Arising from: Y. Zhang et al. Nature 513, 440–443 (2014); doi:10.1038/nature13492
Among mTORC1’s best-studied anabolic actions are enhancing protein synthesis and inhibiting protein degradation by autophagy1,2. When mTORC1 is inactivated during starvation, proteolysis and autophagy increase to provide amino acids for protein synthesis and energy production. However, Zhang et al. recently reported that inhibition of mTORC1 activity for 16 h or more causes a reduction in overall proteolysis by decreasing proteasome expression3. However, their methodology to measure the rates of protein degradation and the rates obtained appear questionable. There is a Reply to this Brief Communication Arising by Zhang, Y. et al. Nature 5xx, http://dx.doi.org/10.1038/nature16473 (2015).
After radiolabelling cell proteins, Zhang et al. observed no degradation of these proteins for 8 h, even in the presence of rapamycin, but then proteolysis accelerated and was reduced by rapamycin after a 16 h delay. Such data are inconsistent with several well-established findings: (1) that newly synthesized proteins are degraded rapidly as cells eliminate mis-folded and short-lived regulatory proteins; (2) that proteolysis by autophagy increases within minutes after rapamycin addition2; and (3) that degradation of pulse-labelled proteins follows exponential kinetics and thus decreases with time4,5.
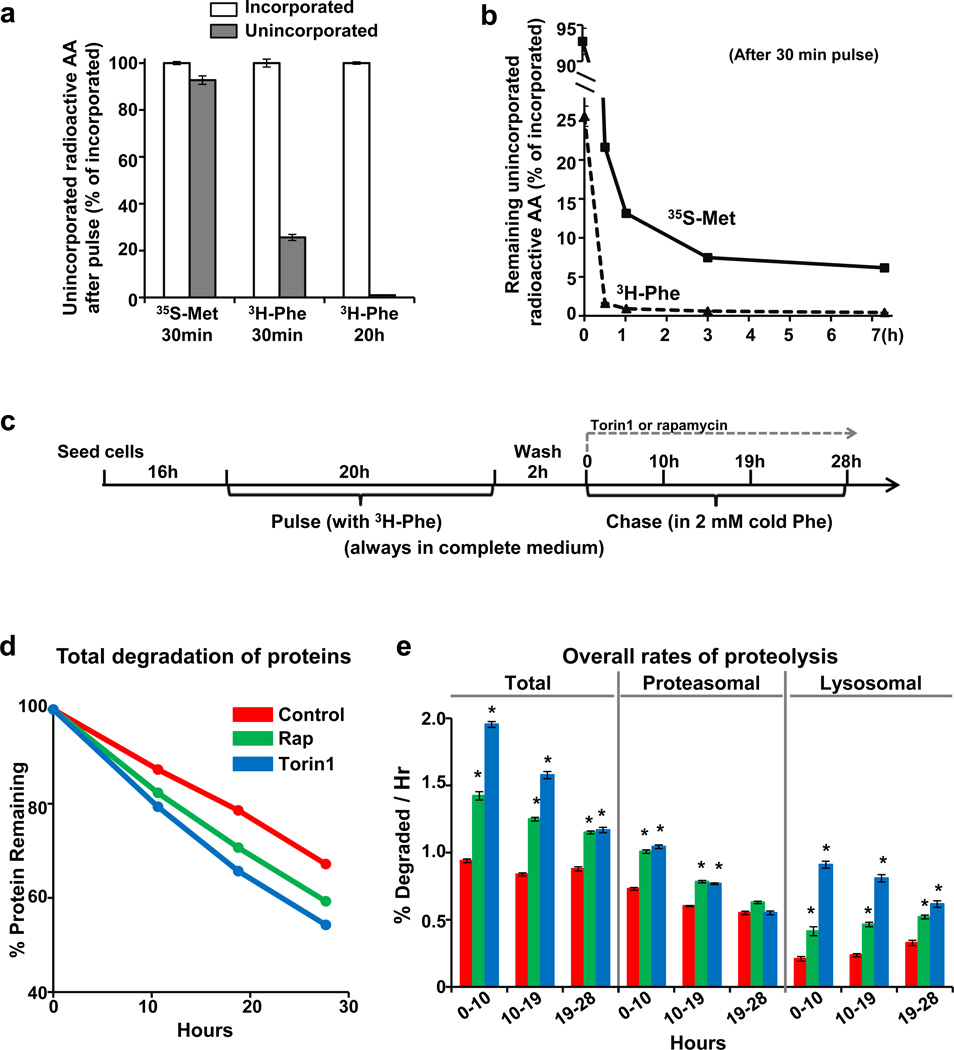
In such pulse–chase studies, it is essential to minimize the continued incorporation of radioactive precursors into proteins during the chase period. If the amount of intracellular unincorporated radioactivity is significant relative to the amount in proteins, then continued incorporation of label can mask the loss of labelled proteins by proteolysis and yield misleadingly slow degradation rates. This effect could account for the reported lack of observed proteolysis for 8 h3. This problem is particularly serious with short pulses and certain precursors. For example, after labelling for 30 min with 35S-Met/Cys in TSC2-null mouse embryonic fibroblasts, the unincorporated radioactivity equalled 92% of the counts in protein, while with 3H-Phe, the unincorporated radioactivity accounted for only 26% (Fig. 1a). In both cases, significant incorporation continues despite chasing with high levels of non-radioactive amino acids. With 35S-Met/Cys, appreciable unincorporated radioactivity remained in the cells for hours, but much less with 3H-Phe (Fig. 1b). To minimize such problems and measure the degradation of long-lived proteins (the bulk of cell proteins), we label with 3H-Phe for 20 h and chase for at least 2 h before measuring degradation (Fig. 1c)6. Consequently, intracellular free 3H-Phe is less than 0.2% of that in proteins, and reincorporation is inconsequential.
