Abstract
Dysregulation of lipid metabolism underlies many chronic diseases such as obesity, diabetes, cardiovascular disease, and cancer. Therefore, understanding enzymatic mechanisms controlling lipid synthesis and degradation is imperative for successful drug discovery for these human diseases. Genes encoding α/β hydrolase fold domain (ABHD) proteins are present in virtually all reported genomes, and conserved structural motifs shared by these proteins predict common roles in lipid synthesis and degradation. However, the physiological substrates and products for these lipid metabolizing enzymes and their broader role in metabolic pathways remain largely uncharacterized. Recently, mutations in several members of the ABHD protein family have been implicated in inherited inborn errors of lipid metabolism. Furthermore, studies in cell and animal models have revealed important roles for ABHD proteins in lipid metabolism, lipid signal transduction, and metabolic disease. The purpose of this review is to provide a comprehensive summary surrounding the current state of knowledge regarding mammalian ABHD protein family members. In particular, we will discuss how ABHD proteins are ideally suited to act at the interface of lipid metabolism and signal transduction. Although, the current state of knowledge regarding mammalian ABHD proteins is still in its infancy, this review highlights the potential for the ABHD enzymes as being attractive targets for novel therapies targeting metabolic disease.
Keywords: Triacylglycerol, Phospholipid, CGI-58, Signal Transduction, Endocannabinoid, Serine Hydrolase
Introduction
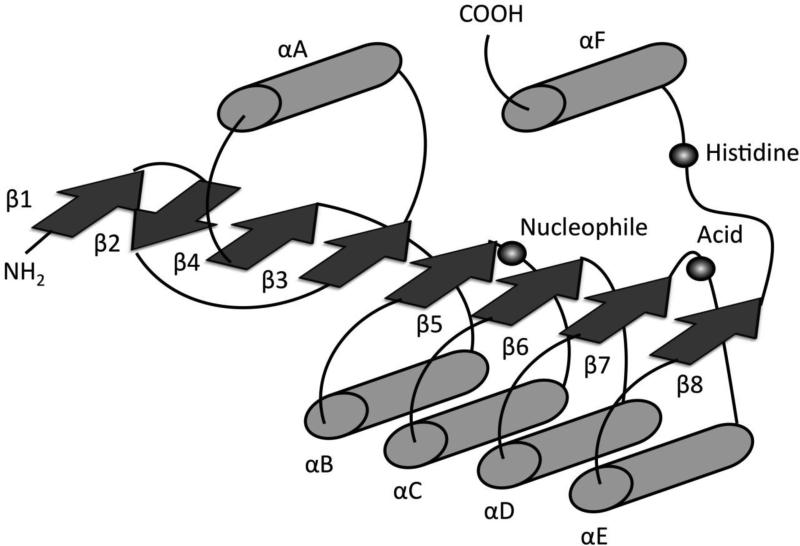
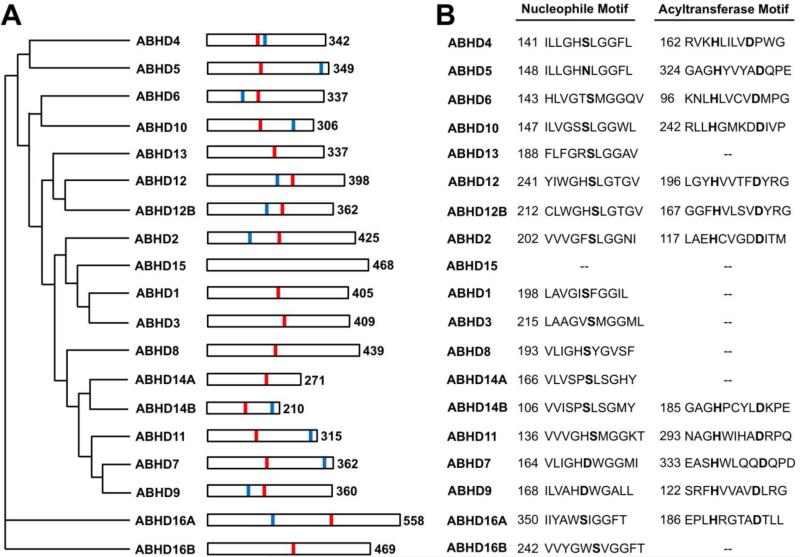
Lipids are essential for life as molecules of energy storage, membrane structure, and signal transduction. Aberrations in lipid signaling and metabolism underlie many human diseases, making it imperative to understand the proteins that regulate these processes. In recent years, the mammalian α/β-hydrolase domain (ABHD) proteins have emerged as novel potential regulators of lipid metabolism and signal transduction. The ABHD family contains at least 19 proteins and is part of a superfamily of proteins that possess an α/β-hydrolase fold [1]. The α/β-hydrolase fold superfamily, which includes proteases, lipases, esterases, dehalogenases, peroxidases, and epoxide hydrolases, is now one of the largest and most diverse protein families known [2]. The canonical α/β hydrolase fold is made up of 8 β-strands, with the second strand being antiparallel (Fig. 1). These β-strands form a core β-sheet which is surrounded by helices and loops connecting the β-strands. Hydrolase activity arises from a catalytic triad composed of nucleophile-acid-histidine residues which are located on loop regions (Fig. 1). The nucleophile (Ser, Cys, or Asp) is always located in a very tight loop, known as the “nucleophilic elbow,” following strand β5 (Fig. 1). The nucleophilic elbow can be identified by a consensus motif Sm-X-Nu-X-Sm, where Sm is a small residue, X is any residue, and Nu is the nucleophile [2]. The corresponding motif in most members of the ABHD family is GXSXG (Fig. 2). The acid residue of the catalytic triad can be either a glutamate or aspartate, usually located after strand β7 (Fig. 1). Finally, the histidine residue is absolutely conserved [2] and is located in a variable loop following the final β strand (Fig. 1). Interestingly, the majority of the ABHD proteins also have a conserved His-XXXX-Asp motif (where X is any residue), which has previously been associated with acyltransferase activity [32,33]. Thus, several ABHD proteins are predicted to possess both hydrolase and acyltransferase activities (Fig. 2). Together, these conserved motifs predict an important role of ABHD proteins in the hydrolysis or synthesis of small molecules involved in lipid metabolism and signal transduction. Although the biochemical substrates and physiological functions of the majority of the ABHD proteins are unknown, there is a growing recognition of the physiological significance of this family in metabolism and disease. In this review, we summarize the current state of knowledge surrounding the functional characterization of the mammalian ABHD proteins, and discuss their emerging importance in lipid metabolism, signal transduction, and metabolic disease. Although all genomes contain many proteins that are predicted to have an α/β-hydrolase fold, this review focuses solely on the mammalian proteins that currently share the ABHD nomenclature (ABHD1-ABHD16B).
Fig 1.
Canonical structure of the α/β hydrolase fold adapted from Nardini and Dijkstra [2]. The α/β Fold is an 8 stranded mostly parallel structure; β-sheets denoted as arrows and α-helices denoted as cylinders. Highly conserved catalytic triad containing a histidine, an acid, and a nucleophile (typically a serine residue in mammalian proteins) are represented by spheres.
Fig 2.
The Human ABHD Family. A. Phylogenetic relationship of the human ABHD proteins based on Clustal W alignment with Poisson correction. The numbers at the right indicate the number of amino acid residues in the full-length human protein. Red lines represent the predicted active site nucleophile, and blue lines indicate the predicted acyltransferase motif (HXXXXD) when present. B. The conserved nucleophilic elbow and acyltransferase motifs. “--“ indicates that the motif is not present in the human protein.
ABHD1: A potential regulator of oxidative stress
Human ABHD1, previously known as lung α/β hydrolase 1 (LABH1) [3], is a 405 residue (45 kDa) protein encoded by 9 exons located on chromosome 2p23.3. ABHD1 is predicted to be a single-pass type II membrane protein, although this has never been experimentally confirmed. Unlike most of the ABHD proteins, ABHD1 lacks a HX4D motif and is not expected to possess acyltransferase activity (Fig. 2). In mice, ABHD1 is ubiquitously expressed, with highest expression in heart and small intestine (Fig. 3). The biochemical function of ABHD1 has not been characterized. However, overexpression of ABHD1 in a renal cell line reduces the generation of reactive oxygen species by nicotinamide adenine dinucleotide phosphate (NADPH) oxidase [4]. Kidney ABHD1 expression is significantly upregulated in a mouse model of oxidative stress-induced hypertension, and it is possible that ABHD1 upregulation may be a protective response to oxidative stress in this model [4]. ABHD1 is also upregulated in a cell model of Huntington's disease, a progressive neurodegenerative disorder [5]. In agreement with a potential anti-oxidant function of ABHD1, the anti-oxidant Nrf2-ARE pathway is upregulated in this model, presumably as a protective response [5]. ABHD1 mRNA levels are also regulated under several other conditions. For example, liver ABHD1 is upregulated in mice challenged with parasitic infection [6]. ABHD1 expression in mouse liver and small intestine is significantly down-regulated by transgenic activation of Notch signaling, which transcriptionally drives cell differentiation [7]. Hippocampal ABHD1 expression is down-regulated by age and up-regulated by exercise in mice [8]. ABHD1 expression is also down-regulated in regenerative neurons in response to spinal cord injury in rats [9]. Together, these studies indicate that ABHD1 is highly transcriptionally controlled and may have an important function in oxidative stress.
