Abstract
We examine the complex evolution of animal nervous systems and discuss the ramifications of this complexity for inferring the nature of early animals. Although reconstructing the origins of nervous systems remains a central challenge in biology, and the phenotypic complexity of early animals remains controversial, a compelling picture is emerging. We now know that the nervous system and other key animal innovations contain a large degree of homoplasy, at least on the molecular level. Conflicting hypotheses about early nervous system evolution are due primarily to differences in the interpretation of this homoplasy. We highlight the need for explicit discussion of assumptions and discuss the limitations of current approaches for inferring ancient phenotypic states.
Homology in the Age of Systems Biology
‘Nothing comes from nothing’ – first attributed to Parmenides.
Homology, since Darwin, has meant similarity due to common descent. A character in two different species is homologous if that character was present in the common ancestor and maintained in the two extant species [1–3]. This definition is clear when the character is well defined: two characters either are, or are not, homologous – there is no gradation [4]. However, complex characters like organs and tissues have many component characters, including proteins, protein networks, and cell types. These parts might not share the same phylogenetic history as the whole character, making homology inference of the whole difficult when there is conflicting signal in the parts [4].
New technologies have begun to unveil the true molecular complexity that underlies organismal phenotypes [5,6]. For evolutionary biologists, these advances provide an unprecedented opportunity to uncover the molecular signatures associated with major evolutionary transitions. For instance, recent studies have used large datasets to investigate the origins of multicellularity [7], the evolution of the bilaterian bauplan [8,9], and the conservation of the neural circuitry underlying social decision-making in vertebrates [10,11].
Such studies are necessarily correlational: they look for coincident state changes in the molecular and phenotypic characters along a phylogeny. This is not necessarily a problem if the connection between molecular and organismal phenotypes is uncontroversial. However, the molecular changes underlying evolutionary novelties are often quite subtle, involving the repurposing of already-existing networks [12]. Because phenotypes of interest have complex molecular underpinnings, their presence in ancestral organisms can be difficult to infer from molecular data alone and rely on often implicit assumptions about the evolutionary linkage between molecular signatures and phenotypes.
Recent work on early nervous system evolution, although it has produced a wealth of interesting data, has encountered such interpretational difficulties, and a consensus view of the origins of nervous systems has been elusive. While this field is interesting in its own right, it also illustrates difficulties common to all complex characters. In particular, it has exposed the degree to which phenotypes and their molecular constituents can become unlinked over deep evolutionary time. Here we consider the conflicting hypotheses and data, put forward a consensus hypothesis on early nervous system evolution, and examine the use and misuse of molecular data for inferring ancestral phenotypes.
Nervous Systems and the Animal Tree
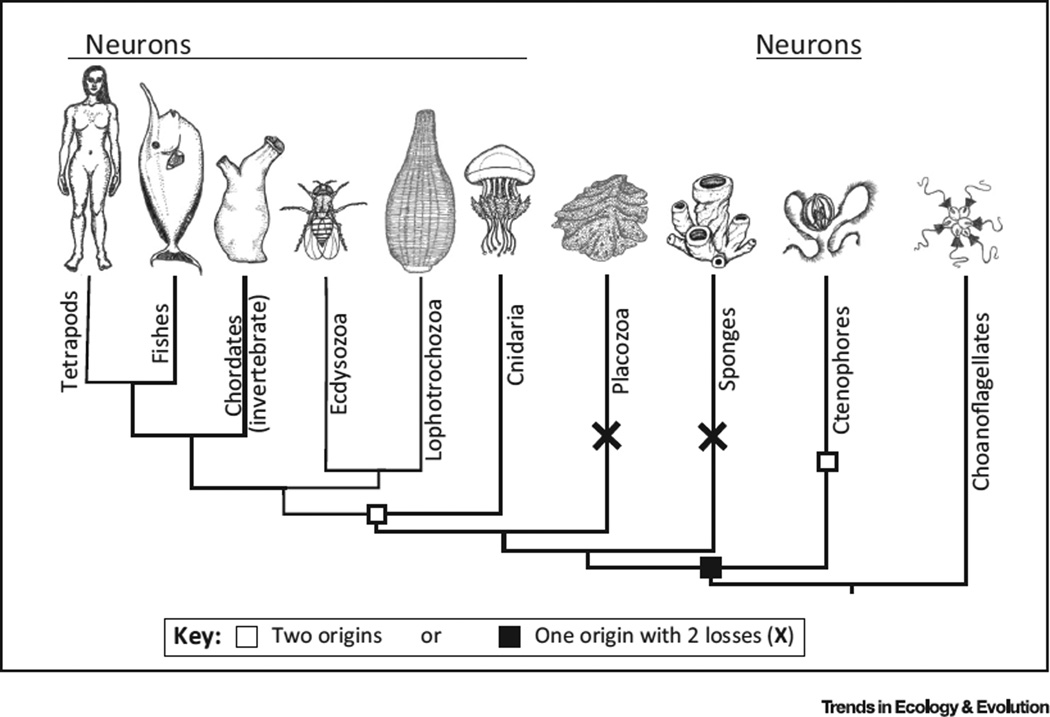
The debate about the evolution of animal nervous systems has focused primarily on whether they have a single origin or were independently derived in ctenophores, the comb jellies, and the common ancestor of all other animals [13–15]. The debate was initiated by phylogenetic work suggesting that the ctenophores are the sister group to all other animals [14–16], a position traditionally ascribed to sponges (Figure 1). Although this placement remains controversial [17], several independent studies have arrived at this conclusion [14–16,18]. With this placement of ctenophores, animals with nervous systems are no longer a monophyletic group. The debate is then whether nervous systems were lost in sponges and placozoans or originated independently in ctenophores and the common ancestor of cnidarians and bilaterians (Figure 1).
