Abstract
Background:
Numerous genes have been reported in relation with gestational diabetes mellitus (GDM), but the findings were not consistently replicated across populations, or there have been no detailed studies on them. Previous literatures suggested that, out of all angiotensin converting enzyme (ACE) gene polymorphisms, only ACE insertion/deletion (I/D) gene polymorphism has a strong association with GDM in Asian Indian women.
Aim:
This study was devoted to evaluate the association of four single nucleotide polymorphisms (SNPs) ACE A240T, C1237T, G2350A and I/D with GDM and Type 2 diabetes mellitus.
Materials and Methods:
This study recruited 105 GDM cases, 119 Type 2 diabetes mellitus subjects and 120 controls. PCR-RFLP was used for identifying genotypes of ACE A240T, C1237T and G2350A and PCR was performed in the case of ACE I/D.
Results:
Significant associations of ACE SNP's, C1237T, and G2350A with GDM were observed. Haplotype analysis revealed the remarkably significant evidence of association with SNP combination ACE A240T, C1237T, G2350A, and I/D with GDM patients (P = 0.024). Individuals possessing haplotype “TTAI” (frequency 30% in GDM and 0 in controls) derived from these SNPs had 185 fold increased risk of developing GDM (95% of confidence interval: 11.13–3102.15), which was highest when compared with other 15 haplotypes.
Conclusion:
Shorter-range haplotypes were also significant, but the only consistently associated alleles were found to be in ACE C1237T, G2350A, and I/D. These results suggested that the variant in close proximity to ACE C1237T, G2350A and/or I/D modulates susceptibility to GDM and noninsulin dependent diabetes mellitus in Indian women.
Keywords: Angiotensin converting enzyme, gestational diabetes mellitus, haplotype, polymorphisms, Type 2 diabetes mellitus
INTRODUCTION
Angiotensin-converting enzyme (ACE) gene (MIM + 1061080) is located on chromosome 17. It produces an enzyme called ACE which plays an important role in renin angiotensin aldosterone system cascade by converting angiotensin I to angiotensin II. Previous studies suggested that ACE polymorphisms may be involved in the etiology of gestational diabetes mellitus (GDM) or Type 2 diabetes mellitus (noninsulin dependent diabetes mellitus [NIDDM (MIM #125853)]). Studies have shown that RAS may play an important role in developing nephropathy in Type 2 diabetes mellitus, but the role of ACE polymorphisms in relation to Type 2 diabetes mellitus poorly understood.[1] Several studies showed that an Alu insertion/deletion (ACE I/D) polymorphism located in intron 16 of ACE gene is associated with the high concentration of ACE levels in plasma.[1] The role of other ACE polymorphisms in relation to patients with GDM or Type 2 diabetes mellitus has not been studied in Indian population till date. GDM women are at an increased risk of developing Type 2 diabetes mellitus later in their lives.[2] Women with the family history of diabetes are at a risk of developing GDM.[3,4] The patho-physiology of GDM is similar to that of Type 2 diabetes mellitus and is characterized by peripheral insulin resistance accompanied by an insulin secretary defect.[5,6,7]
The results of previous studies described the haplotype analysis of ACE gene polymorphisms in different populations, but with hypertension.[8] Studies reported that a linkage disequilibrium (LD) block difference of haplotypes varies in different populations.[8] This suggests that different LD of the polymorphisms located in ACE polymorphisms may be observed in different populations.[8] Keeping these in view, the objective of this study was to analyze the role of these polymorphisms or given haplotypes with GDM and Type 2 diabetes mellitus patients in Indian population. Accordingly, we selected the following four polymorphisms A-240T (ACE 4), ACE C1237T (ACE 6), A2350G (ACE 8), and ACE I/D in our study.
MATERIALS AND METHODS
These are detailed in the following sub-sections.
Study settings
A sample size of 10,000 women reporting to the OPDs of Obstetrics and Gynecology Department, All India Institute of Medical Sciences (AIIMS), New Delhi, India, during the period of May 2010 to June 2013, was screened for meeting the selection criteria. Out of this population, mainly the residents of North India, 344 subjects (120 controls, 105 GDM patients [Group 1], and 119 Type 2 diabetes mellitus [Group 2]) were enrolled in this study. Each subject's consent and the details of her medical and family history were documented through a set of questionnaires. The ethics committee of AIIMS approved the protocol of this study as well as patients' medical/family history questionnaire. A detailed pedigree chart for each subject with family history was drawn and recorded.
