared as previously described [39]. Membranes of P450 4X1 (from 1 L culture) were solubilized in 400 mM potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v), 1.0 mM EDTA, 0.5% sodium CHAPS (w/v), and 1.0 mM imidazole. The mixture was stirred overnight at 4 °C and centrifuged at 105 g for 60 min, and the supernatant was loaded on a Ni-NTA column (6 mL) equilibrated with 400 mM potassium phosphate buffer (pH 7.4) containing 1.0 mM EDTA, 1.0 M KCl, 0.5% CHAPS (w/v), 10 mM β-mercaptomethanol, and 1.0 mM imidazole). The enzyme was eluted with 100 mM potassium phosphate buffer (pH. 7.4) containing 0.5% CHAPS (w/v), 1.0 M KCl, 10 mM β-mercaptomethanol, and a gradient increasing from 50 to 100 mM imidazole. The eluted fractions were pooled and dialyzed four times versus 100 volumes of 200 mM potassium phosphate buffer (pH 7.4) containing 1.0 mM EDTA and 20% glycerol (v/v) at 4 °C. Purified P450 4X1 was stored in small aliquots at −70 °C until used. (Purified 4X1 appeared to be less stable under storage conditions than P450 2W1 [18] and several other recombinant human P450s.)
Real-time PCR Analysis of P450 4X1 Expression
Human polyA+ RNA samples (human adult and fetal liver, kidney, colon, skin, prostate, breast, adult heart and fetal aorta, as well as a number of human brain regions including whole brain, cerebellar hemisphere, basal ganglia, globus pallidus, and amygdala) were obtained from Ambion Inc. (Austin, TX, USA) and Stratagene (La Jolla, CA, USA). Aliqouts of RNAs (1 μg) were reverse-transcribed using a two-step Enhanced Avian™ RT reaction (SigmaAldrich, St. Louis, MO, USA) containing deoxynucleoside triphosphate mix (10 mM dNTP), random nonamers (50 μM in H2O, Enhanced AMV RT ™ (20 U/mL in 200 mM potassium phosphate buffer (pH 7.2) containing 2 mM dithiothreitol, 0.2% Triton X-100 (v/v) and 50% glycerol (v/v)), 10× buffer for AMV RT (500 mM Tris-HCl buffer (pH 8.3) containing 400 mM KCl, 80 mM MgCl2, and 10 mM dithiothreitol) and RNase inhibitor (20 U/μL) in 20 μL volume and used for first strand synthesis (25 °C, 25 min; 42 °C, 50 min) according to the manufacturer's protocol, and 1 μL cDNA was used as template for each PCR. Primers for real-time PCR of human P450 4X1 mRNA were (forward) 5’ CACCGCTTGTACAGTTTGTTGT and (reverse) 5’-AGATACAATAATCCAGGAAAGAAAGAA, adapted from Savas et al. [15], specifically amplifying a 127 bp fragment of the cDNA. GAPDH and 18S RNA qPCR primer assay sets were purchased from SuperArray Bioscience (Frederick, MD, USA).
Quantitative real-time PCR was performed using iQ™ SYBR Green PCR Master Mix™ according to the manufacturer's instructions (Bio-Rad, Hercules, CA, USA). Each cDNA sample was analyzed in triplicate. Real-time RT-PCR reactions (15 μL) were performed with 0.4 μM forward and reverse primers and 1 μL first strand cDNA template (corresponding to 30-50 ng cDNA). The program was set at 95 °C (15 min), followed by 95 °C (30 s), 40 cycles; 55 °C (30 s); and 72 °C (30 s). Real-time PCR was performed on a MyIQ Single-Color Real-Time PCR Detection System ™ (Bio-Rad) in MicroAmp Optical™ 96-well reaction plates (Bio-Rad). P450 4X1 mRNA levels were calculated using the comparative Ct method and normalized to GAPDH expression levels.