Figure 1. Two Alternative Hypotheses of the Origins of Nervous Systems Arising from the Proposed Position of Ctenophores as the Earliest Diverging Animal Phylum.
Either the common ancestor of all animals had a nervous system and neurons were lost twice in placozoa and sponges or the nervous system evolved twice, in ctenophores and in the common ancestor of cnidarians and bilaterians. Animal illustrations by Annika L. Smith.
The position of ctenophores in the animal tree has sometimes been treated as the decisive piece of evidence for determining the origin of nervous systems [17], but this view fails to take into account the possibility that ctenophores and sponges might not resemble stem animals any more than other extant animals do. Indeed, the weight of fossil evidence now suggests that when the first major lineages of animals diverged, nothing resembling extant taxa existed. The first macroscopic (greater than a millimeter in size) animal fossils occur in the middle Ediacaran, a little under 600 million years ago (Mya) [19] – considerably later than the divergence dates of the five main animal lineages (Figure 2) [20–22].
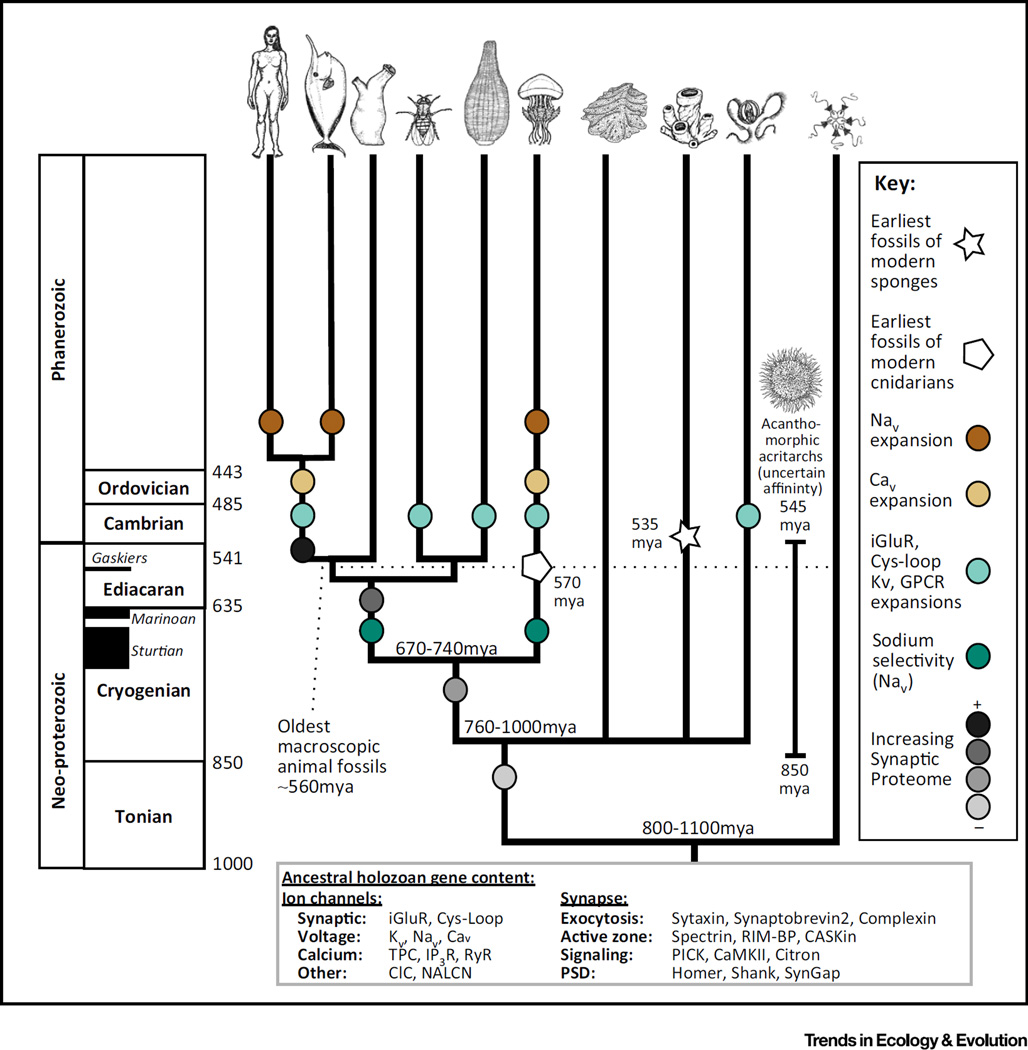
Figure 2. Time Tree of Animal Diversification with Key Events in Nervous System Evolution Annotated on their Respective Branches.