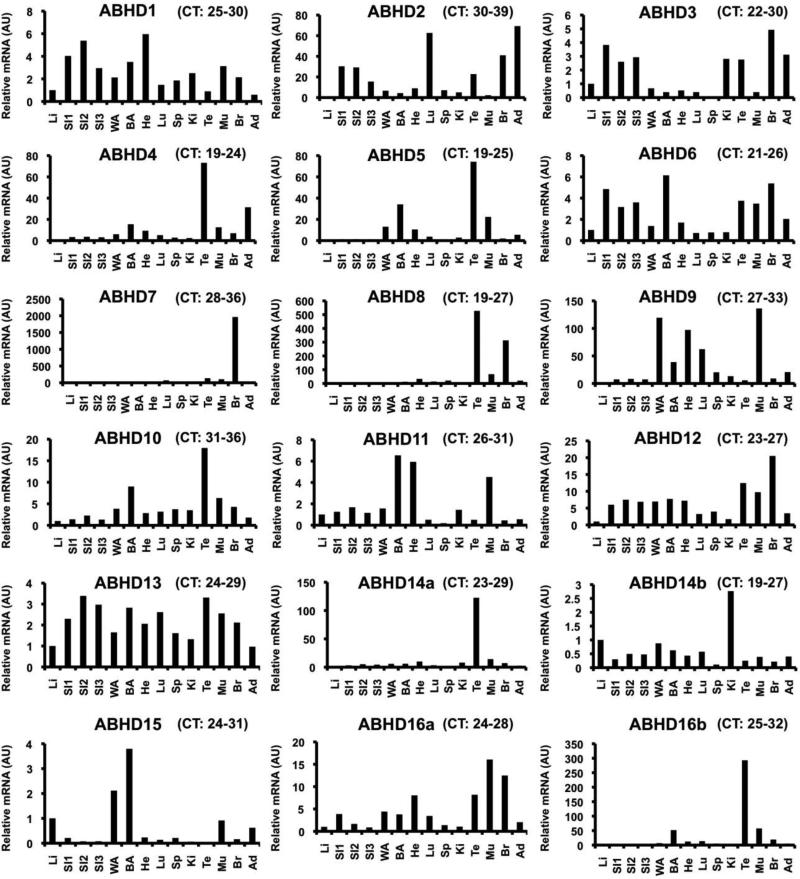
Fig 3.
Tissue Distribution of the ABHD family of enzymes in liver (Li), small intestine (SI1-3), white adipose tissue (WA), brown adipose tissue (BA), heart (He), lung (Lu), spleen (Sp), kidney (Ki), testes (Te), muscle (Mu), brain (Br), and adrenal (Ad). C57/Bl6N male mice were maintained on standard rodent chow until 18 weeks of age. Total RNA was extracted from individual mouse tissues, and an equal amount of total RNA from each sample in each group (n=4) was pooled for each tissue, and relative mRNA abundance was measured using quantitative real-time PCR (normalized to cyclophilin B). The range of threshold cycle (CT) values for each ABHD across tissues are listed above each panel. All PCR amplicons were verfied to correct size by gel electrophoresis and specificity of primer pairs was confirmed by DNA sequencing. The sequence of validated primer pairs are available upon requests.
ABHD2: A suppressor of smooth muscle cell migration and pulmonary emphysema
Human ABHD2, previously known as lung alpha/beta hydrolase 2 (LABH2) [3], is a 425 residue protein (48 kDa) encoded by 11 exons located on chromosome 15q26.1. ABHD2 is expressed in multiple tissues in mice, with highest expression in the lung, adrenal gland, and brain (Fig. 3). ABHD2 is predicted to be a single-pass type II membrane protein, although this has never been experimentally determined. ABHD2 is predicted to possess hydratase catalytic activity [10-13], but its true biochemical function remains unclear. ABHD2 has been previously linked to the migration of vascular smooth muscle cells (SMC), where it is initially expressed in endothelial cells with expression shifting to SMCs to suppress cellular migration and development of intimal hyperplasia [10-12]. ABHD2 is downregulated by lamivudine, a drug used to treat HIV patients, while the level of ABHD2 mRNA increases during monocyte to macrophage differentiation [11, 13]. Suppression of ABHD2 function in mice has been achieved in vivo using both antisense oligonucleotides (ASO) [13] and gene trapping techniques [10]. Knockdown of ABHD2 using ASO treatment in mice has been suggested to block hepatitis B virus propagation in a dose dependent manner without inducing apoptosis, suggesting it plays an important role in the virus's replication process [13, Table 1]. Global deletion of ABHD2 by gene trapping results in a reduction in the number of alveolar type II cells in the lung and a striking accumulation of macrophages in the lungs in aged mice [10, Table 1]. ABHD2 deficiency is also accompanied by an increase in inflammatory cytokines, increased apoptotic cells, reduced surfactant phospholipids, and a protease/anti-protease imbalance in the lung causing age-related emphysema [10]. In addition to its role in the lung, ABHD2 appears to play an important role in macrophage infiltration to atherosclerotic lesions [11]. ABHD2 expression is increased in patients with unstable angina in atherosclerotic lesions, specifically in neointimal lesions where it colocalizes with CD68+ cells [11]. Collectively, ABHD2 seems to play an important role in chronic diseases that involved monocyte/macrophage recruitment (i.e. atherosclerosis and emphysema), yet the true physiological substrates and products of ABHD2 remain unidentified.
Table 1.
qPCR primer design and conditions for tissue distribution analysis.
| Primer Sequence | |
|---|---|
| mABHD1 | F: 5’ – GGG TAC AGA ACG AAC CGA AA – 3’ R: 5’ – AGA GAA CAG GGG TGT GGA TG – 3’ |
| mABHD2 | F: 5’ – TTG ACA TGG CTT TGT GTG GT – 3’ R: 5’ – CGA CAT GGT GAT GAA CTT GC – 3’ |
| mABHD3 | F: 5’ – AGT TTC AGC CGC TTC CTA CA – 3’ R: 5’ – GAG ATC TGT CCT CCG TCT GC – 3’ |
| mABHD4 | F: 5’ – GTT CCA ATC CAC TGG CTG TT – 3’ R: 5’ – AGG ACT CCA TCA TGG CTT TG – 3’ |
| mABHD5 | F: 5’ – TGA CAG TGA TGC GGA AGA AG – 3’ R: 5’ – AGA TCT GGT CGC TCA GGA AA – 3’ |
| mABHD6 | F: 5’ – GAC ACA AGC CAT CCA TCC TT – 3’ R: 5’ – ACT TGC CCC ACT ATG GAC AG – 3’ |
| mABHD7 | F: 5’ – TCC AAA GGG AAT ACC CAC AA – 3’ R: 5’ – CAG GTG CAT CAG ACT CTC CA – 3’ |
| mABHD8 | F: 5’ – GGC TAC ACA TTC GTG GAG GT – 3’ R: 5’ – GTG GGC CAG GAA ACT ACA GA -3’ |
| mABHD9 | F: 5’ – GGC GAG CAT TGC TTT CTA AC – 3’ R: 5’ – TCC ACT TCC TTT GGA GCA TC – 3’ |
| mABHD10 | F: 5’ – CCT ACG CTG ACG ACT GAT GA – 3’ R: 5’ – TGA TGC CTT TTG TCT GCA AG – 3’ |
| mABHD11 | F: 5’ – GCC CTA GGA AGC ACA CTT TG – 3’ R: 5’ – GTC CTG ACT CAT GGC CTC AT – 3’ |
| mABHD12 | F: 5’ – TCA TCC TGA CAG GTG CTG AG – 3’ R: 5’ – ACC AGA TTT GTT GCC ACT CC – 3’ |
| mABHD13 | F: 5’ – AGG CCA ATA TGC TGT GGA AG – 3’ R: 5’ – GGC CAG TAG CCA TCT TTC AA – 3’ |
| mABHD14a | F: 5’ – ACC ACC AGC TAC GTG GAT TC – 3’ R: 5’ – ATC ATG CAG CTT CAC CAC AG – 3’ |
| mABHD14b | F: 5’ – AGG GCC AGA ACC TCT TCT TC – 3’ R: 5’ – ATC GCC CAA GAC CAT TAC AG – 3’ |
| mABHD15 | F: 5’ – TCT TGC TGC ACC AGA AAA TC – 3’ R: 5’ – CGG TGT CCA CTG TGT CCT C – 3’ |
| mABHD16a | F: 5’ – ACA TTG CTG CTG CTA CTT GC – 3’ R: 5’ – GTA CTG GGG GTT GGT CCA G – 3’ |
| mABHD16b | F: 5’ – AGT GCC TCC TCC AGC AGA T – 3’ R: 5’ – TGA GTG GGC CAG TGC ATA G – 3’ |
RNA was extracted from ~100-400mg of tissue in Trizol (Invitrogen). 1μg of RNA was reverse transcribed using omniscript RT (Qiagen) to generate cDNA. Relative mRNA was measured by quantitative real-time PCR using the delta-delta-CT method. Liver was set to 1 for all ABHD profiles.
ABHD3: A medium-chain phosphatidylcholine-specific phospholipase
Human ABHD3, previously known as lung alpha/beta hydrolase 3 (LABH3) [3], is a 409 residue (46 kDa) protein encoded by 9 exons located on chromosome 18q11.2. ABHD3 is predicted to be a single-pass type II membrane protein, yet this has not been experimental confirmed. Like ABHD1, ABHD3 does not possess a HX4D acyltransferase motif (Fig. 2). In mice, ABHD3 is ubiquitously expressed, with highest expression in brain and small intestine (Fig. 3). Based on activity-based protein profiling (ABPP), organophosphorus agents may be useful tools to inhibit ABHD3 serine hydrolase activity [14, 15]. In elegant unbiased metabolomic studies, Long and colleagues [16, Table 1] recently identified that ABHD3 selectively cleaves medium-chain and oxidatively-truncated phospholipids. In these studies, lysates from cells overexpressing ABHD3 had much higher phospholipase activity toward C14-containing phosphatidylcholine (C14-PCs) than cells overexpressing a catalytically dead serine mutant (ABHD3-S220A). In agreement, tissue metabolomics of ABHD3 knockout (ABHD3−/−) mice showed elevated levels of C14-PCs and other C14-phospholipids [16]. ABHD3 may regulate specific pools of PC with potential signaling functions, but further characterization of ABHD3−/− mice is required to determine the function of this activity in physiology and disease [16]. In agreement with a biochemical function in phospholipid metabolism, ABHD3 polymorphisms are significantly associated with the molar percentage of PC 32:2 in human plasma [17, Table 1]. Several additional studies have identified regulation of ABHD3 expression under various conditions. For example, ABHD3 is upregulated in the early response to chemotherapy treatment in human ovarian cancer cell lines [18]. ABHD3 has been identified in a screen for pro-apoptotic genes, being upregulated in microarrays of breast cancer tumors [19]. ABHD3 is also upregulated in a human osteosarcoma cell line overexpressing HIC1 (Hypermethylated in Cancer 1), a tumor suppressor and transcriptional repressor that is silenced in many human tumors [20]. ABHD3 is down-regulated in peripheral blood mononuclear cells (PBMCs) from patients with Crohn's disease, and the promoter contains binding sites for two different transcription factors, T-bet and Early Growth Response (EGRF) [21]. ABHD3 expression in the optic nerve is downregulated by early optic nerve injury in a rat model of glaucoma [22]. Although physiological substrates for ABHD3 have been successfully identified in mice, and diverse modes of transcriptional regulation have been described, the physiological function of ABHD3 in metabolic disease remains unknown.