Study subjects
Active groups were framed following on the characteristics of the diseased conditions namely, GDM (Group 1) and Type 2 diabetes mellitus (Group 2); and healthy women were enrolled to control group. The female subjects who suffered from hypertension, diabetes complications, cardiac diseases, metabolic syndromes (such as, obesity), cancers, HIV, gynecological-, endocrinological-, and neurological-disorders, any other type of diabetes or impaired glucose intolerance were excluded from the study. Each individual group is described in the following sub-sections.
Selection of active subjects
Gestational diabetes mellitus (Group 1)
All pregnant women were screened with Carpenter and Coustan Oral Glucose Tolerance Test (OGTT) (threshold reading for 100 g glucose OGTT were fasting/1 h/2 h/3 h = 95/180/155/140 mg/dl) in 2nd trimester of their gestation, but cases with the strong family history of Type 2 diabetes mellitus were screened in 1st trimester for GDM. Patients who had <2 OGTT values were excluded. The age of the patients in this group ranged between 18 and 42 years.[9]
Type 2 diabetes mellitus (Group 2)
This group consisted of 119 female subjects suffering from Type 2 diabetes mellitus with glucose levels for fasting/2 h ≥ 150/200 mg/dl or those with more than 2 high values of OGTT, for duration of more than a year and age > 43 years (which is the average age of menopause in Indian women).[10]
Controls
Control group subjects were comprised of 120 healthy pregnant women with no reported clinical history of GDM or Type 2 diabetes mellitus. Blood samples from controls (of comparable age to that of GDM patients) were collected after screening with glucose test who had fasting/postprandial (PP) values (after 2 h) <100/140 mg/dl. Patients with impaired glucose intolerance, GDM or Type 1/2 diabetes mellitus were excluded from this group.
Sample collection and DNA extraction
Blood samples were collected after 8 h or overnight fasting form all the subjects included (controls, GDM and Type 2 diabetes mellitus) in this study. Genomic DNA was extracted from leukocytes of whole blood using the salting out method.[11] The yield of extracted DNA was measured at 260/280-nm using a nano-drop (ND1000, Thermo, Delaware, USA) and purity was calculated by taking ratio of readings at 260/280 nm (acceptable ratios ranged between 1.6 and 2.0). The genomic DNA samples were stored at –20°C until further analysis.
Genetic analysis
The ACE gene single nucleotide polymorphisms that is, A-240T (rs4291), ACE C1237T (rs4309), A2350G (rs4343), and ACE I/D (rs4340) were amplified using QB-96 Server Gradient Thermocycler (Quanta Biotech, England), with the help of the previously published primers and methodology.[12] Each PCR product of ACE I/D gene polymorphism, which were reported to be DD genotype, was subjected to a second PCR amplification with insertion-specific primers (FP5'- TGGGACCACAGCGCCCGCCACTAC-3'; RP: 5' TCGCCAGCCCTCCCATGCCCATAA-3') in order to avoid DD mistyping.[13] The sequences of randomly selected samples (6 of each polymorphism) were analyzed through the software ABI3730XI (Applied Biosystem, Texas, USA). The result of this sequence analysis for the above-mentioned polymorphisms (ACE 4, ACE 6, ACE 8, and ACE I/D) confirmed our genotyping results (all PCR and RFLP chemicals were bought from Fermentas, USA).
