Abstract
Background:
Some studies published in recent time revealed that many bacteria from Enterobacteriaceae group are multi-antibiotic-resistant because of the production enzymes carbapenemase particularly New Delhi metallo-beta-lactamase encoded by gene called blaNDM-1. Looking at public health importance of this issue there is a need for studies at other centers to confirm or refute published findings.
Objectives:
This study was designed with the aim of exploring antibiotic resistance in Enterobacteriaceae group of bacteria and also to explore gene and enzyme responsible for it.
Materials and Methods:
Samples of Enterobacteriaceae were collected from wards and outpatient departments. Antibiotic sensitivity was checked by an automated system (VITEK 2 COMPACT). Carbapenemase production was assessed by Modified Hodge Test. Presence of blaNDM-1 was assessed by polymerase chain reaction.
Statistics:
Frequency and percentage were used to describe the data. Frequency of sensitivity was compared between carbapenemase producers and noncarbapenemase producers by Fisher's exact test.
Results:
Forty-seven percent bacteria were found to be producing carbapenemase enzyme. These bacteria were significantly less sensitive to cefoperazone, cefepime, and amikacin. Among carbapenemase-producing organisms, 3% and 6% were resistant to tigecycline and colistin, respectively. Forty percent bacteria were found to be having blaNDM-1 gene. There was a significant difference between blaNDM-1-positive and blaNDM-1-negative for sensitivity toward cefoperazone + sulbactam, imipenem, meropenem, amikacin, tobramycine, ciprofloxacin, and levofloxacin.
Conclusion:
Presence of carbapenemase enzyme and blaNDM-1 gene is associated with high level of resistance in Enterobacteriaceae group of bacteria and only few antibiotics have good sensitivity for these organisms.
Keywords: Antibiotic resistance, blaNDM gene, carbapenemase, Enterobacteriaceae
INTRODUCTION
One of the biggest public health problems at present is antibiotic resistance. Many traditional antibiotics are no longer effective in common infections which are contributing significant morbidity and mortality. The problem is more serious as discovery of new antibiotics is slow and emergences of new resistance in microorganisms are frequent.[1] It had been predicted that, with the present scenario in coming time, there may be very few or no antibiotic which could be used against multidrug-resistant microorganisms.[2,3]
One of the major classes of microorganisms affected by drug resistance is Enterobacteriaceae group of bacteria. In a study by Mulla et al., it was observed that Enterobacteriaceae organism were very less sensitive to amoxicillin + clavulanic acid (13.7%), chloramphenicol (7.6%), cefoperazone (14.4%), cefixime (15.7%), and cefuroxime (17.6). Sensitivity to aztreonam was 32.7%.[4] These bacteria are responsible for various diseases such as urinary tract infections, bacteraemia, septicaemia, hospital, and nosocomial infections, etc. One of the major reasons for resistance in Enterobacteriaceae is production of carbapenemase enzyme. In a study by Kumarasamy et al., it was revealed that Enterobacteriaceae having a gene blaNDM-1 which encodes an enzyme called New Delhi metallo-beta-lactamase (carbapenemase) is resistant to all antibiotics including imipenem and meropenem except colistin and tigecycline.[5,6] This finding is very important and should be a reason for worry as carbapenemase-producing microorganisms are known to cause infections which are associated with high morbidity and mortality. It is observed in a study done by Borer et al. that crude mortality and attributable mortality in the patients of bacteremia by carbapenemase-producing Klebsiella pneumoniae is 71.9% and 50%, respectively.[7] In a similar study by Patel et al., it was observed that mortality among the cases with carbapenemase-resistant K. pneumoniae was significantly more in comparison with control group.[8] It was also observed that even after administrating in vitro sensitive antibiotics there was high mortality and morbidity in patients infected by carbapenamase-producing bacteria.[8] Looking at the public health importance of this it is very important to screen Enterobacteriaceae at various hospitals and institutions for antibiotic sensitivity and carbapenemase production periodically and trends/findings should be conveyed to clinicians, so that they can prescribe antibiotics accordingly.[1,4]
With this idea in mind, this study was designed to evaluate Enterobacteriaceae group of bacteria for their carbapenemase production capacity and also to compare the antibiotic sensitivity of carbapenamase producing bacteria with noncarbapenemase bacteria. It was also necessary to explore the genetic basis of this resistance through evaluating the presence of blaNDM-1 which is considered as a novel gene associated with the production of New Delhi metallo-beta-lactamase enzyme in Enterobacteriaceae. At present, there is a lack of data regarding the antibiotic sensitivity pattern of carbapenemase-producing Enterobacteriaceae and blaNDM-1-producing Enterobacteriaceae in Indian setting. Another aim of this study was to identify antibiotics which shows good sensitivity for carbapenemase-producing and blaNDM-1-positive Enterobacteriaceae, which can be tried in combination for further study to find a suitable antibiotic combination against these bacteria.