The precise timing of these events is mostly unknown but is bounded by the bifurcations, estimates for which are derived from [20,21,78]. The ages of important fossils and the timing of the major glaciation events are also given and are derived from [79,80]. No macroscopic animal fossils are available from the time period when the first animal lineages were diverging. When this is considered along with the length of the terminal branches, it seems more plausible that large-scale convergences of animal tissues might have occurred at the same time as the convergent molecular events. Animal illustrations by Annika L. Smith.
The only earlier fossils suggested to have animal affinities are minute embryos and ‘acritarchs’ [19,23]. No large adult forms are known from this period. It is possible that early macroscopic animals went undetected in the fossil record, but a simpler explanation is that there were no macroscopic adult forms at that time. It might even be possible that some of the larger acritarchs (>1000 cells) are in fact terminal adults. This seems plausible because the massive glaciation events of the Cryogenian would have severely reduced oceanic oxygen, perhaps restricting the size of animals [19]. Stem animals from the ctenophore, sponge, placozoan, cnidarian, and even bilaterian lineages might then have resembled the acritarchs more than any extant animal and characters whose molecular basis was being laid down during the late Cryogenian, such as the number of germ layers, gastrulation, muscle, and nervous systems, might have been present only in nascent form.
Below, we discuss the phyletic distribution of molecular characters associated with various aspects of nervous system function and consider how these distributions inform our understanding of the early evolution of these phenotypes.
Morphogenesis
Ontogenetic origin and morphogenetic gene networks are often considered key evidence for homology. It is becoming clear, however, that the relationship between morphogenes and the characters they encode is complex [24,25]. For instance, many morphogenes and signaling pathways associated with animal development were present in the unicellular ancestor of choanoflagellates and animals [7]. Interestingly, the developmental toolkit of ctenophores more closely resembles that of sponges, while that of placozoans more closely resembles cnidarians and bilaterians [8,26]. Sponges also possess homologs of known neurogenesis genes [27], despite lacking true neurons, and some are even present in choanoflagellates [7]. Genes in the Notch–Delta pathway, for instance, are expressed in the larval globular cells of the sponge Amphimedon queenslandica [28,29]. Notably, these globular cells express many synaptic proteins and have electron-dense vesicles, suggesting a secretory function.
The embryological origin of neurons is also more plastic than previously appreciated. Although most nervous systems are ectodermally derived, neurons originate from non-ectodermal precursors in several species, including hydrozoan cnidarians and echinoderms [30,31], suggesting that non-ectodermal neurons might have evolved at least twice. Thus even the ontogeny of neurons is a plastic character that cannot be used as an unqualified test of homology without comparative and molecular analysis.
No single gene yet found is a perfect marker of nervous systems and even gene networks cannot be used to assess homologous phenotypes without further characterization due to the possibility of independent recruitment into convergent phenotypes, or ‘deep homology’ [12,32]. Perhaps the discovery of more ctenophore- and sponge-specific developmental networks will help reveal the deep roots of neural development.
Chemical Transmission
Synapses are specialized junctions for the rapid diffusion of chemical signals from a secretory surface on the presynaptic cell to a receptive surface on the postsynaptic cell. A large protein scaffold exists in the postsynaptic cell that is the locus of various modulatory pathways [33,34]. Although synapses are often considered to be highly derived, nervous system-specific junctions, they actually depend on many proteins that are not specific to nervous systems or even to metazoans. For instance, the proteins involved in neurotransmitter release are an ancient complex that also manages interorganellar transport and exocytosis, some of it dating back to the last common ancestor of eukaryotes [35]. Also, neurons are not the only cells to make use of this ancient machinery: similar junctions utilizing identical proteins form between lymphocytes and antigen-presenting cells to facilitate the release of lytic granules, creating an ‘immunological synapse’ [36].