Statistical analysis
Demographics and clinical variables between active groups and controls were analyzed with Stata 11.0 (College Station, Texas, USA) for windows. Numerical variables not normally distributed were presented as mean ± standard deviation and median (range) and were compared using ANOVA/Krushkal-Wallis test, followed by Bonferroni correction. Logistic regression analyses were used to test for the association of polymorphisms or haplotypes with GDM and Type 2 diabetes mellitus. Statistical significance was set at P < 0.05. SHEsis software (http://analysis.bio-x.cn; Shi and He, 2005) was used to construct two/three and four locus haplotypes.[14]
RESULTS
Characteristics of study subjects
The statistical analyses of demographic data and clinical profile of 2 active groups and controls showed that there was the insignificant difference in mean age of Group 1 cases (P = 0.16) in comparison with that of controls. However, there was a significant difference in the mean age of Group 2 cases when compared to controls (P < 0.001) and Group 1 cases (P < 0.001). Patients with multigravida were approximately 75% in both controls and Group 1. Group 2 had the maximum number of children (50%) in comparison with that of controls (10%) and Group 1 (5%). Group 1 (GDM) had the strongest family history of NIDDM with mother (P = 0.02), father (P = 0.01) or both (P = 0.001); and highest percentage of GDM occurrence (12.3%) and cesareans (13.33%) in their previous pregnancies. The difference in mean fasting and PP glucose levels in all the groups were statistically significant (P < 0.001).
Allele and genotype frequencies
The genotype frequencies of ACE 4, ACE 6, ACE 8, and ACE I/D are shown in Table 1. Table 1 exhibits that genotype TT of ACE 6 (P = 0.004) and genotype GG of ACE 8 (P < 0.001) gene polymorphisms were significantly associated with GDM. The chances of developing Type 2 diabetes mellitus in postpregnancy of GDM patients with genotype DD of ACE I/D were found to be increased by 2.14 folds (95% confidence interval: 1.14-4.13). Table 2 depicts the allele frequencies of ACE 4, ACE 6, ACE 8, and ACE I/D and each one differed significantly between active groups and controls. According to recessive model,[8] allele T of ACE 6 (P = 0.008) and according to dominant model,[8] allele G of ACE 8 (P < 0.001) were observed to be associated with GDM [Table 1]. Allele G of ACE 8 was also associated with Group 2 cases (P < 0.001).
Table 1.
Genotype and allele frequencies of ACE 4, ACE 6, ACE 8 and ACE I/D polymorphisms
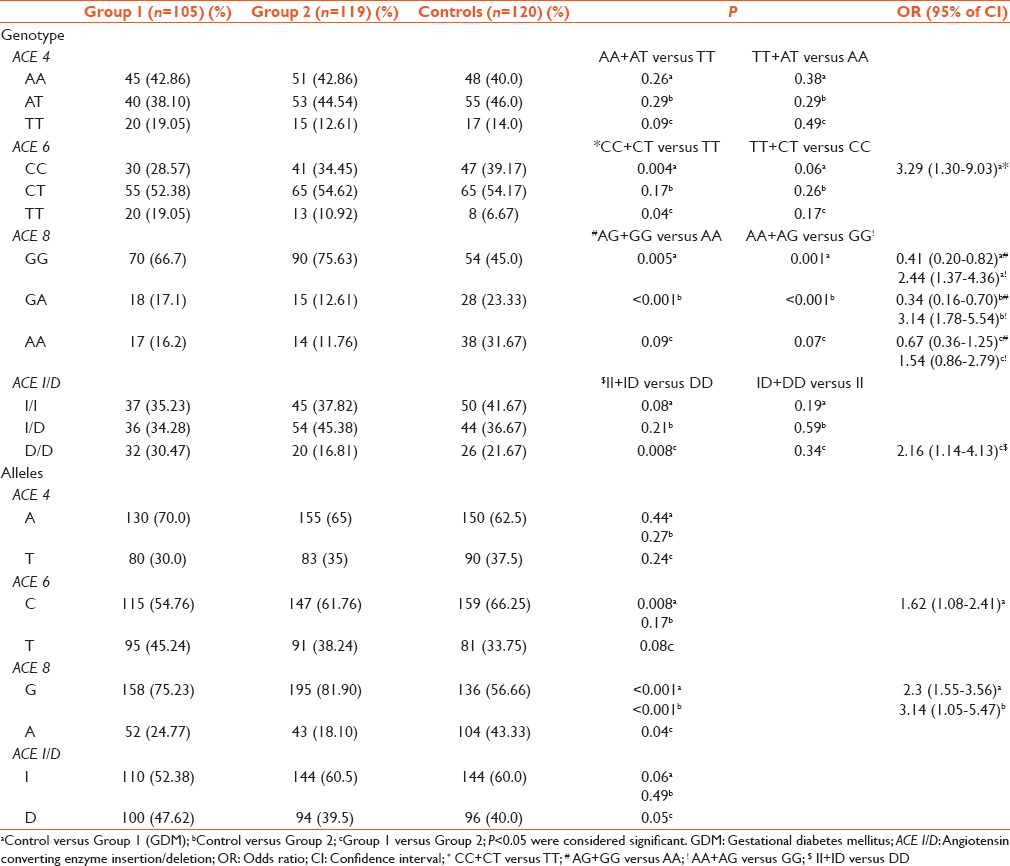
Table 2.
