Abstract
Tetrahymena telomerase holoenzyme subunits p75, p45 and p19 form a subcomplex (7–4–1) peripheral to the catalytic core. We report structures of p45 and p19 and reveal them as the Stn1 and Ten1 subunits of the CST complex, which stimulates telomerase complementary-strand synthesis. 7–4–1 binds telomeric single-stranded DNA, and mutant p19 overexpression causes telomere 3′-overhang elongation. We propose that telomerase-tethered Tetrahymena CST coordinates telomere G-strand and C-strand synthesis.
The telomerase enzyme maintains telomeres and is essential for chromosome stability. In Tetrahymena thermophila telomerase, telomerase reverse transcriptase (TERT), telomerase RNA (TER) and RNA-associated p65 form the catalytic core (TERT–TER–p65), which then assembles the additional telomerase holoenzyme subunits Teb1, p50, p75, p45 and p19 (refs. 1–3 and Fig. 1a). Teb1 is a paralog of the replication protein A (RPA) large subunit RPA70 and has telomeric single-stranded (ss) DNA–binding activity necessary for telomerase recruitment to telomeres3–5. Teb1 and the separate 7–4–1 subcomplex are tethered to the catalytic core by the p50 central hub (Fig. 1a), through associations that stimulate repeat-synthesis activity3,6,7. Cellular depletion of p75, p45 or p19 results in telomere shortening1–3. The structure of 7–4–1 and understanding of how it contributes to telomere maintenance remain largely unknown.
Figure 1.
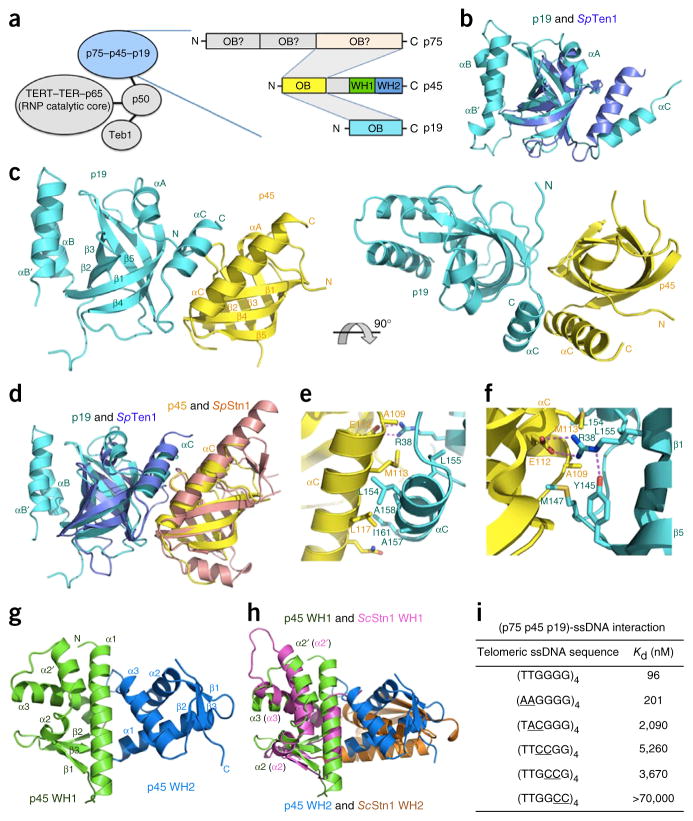
Structural and biochemical analyses of the Tetrahymena 7–4–1 complex. (a) Left, components of Tetrahymena telomerase holoenzyme. Right, domain organization of p75, p45 and p19. The shaded areas indicate domain interactions among p75, p45 and p19. (b) Superposition of p19 and S. pombe (Sp) Ten1. Cyan, p19; light blue, SpTen1. (c) Ribbon diagram of two orthogonal views of the p45N–p19 complex. Yellow, p45N; cyan, p19. (d) Superposition of the p45N–p19 complex and the S. pombe Stn1N–Ten1 complex. Yellow, p45N; cyan, p19; brown, SpStn1N; light blue, SpTen1. The superposition is based on the structures of p45N and SpStn1N. (e) The hydrophobic interface in the p45N–p19 complex. Hydrophobic residues from the αC helices of both p45N and p19 form the core of the interface. (f) Electrostatic interactions at the p45N-p19 interface. The salt bridge and hydrogen-bonding interactions are shown as dashed magenta lines. (g) Ribbon diagram of p45C. Green, WH1; blue, WH2. (h) Superposition of p45C and S. cerevisiae (Sc) Stn1C. The two WH motifs of p45C are colored as in g, and the two WH motifs of ScStn1C are in magenta and orange, respectively. The superposition is based on only the structures of the WH1 motifs of p45C and ScStn1C. (i) In vitro MST binding data of interactions between 7–4–1 and wild-type and mutant telomeric ssDNAs. The substituted nucleotides are underlined.