MATERIALS AND METHODS
This study was done at Department of Microbiology of Government Medical College and New Civil Hospital, Surat (Gujrat) during a period of January 2013 to March 2013. The study was approved by Institutional Ethics Committee. Informed consent was waived because the data were collected as a part of routine laboratory tests for the patients, and there was no additional risk possessed to the patients. Identity of samples was decoded. All measures were taken to keep the data safe and secure, so that identification of patients could not be revealed.
All clinical samples coming to microbiology department for bacterial culture and sensitivity were screened for isolation of Enterobacteriaceae and identified by standard biochemical reactions. Antibiotic sensitivity was checked by an automated system (VITEK 2 Compact). Carbapenemase production was assessed by Modified Hodge Test with imipenem disk (10 mg) (HiMedia, Mumbai, India) and compared with known carbapenemase-producing QC strains of K. pneumoniae ATCC BAA-1705 and known carbapenemase-negative QC strain of K. pneumoniae ATCC BAA-1706 for phenotypic detection of carbapenamase production as per Clinical and Laboratory Standards Institute guidelines 2012.[9] Presence of blaNDM-1 was assessed by polymerase chain reaction (Applied Biosystems 7300, DNA Analyzer, Perkin Elmer, USA). Assay used were TaqMan® gene expression assay. The oligonucleotide primers used were 5'-CTGGTTCGACAACGCATTGG-3', reverse primer 3'-GACAAGATGGGCGGTATGGA-5', and reporter 1 sequence was ATAAGTCGCAATCCCC with reporter 1 dye FAM and reporter 1 quencher NFQ primer sequence. The primers and probes were designed from alignments of available sequences obtained from the GenBank nucleotide sequence database.
Descriptive statistics in the form of frequency and percentage was used to describe the data. Frequency of sensitivity was compared between carbapenemase producers and noncarbapenemase producers by Fisher's exact test with the help of OpenEpi software. P <0.05 was considered significant.
RESULTS
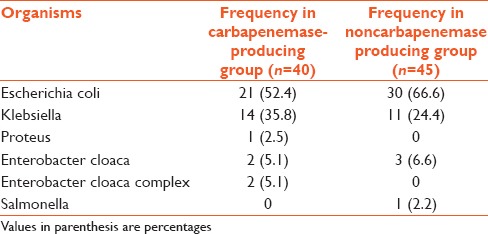
Out of 85 Enterobacteriaceae sample isolated, 40 (47%) were found to be carbapenemase producers on the basis of Modified Hodge Test. The majority of samples were urine sample and majority of organisms isolated were Escherichia coli and Klebsiella [Tables 1 and 2].
Table 1.
Type of samples from which Enterobacteriaceae were isolated
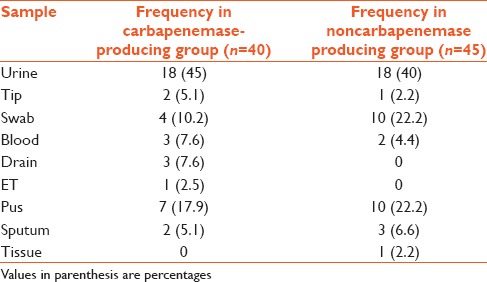
Table 2.