LC-Mass Spectrometry (MS)/MS Analysis
LC-MS/MS analysis was performed on a Waters Acquity UPLC system (Waters, Milford, MA, USA) connected to a ThermoFinnigan LTQ mass spectrometer (ThermoFisher, Watham, MA, USA). Analysis was performed in the electrospray ionization (ESI) positive or negative ion mode using an Acquity UPLC BEH octadecylsilane (C18) column (1.7 μm, 1.0 mm × 100 mm). All analysis was performed using a gradient from Buffer A (10 mM NH4CH3CO2 in 5% CH3CN, v/v) to Buffer B (10 mM NH4CH3CO2 plus 95% CH3CN, v/v). The following gradient program was used with a flow rate of 100 μL/min. Sample (15 μL of a total of 90 μL) was injected on the column using an autosampler system using solvent mixture A:B/95:5 (v/v) for 0-3 min; A:B/80:20 (v/v) for 3-6 min; A:B/60:40 (v/v) for 6-9 min; A:B/0:100 (v/v) for 9-10 min. The temperature of the column was maintained at 55-60°C. ESI conditions were as follow: source voltage 4 kV, source current 100 μA, auxiliary gas flow rate setting 20, sweep gas flow rate setting 5, sheath gas flow setting 34, capillary voltage −49 V; capillary temperature 350 °C, tube lens voltage −90 V. MS/MS conditions were as follow: normalized collision energy 35%; activation Q 0.250; activation time 30 ms.
Data were acquired in positive and negative ion modes using the Xcalibur software package (ThermoElectron) with one full scan from m/z 100 to 500 followed by data-dependent MS/MS scans of putative mono- and dioxygenated products (Supplementary material Table S3). Anandamide, 2-AG, arachidonic acid, docosahexaenoic acid, eicosapentaenoic acid, eicosatrienoic acid, prostaglandin E2, and two stable analogs of anandamide and Δ9-tetrahydrocannabinol, CB-25 and CB-52, were purchased from Cayman Chemicals (Ann Arbor, MI, USA). Dopamine-HCl, tyramine-HCl, loratadine, clotrimazole, and terfenadine were purchased from SigmaAldrich.
Search for Putative Substrates using Bicistronic P450 4X1 Protein
A number of potential substrates (100 μM) (Supplementary material Table S3 and Figs. S3-S6) were incubated in 100 mM potassium phosphate buffer (pH.7.4) with bicistronic membranes containing P450 4X1 protein and human NADPH-P450 reductase (0.3 μM) in a total volume of 0.5 mL. All samples had two controls, one without the addition of the NADPH-generating system and one without protein. The reactions were carried out at 37 °C (30 min) and initiated by the addition of an NADPH-regenerating system [40]). The reactions were terminated by the addition of 1.0 mL of ethyl acetate and extracted (3 times, with separation each time by centrifugation at 3 × 103 g for 10 min); the combined extracts were dried under an N2 stream and the residue was dissolved in a 50:50 mixture of CH3CN/H2O (v/v). Similar incubation procedures were carried out with all test substrates.
For steady-state analysis of the anandamide oxidation reaction, bicistronic P450 4X1 protein (with NADPH-P450 reductase) was used at a final concentration of 0.38 μM with incubation (37 °C) for 1, 5, 10, 15, 30, 45, 60, and 120 min. Different concentrations of bicistronic P450 4X1 protein were used (0.075, 0.38, 0.75, 1.13, 1.50 μM) with incubation for 30 min at 37 °C. In the same studies, the enzyme was preincubated with the mechanism-based inhibitor ABT (20 μM). ABT was incubated in the presence and absence of the NAPDH-generating system for 10 min prior to the addition of anandamide.
In assays using purified P450 enzymes, P450 4X1 (0.1 μM) was mixed with purified recombinant (E. coli) rat NADPH-P450 reductase [41] (0.5 μM), 30 μM L-α-dilaurolyl-snglycero-3-phosphocholine, and substrate in 100 mM potassium phosphate buffer (pH 7.4) and incubated for 5 min at room temperature (total volume of 0.5 mL). Reactions were started after 5 min of pre-incubation at 37 °C with the addition of an NADPH-generating system [35]. The reactions were terminated by addition of two volumes of ethyl acetate and analyzed as described above.
Assay of Cholesterol Oxidation
Assays of cholesterol oxidation were performed using a general procedure described elsewhere [17].
Other Assays and Methods
Concentrations of P450s were estimated using the CO-difference spectra assay [42] with an OLIS/Aminco DW2a spectrophotometer (On-Line Instrument System, Bogart, GA, USA). SDS-polyacrylamide gel electrophoresis was performed according to Laemmli [43] and staining was done using an ammoniacal silver method [44].
Data Analysis
All kinetic data were analyzed by analysis of variance (one-way ANOVA) followed by multiple comparisons using Kolmogorov–Smirnov's test for normality, Dunnet's test for comparison of groups against control groups, and Student-Newman–Keul's test for comparison of all groups’ pair-wise. A Kruskal-Wallis test was used for non-parametric data. SPSS version 13 software for Windows (SSPS, Chicago, IL, USA) was used. Results are expressed as means ± SEM. The computer program GraphPad Prism for Windows 5.0 (GraphPad Prism Software, San Diego, CA, USA) was used to create graphs. Values of p <0.05 were considered to be significant.