ABHD4: An enzymatic regulator of endocannabinoid signaling and suppressor of tumor growth
Human ABHD4 is a 342 residue protein (39 kDa) encoded by 7 exons located on chromosome 14q11.2. Murine ABHD4 is ubiquitously expressed in multiple tissues, with highest expression in brain, small intestine, kidney and testis (Fig. 3), yet the subcellular localization of ABHD4 remains uncharacterized. Although vastly understudied, ABHD4 has recently been suggested to play a role in tumor suppression through limiting cell proliferation and cell cycling [23]. ABHD4 has also been described as a lyso-N-acyl phosphatidylethanolamine lipase, capable of hydrolyzing both N-acyl phosphatidylethanolamine (NAPE) and lysoNAPE to generate glycerophosphoarachidonyl ethanolamine (Gp-AEA) [23-25]. ABHD4 shows no enzymatic activity towards other lysophospholipid substrates tested, but rather with just a wide range of lyso-NAPE substrates, including the anandamide (AEA) precursor N-arachidonoyl lyso-NAPE [23, 24]. Lipopolysaccharide (LPS) has previously been shown to induce AEA synthesis, yet knockdown of ABHD4 does not affect LPS-induced AEA synthesis [25]. Cells treated with LPS for 90 minutes results in only a modest 27% increase in ABHD4 mRNA, suggesting that ABHD4 may be dominant in only the long term synthesis of AEA rather than the acute generation of the endocannabinoid in response to a LPS stimulus [25]. More recently, ABHD4 has also been suggested to play an important role in cancer metastasis through a mechanism that does not involve its role in AEA synthesis [26]. ABHD4 knockdown through the use of short-hairpin RNA has been shown to render cells resistant to anoikis, a process in which anchorage-dependent cells undergo cell death upon detachment from the extracellular matrix [26]. Knockdown of ABHD4 using the short hairpin RNA also results in more moderate tumor growth compared to controls, suggesting it may be a target gene of the tumor suppressor gene, p53 [23]. ABHD4 has also been shown to be upregulated in in vitro matured (IVM) oocytes, again supporting its proposed role in tumor growth suppression [27]. While the physiological substrates for ABHD4 have been successfully identified as NAPE and lysoNAPE, and ABHD4 seems to play a role in cancer pathogenesis, its role in metabolism remains poorly understood.
ABHD5: Critical integrator of phospholipid and triacylglycerol metabolism & guardian against Chanarin-Dorfman syndrome (CDS)
Human ABHD5, also known as Comparative Gene Identification 58 (CGI-58) is the most well characterized member of the ABHD family of protein. ABHD5 is a 349 (39 kDa) protein encoded by 7 exons located on chromosome 3p21.33. In mice, ABHD5 is widely expressed, with highest expression in testis and adipose tissue (Fig. 3). ABHD5 is the causative gene mutated in human Chanarin-Dorfman Syndrome, also known as Neutral Lipid Storage Disease with Ichthyosis (NLSDI), a rare, autosomal recessive non-lysosomal disorder of ectopic triacylglycerol accumulation [28, 29]. All identified ABHD5 mutations result in ichthyosis, often accompanied by hepatomegaly, hepatic steatosis, and various neurological disabilities [30]. Because NLSDI implicates ABHD5 as an important protein in triacylglycerol metabolism, ABHD5 has been highly studied and is the most well known member of the ABHD family. In recent years, several key in vitro and cell-based studies have shed light on the function of ABHD5. The nucleophilic serine is replaced by an asparagine residue in ABHD5 (Fig. 2). As a result, ABHD5 does not possess intrinsic hydrolase activity against triacylglycerol. Instead, ABHD5 has been shown to specifically co-activate Adipose Triacylglycerol Lipase (ATGL), the rate-limiting triacylglycerol hydrolase in adipose tissue, through an unknown mechanism [31]. In addition, ABHD5 has been reported to be involved in glycerophospholipid metabolism given that it possesses intrinsic lysophosphatidic acid acyltransferase (LPAAT) activity to generate the key signaling lipid phosphatidic acid [32, 33]. In adipocytes, ABHD5 is localized to the lipid droplet through interaction with perilipin-1 (Plin-1) [34-36]. Following Plin-1 phosphorylation by cyclic AMP-activated protein kinase A (PKA) during lipolytic hormone stimulation [34, 36], ABHD5 is released and a large portion disperses to the cytoplasm while a small portion remains at the lipid droplet, presumably interacting with ATGL [37]. The physiological function of ABHD5 has proven challenging to study in mouse models. ABHD5 total body knockout mice (ABHD5−/−) die shortly after birth due to a skin barrier defect, which closely resembles ichthyosis in human patients [38, Table 1]. Because of this, the function of ABHD5 in obesity and metabolism cannot be studied in ABHD5−/− mice, and although several tissue-specific ABHD5 knockout mouse models have been generated, associated phenotypes have not yet been published. As an alternative approach, ABHD5 knockdown in adult mice using antisense oligonucleotides has been useful to study the role of ABHD5 in metabolism and disease. Unexpectedly, knockdown of ABHD5 in adipose tissue results in lower fat mass and completely prevents high fat diet-induced obesity [39, Table 1], in contrast to mild obesity in ATGL−/− mice [40]. The phenotype of ABHD5 knockdown suggests that co-activation of ATGL-mediated triacylglycerol hydrolysis may not be the primary physiological function of ABHD5 in adipose tissue. Interestingly, adipose-selective overexpression of ABHD5 in mice has no effect on body weight and does not increase lipolysis, suggesting that endogenous levels of ABHD5 are not rate-limiting for ATGL-mediated lipolysis in vivo [41, Table 1]. In the liver, ABHD5 knockdown results in severe steatosis, yet paradoxically improves systemic glucose tolerance and hepatic insulin signaling [39, 42]. Whereas hepatic insulin signaling is improved, inflammatory signaling pathways in the liver are dramatically blunted by ABHD5 knockdown [42]. Lipidomic analysis of liver lipids revealed significant changes in several glycerophospholipid species with signaling potential, suggesting that ABHD5 functions to generate inflammatory signaling lipids that can promote insulin resistance [42]. Interestingly, triacylglycerol accumulation in NLDSI skin fibroblasts has previously been determined to be secondary to a defect in phospholipid metabolism [43, 44]. Although much progress has been made in understanding the biochemical function of ABHD5, it will be important to clarify physiological roles of ABHD5 in lipid metabolism and signal transduction that may be distinct from ATGL co-activation [45].
ABHD6: A physiological regulator of endocannabinoid signaling
Human ABHD6, also known as 2-arachidonylglycerol hydrolase, is a 337 residue protein (38 kDa) encoded by 10 exons on chromosome 3p21.1. It is ubiquitously expressed in mouse with highest expression in brown adipose tissue (BAT), small intestine and brain (Fig. 3). ABHD6 is predicted to have a single N-terminal transmembrane region, and based on N-linked glycosidase digestion studies ABHD6 is predicted to have a cytosolic facing orientation. Currently, there are no known mutations in ABHD6 associated with human disease. ABHD6 has been proposed as a possible target gene for Epstein-Barr virus (EBV) nuclear antigen 2 [46]. Furthermore, ABHD6 expression might be linked to the pathogenesis of EBV-related disorders such as: endemic Burkitt's lymphoma, Hodgkin's Lymphoma, and Post-Transplant Lymphoma [46, 47]. The first physiological substrate identified for ABHD6 was 2-arachidonylglycerol (2-AG) [48-50], an endocannabinoid signaling lipid that plays key roles in neurotransmission and metabolic disease. Although monoacylglycerol lipase (MAGL) was long thought to be the only mechanism of 2-AG hydrolysis, recently both ABHD6 and ABHD12 have also been shown to hydrolyze this key signaling lipid [48-52]. When ABHD6 is inhibited it does not affect MAGL activity, the primary enzyme associated with 2-AG hydrolysis in the brain, and ABHD6 controls the stimulated accumulation of 2-AG in intact neurons independently of MAGL [48, 51-53]. In BV-2 cell homogenates and intact BV-2 cells, inhibition of ABHD6 reduces 2-AG hydrolysis by ~50% [51]. It has also been shown to control the efficacy of 2-AG at CB receptors in neurons, suggesting ABHD6 enzymatic activity is key in controlling the bioactivity of this lipid transmitter [51]. Given that MAGL, ABHD6, and ABHD12 display distinct subcellular localization, it has been postulated that each enzyme acts to control specific subcellular pools of 2-AG [48, 51]. Since ABHD6 is found post-synaptically as opposed to MAGL, which is found presynaptically, it does not appear to play an active role in depolarization-induced suppression of excitation (DSE) [50, 52]. As well as its role in endocannabinoid metabolism, ABDH6 is differentially expressed among 7 tumor cell lines with highest expression in bone, prostate, and leukocyte tumor cell lines [46]. ABHD6 displayed lowest expression in liver and ovary tumor cells lines, but was not expressed in brain or cervical tumor cell lines [46]. ABHD6 shows exceptional high expression levels in Ewing family tumors, but not in other related sarcomas, suggesting that ABHD6 may be regulated by onco-fusion proteins in Ewing family tumors [54]. ABHD6 expression has been shown to be highly correlated with an Ewing family tumor-associated gene, aristaless [54]. However, tumor growth velocity, apoptosis rate and cell morphology are unaffected with ABHD6 knockdown [54]. Although one physiological substrate for ABHD6 has been successfully identified as 2-AG, it remains unclear whether ABHD6 regulates endocannabinoid signaling in peripheral tissues, and whether this signaling role is involved in cancer pathogenesis.