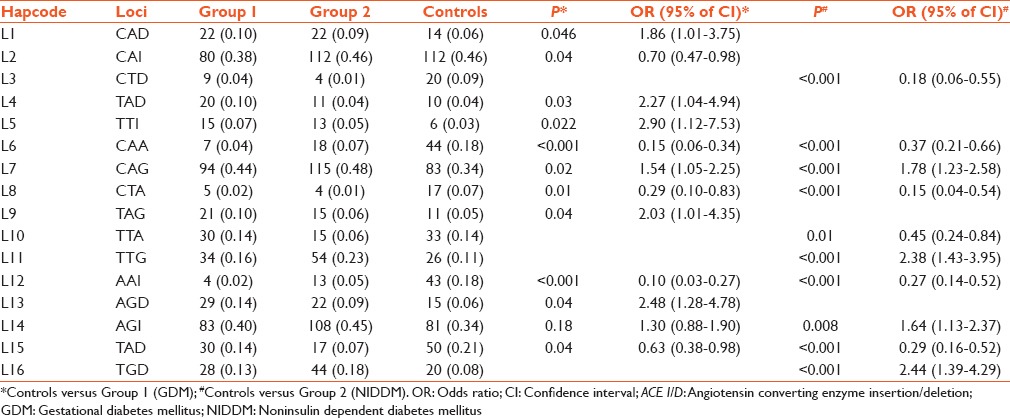
Observed 3 locus haplotypes of ACE 4, ACE 6, ACE 8 and ACE I/D polymorphisms and haplotype frequency analysis in groups and controls
Haplotype frequencies
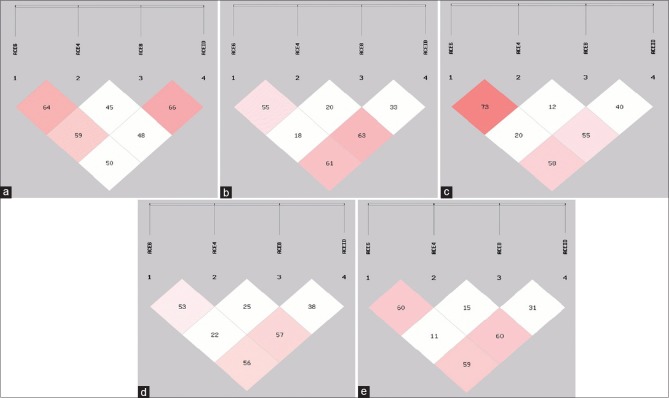
The LD analysis was made of four ACE polymorphisms in individuals and combined groups Figure 1a–e]. Haplotype analysis of ACE 4, ACE 6, ACE 8, and ACE I/D gene polymorphisms were carried out by combining them in the loci of two, three, and four respectively. A total of 24 haplotype combinations were formed both in two-locus and three locus haplotypes of which sixteen haplotypes were significant [Tables 2 and 3]. In four loci haplotype analyses only eight out of sixteen haplotype combinations were found to be significant [Table 4]. All polymorphisms (ACE 4, ACE 6, ACE 8, and ACE I/D) with high LD (D' values = 0.9 and r2 values = 0.8) were reanalyzed.
Figure 1.
Graphical overview of pair-wise linkage disequilibrium analysis of the investigated polymorphisms. (a) Controls; (b) Group 1; (c) Group 2; (d) Combined data of Group 1 and controls; (e) Combined data of Group 2 and controls
Table 3.