Some postsynaptic proteins can also be traced back to our protist ancestors (Figure 2) [37,38], but vertebrate synapses contain thousands of proteins, many of which arose more recently [34], and vertebrates appear to have the most complex synapses [33]. However, the comparative genomics of synapses has been largely guided by proteomic surveys of vertebrates [39,40], so much of this pattern is probably due to vertebrate-specific ascertainment bias, and it does not follow that non-vertebrates have ‘simpler’ synapses or that their synapses are more similar to those of the first animals.
Ctenophore and cnidarian synapses, and non-vertebrate synapses in general, have diverse morphologies that differ both from vertebrates and from one another [41]. One key difference is that, while vertebrate synapses tend to be unidirectional and have distinct pre- and postsynaptic specializations, these specializations are often less clear in non-vertebrates [41]. In cnidarians and ctenophores, many synapses are fully bidirectional, especially in the motor nerve nets where coordination is central and the direction of signal flow probably does not hold much information [42]. It is likely that these bidirectional synapses have as-yet-unidentified functional complexity.
Placozoans and sponges, although lacking true synapses, have many protein families associated with synapses, including postsynaptic density proteins and neuropeptides [27,43–46], although these are not coexpressed in sponges as they are in animals with nervous systems [47]. However, sponges do have contractile behaviors that depend on well-known neurotransmitters [48,49]. Even some choanoflagellates have intercellular bridges with occluding electron densities [50] and genes for the synthesis of some neuropeptides [51]. Which, if any, of these structures are homologous to bona fide synapses? Vertebrate-specific acquisition bias and missing data from nervous system-lacking organisms and ctenophores make it difficult to judge, but the groundwork is clearly laid for an investigation of this question.
Electrical Properties
Regulation of ionic gradients by pumps and channels is a ubiquitous property of cellular life forms. Neurons use the potential energy in these transmembrane ionic potentials to drive regenerative traveling electrical signals called action potentials (APs) that carry signals down axons. Many other organisms fire APs for signaling purposes, just as animals do [52].
APs are generated by proteins in the family of voltage-gated ion channels, found across eukaryotes [52,53]. APs in non-animals typically deliver a rapid burst of ions that directly affect cellular physiology. Calcium channels in animals also trigger intracellular pathways for various purposes, but the APs along axons are primarily carried by sodium ions, which do not trigger intracellular pathways. The advent of sodium channels has been hypothesized to represent a key transition in the origin of nervous systems because they allowed constant firing without building up toxic intracellular calcium levels [52,54]. In fact, this transition did not happen just once, but twice, via convergent amino acid substitutions in cnidarians and bilaterians [55–57].
Most of the information in the neural code is in the frequency and duration of AP bursts, both of which are controlled by voltage-gated potassium channels (Kvs). Kvs have also undergone convergent changes at the biophysical level in cnidarians and bilaterians [58]. Additionally, Kvs have experienced large and independent gene family expansions in ctenophores, cnidarians, and bilaterians, suggesting independent gains in electrical code complexity in these lineages [59,60].
The ion channel families that transduce the chemical signals in synapses have undergone even larger expansions. Interestingly, these expansions occur on the same branches as the Kv expansions [60]. Synaptic channels have radiated independently in ctenophores, cnidarians, and bilaterians, causing these different lineages to converge toward a similar genomic ion channel complement [60]. These results are not dependent on the choice of species tree, as has been claimed ([17], see SI text 61). There have also been reported cases of convergence to glutamate sensitivity in a number of bilaterian synaptic channel families [61,62].
Origin of Nervous Systems
Despite the complex phyletic patterns of nervous system-associated genes discussed above, we will put forward a consensus view of early nervous system evolution. For over 300 My, from the common ancestor of animals until the Ediacaran biota, animals were probably very small, perhaps microscopic. Yet they, and even their common ancestor with choanoflagellates, possessed many of the genes that power modern neurons. It seems likely, given their extensive molecular toolkit, that these early animals were capable of intercellular communication and coordinated behavior. Did these early structures constitute a nervous system? This question might come down to minor distinctions. For instance, if a nervous system means electrical impulses coupled to intercellular chemical signals, the first animals almost certainly had nervous systems. However, under this definition we might have to say that choanoflagellates also have nervous systems. On the other hand, if nervous systems must have specialized axons and dendrites and maintain a complex electrical code, they might have originated much later, and perhaps more than twice.
Conversely, animals without nervous systems display complex behaviors that rival those of their nervous system-bearing relatives. Placozoans, sponge larvae, and cnidarian larvae without nervous systems have motility and taxis no less complex than cnidarian larvae with neurons [63,64]. Several animals, all of which are small in size [65,66], have even lost core features of neurons, like sodium-based APs. Perhaps a bona fide nervous system provided little advantage to early microscopic animals, and animal size, rather than behavioral complexity, was the biggest driver of nervous system elaboration. It is therefore possible that, whatever definition of the nervous system one chooses, its early evolution was characterized by many origins, dead ends, and reversals.