ABHD7: A brain restricted epoxide hydrolase
Human ABHD7, also known as epoxide hydrolase 4 (EPHX4) or epoxide hydrolase-related protein (EPHXRP), is a 362 residue (42 kDa) protein encoded by 7 exons located on chromosome 1p22.1. ABHD7 is predicted to be a single-pass type II membrane protein, but this need to be confirmed experimentally. ABHD7 possesses an aspartate nucleophile rather than a serine (Fig. 2), and therefore is expected to function as an epoxide hydrolase with activity toward epoxide-containing lipid substrates. Epoxide-containing lipids have potent bioactive effects in host defense, development, blood pressure, inflammation, and pain [55]. Epoxide hydrolases reduce the bioactivity of these lipids through conversion to 1,2-diols [55]. Although the specific substrates are unknown, ABHD7 activity is predicted to negatively regulate the bioactive effects of some epoxide lipids. In mice, ABHD7 is much more highly expressed in brain compared to other tissues (Fig. 3). ABHD7 has been identified as a direct target gene of the oncogene ZNF217, a transcriptional repressor overexpressed in many human tumors [56]. Based on this, it is possible that ABHD7 downregulation may contribute to the pathogenesis of some cancers. On the other hand, ABHD7 has been found to be hypomethylated in human colorectal cancer [57]. In short, very little is known about this brain restricted ABHD enzyme.
ABHD8: A member of the ABHD family with no known function
Human ABHD8 is 385 residue protein (47 kDa) encoded by 5 exons on chromosome 19p13.12. Subcellular localization, membrane topology, and physiological substrates of ABHD8 are currently unknown. It is most highly expressed in testes and brain in mouse, yet ABHD8 mRNA is quite abundant in all tissues examined (Fig. 3). Currently there are no published data about regulation, biochemical function, or physiological function of ABHD8.
ABHD9: A predicted epoxide hydrolase linked to cancer pathogenesis
Human ABHD9, also known as epoxide hydrolase 3 (EPHX3), is a 360 residue (41 kDa) protein encoded by 7 exons on chromosome 19p13.12. Like ABHD7, ABHD9 is annotated as an epoxide hydrolase, based on the presence of a nucleophilic aspartate in place of a serine (Fig. 2). In mice, we found ABHD9 to be ubiquitously expressed, with highest expression in skeletal muscle, white adipose tissue, and heart (Fig. 3). In contrast to our findings (Fig. 3), a recent report found low levels of ABHD9 mRNA in skeletal muscle [58]. This discrepancy might be due to detection of different ABHD9 variants or perhaps varying expression in different skeletal muscle sites (data in Figure 3 represents expression in quadricep muscle). Epoxyeicosatrienoic acids, angiogenic bioactive lipids, have recently been identified as endogenous substrates of ABHD9, suggesting that ABHD9 may negatively regulate angiogenesis [58]. Interestingly, epigenetic silencing of ABHD9 has been identified in several types of cancer. For example, ABHD9 is hypermethylated in human malignant melanomas [59], gastric cancers [60], adenoid cystic carcinomas of salivary glands [61], and colorectal carcinomas [62]. Hypermethylation of ABHD9 has been associated with prostate cancer recurrence [63, 64], and AHBD9 expression is also deregulated in some B cell malignancies [65]. Based on these studies, it is possible that ABHD9 substrates may contribute to cancer development. The chromosome 19p13.12 region, which includes ABHD9, is a frequent region of copy number gain in ovarian cancer, but ABHD9 has not been specifically implicated [66]. Interestingly, ABHD9 mutations are also predicted to cause human autosomal recessive nonlamellar, non-erythrodermic congenital ichthyosis [67]. ABHD9 also possesses activity toward leukotoxin to produce metabolites that mediate acute respiratory distress syndrome (ARDS) [58]. Based on these studies, ABHD9 may be a potential drug target to regulate bioactive epoxide lipids that are important in physiology and several diseases.
ABHD10: An enzymatic degrader of mycophenolic acid acyl-glucuronide
Human ABHD10 is a 306 residue protein (34 kDa) encoded by 5 exons on chromosome 3q13.2. ABHD10 is ubiquitously expressed in mouse tissues with highest expression found in testes and brown adipose tissue (Fig. 3). It is predicted to localize to mitochondria, although this has never been confirmed. In a study that sought to identify the targets of hexadecyl fluorophosphonate (HDFP), a candidate probe for depalmitoylating enzymes, ABHD10 was identified as a serine hydrolase that interacted with that probe [68]. ABHD10 has also been identified as the enzyme responsible for acyl glucuronide deglucoronidation of mycophenolic acid, the active metabolite of the immunosuppressant mycophenolate mofetil [69, 70]. Acyl-glucuronides are known to be toxic to the liver, and therefore ABHD10 may play a key role in the liver as a detoxification enzyme [69]. Recently, competitive activity-based protein profiling has identified a small molecule inhibitor that is a very potent and selective towards ABHD10 inactivation in vitro and in vivo, ABL303 [71]. Advancements with inhibitors as well as genetically modified animal models should continue to shed light on the biochemical and physiological function of ABHD10 in the coming years.
ABHD11: An enzymatic link to Williams-Beuren syndrome
Human ABHD11, previously named Williams-Beuren syndrome chromosomal region 21 protein (WBSCR21) or PP1226, is a 315 residue (35 kDa) protein encoded by 6 exons located on chromosome 7q11.23. Isoforms shorter than 325 amino acids are predicted from truncated mRNAs, but this has yet to be confirmed at the protein level. ABHD11 is predicted to be a mitochondrial protein, and has been detected in human skeletal muscle mitochondria [72]. ABHD11 is also enriched in the mitochondrial fraction of BV-2 cells, a murine microglial cell line [51]. In mice, ABHD11 is ubiquitously expressed, with highest expression in brown adipose tissue, heart, and skeletal muscle (Fig. 3). ABHD11 is one of several genes deleted in Williams-Beuren syndrome, a contiguous gene disorder caused by hemizygous deletion of a critical region on chromosome 7q11.23. WBS patients present with diverse neurodevelopmental features, including cardiovascular disease, mental retardation, opthalmic defects, dysmorphic facial features, premature aging of the skin, and unique cognitive and behavioral disabilities [73]. It is unknown whether any of these phenotypic features result specifically from ABHD11 deletion. In mouse white adipose tissue, high fat diet feeding decreases ABHD11 expression, while rosiglitazone treatment increases ABHD11 expression [74]. In addition, ABHD11 expression in brown adipose tissue is significantly reduced in hormone-sensitive lipase (HSL) knockout mice [74]. Although the endogenous substrates are unknown, recently identified inhibitors could be useful in characterizing the biochemical and physiological function of ABHD11. For example, a potent and selective carbamate inhibitor of ABHD11has been identified [75]. A 1,2,3-triazole urea inhibitor of ABHD11 has also been identified which results in covalent, irreversible inhibition [76]. Activity-based proteomics have identified ABHD11 serine hydrolase activity as a potential biomarker in human lung adenocarcinoma, since increased ABHD11 activity in lung adenocarcinomas significantly predicted the development of distant metastases [77]. Identification of the substrates and physiological function of ABHD11 could lead to better understanding of the pathogenesis of Williams-Beuren syndrome.
ABHD12: Regulator of endocannabinoid signaling and protector against PHARC development
Human ABHD12, also known as ABHD12A, c20orf22, and 2-arachidonoylglerol hydrolase, is a 398 residue protein (45 kDa) encoded by 13 exons on chromosome 20p11.21. Isoforms shorter than 398 amino acids are predicted from truncated mRNAs, but this has yet to be confirmed at the protein level. ABHD12 is predicted to be a single-pass integral membrane protein with its active site facing the ER lumen or extracellular space based on N-linked glycosidase digestion studies [48,49]. ABHD12 is ubiquitously expressed in mouse tissue with highest expression in the brain (Fig. 3), where it accounts for ~9% of total hydrolase activity toward the endocannabinoid 2-AG, along with monoacylglycerol lipase (MAGL) and ABHD6 [48,49,51]. In vivo inhibitor profiling demonstrates substrate specificity differences between ABHD12 and ABHD6, where ABHD12 prefers 1 (3)- and 2-isomers of arachidonoylglycerol [78]. In various brain cell types, specifically the microglia, ABHD12 transcripts are highly expressed [49]. ABHD12 is also abundant in other cell types such as macrophages and osteoclasts [49], and it is important to note that ABHD12 mRNA is very abundant and ubiquitiously expressed in all mouse tissue examined (Fig. 3). Much like ABHD6, ABHD12 also has no effect of the recovery of DSE, yet it does appears to traffic throughout the neuron [50]. Interestingly, mutations in ABHD12 have been shown to be causative of a neurodegenerative disease called polyneuropathy, hearing loss, ataxia, retinitis pigmentosa and cataract (PHARC) in humans [79, Table 1]. This disorder is a slow, progressive neurologic disorder that displays a phenotype resembling Refsum disease, which is characterized by a biochemical defect in the degradation of phytanic acid and plasmalogen lipids [79]. Four different ABHD12 mutations have been associated with PHARC development suggesting a causal genotype-phenotype relationship [79]. It has been suggested that ABHD12 defects leads to PHARC based on its enzymatic ability to hydrolyze 2-AG, since the endocannabinoid has important functions in synaptic plasticity and neuroinflammation [79]. However, careful studies examining whether ABHD12's ability to hydrolyze 2-AG is involved in PHARC development are still necessary. A nonsense mutation in ABHD12 has also been associated with an autosomal recessive genetically heterogeneous disorder called Usher Syndrome, specifically Usher Syndrome 3 [80]. Usher syndrome 3 is associated with congenital sensorineural hearing impairment, retinitis pigmentosa, hearing loss and cataracts representing a variant of PHARC [80]. In addition, a genome-wide association study identified ABHD12 as a candidate gene associated with concentrations of liver enzymes in plasma, a widely used indicator of liver disease [81]. Finally, ABHD12 gene expression has also been associated with colorectal cancer development [82]. Although one physiological substrate for ABHD12 has been successfully identified as 2-AG, the physiological role of this activity remains unclear.
ABHD12B: An ABHD with unknown function
Human ABHD12B, previously known as C14orf29, is a 362 residue (41kDa) protein encoded by 13 exons located on chromosome 14q22.1. Isoforms shorter than 362 amino acids are predicted from truncated mRNAs, but this has yet to be confirmed at the protein level. Although this gene shares a similar nomenclature and predicted structural homology with ABHD12, it is encoded by a completely separate gene. The subcellular localization of and the biochemical function of ABHD12B has not been determined. In addition, ABHD12B expression has not yet been characterized in mice. To our knowledge, the only report of ABHD12B expression in humans is significant downregulation of ABHD12B mRNA in skin tumors that result from mutations in the tumor suppressor gene CYLD [83]. Although ABHD12B is predicted to have both hydrolase and acyltransferase activity (Figure 2), this enzyme has not yet been characterized.