Observed 2 locus haplotypes of ACE 4, ACE 6, ACE 8 and ACE I/D polymorphisms and haplotype frequency analysis in groups and controls
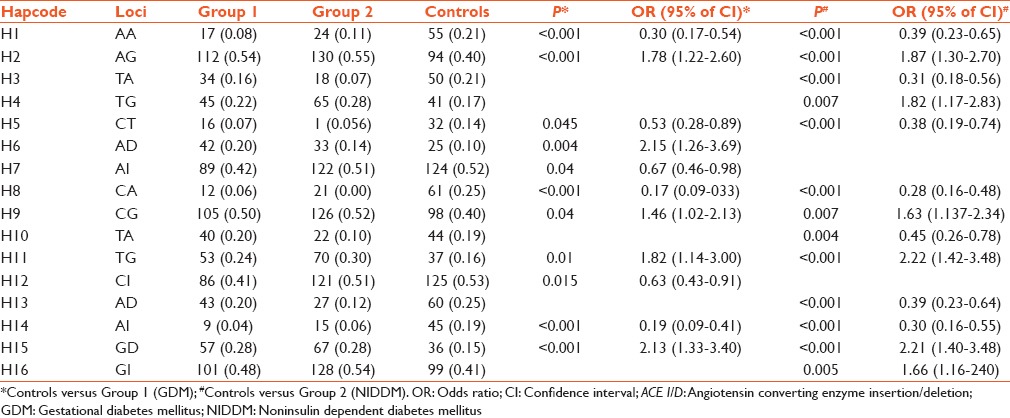
Table 4.
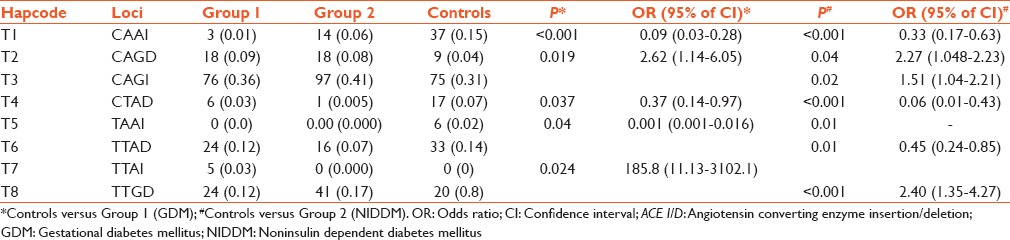
Observed 4 locus haplotypes of ACE 4, ACE 6, ACE 8 and ACE I/D polymorphisms and haplotype frequency analysis in groups and controls
Haplotype analyses of two-locus haplotypes
From Table 3, it appears that two loci haplotypes H1, H5, H8, and H14 were the protected ones in both active groups but haplotypes H2, H9, H11, and H15 were significantly associated as risk ones in these groups. In an individual group, the situation is different, and it is stated as follows. Haplotype H6 was found to be associated (risk) and haplotypes H7 and H12 were observed to be protective in Group 1 (GDM). In Group 2 haplotypes, H3, H10, and H13 were protective and H4 and H16 were risk haplotypes.
Three locus haplotype analysis
Table 2 shows that haplotype L2 was protective in Group 1 whereas haplotypes L1, L4, L5, L9, and L13 were strongly associated with Group 1. In Group 2, haplotypes L3 and L10 were protective but L11 and L16 was only risk haplotypes. Haplotypes L6, L8, L12, and L15 were protected in both groups. Strong association of haplotypes L7 and L14 with both groups were observed.
Haplotype analysis of four locus haplotypes
Table 4 shows four loci haplotypes depicting T1 and T4 to be protective in both groups. Haplotypes T2 and T5 were strongly associated with both active groups. In Group 1, haplotype T7 was statistically significant. In Group 2, haplotypes T3 and T8 may be a disease causing haplotypes, whereas T6 may play a protective role.