Around 600 Mya, after all five major lineages (ctenophores, sponges, placozoans, cnidarians, and bilaterians) had diverged, there was a sudden change. Large expansions of the gene families associated with synaptic and electrical complexity occurred together with profound changes on the biophysical level [34,38,57,58,60]. These changes occurred convergently in the stem lineages of ctenophores, cnidarians, and bilaterians (Figure 2). Although many of these genomic events cannot yet be dated precisely, they most likely followed the end of the worldwide glaciation events, contemporaneously with the rise of oceanic oxygen and macroscopic animal forms. The rise of inter-animal predation in the early Cambrian probably provided selective pressure to evolve complex behaviors, neural organization [67], and musculature [68]. It is perhaps not meaningful, therefore, to assign a single date to the origin of nervous systems, especially when one considers the genomic continuity between sensu stricto neurons and proto-neurons, and the large degree of homoplasy in animal nervous systems as a whole. Furthermore, the relatively late appearance of many aspects of nervous system complexity suggests that the precise branching order of the animal tree will not resolve the difficulties described above.
Concluding Remarks
Advances in genomic techniques make it possible to pull phenotypes apart into their constituent mechanisms. Putting them back together for evolutionary inference is difficult. In reviewing the recent work on the early evolution of the nervous system, we observe several repeated and interrelated difficulties that will be common to any study on the molecular basis for phenotype evolution. How can molecular data best be used to inform studies of ancestral phenotypes?
Disagreements about the homology status of complex organs are often the result of differing assumptions about how strongly certain molecular states predict organismal phenotypes. However, the evolutionary link between mechanisms and phenotypes is rarely, if ever, one to one, as illustrated by the genomic data we have reviewed above. For instance, shared molecular structures in similar extant phenotypes can be the product of convergence [12] or homology. Conversely, molecular differences between similar phenotypes are either due to systems drift [24] if the phenotypes are homologous or simply the expected differences due to non-homology. We submit that these possibilities should always be considered when molecular phenotypes are put forward as signatures of organismal phenotypes. They are too often ignored in favor of the understandable but overly simplistic assumption that cases of convergence and systems drift are too rare to be taken into account. Below, we make several suggestions for how to prioritize research in situations where inference is difficult.
Ideally, the evolutionary linkage between molecular and organismal phenotypes would be captured in a probabilistic framework (see Outstanding Questions). Such methods exist only for very simple systems [69] but future advances in modeling the coevolution of phenotypes and mechanisms are expected. Even with these possibilities, systems drift and homoplasy – based on subtle rearrangement of ancient mechanisms – will often make it difficult to reconstruct ancestral phenotypes. The ability to reduce systems to their molecular components does not necessarily imply the ability to infer the evolution of systems from the evolution of those components.
Gene family origins and the presence or absence of a gene in a genome are not strong or reliable signals of key phenotypic changes. Most metazoan gene families predate the phenotype that they currently mediate and many are highly pleiotropic. Rather, greater attention should be paid to changes at the systems level, such as gene family expansions [59,60] or coexpression [47], or at the level of physiology and biophysics, such as switches in potassium channel gating [58], glutamate reception [61,62], or sodium selectivity [55,57]. However, such changes can be difficult to detect from sequence data alone. Subtle changes in morphogenetic gene networks can be particularly elusive [12,32]. More work on systems-level approaches to evolution will help to identify evolutionarily pertinent characters.
As noted by Dunn and Ryan (2015), genome annotation is biased toward bilaterians, which results in underestimation of the complexity of non-bilaterians [70]. Increased focus on the biology of choanoflagellates, ctenophores, sponges, and placozoans is necessary before the full diversity of animal nervous systems can be appreciated.
Models of phenotype evolution are often insufficient [71] and our intuitions about the parsimonious nature of major animal innovations are being increasingly challenged [72,73]. A shift in our assumptions might therefore be warranted (see Outstanding Questions). Although it is sometimes desirable to find apomorphic characters, an alternative way to organize and understand the evolution of animal forms is as a set of stable states toward which the different lineages have been pulled. It is increasingly clear that animal diversity is dominated by these sorts of patterns and not by divisions between living fossils and derived forms.
Outstanding Questions.
How can we best harness molecular data for inferring historical phenotypic changes? More specifically, what kinds of molecular phenotypes are the best ‘signatures’ of which kinds of phenotypic changes?
How should we model the coevolution of molecular and organismal phenotypes, given that molecular signatures of past phenotypic changes are commonly inferred assuming parsimony, whereas phenotypes are modeled with fairly simple yet often inaccurate models (e.g., Brownian motion)?