Supplementary Material
Acknowledgments
This work was supported in part by the Henning and Johan Trone Holst stiftelse (to K. S), Svenska Läkaresällskapet och Apotekarsocietetén (to K. S), and U. S. Public Health Service grants R37 CA090426 and P30 ES000267 (to F. P. G.). We thank M. V. Martin for technical assistance and D. L. Hachey and M. W. Calcutt of the Vanderbilt Mass Spectrometry Facility Core for technical assistance and discussions.
Abbreviations
- ABT
1-aminobenzotriazole
- CB-25
N-cyclopropyl-11-(3-hydroxy-5-pentylphenoxy)-undecanamide
- CB-52
N-cyclopropyl-11-(2-hydroxy-5-pentylphenoxy)-undecanamide
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- EET
eicosatrienoic
- ESI
electrospray ionization
- EST
expressed sequence tag
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HETE
hydroxyeicosatetraenoic acid
- LB
Luria-Bertani (medium)
- MS
mass spectrometry
- NTA
nitrilotriacetic acid
- P450
cytochrome P450, also termed “heme thiolate P450” [1]
- PBS
phosphate-buffered saline (15 mM potassium phosphate buffer, pH.7.4, containing 150 mM NaCl)
- PCA
polymerase chain assembly
- PCR
polymerase chain reaction
- PPAR
peroxisome proliferator activated receptor
- RT
reverse transcriptase
- SDS
sodium dodecyl sulfate
- UPLC
ultra performance liquid chromatography
Footnotes
Enzymes
Cytochrome P450, unspecific monooxyygenase, EC 1.14.14.1; NADPH-P450 reductase, NADPH:hemoprotein oxidoreductase, EC 1.6.2.4
Subdivision
Enzymology
References
- 1.Palmer G, Reedijk J. Nomenclature of electron-transfer proteins. Recommendations 1989. J Biol Chem. 1992;267:665–677. [PubMed] [Google Scholar]
- 2.Capdevila J, Holla VR, Falck JR. Cytochrome P450 and the metabolism and bioactivation of arachidonic acid and eicosanoids. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 531–551. [Google Scholar]
- 3.Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 377–530. [Google Scholar]
- 4.Kalsotra A, Strobel HW. Cytochrome P450 4F subfamily: at the crossroads of eicosanoid and drug metabolism. Pharmacol Ther. 2006;112:589–611. doi: 10.1016/j.pharmthera.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Qu W, Bradbury JA, Tsao CC, Maronpot R, Harry GJ, Parker CE, Davis LS, Breyer MD, Waalkes MP, Falck JR, Chen J, Rosenberg RL, Zeldin DC. Cytochrome P450 CYP2J9, a new mouse arachidonic acid omega-1 hydroxylase predominantly expressed in brain. J Biol Chem. 2001;276:25467–25479. doi: 10.1074/jbc.M100545200. [DOI] [PubMed] [Google Scholar]
- 6.Bornheim LM, Kim KY, Chen B, Correia MA. Microsomal cytochrome P450-mediated liver and brain anandamide metabolism. Biochem Pharmacol. 1995;50:677–686. doi: 10.1016/0006-2952(95)00177-2. [DOI] [PubMed] [Google Scholar]
- 7.Kozak KR, Marnett LJ. Oxidative metabolism of endocannabinoids. Prostaglandins Leukot Essent Fatty Acids. 2002;66:211–220. doi: 10.1054/plef.2001.0359. [DOI] [PubMed] [Google Scholar]
- 8.Palmer SL, Khanolkar AD, Makriyannis A. Natural and synthetic endocannabinoids and their structure-activity relationships. Curr Phar Des. 2000;6:1381–1397. doi: 10.2174/1381612003399419. [DOI] [PubMed] [Google Scholar]
- 9.Hanus L, Abu-Lafi S, Fride E, Breuer A, Vogel Z, Shalev DE, Kustanovich I, Mechoulam R. 2-Arachidonyl glyceryl ether, an endogenous agonist of the cannabinoid CB1 receptor. Proc Natl Acad Sci USA. 2001;98:3662–3665. doi: 10.1073/pnas.061029898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 11.Snider NT, Kornilov AM, Kent UM, Hollenberg PF. Anandamide metabolism by human liver and kidney microsomal cytochrome P450 enzymes to form hydroxyeicosatetraenoic and epoxyeicosatrienoic acid ethanolamides. J Pharmacol Exp Ther. 2007;321:590–597. doi: 10.1124/jpet.107.119321. [DOI] [PubMed] [Google Scholar]
- 12.Stark K, Guengerich FP. The orphan human cytochrome P450: what is left to learn? Drug Metab Rev. 2007;39:627–637. doi: 10.1080/03602530701467708. [DOI] [PubMed] [Google Scholar]