DISCUSSION
GDM is defined as any glucose intolerance detected during pregnancy.[15] Epidemiological studies revealed that GDM may be associated with increased feto-maternal morbidity and long-term complications in mothers and off-spring.[15] The etiologic role of genes implicated in RAS has been widely studied. Several clinical studies have reported a possible association between ACE polymorphisms and pathological phenotypes.[14,16,17,18,19,20,21,22,23,24] The present study shows the relation between four ACE polymorphisms in a group of North Indian pregnant women with or without GDM. Two out of four polymorphisms (ACE 6 and ACE 8) were found to be associated with GDM patients. These associations were independent of other diabetes-associated risk factors. ACE 4 and ACE I/D were associated neither with GDM nor with Type 2 diabetes mellitus patients. Of the studied polymorphisms, only ACE I/D were previously studied in patients with GDM and Type 2 diabetes mellitus in various populations.[25,26] The association of ACE I/D with GDM were contradictory, as in Asian Indian population DD genotype was strongly associated with GDM, but in Czechoslovakian population it was reported to be absent.[25,26]
Linkage disequilibrium
According to LD analysis, 4 polymorphisms (ACE 4, ACE 6, ACE 8, and ACE I/D) were in LD with statistical significance [Figure 1a–e] and constructed various haplotypes: One of them associated with highest risk (T7: TTAI) and the other was significantly protective (T5: TAAI) for GDM. To the best of our knowledge, haplotype studies on ACE polymorphism, besides hypertension or Alzheimer disease, and diabetes complications are not reported till date. Zhu et al. studied 10 polymorphisms in hypertensive patients from African-American and European-American populations.[27] The authors reported a haplotype with three polymorphisms associated with hypertension and the haplotype association was different in African-American and European-American subjects.[27] In the African-Americans, the haplotype associated included rs4343, rs4353, and rs4363 polymorphisms (AAA haplotype), and in European-Americans the haplotype included the rs4335, rs4343, and rs4344 polymorphisms (GGG haplotype). The risk haplotype in the Mexican Mestizo population includes rs4335 G and rs4344 G alleles. On the other hand, rs4353 A allele was detected in African-American as well as in European-American population.
The most studied ACE gene polymorphism, in this case, is I/D. In a study by Zhu et al.,[12] two polymorphisms (ACE 8 and ACE 4) accounted for the variation of ACE concentrations. ACE 8 had the most significant effect, and accounted for 19% of the total variation in ACE levels, while ACE 4 located in the 5' promoter region of the gene accounted for 6% of the variation. After adjustment for the effect of ACE 4, I/D polymorphism was found to be no longer associated with ACE levels, indicating that it is in LD with ACE 8 and unlikely to be a functional polymorphism.[12] The association of other ACE polymorphisms with ACE levels was reported to be contradictory with positive and negative associations.[28,29,30] The association ACE 4 polymorphism was absent in hypertensive Mexican Mestizo population.[8] In this study too, the association ACE 4 with GDM and Type 2 diabetes mellitus was observed to be absent.
Limitations of the study
The limitations of our study were inherited in the fact that ACE levels were not measured, and the study sample size was restricted to n = 344 comprising of 3 groups. The ACE levels could not be estimated in GDM patients because blood samples collecting varied at months of pregnancy for all patients (as some patients developed GDM in different trimesters of pregnancy).[25] Moreover, the T2DM patients were already in ACE-inhibitor drugs, which could have misled the estimated level. This pilot study has indicated the plausibly valuable associations of the polymorphism with the diseased conditions of a specific population. Hence, this study may be extended to other ethnic groups in establishing different regulatory regions in ACE gene along with ACE levels.
CONCLUSIONS
In summary, our results suggested that the ACE 4, ACE 6 ACE 8 and ACE I/D ACE gene polymorphisms might play an important role in the development of GDM and NIDDM in North Indian women population. In our study, it was possible to identify risk haplotypes and protective haplotypes for GDM and NIDDM. Individuals with the risk haplotype showed increased Glucose levels as compared to individuals with the protective haplotype.