Organisms isolated from both groups
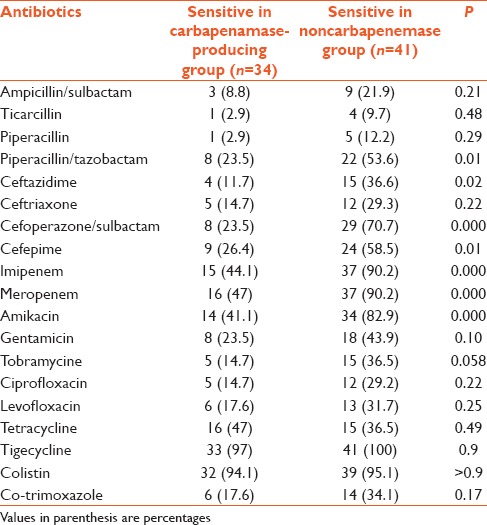
There was a significant difference between two groups for the sensitivity of few antibiotics like (piperacillin + tazobactam, ceftazidime, cefoperazone + sulbactam, cefepime, meropenem, and amikacin). After bonferroni correction, there was a significant difference for cefoperazone, cefepime, and amikacin [Table 3]. Antibiotics for which at least 50% organism were sensitive in carbapenemase nonproducing group are piperacillin + tazobactam, cefoperazone + sulbactam, cefepime, imipenem, meropenem, amikacin, tigecycline, and colistin. There was 100% sensitivity for tigecycline and 95.5% for colistin [Table 3]. Antibiotics for which at least 50% organisms were sensitive in the carbapenemase-producing group were imipenem, meropenem, colistin, and tigecycline. Around 15% bacteria were resistant to tigecycline and around 13% were resistant to colistin [Table 3].
Table 3.
Antibiotic sensitivity in carbapenemase-producing bacteria and noncarbapenemase producing bacteria
There was a significant difference between blaNDM-1-positive and blaNDM-1-negative group for piperacillin + tazobactam, ceftazidime, ceftriaxone, cefoperazone + sulbactam, cefepime, imipenem, meropenem, amikacin, gentamicin, tobramycine, ciprofloxacin, and levofloxacin. After bonferroni correction, significant difference was obtained in cefoperazone + sulbactam, imipenem, meropenem, amikacin, tobramycine, ciprofloxacin, and levofloxacin [Table 4]. Antibiotics for which at least 50% blaNDM-1-positive organisms were sensitive were imipenem (58.8%), meropenem (52.9%), tigecycline (70.4%), and colistin (94.1%) [Table 4].
Table 4.
Comparative antibiotic sensitivity in blaNDM-1-positive bacteria and blaNDM-1-negative bacteria
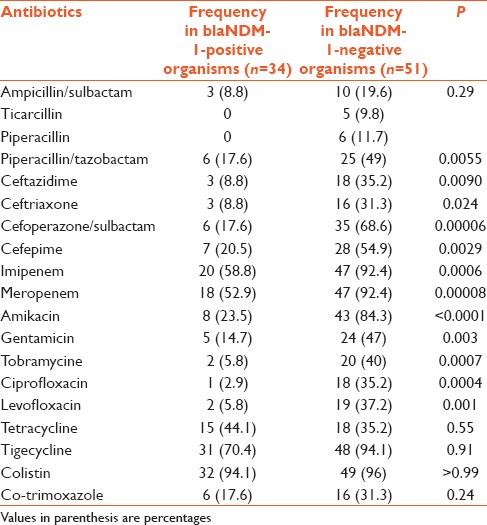
Antibiotic for which at least 50% blaNDM-1-negative organisms were sensitive were cefoperazone + sulbactam (68.6%), cefepime (54.9%), imipenem (92.4%), meropenem (92.4), amikacin (84.3), tigecycline (94.1), and colistin (96).
DISCUSSION
This study revealed that carbapenemase-producing Enterobacteriaceae are very common in hospital setting which are not much sensitive to majority of antibiotics except imipenam, meropenam, colistin, and tigecycline. These organisms are different from carbapenemase nonproducers in sensitivity to different antibiotics such as piperacillin + tazobactum, ceftazidime, cefoperazone + sulbactam, cefepime, meropenem, and amikacin. Colistin and tigecycline resistance was also observed in carbapenemase-producing Enterobacteriaceae. Surprisingly, small portion of carbapenemase producers was also resistant to colistin and tigecycline. Similarly, for specific carbapenemase “NDM-1” producing Enterobacteriaceae sensitivity for majority of antibiotics were very low and resistance was observed for most effective drugs like tigecycline and colistin.