- 13.Bylund J, Zhang C, Harder DR. Identification of a novel cytochrome P450, CYP4X1, with unique localization specific to the brain. Biochem Biophys Res Commun. 2002;296:677–684. doi: 10.1016/s0006-291x(02)00918-x. [DOI] [PubMed] [Google Scholar]
- 14.Al-Anizy M, Horley NJ, Kuo CW, Gillett LC, Laughton CA, Kendall D, Barrett DA, Parker T, Bell DR. Cytochrome P450 Cyp4x1 is a major P450 protein in mouse brain. FEBS J. 2006;273:936–947. doi: 10.1111/j.1742-4658.2006.05119.x. [DOI] [PubMed] [Google Scholar]
- 15.Savas U, Hsu MH, Griffin KJ, Bell DR, Johnson EF. Conditional regulation of the human CYP4X1 and CYP4Z1 genes. Arch Biochem Biophys. 2005;436:377–385. doi: 10.1016/j.abb.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Expression patterns of mouse and human CYP orthologs (families 1-4) during development and in different adult tissues. Arch Biochem Biophys. 2005;436:50–61. doi: 10.1016/j.abb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z-L, Bartleson CJ, Ham A-JL, Guengerich FP. Heterologous expression, purification, and properties of human cytochrome P450 27C1. Arch Biochem Biophys. 2006;445:138–146. doi: 10.1016/j.abb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z-L, Sohl CD, Shimada T, Guengerich FP. Recombinant enzymes over-expressed in bacteria show broad catalytic specificity of human cytochrome P450 2W1 and limited activity of human cytochrome P450 2S1. Mol Pharmacol. 2006;69:2007–2014. doi: 10.1124/mol.106.023648. [DOI] [PubMed] [Google Scholar]
- 19.Parikh A, Gillam EMJ, Guengerich FP. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nature Biotechnol. 1997;15:784–788. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- 20.Cheesman MJ, Baer BR, Zheng YM, Gillam EM, Rettie AE. Rabbit CYP4B1 engineered for high-level expression in Escherichia coli: ligand stabilization and processing of the N-terminus and heme prosthetic group. Arch Biochem Biophys. 2003;416:17–24. doi: 10.1016/s0003-9861(03)00278-9. [DOI] [PubMed] [Google Scholar]
- 21.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson TH, Hsu MH, Kronbach T, Barnes HJ, Chan G, Waterman MR, Kemper B, Johnson EF. Purification and characterization of recombinant-expressed cytochrome P450 2C3 from Escherichia coli: 2C3 encodes the 6β-hydroxylase deficient form of P450 3b. Arch Biochem Biophys. 1993;300:510–516. doi: 10.1006/abbi.1993.1069. [DOI] [PubMed] [Google Scholar]
- 23.Licad-Coles E, He K, Yin H, Correia MA. Cytochrome P450 2C11: Escherichia coli expression, purification, functional characterization, and mechanism-based inactivation of the enzyme. Arch Biochem Biophys. 1997;338:35–42. doi: 10.1006/abbi.1996.9795. [DOI] [PubMed] [Google Scholar]
- 24.Rouzer CA, Marnett LJ. Non-redundant functions of cyclooxygenases: Oxygenation of endocannabinoids. J Biol Chem. 2008;283:8065–8069. doi: 10.1074/jbc.R800005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emoto C, Murase S, Sawada Y, Iwasaki K. In vitro inhibitory effect of 1-aminobenzotriazole on drug oxidations in human liver microsomes: a comparison with SKF-525A. Drug Metab Pharmacokinet. 2005;20:351–357. doi: 10.2133/dmpk.20.351. [DOI] [PubMed] [Google Scholar]
- 26.Brizzi A, Brizzi V, Cascio MG, Bisogno T, Sirianni R, Di Marzo V. Design, synthesis, and binding studies of new potent ligands of cannabinoid receptors. J Med Chem. 2005;48:7343–7350. doi: 10.1021/jm0501533. [DOI] [PubMed] [Google Scholar]
- 27.Fowler CJ. The pharmacology of the cannabinoid system--a question of efficacy and selectivity. Mol Neurobiol. 2007;36:15–25. doi: 10.1007/s12035-007-0001-6. [DOI] [PubMed] [Google Scholar]
- 28.De Petrocellis L, Bisogno T, Maccarrone M, Davis JB, Finazzi-Agro A, Di Marzo V. The activity of anandamide at vanilloid VR1 receptors requires facilitated transport across the cell membrane and is limited by intracellular metabolism. J Biol Chem. 2001;276:12856–12863. doi: 10.1074/jbc.M008555200. [DOI] [PubMed] [Google Scholar]