Financial support and sponsorship
There was no external financial support to carry out this research activities.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Grzeszczak W, Zychma MJ, Lacka B, Zukowska-Szczechowska E. Angiotensin I-converting enzyme gene polymorphisms: Relationship to nephropathy in patients with non-insulin dependent diabetes mellitus. J Am Soc Nephrol. 1998;9:1664–9. doi: 10.1681/ASN.V991664. [DOI] [PubMed] [Google Scholar]
- 2.Linné Y, Barkeling B, Rössner S. Natural course of gestational diabetes mellitus: Long term follow up of women in the SPAWN study. BJOG. 2002;109:1227–31. doi: 10.1016/s1470-0328(02)01973-0. [DOI] [PubMed] [Google Scholar]
- 3.Williams MA, Qiu C, Dempsey JC, Luthy DA. Familial aggregation of type 2 diabetes and chronic hypertension in women with gestational diabetes mellitus. J Reprod Med. 2003;48:955–62. [PubMed] [Google Scholar]
- 4.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: A systematic review. Diabetes Care. 2002;25:1862–8. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan TA. Pancreatic B-cell defects in gestational diabetes: Implications for the pathogenesis and prevention of type 2 diabetes. J Clin Endocrinol Metab. 2001;86:989–93. doi: 10.1210/jcem.86.3.7339. [DOI] [PubMed] [Google Scholar]
- 6.Kautzky-Willer A, Prager R, Waldhausl W, Pacini G, Thomaseth K, Wagner OF, et al. Pronounced insulin resistance and inadequate beta-cell secretion characterize lean gestational diabetes during and after pregnancy. Diabetes Care. 1997;20:1717–23. doi: 10.2337/diacare.20.11.1717. [DOI] [PubMed] [Google Scholar]
- 7.Ryan EA, Imes S, Liu D, McManus R, Finegood DT, Polonsky KS, et al. Defects in insulin secretion and action in women with a history of gestational diabetes. Diabetes. 1995;44:506–12. doi: 10.2337/diab.44.5.506. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Rodríguez N, Posadas-Romero C, Villarreal-Molina T, Vallejo M, Del-Valle-Mondragón L, Ramírez-Bello J, et al. Single nucleotide polymorphisms of the angiotensin-converting enzyme (ACE) gene are associated with essential hypertension and increased ACE enzyme levels in Mexican individuals. PLoS One. 2013;8:e65700. doi: 10.1371/journal.pone.0065700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter MW, Coustan DR. Criteria for screening tests for gestational diabetes. Am J Obstet Gynecol. 1982;144:768–73. doi: 10.1016/0002-9378(82)90349-0. [DOI] [PubMed] [Google Scholar]
- 10.Govil D. Health Needs of Middle Aged Population: An Unaddressed Link. EAPS; European Population Conference Vienna; Austria. 2010. [Google Scholar]
- 11.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Bouzekri N, Southam L, Cooper RS, Adeyemo A, McKenzie CA, et al. Linkage and association analysis of angiotensin I-converting enzyme (ACE)-gene polymorphisms with ACE concentration and blood pressure. Am J Hum Genet. 2001;68:1139–48. doi: 10.1086/320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerousse O, Danilov S, Alhenc-Gelas F. Genetics of angiotensin I-converting enzyme. Clin Exp Hypertens. 1997;19:659–69. doi: 10.3109/10641969709083177. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XL, Wu LQ, Liu X, Yang YQ, Tan HW, Wang XH, et al. Association of angiotensin-converting enzyme gene I/D and CYP11B2 gene -344T/C polymorphisms with lone atrial fibrillation and its recurrence after catheter ablation. Exp Ther Med. 2012;4:741–747. doi: 10.3892/etm.2012.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Diabetes Association. Gestational diabetes mellitus (position statement) Diabetes Care. 2004;27(Suppl 2):S88–90. doi: 10.2337/diacare.27.2007.s88. [DOI] [PubMed] [Google Scholar]
- 16.Kabadou IA, Soualmia H, Jemaa R, Feki M, Kallel A, Souheil O, et al. G protein beta3 subunit gene C825T and angiotensin converting enzyme gene insertion/deletion polymorphisms in hypertensive Tunisian population. Clin Lab. 2013;59:85–92. doi: 10.7754/clin.lab.2013.111105. [DOI] [PubMed] [Google Scholar]