Nervous systems and several other complex animal phenotypes show a large degree of homoplasy, suggesting general principles underlying tissue function. How can we use comparative data for both evolutionary and translational studies to gain insight into these fundamental functional properties?
Genome annotation and functional analyses in early-branching animal lineages (e.g., ctenophores, sponges) and choanoflagellates, the sister taxon of all animals, are biased by our knowledge of bilaterians. How will in-depth studies of these important organisms inform our understanding of early nervous system evolution?
How much convergence should we expect in complex animal phenotypes? Instances of convergence in nervous systems are common [74]. Classic cases include the independent origin of camera eyes and of electric organs in fishes [75–77]. If these examples can occur over shorter timescales, surely more radical convergence is possible over longer periods.
The level of phenotypic plasticity in animal evolution also provides an opportunity to better understand the principles of phenotypes. Cases of convergence, loss, and repurposing can provide useful information that is not available within a single model system (see Outstanding Questions). For instance, selectivity for sodium in cnidarian and bilaterian voltage-gated ion channels [55,57] and sensitivity to glutamate in non-vertebrate Cys-loop receptors [61,62] both evolved by convergence toward similar geometries of the pore and binding pockets, respectively.
The diversity of animals, which is characterized by widespread homoplasy and plasticity, is therefore a challenge and an opportunity. A successful comparative approach will take advantage of this plasticity and not try to abstract from it. We are now well poised to take advantage of a fully comparative framework that includes the entire animal tree, but we must be cognizant of the phenotypic patterns that occur there [72]. A diversity-based approach will enable an understanding of nervous systems that goes beyond the merely descriptive.
Trends.
New phylogenetic evidence suggests that nervous systems are not monophyletic. There is a debate about whether this indicates a single origin of nervous systems with several losses or multiple independent origins of neurons.
Comparative genomics studies have found that many of the gene families associated with extant nervous system function were present before the origin of animals. However, changes at the biophysical level and gene family expansions occurred independently in several animal lineages, suggesting widespread homoplasy in nervous systems.
Fossils of the first animals are microscopic, about the same size as the larvae of various extant marine species, which display complex behavior although only some have nervous systems. Together with colonial choanoflagellates, these larvae may provide the best model systems for understanding behavior and the origin of nervous systems in early animals.
Acknowledgments
This opinion article emerged from discussions originating from B.J.L.’s thesis committee. B.J.L. is supported by NIH NRSA 1F32GM112504-01A1. H.H.Z. (IOS-1443637) and H.A.H. (IOS-0843712, IOS-1354942, IOS-1501704) are supported by the US National Science Foundation.
References
- 1.Bock WJ. Philosophical foundations of classical evolutionary classification. Syst. Zool. 1973;22:375–392. [Google Scholar]
- 2.Szucsich NU, Wirkner CS. Homology: a synthetic concept of evolutionary robustness of patterns. Zool. Scr. 2007;36:281–289. [Google Scholar]
- 3.Wagner GP. The biological homology concept. Annu. Rev. Ecol. Syst. 1989;20:51–69. [Google Scholar]
- 4.Hall BK. Homology: The Hierarchial Basis of Comparative Biology. Academic Press; 1994. [Google Scholar]
- 5.Wan C, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soon WW, et al. High-throughput sequencing for biology and medicine. Mol. Syst. Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter DJ, King N. The genomic and cellular foundations of animal origins. Annu. Rev. Genet. 2013;47:509–537. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 8.Ryan J, et al. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo. 2010;1:9. doi: 10.1186/2041-9139-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauri A, et al. Development of the annelid axochord: insights into notochord evolution. Science. 2014;345:1365–1368. doi: 10.1126/science.1253396. [DOI] [PubMed] [Google Scholar]
- 10.O’Connell LA, Hofmann HA. Evolution of a vertebrate social decision-making network. Science. 2012;336:1154–1157. doi: 10.1126/science.1218889. [DOI] [PubMed] [Google Scholar]
- 11.O’Connell LA, Hofmann HA. The vertebrate mesolimbic reward system and social behavior network: a comparative synthesis. J. Comp. Neurol. 2011;519:3599–3639. doi: 10.1002/cne.22735. [DOI] [PubMed] [Google Scholar]