To initiate the study of 7–4–1, we determined the crystal structure of p19 at a resolution of 1.7 Å (Supplementary Table 1). It revealed a classical oligonucleotide- and oligosaccharide-binding (OB)-fold architecture with a large two-helix insertion (Fig. 1b). An unbiased structural homology search revealed that the OB fold of p19 is closely related to those of Stn1 and Ten1 of the CST complex (refs. 8–10 and structural superpositions in Fig. 1b and Supplementary Fig. 1). The CST complex, composed of the OB fold–containing proteins Cdc13, Stn1 and Ten1 in budding yeast, or Ctc1, Stn1 and Ten1 in vertebrates and plants, has important roles in generating telomeric 3′-G overhangs and providing chromosome-end protection through the recruitment and stimulation of DNA polymerase α (Polα)–primase11–13. The structure of p19 led us to hypothesize that 7–4–1 might be the Tetrahymena CST, in which p75 is the Ctc1-like component, p45 is Stn1, and p19 is Ten1. Consistently with this idea, yeast two-hybrid analysis revealed that, similarly to the interaction between Stn1 and Ten1 in CST8,9, the N-terminal fragment of p45 (p45N) is necessary and sufficient to mediate p19 interaction (Supplementary Fig. 2a). To extend the comparison, we reconstituted the complex between p19 and p45N and determined its structure to 2.2-Å resolution (Supplementary Table 2).
The p45N–p19 structure revealed that p45N is an OB fold closely related to that of fission yeast Schizosaccharomyces pombe Stn1 (ref. 8), with a Cα r.m.s. deviation of 1.9 Å (Fig. 1c,d). In addition to similarities between individual components, the p45N–p19 complex adopts a three-dimensional architecture similar to that of the Stn1 N-terminal region (Stn1N)–Ten1 complex; the two subunits pack against each other mainly through hydrophobic contacts mediated by the two C-terminal helices (Fig. 1d,e and Supplementary Fig. 2b,c). In addition, at the N terminus of the p45N αC helix, the side chain of E112 mediates two salt-bridge contacts with R38 of p19, which also accepts an intramolecular hydrogen bond from Y145 of p19 (Fig. 1f). This electrostatic-interaction network extends the contact interface area and helps stabilize the relative orientation of p45N and p19.
The C-terminal region of Stn1 (Stn1C) contains a globular domain with two adjacent winged helix-turn-helix (WH) motifs8–10. In contrast, the paralogous subunit of RPA, RPA32, has only one WH motif11,12,14. To examine whether the structural similarity between p45 and Stn1 could be extended to their C-terminal regions, we determined the structure of the C-terminal domain of p45 (p45C) at a resolution of 2.3 Å (Fig. 1g and Supplementary Table 3). The structure shows that p45C is indeed composed of two WH motifs (Fig. 1g). The first WH motif is closely related to S. cerevisiae Stn1C WH1; an extra helix, α2′, between helices α2 and α3 in ScStn1C WH1, which is not observed in other WH motifs, is conserved in p45C WH1 (refs. 8,10 and structural superposition in Fig. 1h). Notably, the second WH motif of p45C exhibits a striking structural resemblance to human Stn1C WH2 (ref. 9), with a Cα r.m.s. deviation of 1.5 Å (Fig. 1h and Supplementary Fig. 3). Together, our results demonstrate that p45 and p19 are structural homologs of Stn1 and Ten1 proteins. The subunit order of p75–p45–p19 (refs. 11,12 and Supplementary Figs. 2 and 4) places p75 in a position analogous to that of the evolutionarily variable large subunit of CST complexes.