ABHD13: Another understudied ABHD enzyme
Human ABHD13, previously known as C13orf6, is a 337 residue (39 kDa) protein encoded by 2 exons located on chromosome 13q33.3. ABHD13 is predicted to be a single-pass type II membrane protein, but this has never been confirmed experimentally. ABHD13 lacks a HX4D motif and is not expected to possess acyltransferase activity, but has a conserved catalytic triad predictive of esterase activity (Fig. 2). In mice, ABHD13 mRNA is very abundantly and ubiquitously expressed, with highest levels found in testes and the distal small intestine (Fig. 3). Very little is known about the regulation of ABHD13 expression. ABHD13 is upregulated in the early response to chemotherapy treatment in human ovarian cancer cell lines [18]. It has also been reported that ABHD13 is regulated by circadian rhythm in rat liver [84]. Currently, there is no published information on the biochemical or physiological function of ABHD13.
ABHD14A: An enzymatic link to autism spectrum disorder
Human ABHD14A, also known as Dorz1, is a 271 residue (30 kDa) protein encoded by 7 exons located on chromosome 3p21.2. ABHD14a is predicted to be a single-pass type II membrane protein, but this has yet to be experimentally verified. ABHD14a is not expected to possess acyltransferase activity, but possesses a conserved catalytic triad predicting esterase activity (Fig. 2). In mice, ABHD14A mRNA in very abundantly and ubiquitously expressed, with higher expression in testis compared other tissues (Fig. 3). The biochemical function and substrates of ABHD14A are unknown. Like ABHD11, ABHD14A may be involved in the pathophysiology of Williams-Beuren syndrome (WBS). In contrast to the deletion of ABHD11 in WBS, ABHD14A expression is higher in skin fibroblasts from WBS patients [85]. In addition, ABHD14A is also a candidate gene associated with autism spectrum disorder, a genetically complex disorder [86]. In mice, ABHD14A expression correlates strongly with neuron development in the cerebellum, suggesting that ABHD14A may play a role in the differentiation or proliferation of neuron precursors [87]. It has also been reported that ABHD14A expression is significantly down-regulated in mouse bone marrow-derived stem cells exposed to intermittent hypoxia [88]. It will be interesting to determine the enzymatic function of ABHD14A, and whether this is related to the development of either autism spectrum disorder or Williams-Beuren syndrome in humans.
ABHD14B: The smallest ABHD enzyme with large potential
ABHD14B, also known as CCG1-interacting factor B (CIB), is a small 210 residue protein (22 kDa) encoded by 4 exons on chromosome 3p21.2. Isoforms shorter than 210 amino acids are predicted from truncated mRNAs, but this has yet to be confirmed at the protein level. ABHD14b-GFP fusion proteins have been shown to localize to both the cytoplasm and nucleus in transfected COS cells [89]. ABHD14B is abundantly and ubiquitously expressed in most mouse tissues, with highest expression in the kidney and liver (Fig. 3). In vitro, ABHD14B has been shown to have hydrolase activity towards the common lipase substrate p-nitrophenyl butyrate in a dose dependent manner [89]. However, true physiological substrates have yet to be identified in mammalian systems. ABHD14B has also been speculated to be a eukaryotic transcription factor [89, 90], but additional work is needed to confirm it nuclear role. ABHD14B gene expression has been shown to be upregulated in metastastic neuroendocrine tumors, and if upregulated >4 fold it would indicate the primary tumor is located in the stomach [90]. Therefore, ABHD14B is a valuable gene that can be used to identify the site of primary neuroendocrine tumor origin from its metastatic products [90]. Collectively, very little is know regarding the biochemical and physiological role of ABHD14B.
ABHD15: Novel target of insulin-stimulated Akt phosphorylation
Human ABHD15 is a 468 residue (52kDa) protein encoded by 2 exons located on chromosome 17q11.2. ABHD15 shares most sequence similarity with ABHDs 1, 2, and 3 (Fig. 2). However, ABHD15 lacks both a nucleophilic elbow and a HX4D motif, making biochemical activity prediction difficult. ABHD15 is actually predicted to be a secreted protein, although this has not been confirmed experimentally. In mice, ABHD15 is widely expressed, with highest expression in adipose tissue, liver, and skeletal muscle (Fig. 3). This tissue distribution is quite interesting given that these tissues are primary sites of postprandial insulin action, and ABHD15 has recently been described as a potential novel player in insulin signaling [91,92]. In mouse 3T3-L1 adipocytes, ABHD15 has been identified as a phosphorylation substrate of protein kinase Akt/PKB, with at least three insulin-induced phosphorylation sites (T142, S426, and S442) [91]. ABHD15 also forms a complex with cyclic AMP phosphodiesterase 3B (PDE3B) in 3T3-L1 adipocytes, and PDE3B protein expression is dependent on ABHD15 interaction [92]. This finding suggests that ABHD15 may stabilize PDE3B to negatively regulate cAMP levels. PDE3B itself is activated by Akt phosphorylation downstream of insulin signaling [93]. Under conditions of insulin stimulation, PDE3B reduces the cellular pool of cAMP, diminishing protein kinase A activation and hormone-stimulated lipolysis in adipocytes [93]. It is unclear how protein-protein interaction with ABHD15 increases PDE3B protein levels and activity. However, ABHD15 appears to be critical for the suppression of adipocyte lipolysis in response to insulin. The high expression of ABHD15 in white adipose tissue in mice (Fig. 3) suggests that ABHD15 may be physiologically important in adipocytes in vivo. It is interesting to note the contrast between ABHD15 and ABHD5. Both ABHD proteins are highly expressed in adipose tissue and have been implicated in the regulation of lipolysis, but with opposing effects. Since ABHD15 does not possess a conserved hydrolase or acyltransferase motif (Fig. 2), further studies will be needed to determine how ABHD15 regulates insulin action and nutrient metabolism.
ABHD16A: Potential enzymatic regulator of immunity
Human ABHD16A, also known as Human Lymphocyte Antigen B-associated transcript 5 (BAT5), is a 558 residue (63 kDa) protein encoded by 20 exons located on chromosome 6p21.33. ABHD16A is predicted to be a multi-pass membrane protein, and possesses both an esterase catalytic triad and an acyltransferase domain (Fig 2). In mice, ABHD16A mRNA is abundantly and ubiquitously expressed with highest expression in skeletal muscle and brain (Fig. 3). It has been reported that ABHD16A is palmitoylated, although the function of this modification is unknown [94]. Activity-based protein profiling has identified ABHD16A as a serine hydrolase with unknown substrates and function [95]. ABHD16A is part of a cluster of genes within the human major histocompatability complex (MHC) class III, suggesting ABHD16A may be involved in immunity [96, 97]. Interestingly, ABHD16A polymorphisms are significantly associated with susceptibility to coronary artery aneurysm formation in Kawasaki disease, an autoimmune disease of vascular inflammation [97]. More recently, single nucleotide polymorphisms in ABHD16A have been associated with backfat thickness in Italian Large White pigs [98], potentially linking ABHD16A function to adipose tissue function. Proteomics show ABHD16A to be localized to the plasma membrane in human platelets and mouse megakaryocytes [99]. It has also been reported that ABHD16A mRNA is regulated by miR-155 in activated B cells [100,101]. In a high-throughput yeast two-hybrid screen for protein interactions, ABHD16A was found to interact with several proteins, including membrane receptors and chaperone/processing proteins [102]. However, it is unclear how ABHD16A functions in these interactions. Identification of the substrates and biochemical function of ABHD16a's predicted hydrolase and acyltransferase activities could shed light on the putative function of ABHD16a in immunity.
ABHD16B: Another understudied member of the ABHD family
ABHD16B is a 469 residue protein (53 kDa) encoded by 1 exon on chromosome 20q13.33. Subcellular location of ABHD16B is currently unknown. ABHD16B possesses a canonical esterase catalytic triad, but lacks the acyltransferase motif shared by it ABHD relatives. It is expressed in most tissues with highest expression found in the testes, skeletal muscle, and brown adipose tissue (Fig. 3). Currently there are no published data surrounding ABHD16B's mRNA regulation, biochemical function, or physiological function.
A putative common role for ABHD enzymes: Integration of glycerophospholipid metabolism and lipid signal transduction
Although it is unlikely that all members of the ABHD protein family share a common biological role, there is early evidence to support this concept. In particular, of the ABHD enzymes with known substrates, most of them share the ability to either hydrolyze or synthesize different glycerophospholipid species [16,24,25,32,33,48,51]. For example ABHD3, ABHD4, and ABHD6 hydrolyze the glycerophospholipids C14-lysophosphatidylcholine, N-acyl phosphatidylethanolamine (NAPE), and 2-arachidonoylglycerol, respectively [16,24,48]. While, ABHD5 has recently been shown to acylate lysophosphatidic acid to form phosphatidic acid [32,33]. In further support of the ABHD's role in glycerophospholipid metabolism, there is a significant decrease in phosphatidylcholine in the bronchoalveolar lavage of ABHD2 knockout mice [10], and more recently ABHD3 reached genome-wide significance as a predictor of plasma phospholipid levels in humans [17]. Although much additional work is needed to identify physiological substrates for the remaining ABHD family members, it will be important to consider glycerophospholipids as clear potential substrates. It is well known that glycerophospholipids serve as key structural elements for the cell membrane, but many also acts a key intermediates in neurotransmission and cellular signaling cascades. In line with this, another shared feature of the ABHD family is that several members have been directly implicated in signaling pathways that involve enzymatic synthesis or degradation of key signaling lipids. For instance, both ABHD6 and ABHD12 have been identified and 2-AG hydrolases [48], linking these enzymes to the termination of acute endocannabinoid signaling [48,49,51,78]. In contrast, ABHD4 has been implicated as a NAPE and lysoNAPE lipase, implicating ABHD4 in endocannabinoid synthesis [24,25]. Further, ABHD5 has recently been shown to be necessary for the generation of phosphatidic acid and other signaling lipids in response to inflammatory stimuli [42], which then has many secondary effects on both insulin signaling and energy metabolism. Although this still needs to be experimentally tested, ABHD3's ability to hydrolyze C14-lysophosphatidylcholine is likely to alter cellular signaling, given that lysophosphatidylcholine species are potent regulators of GPCR and TRPC5 signaling [103,104]. Collectively, several members of the ABHD protein family share the ability to metabolize diverse glycerophospholipid species that are intimately involved in cellular signaling. These shared features of the known ABHDs will likely provide important clues as progress is made with the characterization of the rest of the ABHD protein family.