- 12.Shubin N, et al. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 13.Moroz LL. On the independent origins of complex brains and neurons. Brain Behav. Evol. 2009;74:177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan JF, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 17.Pisani D, et al. Genomic data do not support comb jellies as the sister group to all other animals. Proc. Natl. Acad. Sci. U.S.A. 2015 doi: 10.1073/pnas.1518127112. Published online November 2, 2015. http://dx.doi.org/10.1073/pnas.1518127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whelan NV, et al. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5773–5778. doi: 10.1073/pnas.1503453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budd GE. The earliest fossil record of the animals and its significance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:1425–1434. doi: 10.1098/rstb.2007.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson KJ, et al. The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:1435–1443. doi: 10.1098/rstb.2007.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgecombe GD, et al. Higher-level metazoan relationships: recent progress and remaining questions. Org. Divers. Evol. 2011;11:151–172. [Google Scholar]
- 22.dos Reis M, et al. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr. Biol. 2015;25:2939–2950. doi: 10.1016/j.cub.2015.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagadorn JW, et al. Cellular and subcellular structure of Neoproterozoic animal embryos. Science. 2006;314:291–294. doi: 10.1126/science.1133129. [DOI] [PubMed] [Google Scholar]
- 24.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 25.Young RL, Wagner GP. Why ontogenetic homology criteria can be misleading: lessons from digit identity transformations. J. Exp. Zool. B Mol. Dev. Evol. 2011;316B:165–170. doi: 10.1002/jez.b.21396. [DOI] [PubMed] [Google Scholar]
- 26.Pang K, et al. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. Evodevo. 2010;1:10. doi: 10.1186/2041-9139-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards GS, et al. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 29.Richards GS, Degnan BM. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. Evodevo. 2012;3:15. doi: 10.1186/2041-9139-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Z, et al. Direct development of neurons within foregut endoderm of sea urchin embryos. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9143–9147. doi: 10.1073/pnas.1018513108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartenstein V, Stollewerk A. The evolution of early neurogenesis. Dev. Cell. 2015;32:390–407. doi: 10.1016/j.devcel.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hejnol A, Lowe CJ. Animal evolution: stiff or squishy notochord origins? Curr. Biol. 2014;24:R1131–R1133. doi: 10.1016/j.cub.2014.10.059. [DOI] [PubMed] [Google Scholar]
- 33.Emes RD, et al. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci. 2008;11:799–806. doi: 10.1038/nn.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emes RD, Grant SGN. Evolution of synapse complexity and diversity. Annu. Rev. Neurosci. 2012;35:111–131. doi: 10.1146/annurev-neuro-062111-150433. [DOI] [PubMed] [Google Scholar]
- 35.Kloepper TH, et al. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angus KL, Griffiths GM. Cell polarisation and the immunological synapse. Curr. Opin. Cell Biol. 2013;25:85–91. doi: 10.1016/j.ceb.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkhardt P, et al. Evolutionary insights into premetazoan functions of the neuronal protein Homer. Mol. Biol. Evol. 2014;31:2342–2355. doi: 10.1093/molbev/msu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Husi H, et al. Proteomic analysis of NMDA receptor–adhesion protein signaling complexes. Nat. Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 40.Collins MO, et al. Molecular characterization and comparison of the components and multiprotein complexes in the post-synaptic proteome. J. Neurochem. 2006;97(Suppl. 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 41.Cobb JLS, Pentreath VW. Comparison of the morphology of synapses in invertebrate and vertebrate nervous systems: analysis of the significance of the anatomical differences and interpretation of the morphological specializations. Prog. Neurobiol. 1978;10:231–252. doi: 10.1016/0301-0082(78)90004-7. [DOI] [PubMed] [Google Scholar]
- 42.Anderson PA. Physiology of a bidirectional, excitatory, chemical synapse. J. Neurophysiol. 1985;53:821–835. doi: 10.1152/jn.1985.53.3.821. [DOI] [PubMed] [Google Scholar]
- 43.Smith CL, et al. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 2014;24:1565–1572. doi: 10.1016/j.cub.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakarya O, et al. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE. 2007;2:e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivera AS, et al. Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J. Exp. Biol. 2012;215:1278–1286. doi: 10.1242/jeb.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishnan A, et al. The GPCR repertoire in the demosponge Amphimedon queenslandica: insights into the GPCR system at the early divergence of animals. BMC Evol. Biol. 2014;14:270. doi: 10.1186/s12862-014-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conaco C, et al. Functionalization of a protosynaptic gene expression network. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10612–10618. doi: 10.1073/pnas.1201890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellwanger K, et al. GABA and glutamate specifically induce contractions in the sponge Tethya wilhelma. J. Comp. Physiol. A. 2007;193:1–11. doi: 10.1007/s00359-006-0165-y. [DOI] [PubMed] [Google Scholar]
- 49.Elliott GRD, Leys SP. Evidence for glutamate, GABA and NO in coordinating behaviour in the sponge, Ephydatia muelleri (Demospongiae Spongillidae) J. Exp. Biol. 2010;213:2310–2321. doi: 10.1242/jeb.039859. [DOI] [PubMed] [Google Scholar]