To extend the comparison of 7–4–1 to CST, we tested binding of 7–4–1 to ssDNA with Tetrahymena telomeric repeats (T1T2G3G4G5G6). Microscale thermophoresis (MST) assays showed that 7–4–1 bound to the four-repeat ssDNA (T1T2G3G4G5G6)4 with a Kd of 96 nM (Fig. 1i and Supplementary Fig. 5). Oligonucleotides with only two or three telomeric repeats bound 7–4–1 with reduced affinities (Supplementary Fig. 5). To identify nucleotides critical for interaction with 7–4–1, we evaluated ssDNAs containing double-nucleotide substitutions. Substitution of the two thymine bases had a marginal effect on binding (Fig. 1i and Supplementary Fig. 5). In contrast, substitutions of guanine bases reduced the binding of 7–4–1. Among the guanines, G6 was the most important position (Fig. 1i and Supplementary Fig. 5). Therefore, the telomeric repeat DNA–binding preference of 7–4–1 parallels those of yeast and human CST complexes, all of which are distinct from the nonspecific ssDNA binding of RPA.
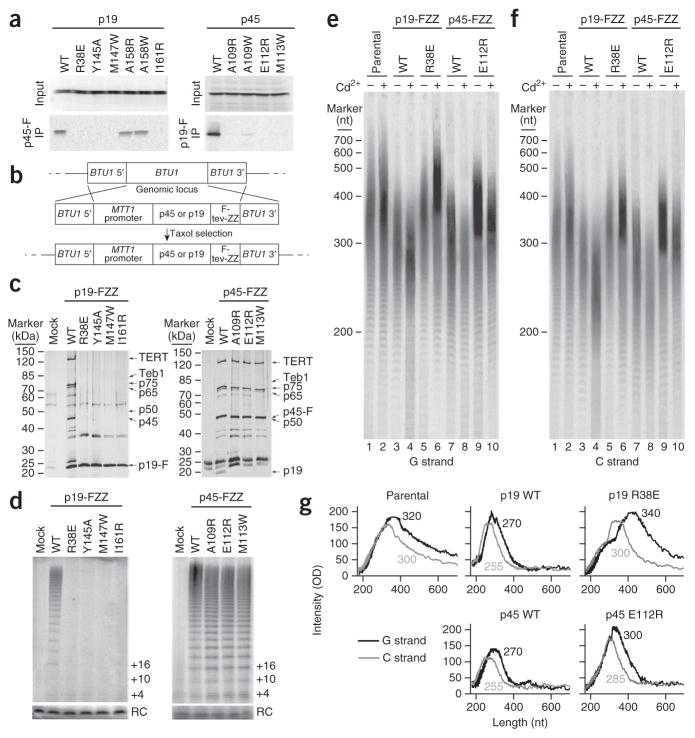
To corroborate our structural analysis, we expressed wild-type or mutant p45 and p19 proteins deficient in p45-p19 interaction in rabbit reticulocyte lysate and examined their interaction with a triple FLAG–tagged wild-type partner5. Concordantly with the crystal structure, single–amino acid substitutions of hydrophobic residues of p19 (Y145A, M147W and I161R) on the interface were sufficient to abrogate interaction of p19 with p45 (Fig. 2a and Supplementary Data Set 1). Moreover, p45 mutations (A109R, A109W and M113W) on the other side of the interface also abrogated the interaction (Fig. 2a and Supplementary Data Set 1). Notably, disruption of the salt-bridge interactions between E112 of p45 and R38 of p19 abolished the p45-p19 interaction (Fig. 2a and Supplementary Data Set 1). Thus, both the hydrophobic and the electrostatic contacts observed in the crystal structure are important for p45N-p19 interaction.