Figure 1. Pulse–chase methodology using 3H-Phe allows valid measurements of degradative rates and shows that mTOR inhibition causes rapid and sustained increases in protein degradation by both proteasomal and lysosomal pathways in TSC2-null MEFs.
a, After a 30-min pulse of radioactive amino acids and repeated washing, large amounts of free (trichloroacetic acid (TCA)-soluble) labelled precursors remain inside cells not incorporated into protein. Cell proteins were labelled by incubation with 35S-Met/Cys for 30 min, with 3H-Phe for 30 min, or with 3H-Phe for 20 h. After three quick washes with chase medium containing 2 mM non-radioactive Met/Cys or Phe, 10% TCA was added to cells to precipitate proteins. TCA-soluble radioactivity (unincorporated precursors) was measured by scintillation counting and expressed relative to the amount of radioactivity incorporated into TCA-precipitable proteins. AA, amino acids. b, After a 30-min pulse, much higher levels of unincorporated 35S-Met/Cys remain in cells for several hours compared to 3H-Phe. After chasing for the indicated times, the medium was removed, and cell proteins were precipitated with TCA. Unincorporated counts at different time points were plotted relative to the initial incorporated radioactivity. c, Experimental protocol for Fig. 1d, e (always in normal medium). mTOR inhibitors (100 nM Torin1, 300 nM rapamycin) were added at time 0 of chase. d, mTOR inhibition with Torin1 or rapamycin causes rapid and sustained increases in overall protein degradation. Levels of TCA-insoluble pre-labelled proteins remaining at different times were plotted against the initial total incorporated radioactivity. e, Treatment with Torin1 or rapamycin increased both proteasomal and lysosomal proteolysis for at least 19–28 h. Alternatively, the average rates of total proteolysis during the indicated periods were measured by the release of 3H-Phe into medium. The inclusion of concanamycin A (100 nM) to block lysosomal proteolysis allowed specific measurement of proteasomal (concanamycin-A-resistant) and lysosomal (concanamycin-A-sensitive) degradation. Error bars represent s.e.m. (n = 4 biological replicates). *P < 0.05 (by Student’s t-test).
By avoiding this recycling problem and conducting our assays in complete medium (without the prolonged serum deprivation used by Zhang et al.3), we found that: (1) mTOR inhibition with rapamycin or Torin1 actually enhanced overall proteolysis immediately in various cells tested, including TSC2-null mouse embryonic fibroblasts used by Zhang et al.3 (Fig. 1d); (2) with rapamycin treatment, degradation rates remained increased for 28 h (measured by determining the radioactivity remaining in cell proteins with time (Fig. 1d)), in contrast to the delayed reduction reported after 16 h3; (3) proteolysis was also found to increase when we measured rates of release of 3H-Phe from proteins into medium6–9 at different times (Fig. 1e); and (4) using selective inhibitors to determine which degradation pathway was activated, we found that mTOR inhibition stimulated proteolysis by both proteasomes and autophagy (lysosomes) (Fig. 1e), as we demonstrated elsewhere.
Thus, we were unable to detect any decrease in proteolysis within 24 h of mTORC1 inhibition as reported by Zhang et al.3, but instead demonstrate a prolonged enhancement of proteasome-mediated degradation, which in nutrient-deprived cells must complement the activation of autophagy in providing an endogenous source of amino acids. The major differences between our pulse–chase approach and that of Zhang et al. is that we always compare the degradation of the exact same pool of labelled proteins and conducted our assays in complete medium, while they used prolonged serum deprivation of TSC2-null cells. This approach is problematic for pulse–chase studies of mTORC1’s effects on proteolysis since serum deprivation itself stimulates protein breakdown and mTORC1 alters patterns of synthesis. These differences in methodology may also help explain why Zhang et al. did not observe increased proteolysis upon mTORC1 inhibition3 (as shown in Fig. 1 and elsewhere).
Footnotes
Author contributions:
J.Z. and A.L.G. designed experiments, analysed data and prepared this manuscript; J.Z. and G.A.G. performed these experiments.
Competing Financial Interests: Declared none.
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg AL, St John AC. Intracellular protein degradation in mammalian and bacterial cells: part 2. Annu. Rev. Biochem. 1976;45:747–804. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- 5.Rock KL, Farfan-Arribas DJ, Colbert JD, Goldberg AL. Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 2014;35:144–152. doi: 10.1016/j.it.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor γ coactivator 1α or 1β overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J. Biol. Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piccirillo R, Goldberg AL. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J. 2012;31:3334–3350. doi: 10.1038/emboj.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad. Sci. USA. 2015 doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]