Conclusions and Future Perspectives
Although the functional annotation the ABHD protein family is in its infancy, ongoing studies have great promise in advancing our fundamental understanding of lipid metabolism in human disease. The ABHD enzyme family shares commons features that predict roles in lipid metabolism, signal transduction, and metabolic disease. More importantly, several ABHD family members have been associated with several human diseases of altered lipid metabolism. Given that serine hydrolases are easily amenable to chemical inhibition, development of small molecular inhibitors will become instrumental in defining the biology associated with each ABHD enzyme. Furthermore, the generation of global and conditional knockout mouse models will be key in defining biochemical and physiological roles of ABHD enzymes in vivo. Progress on these fronts has important implications for targeting ABHD enzymes for the treatment or prevention of lipid metabolic disease and diseases of altered signal transduction.
Table 2.
Identified human ABHD mutations, genetic variants, and mouse models of ABHD expression.
| Protein | Human Mutations | Human Polymorphisms | Mouse Models |
|---|---|---|---|
| ABHD2 | Global deletion: Progressive emphysema characterized by reduced surfactant phospholipids and increased inflammation, apoptosis, and macrophage accumulation in lung[10]. Antisense oligonucleotide-mediated knockdown: Prevention of hepatitis B virus propagation [13]. |
||
| ABHD3 | Association with the molar percentage of PC 32:2 in plasma[17]. | Global deletion: Elevated levels of C14-PCs and other C14 phospholipids in tissues [16]. | |
| ABHD5 | Neutral Lipid Storage Disease with Ichthyosis (also known as Chanarin-Dorfman Syndrome), a disease of ectopic triacylglycerol accumulation. Further characteristics: Jordan anomaly, hepatomegaly, hepatic steatosis, various neurological disabilities [28,29]. | Global deletion: Premature death due to skin barrier defect, elevated hepatic triacylglycerol [38]. Antisense oligonucleotide-mediated knockdown: Hepatic steatosis, prevention of diet-induced obesity, decreased fat mass, improved hepatic insulin signaling, attenuated hepatic inflammation [39,42]. Adipose-selective overexpression: No change in fat mass or lipolysis [41]. |
|
| ABHD12 | PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa and cataract), a neurodegenerative disease[79]. Usher syndrome type 3 Congenital hearing loss and retinitis pigmentosa [80] |
Highlights.
α/β-hydrolase domain (ABHD) enzymes regulate glycerophospholipid metabolism.
Several mutations in ABHD enzymes have been implicated in human diseases.
ABHD enzymes synthesize or degrade lipids involved in cellular signal transduction.
ABHD enzymes are attractive targets for new therapies targeting metabolic diseases.
Acknowledgements
This work was supported in part by the National Heart, Lung, and Blood Institute (R00-HL096166 to J.M.B) and the American Heart Association (11BGIA-7840072 to J.M.B and 12PRE11910081 to C.C.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 2.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 3.Edgar AJ, Polak JM. Cloning and tissue distribution of three murine alpha/beta hydrolase fold protein cDNAs. Biochem Biophys Res Commun. 2002;292:617–625. doi: 10.1006/bbrc.2002.6692. [DOI] [PubMed] [Google Scholar]
- 4.Stoelting M, Geyer M, Reuter S, Reichelt R, Bek MJ, Pavenstadt H. Alpha/beta hydrolase 1 is upregulated in D5 dopamine receptor knockout mice and reduces O2- production of NADPH oxidase. Biochem Biophys Res Commun. 2009;379:81–85. doi: 10.1016/j.bbrc.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 5.van Roon-Mom WM, Pepers BA, t Hoen PA, Verwijmeren CA, den Dunnen JT, Dorsman JC, van Ommen GB. Mutant huntingtin activates Nrf2-responsive genes and impairs dopamine synthesis in a PC12 model of Huntington's disease. BMC Mol Biol. 2008;9:84. doi: 10.1186/1471-2199-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kierstein S, Noyes H, Naessens J, Nakamura Y, Pritchard C, Gibson J, Kemp S, Brass A. Gene expression profiling in a mouse model for African trypanosomiasis. Genes Immun. 2006;7:667–679. doi: 10.1038/sj.gene.6364345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler JC, Zecchini VR, Jones PH. Intestinal activation of Notch signaling induces rapid onset hepatic steatosis and insulin resistance. PLoS One. 2011;6:e20767. doi: 10.1371/journal.pone.0020767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohman RA, Rodriguez-Zas SL, Southey BR, Kelley KW, Dantzer R, Rhodes JS. Voluntary wheel running reverses age-induced changes in hippocampal gene expression. PLoS One. 2011;6:e22654. doi: 10.1371/journal.pone.0022654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siebert JR, Middelton FA, Stelzner DJ. Intrinsic response of thoracic propriospinal neurons to axotomy. BMC Neurosci. 2010;11:69. doi: 10.1186/1471-2202-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin S, Zhao G, Li Z, Nishimoto Y, Isohama Y, Shen J, Ito T, Takeya M, Araki K, He P, Yamamura K. Age-related pulmonary emphysema in mice lacking alpha/beta hydrolase domain containing 2 gene. Biochem Biophys Res Commun. 2009;380:419–424. doi: 10.1016/j.bbrc.2009.01.098. [DOI] [PubMed] [Google Scholar]
- 11.Miyata K, Nakayama M, Mizuta S, Hokimoto S, Sugamura K, Oshima S, Oike Y, Sugiyama S, Ogawa H, Yamamura K. Elevated mature macrophage expression of human ABHD2 gene in vulnerable plaque. Biochem Biophys Res Commun. 2008;365:207–213. doi: 10.1016/j.bbrc.2007.10.127. [DOI] [PubMed] [Google Scholar]
- 12.Miyata K, Oike Y, Hoshii T, Maekawa H, Ogawa H, Suda T, Araki K, Yamamura K. Increase of smooth muscle cell migration and of intimal hyperplasia in mice lacking the alpha/beta hydrolase domain containing 2 gene. Biochem Biophys Res Commun. 2005;329:296–304. doi: 10.1016/j.bbrc.2005.01.127. [DOI] [PubMed] [Google Scholar]
- 13.Ding X, Yang J, Wang S. Antisense oligonucleotides targeting abhydrolase domain containing 2 block human hepatitis B virus propagation. Oligonucleotides. 2011;21:77–84. doi: 10.1089/oli.2011.0280. [DOI] [PubMed] [Google Scholar]
- 14.Casida JE, Nomura DK, Vose SC, Fujioka K. Organophosphate-sensitive lipases modulate brain lysophospholipids, ether lipids and endocannabinoids. Chem Biol Interact. 2008;175:355–364. doi: 10.1016/j.cbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat Chem Biol. 2008;4:373–378. doi: 10.1038/nchembio.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long JZ, Cisar JS, Milliken D, Niessen S, Wang C, Trauger SA, Siuzdak G, Cravatt BF. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat Chem Biol. 2011;7:763–765. doi: 10.1038/nchembio.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, Johansson A, Rudan I, Aulchenko YS, Kirichenko AV, Janssens AC, Jansen RC, Gnewuch C, Domingues FS, Pattaro C, Wild SH, Jonasson I, Polasek O, Zorkoltseva IV, Hofman A, Karssen LC, Struchalin M, Floyd J, Igl W, Biloglav Z, Broer L, Pfeufer A, Pichler I, Campbell S, Zaboli G, Kolcic I, Rivadeneira F, Huffman J, Hastie ND, Uitterlinden A, Franke L, Franklin CS, Vitart V, Nelson CP, Preuss M, Bis JC, O'Donnell CJ, Franceschini N, Witteman JC, Axenovich T, Oostra BA, Meitinger T, Hicks AA, Hayward C, Wright AF, Gyllensten U, Campbell H, Schmitz G. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L'Esperance S, Bachvarova M, Tetu B, Mes-Masson AM, Bachvarov D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genomics. 2008;9:99. doi: 10.1186/1471-2164-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin B, Huntley D, Abuali G, Langley SR, Sindelar G, Petretto E, Butcher S, Grimm S. Determining signalling nodes for apoptosis by a genetic high-throughput screen. PLoS One. 2011;6:e25023. doi: 10.1371/journal.pone.0025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guerardel C, Ramsey K, Monte D, Begue A, Tschan MP, Stephan DA, Leprince D. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1) J Biol Chem. 2009;284:20927–20935. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Becker NQ, Moss AC. In silico Analysis of T-bet Activity in Peripheral Blood Mononuclear Cells in Patients with Inflammatory Bowel Disease (IBD) In Silico Biol. 2009;9:355–363. doi: 10.3233/ISB-2009-0410. [DOI] [PubMed] [Google Scholar]
- 22.Johnson EC, Doser TA, Cepurna WO, Dyck JA, Jia L, Guo Y, Lambert WS, Morrison JC. Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci. 2011;52:504–518. doi: 10.1167/iovs.10-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, Karnezis AN, Attardi LD. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J Biol Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Wang L, Harvey-White J, Huang BX, Kim HY, Luquet S, Palmiter RD, Krystal G, Rai R, Mahadevan A, Razdan RK, Kunos G. Multiple pathways involved in the biosynthesis of anandamide. Neuropharmacology. 2008;54:1–7. doi: 10.1016/j.neuropharm.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson CD, Hurren R, Kasimer D, MacLean N, Eberhard Y, Ketela T, Moffat J, Schimmer AD. A genome wide shRNA screen identifies alpha/beta hydrolase domain containing 4 (ABHD4) as a novel regulator of anoikis resistance. Apoptosis. 2012;17:666–678. doi: 10.1007/s10495-012-0723-4. [DOI] [PubMed] [Google Scholar]
- 27.Katz-Jaffe MG, McCallie BR, Preis KA, Filipovits J, Gardner DK. Transcriptome analysis of in vivo and in vitro matured bovine MII oocytes. Theriogenology. 2009;71:939–946. doi: 10.1016/j.theriogenology.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Igal RA, Rhoads JM, Coleman RA. Neutral lipid storage disease with fatty liver and cholestasis. J Pediatr Gastroenterol Nutr. 1997;25:541–547. doi: 10.1097/00005176-199711000-00011. [DOI] [PubMed] [Google Scholar]