These findings supports the study done by Kumarasamy et al., and Deshpande et al., which reported the existence of such multi-drug resistance organisms in Indian hospitals.[3,5] Few more studies were done with similar objectives also confirm the existence of such organisms.[10,11,12,13] The antibiotic sensitivity pattern observed in this study is very alarming. Sensitivity for imipenem and meropenem was found to be around 50%, and there was also resistance for reserved drugs like colistin and tigecycline. In few studies done in Indian setting, it was observed that there was no resistance for tigecycline in NDM-1 producer organisms but in this study resistance was found for tigecycline and colistin both.[3,14] Here it is important to note that resistance to NDM-1 producers if more problematic because it is not confined to the single strain of the bacteria and can spread to nonclonal strain also. Antibiotic sensitivity pattern observed in this study and also in other studies proves that majority of such bacteria are multidrug resistant and this can be explained on the basis of presence of genes responsible for resistance to other antibiotics on same plasmid which has blaNDM-1.[15]
In the light of these findings, there is a need of using various measures to prevent antibiotic resistance so that these kinds of resistance to antibiotics can be prevented. There is a need of widespread screening of Enterobacteriaceae group of bacteria for antibiotic sensitivity and present of blaNDM-1, so that the exact prevalence of such organism can be assessed.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We would like to thank Dr. Ambuj Kumar, Associate Professor, Centre for EBM, University of South Florida, for help in manuscript writing during the workshop of manuscript writing in Surat (Fogarty training grant - #1D43TW006793-01A2-AITRP).
REFERENCES
- 1.Charan J, Mulla S, Ryavanki S, Kantharia N. New Delhi metallo-beta lactamase-1 containing Enterobacteriaceae: Origin, diagnosis, treatment and public health concern. Pan Afr Med J. 2012;11:22. [PMC free article] [PubMed] [Google Scholar]
- 2.Abdul Ghafur K. An obituary - On the death of antibiotics! J Assoc Physicians India. 2010;58:143–4. [PubMed] [Google Scholar]
- 3.Deshpande P, Rodrigues C, Shetty A, Kapadia F, Hedge A, Soman R. New Delhi metallo-beta lactamase (NDM-1) in Enterobacteriaceae: Treatment options with carbapenems compromised. J Assoc Physicians India. 2010;58:147–9. [PubMed] [Google Scholar]
- 4.Mulla S, Charan J, Panvala T. Antibiotic sensitivity of Enterobacteriaceae at a tertiary care center in India. Chron Young Sci. 2011;2:214–8. [Google Scholar]
- 5.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect Dis. 2011;11:355–62. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 7.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, et al. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol. 2009;30:972–6. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 8.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 9.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 10.Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y. Prevalence of New Delhi metallo-ß-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents. 2012;39:265–6. doi: 10.1016/j.ijantimicag.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 11.Khan AU, Nordmann P. NDM-1-producing Enterobacter cloacae and Klebsiella pneumoniae from diabetic foot ulcers in India. J Med Microbiol. 2012;61:454–6. doi: 10.1099/jmm.0.039008-0. [DOI] [PubMed] [Google Scholar]
- 12.Seema K, Ranjan Sen M, Upadhyay S, Bhattacharjee A. Dissemination of the New Delhi metallo-ß-lactamase-1 (NDM-1) among Enterobacteriaceae in a tertiary referral hospital in north India. J Antimicrob Chemother. 2011;66:1646–7. doi: 10.1093/jac/dkr180. [DOI] [PubMed] [Google Scholar]
- 13.Datta S, Roy S, Chatterjee S, Saha A, Sen B, Pal T, et al. A five-year experience of carbapenem resistance in Enterobacteriaceae causing neonatal septicaemia: Predominance of NDM-1. PLoS One. 2014;9:e112101. doi: 10.1371/journal.pone.0112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fomda BA, Khan A, Zahoor D. NDM-1 (New Delhi metallo beta lactamase-1) producing Gram-negative bacilli: Emergence and clinical implications. Indian J Med Res. 2014;140:672–8. [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin C, Liolios L, Peleg AY. Phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. J Clin Microbiol. 2006;44:3139–44. doi: 10.1128/JCM.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