- 17.Yadav S, Hasan N, Marjot T, Khan MS, Prasad K, Bentley P, et al. Detailed analysis of gene polymorphisms associated with ischemic stroke in South Asians. PLoS One. 2013;8:e57305. doi: 10.1371/journal.pone.0057305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai T, Shikishima K, Matsushima M, Tsuneoka H. Genetic polymorphisms associated with endothelial function in nonarteritic anterior ischemic optic neuropathy. Mol Vis. 2013;19:213–9. [PMC free article] [PubMed] [Google Scholar]
- 19.Yang SJ, Kim S, Park H, Kim SM, Choi KM, Lim Y, et al. Sex-dependent association between angiotensin-converting enzyme insertion/deletion polymorphism and obesity in relation to sodium intake in children. Nutrition. 2013;29:525–30. doi: 10.1016/j.nut.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Jing Q, Wang X, Ma Y, Yang M, Huang G, Zhao X, et al. Angiotensin-converting enzyme I/D polymorphism and the risk of thoracic aortic dissection in Chinese Han population. Mol Biol Rep. 2013;40:1249–54. doi: 10.1007/s11033-012-2167-x. [DOI] [PubMed] [Google Scholar]
- 21.Rasyid H, Bakri S, Yusuf I. Angiotensin-converting enzyme gene polymorphisms, blood pressure and pulse pressure in subjects with essential hypertension in a South Sulawesi Indonesian population. Acta Med Indones. 2012;44:280–3. [PubMed] [Google Scholar]
- 22.Dhar S, Ray S, Dutta A, Sengupta B, Chakrabarti S. Polymorphism of ACE gene as the genetic predisposition of coronary artery disease in Eastern India. Indian Heart J. 2012;64:576–81. doi: 10.1016/j.ihj.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Dong X, Li B, Zhang XQ, Zeng Y, Wei YP, et al. Correlation of angiotensin converting enzyme gene polymorphism with perioperative myocardial protection under extracorporeal circulation. Asian Pac J Trop Med. 2012;5:995–9. doi: 10.1016/S1995-7645(12)60189-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Xu G, Liu D, Fan X, Zhu W, Liu X. Angiotensin-converting enzyme insertion/deletion polymorphism contributes to ischemic stroke risk: A meta-analysis of 50 case-control studies. PLoS One. 2012;7:e46495. doi: 10.1371/journal.pone.0046495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan IA, Jahan P, Hasan Q, Rao P. Angiotensin-converting enzyme gene insertion/deletion polymorphism studies in Asian Indian pregnant women biochemically identifies gestational diabetes mellitus. J Renin Angiotensin Aldosterone Syst. 2014;15:566–71. doi: 10.1177/1470320313502106. [DOI] [PubMed] [Google Scholar]
- 26.Dostálová Z, Bienertová-Vasku AJ, Vasku A, Gerychová R, Unzeitig V. Insertion-deletion polymorphism in the gene for angiotensin-converning enzyme (I/D ACE) in pregnant women with gestational diabetes. Ceska Gynekol. 2006;71:369–73. [PubMed] [Google Scholar]
- 27.Zhu X, Chang YP, Yan D, Weder A, Cooper R, Luke A, et al. Associations between hypertension and genes in the renin-angiotensin system. Hypertension. 2003;41:1027–34. doi: 10.1161/01.HYP.0000068681.69874.CB. [DOI] [PubMed] [Google Scholar]
- 28.Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, Soubrier F. Identification of new polymorphisms of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two-QTL segregation-linkage analysis. Am J Hum Genet. 1996;58:1268–78. [PMC free article] [PubMed] [Google Scholar]
- 29.Keavney B, McKenzie CA, Connell JM, Julier C, Ratcliffe PJ, Sobel E, et al. Measured haplotype analysis of the angiotensin-I converting enzyme gene. Hum Mol Genet. 1998;7:1745–51. doi: 10.1093/hmg/7.11.1745. [DOI] [PubMed] [Google Scholar]
- 30.Baghai TC, Binder EB, Schule C, Salyakina D, Eser D, Lucae S, et al. Polymorphisms in the angiotensin-converting enzyme gene are associated with unipolar depression, ACE activity and hypercortisolism. Mol Psychiatry. 2006;11:1003–15. doi: 10.1038/sj.mp.4001884. [DOI] [PubMed] [Google Scholar]