- 29.Herradon E, Martin MI, Lopez-Miranda V. Characterization of the vasorelaxant mechanisms of the endocannabinoid anandamide in rat aorta. Br J Pharmacol. 2007;152:699–708. doi: 10.1038/sj.bjp.0707404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa B, Parolaro D, Colleoni M. Chronic treatment with the endocannabinoid anandamide increases cytochrome P450 metabolizing system in the rat. Eur J Pharmacol. 2002;449:61–69. doi: 10.1016/s0014-2999(02)01994-5. [DOI] [PubMed] [Google Scholar]
- 31.Felder CC, Nielsen A, Briley EM, Palkovits M, Priller J, Axelrod J, Nguyen DN, Richardson JM, Riggin RM, Koppel GA, Paul SM, Becker GW. Isolation and measurement of the endogenous receptor agonist, anandamide, in brain and peripheral tissues of human and rat. FEBS Lett. 1996;393:231–235. doi: 10.1016/0014-5793(96)00891-5. [DOI] [PubMed] [Google Scholar]
- 32.Zygmunt PM, Julius I, Di Marzo I, Högestätt ED. Anandamide—the other side of the coin. Trends Pharmacol Sci. 2000;21:43–44. doi: 10.1016/s0165-6147(99)01430-3. [DOI] [PubMed] [Google Scholar]
- 33.Darmani NA, Izzo AA, Degenhardt B, Valenti M, Scaglione G, Capasso R, Sorrentini I, Di Marzo Y. Involvement of the cannabimimetic compound, N-palmitoyl-ethanolamine, in inflammatory and neuropathic condiditons: Review of the available pre-clinical data, and first human studies. Neuropharmacology. 2005;48:1154–1163. doi: 10.1016/j.neuropharm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Kalsotra A, Turman CM, Kikuta Y, Strobel HW. Expression and characterization of human cytochrome P450 4F11: Putative role in the metabolism of therapeutic drugs and eicosanoids. Toxicol Appl Pharmacol. 2004;199:295–304. doi: 10.1016/j.taap.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 35.Schenkman JB, Remmer H, Estabrook RW. Spectral studies of drug interaction with hepatic microsomal cytochrome P-450. Mol Pharmacol. 1967;3:113–123. [PubMed] [Google Scholar]
- 36.Guengerich FP. Oxidation-reduction properties of rat liver cytochrome P-450 and NADPH-cytochrome P-450 reductase related to catalysis in reconstituted systems. Biochemistry. 1983;22:2811–2820. doi: 10.1021/bi00281a007. [DOI] [PubMed] [Google Scholar]
- 37.Cui X, Kawashima H, Barclay TB, Peters JM, Gonzalez FJ, Morgan ET, Strobel HW. Molecular cloning and regulation of expression of two novel mouse CYP4F genes: expression in peroxisome proliferator-activated receptor α-deficient mice upon lipopolysaccharide and clofibrate challenges. J Pharmacol Exp Ther. 2001;296:547–555. [PubMed] [Google Scholar]
- 38.Sandhu P, Baba T, Guengerich FP. Expression of modified cytochrome P450 2C10 (2C9) in Escherichia coli, purification, and reconstitution of catalytic activity. Arch Biochem Biophys. 1993;306:443–450. doi: 10.1006/abbi.1993.1536. [DOI] [PubMed] [Google Scholar]
- 39.Gillam EMJ, Baba T, Kim B-R, Ohmori S, Guengerich FP. Expression of modified human cytochrome P450 3A4 in Escherichia coli and purification and reconstitution of the enzyme. Arch Biochem Biophys. 1993;305:123–131. doi: 10.1006/abbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- 40.Guengerich FP, Bartleson CJ. Analysis and characterization of enzymes and nucleic acids. In: Hayes AW, editor. Principles and Methods of Toxicology. CRC Press; Boca Raton, FL: 2007. pp. 1981–2048. [Google Scholar]
- 41.Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. Role of the alanine at position 363 of cytochrome P450 2B2 in influencing the NADPH- and hydroperoxide-supported activities. Arch Biochem Biophys. 1998;350:324–332. doi: 10.1006/abbi.1997.0534. [DOI] [PubMed] [Google Scholar]
- 42.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 43.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.