- 29.Lefevre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret JL, Weissenbach J, Ozguc M, Lathrop M, Prud'homme JF, Fischer J. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69:1002–1012. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 31.Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh AK, Ramakrishnan G, Chandramohan C, Rajasekharan R. CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J Biol Chem. 2008;283:24525–24533. doi: 10.1074/jbc.M801783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montero-Moran G, Caviglia JM, McMahon D, Rothenberg A, Subramanian V, Xu Z, Lara-Gonzalez S, Storch J, Carman GM, Brasaemle DL. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J Lipid Res. 2009 doi: 10.1194/jlr.M001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi T, Omatsu N, Matsushita S, Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem. 2004;279:30490–30497. doi: 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi T, Omatsu N, Morimoto E, Nakashima H, Ueno K, Tanaka T, Satouchi K, Hirose F, Osumi T. CGI-58 facilitates lipolysis on lipid droplets but is not involved in the vesiculation of lipid droplets caused by hormonal stimulation. J Lipid Res. 2007;48:1078–1089. doi: 10.1194/jlr.M600493-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem. 2007;282:5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- 38.Radner FP, Streith IE, Schoiswohl G, Schweiger M, Kumari M, Eichmann TO, Rechberger G, Koefeler HC, Eder S, Schauer S, Theussl HC, Preiss-Landl K, Lass A, Zimmermann R, Hoefler G, Zechner R, Haemmerle G. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) J Biol Chem. 2009 doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown JM, Betters JL, Lord C, Ma Y, Han X, Yang K, Alger HM, Melchior J, Sawyer J, Shah R, Wilson MD, Liu X, Graham MJ, Lee R, Crooke R, Shulman GI, Xue B, Shi H, Yu L. CGI-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. J Lipid Res. 2010;51:3306–3315. doi: 10.1194/jlr.M010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, Wagner EF, Klingenspor M, Hoefler G, Zechner R. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 41.Caviglia JM, Betters JL, Dapito DH, Lord CC, Sullivan S, Chua S, Yin T, Sekowski A, Mu H, Shapiro L, Brown JM, Brasaemle DL. Adipose-selective overexpression of ABHD5/CGI-58 does not increase lipolysis or protect against diet-induced obesity. J Lipid Res. 2011;52:2032–2042. doi: 10.1194/jlr.M019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord CC, Betters JL, Ivanova PT, Milne SB, Myers DS, Madenspacher J, Thomas G, Chung S, Liu M, Davis MA, Lee RG, Crooke RM, Graham MJ, Parks JS, Brasaemle DL, Fessler MB, Brown HA, Brown JM. CGI-58/ABHD5-Derived Signaling Lipids Regulate Systemic Inflammation and Insulin Action. Diabetes. 2012;61:355–363. doi: 10.2337/db11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Igal RA, Coleman RA. Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in neutral lipid storage disease fibroblasts. J Biol Chem. 1996;271:16644–16651. doi: 10.1074/jbc.271.28.16644. [DOI] [PubMed] [Google Scholar]
- 44.Igal RA, Coleman RA. Neutral lipid storage disease: a genetic disorder with abnormalities in the regulation of phospholipid metabolism. J Lipid Res. 1998;39:31–43. [PubMed] [Google Scholar]
- 45.Lord CC, Brown JM. Distinct roles for alpha-beta hydrolase domain 5 (ABHD5/CGI-58) and adipose triglyceride lipase (ATGL/PNPLA2) in lipid metabolism and signaling. Adipocyte. 2012;1 doi: 10.4161/adip.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F, Fei X, Xu J, Ji C. An unannotated alpha/beta hydrolase superfamily member, ABHD6 differentially expressed among cancer cell lines. Mol Biol Rep. 2009;36:691–696. doi: 10.1007/s11033-008-9230-7. [DOI] [PubMed] [Google Scholar]
- 47.Maier S, Staffler G, Hartmann A, Hock J, Henning K, Grabusic K, Mailhammer R, Hoffmann R, Wilmanns M, Lang R, Mages J, Kempkes B. Cellular target genes of Epstein-Barr virus nuclear antigen 2. J Virol. 2006;80:9761–9771. doi: 10.1128/JVI.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savinainen JR, Saario SM, Laitinen JT. The serine hydrolases MAGL, ABHD6 and ABHD12 as guardians of 2-arachidonoylglycerol signalling through cannabinoid receptors. Acta Physiol (Oxf) 2012;204:267–276. doi: 10.1111/j.1748-1716.2011.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straiker A, Wager-Miller J, Hu SS, Blankman JL, Cravatt BF, Mackie K. COX-2 and fatty acid amide hydrolase can regulate the time course of depolarization-induced suppression of excitation. Br J Pharmacol. 2011;164:1672–1683. doi: 10.1111/j.1476-5381.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marrs WR, Blankman JL, Horne EA, Thomazeau A, Lin YH, Coy J, Bodor AL, Muccioli GG, Hu SS, Woodruff G, Fung S, Lafourcade M, Alexander JP, Long JZ, Li W, Xu C, Moller T, Mackie K, Manzoni OJ, Cravatt BF, Stella N. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat Neurosci. 2010;13:951–957. doi: 10.1038/nn.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong P, Pan B, Gao XP, Blankman JL, Cravatt BF, Liu QS. Genetic deletion of monoacylglycerol lipase alters endocannabinoid-mediated retrograde synaptic depression in the cerebellum. J Physiol. 2011;589:4847–4855. doi: 10.1113/jphysiol.2011.215509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marrs WR, Horne EA, Ortega-Gutierrez S, Cisneros JA, Xu C, Lin YH, Muccioli GG, Lopez-Rodriguez ML, Stella N. Dual inhibition of alpha/beta-hydrolase domain 6 and fatty acid amide hydrolase increases endocannabinoid levels in neurons. J Biol Chem. 2011;286:28723–28728. doi: 10.1074/jbc.M110.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Max D, Hesse M, Volkmer I, Staege MS. High expression of the evolutionarily conserved alpha/beta hydrolase domain containing 6 (ABHD6) in Ewing tumors. Cancer Sci. 2009;100:2383–2389. doi: 10.1111/j.1349-7006.2009.01347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morisseau C. Role of epoxide hydrolases in lipid metabolism. Biochimie. 2012 doi: 10.1016/j.biochi.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krig SR, Jin VX, Bieda MC, O'Geen H, Yaswen P, Green R, Farnham PJ. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnekenburger M, Diederich M. Epigenetics Offer New Horizons for Colorectal Cancer Prevention. Curr Colorectal Cancer Rep. 2012;8:66–81. doi: 10.1007/s11888-011-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Decker M, Adamska M, Cronin A, DiGiallonardo F, Burgener J, Marowsky A, Falck JR, Hammock C, Gruzdev A, Zeldin DC, Arand M. EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J Lipid Res. 2012;53:2038–2045. doi: 10.1194/jlr.M024448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furuta J, Nobeyama Y, Umebayashi Y, Otsuka F, Kikuchi K, Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2'-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bell A, Bell D, Weber RS, El-Naggar AK. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer. 2011;117:2898–2909. doi: 10.1002/cncr.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oster B, Thorsen K, Lamy P, Wojdacz TK, Hansen LL, Birkenkamp-Demtroder K, Sorensen KD, Laurberg S, Orntoft TF, Andersen CL. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int J Cancer. 2011;129:2855–2866. doi: 10.1002/ijc.25951. [DOI] [PubMed] [Google Scholar]
- 63.Cottrell S, Jung K, Kristiansen G, Eltze E, Semjonow A, Ittmann M, Hartmann A, Stamey T, Haefliger C, Weiss G. Discovery and validation of 3 novel DNA methylation markers of prostate cancer prognosis. J Urol. 2007;177:1753–1758. doi: 10.1016/j.juro.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 64.Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, Ittmann M, Haefliger C, Lesche R, Hartmann A, Corman J, Wheeler T. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol. 2009;181:1678–1685. doi: 10.1016/j.juro.2008.11.120. [DOI] [PubMed] [Google Scholar]
- 65.Micci F, Panagopoulos I, Tjonnfjord GE, Kolstad A, Delabie J, Beiske K, Heim S. Molecular cytogenetic characterization of t(14;19)(q32;p13), a new recurrent translocation in B cell malignancies. Virchows Arch. 2007;450:559–565. doi: 10.1007/s00428-007-0407-6. [DOI] [PubMed] [Google Scholar]
- 66.Gorringe KL, George J, Anglesio MS, Ramakrishna M, Etemadmoghadam D, Cowin P, Sridhar A, Williams LH, Boyle SE, Yanaihara N, Okamoto A, Urashima M, Smyth GK, Campbell IG, Bowtell DD. Copy number analysis identifies novel interactions between genomic loci in ovarian cancer. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ala U, Piro RM, Grassi E, Damasco C, Silengo L, Oti M, Provero P, Di Cunto F. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput Biol. 2008;4:e1000043. doi: 10.1371/journal.pcbi.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukami T, Yokoi T. The Emerging Role of Human Esterases. Drug Metab Pharmacokinet. 2012 doi: 10.2133/dmpk.dmpk-12-rv-042. [DOI] [PubMed] [Google Scholar]