- 50.Dayel MJ, et al. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fairclough SR, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hille B. Ion Channels of Excitable Membranes. 3rd. Sinauer Associates; 2001. [Google Scholar]
- 53.Verret F, et al. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 2010;187:23–43. doi: 10.1111/j.1469-8137.2010.03271.x. [DOI] [PubMed] [Google Scholar]
- 54.Hille B. Ionic channels: evolutionary origins and modern roles. Exp. Physiol. 1989;74:785–804. doi: 10.1113/expphysiol.1989.sp003349. [DOI] [PubMed] [Google Scholar]
- 55.Liebeskind BJ, et al. Evolution of sodium channels pre-dates the origin of nervous systems in animals. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liebeskind BJ. Evolution of sodium channels and the new view of early nervous system evolution. Commun. Integr. Biol. 2011;4:679–683. doi: 10.4161/cib.17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gur Barzilai M, et al. Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep. 2012;2:242–248. doi: 10.1016/j.celrep.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinson AS, et al. Functional evolution of Erg potassium channel gating reveals an ancient origin for IKr. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5712–5717. doi: 10.1073/pnas.1321716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X, et al. Major diversification of voltage-gated K+ channels occurred in ancestral parahoxozoans. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E1010–E1019. doi: 10.1073/pnas.1422941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liebeskind BJ, et al. Convergence of ion channel genome content in early animal evolution. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E846–E851. doi: 10.1073/pnas.1501195112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kehoe J, et al. Aplysia Cys-loop glutamate-gated chloride channels reveal convergent evolution of ligand specificity. J. Mol. Evol. 2009;69:125–141. doi: 10.1007/s00239-009-9256-z. [DOI] [PubMed] [Google Scholar]
- 62.Lynagh T, et al. Molecular basis for convergent evolution of glutamate recognition by pentameric ligand-gated ion channels. Sci. Rep. 2015;5:8558. doi: 10.1038/srep08558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ludeman DA, et al. Evolutionary origins of sensation in metazoans: functional evidence for a new sensory organ in sponges. BMC Evol. Biol. 2014;14:3. doi: 10.1186/1471-2148-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maldonado M, et al. The cellular basis of photobehavior in the tufted parenchymella larva of demosponges. Mar. Biol. 2003;143:427–441. [Google Scholar]
- 65.Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 66.Nordström K, et al. A simple visual system without neurons in jellyfish larvae. Proc. Biol. Sci. 2003;270:2349–2354. doi: 10.1098/rspb.2003.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monk T, Paulin MG. Predation and the origin of neurones. Brain Behav. Evol. 2014;84:246–261. doi: 10.1159/000368177. [DOI] [PubMed] [Google Scholar]
- 68.Steinmetz PRH, et al. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lartillot N, Poujol R. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol. Biol. Evol. 2011;28:729–744. doi: 10.1093/molbev/msq244. [DOI] [PubMed] [Google Scholar]
- 70.Dunn CW, et al. The hidden biology of sponges and ctenophores. Trends Ecol. Evol. 2015;30:282–291. doi: 10.1016/j.tree.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Pennell MW, et al. Model adequacy and the macroevolution of angiosperm functional traits. Am. Nat. 2014 doi: 10.1086/682022. Published online April 7, 2014. http://dx.doi.org/10.1101/004002. [DOI] [PubMed] [Google Scholar]
- 72.Dunn CW, et al. Animal phylogeny and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 2014;45:371–395. [Google Scholar]
- 73.Halanych KM. The ctenophore lineage is older than sponges? That cannot be right! Or can it? J. Exp. Biol. 2015;218:592–597. doi: 10.1242/jeb.111872. [DOI] [PubMed] [Google Scholar]
- 74.Nishikawa KC. Evolutionary convergence in nervous systems: insights from comparative phylogenetic studies. Brain Behav. Evol. 2002;59:240. doi: 10.1159/000063561. [DOI] [PubMed] [Google Scholar]
- 75.Arnegard ME, et al. Old gene duplication facilitates origin and diversification of an innovative communication system – twice. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22172–22177. doi: 10.1073/pnas.1011803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thompson A, et al. Expression evolution facilitated the convergent neofunctionalization of a sodium channel gene. Mol. Biol. Evol. 2014;31:1941–1955. doi: 10.1093/molbev/msu145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gallant JR, et al. Genomic basis for the convergent evolution of electric organs. Science. 2014;344:1522–1525. doi: 10.1126/science.1254432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peterson KJ, Butterfield NJ. Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9547–9552. doi: 10.1073/pnas.0503660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antcliffe JB, et al. Giving the early fossil record of sponges a squeeze. Biol. Rev. 2014;89:972–1004. doi: 10.1111/brv.12090. [DOI] [PubMed] [Google Scholar]
- 80.Liu AG, et al. Haootia quadriformis n. gen., n. sp., interpreted as a muscular cnidarian impression from the Late Ediacaran period (approx 560 Ma) Proc. Biol. Sci. 2014;281:20141202. doi: 10.1098/rspb.2014.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]