- 70.Iwamura A, Fukami T, Higuchi R, Nakajima M, Yokoi T. Human alpha/beta hydrolase domain containing 10 (ABHD10) is responsible enzyme for deglucuronidation of mycophenolic acid acyl-glucuronide in liver. J Biol Chem. 2012;287:9240–9249. doi: 10.1074/jbc.M111.271288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuhl AM, Mohr JT, Bachovchin DA, Niessen S, Hsu KL, Berlin JM, Dochnahl M, Lopez-Alberca MP, Fu GC, Cravatt BF. Competitive activity-based protein profiling identifies aza-beta-lactams as a versatile chemotype for serine hydrolase inhibition. J Am Chem Soc. 2012;134:5068–5071. doi: 10.1021/ja300799t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lefort N, Yi Z, Bowen B, Glancy B, De Filippis EA, Mapes R, Hwang H, Flynn CR, Willis WT, Civitarese A, Hojlund K, Mandarino LJ. Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J Proteomics. 2009;72:1046–1060. doi: 10.1016/j.jprot.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet. 2002;110:429–438. doi: 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- 74.Shen WJ, Patel S, Yu Z, Jue D, Kraemer FB. Effects of rosiglitazone and high fat diet on lipase/esterase expression in adipose tissue. Biochim Biophys Acta. 2007;1771:177–184. doi: 10.1016/j.bbalip.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wiedl T, Arni S, Roschitzki B, Grossmann J, Collaud S, Soltermann A, Hillinger S, Aebersold R, Weder W. Activity-based proteomics: identification of ABHD11 and ESD activities as potential biomarkers for human lung adenocarcinoma. J Proteomics. 2011;74:1884–1894. doi: 10.1016/j.jprot.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 78.Navia-Paldanius D, Savinainen JR, Laitinen JT. Biochemical and pharmacological characterization of human a/b-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12) J Lipid Res. 2012;53:2413–2424. doi: 10.1194/jlr.M030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fiskerstrand T, H'Mida-Ben Brahim D, Johansson S, M'Zahem A, Haukanes BI, Drouot N, Zimmermann J, Cole AJ, Vedeler C, Bredrup C, Assoum M, Tazir M, Klockgether T, Hamri A, Steen VM, Boman H, Bindoff LA, Koenig M, Knappskog PM. Mutations in ABHD12 cause the neurodegenerative disease PHARC: An inborn error of endocannabinoid metabolism. Am J Hum Genet. 2010;87:410–417. doi: 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eisenberger T, Slim R, Mansour A, Nauck M, Numberg G, Decker C, Dafinger C, Ebermann I, Bergmann C, Bolz HJ. Targeted next-generation sequencing identifies a homozygous nonsense mutation in ABHD12, the gene underlying PHARC in a family clinically diagnosed with Usher syndrome type 3. Orphanet J Rare Dis. 2012;7:59. doi: 10.1186/1750-1172-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Gieger C, Heard-Costa NL, Hottenga J-J, Kühnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Leach IM, O'Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJC, Dixon AL, Döring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen A-L, Heath S, Hirschfield GM, Hofman A, Homuth G, Hyppönen E, Janssen HLA, Johnson T, Kangas AJ, Kema IP, Kühn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin J, Loos RJF, Martin NG, Moffatt MF, Montgomery GW, Munroe PB, Musunuru K, Nakamura Y, O'Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JNL, Seedorf U, Sen-Chowdhry S, Siminovitch KA, Smit JH, Spector TD, Tan W, Teslovich TM, Tukiainen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann H-E, Willemsen G, Würtz P, Xu C, Yerges-Armstrong LM, Alcohol Genome-wide Association (AlcGen) Consortium. Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study. Genetic Investigation of Anthropometric Traits (GIANT) Consortium. Global Lipids Genetics Consortium. Genetics of Liver Disease (GOLD) Consortium. International Consortium for Blood Pressure (ICBP-GWAS) Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietiläinen KH, Schumann G, Snieder H, Sternberg MJE, Stolk RP, Thomas HC, Thorsteinsdottir U, Uda M, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JCM, Wolffenbuttel BHR, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Völzke H, Schadt EE, Scott J, Järvelin M-R, Elliott P, Kooner JS. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshida T, Kobayashi T, Itoda M, Muto T, Miyaguchi K, Mogushi K, Shoji S, Shimokawa K, Iida S, Uetake H, Ishikawa T, Sugihara K, Mizushima H, Tanaka H. Clinical omics analysis of colorectal cancer incorporating copy number aberrations and gene expression data. Cancer Inform. 2010;9:147–161. doi: 10.4137/cin.s3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajan N, Elliott R, Clewes O, Mackay A, Reis-Filho JS, Burn J, Langtry J, Sieber-Blum M, Lord CJ, Ashworth A. Dysregulated TRK signalling is a therapeutic target in CYLD defective tumours. Oncogene. 2011;30:4243–4260. doi: 10.1038/onc.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Almon RR, Yang E, Lai W, Androulakis IP, DuBois DC, Jusko WJ. Circadian variations in rat liver gene expression: relationships to drug actions. J Pharmacol Exp Ther. 2008;326:700–716. doi: 10.1124/jpet.108.140186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henrichsen CN, Csardi G, Zabot MT, Fusco C, Bergmann S, Merla G, Reymond A. Using transcription modules to identify expression clusters perturbed in Williams-Beuren syndrome. PLoS Comput Biol. 2011;7:e1001054. doi: 10.1371/journal.pcbi.1001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, Shields DC, Abrahams BS, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolton PF, Bourgeron T, Brennan S, Cali P, Correia C, Corsello C, Coutanche M, Dawson G, de Jonge M, Delorme R, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Foley S, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Green J, Guter SJ, Hakonarson H, Holt R, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Lamb JA, Leboyer M, Le Couteur A, Leventhal BL, Lord C, Lund SC, Maestrini E, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Miller J, Minopoli F, Mirza GK, Munson J, Nelson SF, Nygren G, Oliveira G, Pagnamenta AT, Papanikolaou K, Parr JR, Parrini B, Pickles A, Pinto D, Piven J, Posey DJ, Poustka A, Poustka F, Ragoussis J, Roge B, Rutter ML, Sequeira AF, Soorya L, Sousa I, Sykes N, Stoppioni V, Tancredi R, Tauber M, Thompson AP, Thomson S, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Vorstman JA, Wallace S, Wang K, Wassink TH, White K, Wing K, Wittemeyer K, Yaspan BL, Zwaigenbaum L, Betancur C, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Geschwind DH, Haines JL, Hallmayer J, Monaco AP, Nurnberger JI, Jr., Pericak-Vance MA, Schellenberg GD, Scherer SW, Sutcliffe JS, Szatmari P, Vieland VJ, Wijsman EM, Green A, Gill M, Gallagher L, Vicente A, Ennis S. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131:565–579. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoshino J, Aruga J, Ishiguro A, Mikoshiba K. Dorz1, a novel gene expressed in differentiating cerebellar granule neurons, is down-regulated in Zic1-deficient mouse. Brain Res Mol Brain Res. 2003;120:57–64. doi: 10.1016/j.molbrainres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 88.Gharib SA, Dayyat EA, Khalyfa A, Kim J, Clair HB, Kucia M, Gozal D. Intermittent hypoxia mobilizes bone marrow-derived very small embryonic-like stem cells and activates developmental transcriptional programs in mice. Sleep. 2010;33:1439–1446. doi: 10.1093/sleep/33.11.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Padmanabhan B, Kuzuhara T, Adachi N, Horikoshi M. The crystal structure of CCG1/TAF(II)250-interacting factor B (CIB) J Biol Chem. 2004;279:9615–9624. doi: 10.1074/jbc.M312165200. [DOI] [PubMed] [Google Scholar]
- 90.Posorski N, Kaemmerer D, Ernst G, Grabowski P, Hoersch D, Hommann M, von Eggeling F. Localization of sporadic neuroendocrine tumors by gene expression analysis of their metastases. Clin Exp Metastasis. 2011;28:637–647. doi: 10.1007/s10585-011-9397-5. [DOI] [PubMed] [Google Scholar]
- 91.Gridley S, Lane WS, Garner CW, Lienhard GE. Novel insulin-elicited phosphoproteins in adipocytes. Cell Signal. 2005;17:59–66. doi: 10.1016/j.cellsig.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 92.Chavez JA, Gridley S, Sano H, Lane WS, Lienhard GE. The 47kDa Akt substrate associates with phosphodiesterase 3B and regulates its level in adipocytes. Biochem Biophys Res Commun. 2006;342:1218–1222. doi: 10.1016/j.bbrc.2006.02.091. [DOI] [PubMed] [Google Scholar]
- 93.Omar B, Zmuda-Trzebiatowska E, Manganiello V, Goransson O, Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal. 2009;21:760–766. doi: 10.1016/j.cellsig.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg Med Chem Lett. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spies T, Blanck G, Bresnahan M, Sands J, Strominger JL. A new cluster of genes within the human major histocompatibility complex. Science. 1989;243:214–217. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- 97.Spies T, Bresnahan M, Strominger JL. Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B. Proc Natl Acad Sci U S A. 1989;86:8955–8958. doi: 10.1073/pnas.86.22.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsieh YY, Lin YJ, Chang CC, Chen DY, Hsu CM, Wang YK, Hsu KH, Tsai FJ. Human lymphocyte antigen B-associated transcript 2, 3, and 5 polymorphisms and haplotypes are associated with susceptibility of Kawasaki disease and coronary artery aneurysm. J Clin Lab Anal. 2010;24:262–268. doi: 10.1002/jcla.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]; 94 Fontanesi L, Galimberti G, Calo DG, Fronza R, Martelli PL, Scott E, Columbo M, Schiavo G, Casadio R, Buttazzoni L, Russo V. Identification and association analysis of several hundred single nucleotide polymorphisms within candidate genes for backfat thickness in Italian Large White pigs using a selective genotyping approach. J Anim Sci. 2012 doi: 10.2527/jas.2011-4797. (in press: PMID 22367074) [DOI] [PubMed] [Google Scholar]
- 99.Senis YA, Tomlinson MG, Garcia A, Dumon S, Heath VL, Herbert J, Cobbold SP, Spalton JC, Ayman S, Antrobus R, Zitzmann N, Bicknell R, Frampton J, Authi KS, Martin A, Wakelam MJ, Watson SP. A comprehensive proteomics and genomics analysis reveals novel transmembrane proteins in human platelets and mouse megakaryocytes including G6b-B, a novel immunoreceptor tyrosine-based inhibitory motif protein. Mol Cell Proteomics. 2007;6:548–564. doi: 10.1074/mcp.D600007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KG, Enright AJ, Gait MJ, Vigorito E. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 103.Fleming PK, Dedman AM, Xu SZ, Li J, Zeng F, Naylor J, Benham CD, Bateson AN, Muraki K, Beech DJ. Sensing of lysophospholipids by TRPC4 calcium channel. J Biol Chem. 2006;281:4977–4982. doi: 10.1074/jbc.M510301200. [DOI] [PubMed] [Google Scholar]
- 104.Murakami N, Yokomizo T, Okuno T, Shimizu T. G2A is a proton-sensing G-protein-coupled receptor antagonized by lysophosphatidylcholine. J Biol Chem. 2004;279:42484–42491. doi: 10.1074/jbc.M406561200. [DOI] [PubMed] [Google Scholar]