Figure 2.
Disruption of the p45N-p19 interaction affects holoenzyme assembly and telomere synthesis. (a) Untagged wild-type (WT) and mutant p19 and p45 proteins, expressed with radiolabeling in rabbit reticulocyte lysate (input samples) and assayed for physical interaction with FLAG (F)-tagged unlabeled p45 or p19 by anti-FLAG–resin binding and elution (IP samples)5. (b) Schematic diagram of the experimental design for protein overexpression from the MTT1 promoter by transgene integration at BTU1. (c,d) Affinity purifications from equivalent amounts of cells overexpressing the indicated proteins, assayed by SDS-PAGE and silver staining (c) or direct primer-extension assay for telomerase activity (d). In c, holoenzymesubunit migration positions are indicated. FZZ, FLAG-tev-ZZ tag (described in Online Methods). In d, RC indicates precipitation recovery control, and primer-extension products are indicated by nucleotides added to the primer. (e,f) Denaturing gel electrophoresis and Southern blotting detecting G-strand (e) and C-strand (f) telomeric repeat–tract lengths in cell lines overexpressing the indicated protein or in the parental CU522 cell line. G-strand and C-strand hybridization were performed by sequential probing of the same blot with end-labeled oligonucleotides. The migration of end-labeled DNA ladder run on the same gel is indicated. (g) Quantification of G-strand (black line) and C-strand (gray line) lengths from blots shown in e and f in cells treated with cadmium. Data shown were derived from a total of two replicates from independently grown cell cultures with the same conditions of induced protein overexpression (mid-log-phase cells ± cadmium addition for 4 h).
To investigate the role of p45-p19 interaction in telomerase holoenzyme assembly, we constructed transgenic strains to place wild-type or mutant p19 or p45 expression under the control of the cadmium-inducible MTT1 promoter in Tetrahymena (Fig. 2b). Basal transcription from the MTT1 promoter generated modest overexpression, and addition of cadmium induced high-level protein overexpression3. As expected, wild-type p19 but not the p45 binding–deficient mutants efficiently pulled down other telomerase holoenzyme components (Fig. 2c and Supplementary Data Set 1). In contrast, both wild-type and mutant p45 proteins were able to pull down other holoenzyme subunits, except for p19, which was associated with only wild-type p45 (Fig. 2c and Supplementary Data Set 1). Furthermore, telomerase RNP activity purified through tagged p19 but not tagged p45 required p45-p19 interaction (Fig. 2d and Supplementary Data Set 1). Collectively, these results support the connection of p19 to the rest of telomerase holoenzyme through interaction with p45.
Next, we assessed the role of p45-p19 interaction in telomere-length regulation (Fig. 2e,f and Supplementary Data Set 1). Telomere DNA-strand and 3′-overhang lengths matched those expected from previous studies15 for the parental strain CU522 without protein overexpression (Fig. 2g). Overexpression of wild-type p19 or p45 reduced telomere length, and we observed some shortening associated with basal-level transgene expression and rapid additional telomere shortening induced by cadmium addition for the 4-h window of one cell cycle (Fig. 2e–g and Supplementary Data Set 1). This reflects an induced telomere-replication defect, because the loss of telomere length far exceeds that from shutoff of telomerase function alone16,17. In contrast to wild-type p45 and p19, overexpressed mutant p19 and p45 proteins deficient in p45-p19 interaction did not induce rapid telomere shortening (Fig. 2e,f and Supplementary Data Set 1). Cadmium-induced high-level overexpression of mutant p45 shortened both strands by at most two or three repeats (Fig. 2e–g and Supplementary Data Set 1). High-level overexpression of mutant p19 did not change C-strand length but caused overelongation of the G strand (Fig. 2e–g and Supplementary Data Set 1). This altered G-overhang phenotype is previously undescribed in Tetrahymena; the depletion of telomerase subunits or Tetrahymena telomere proteins has surprisingly little effect on overhang structure3,17,18. We suggest that because overexpression of p19 R38E does not compromise general telomere replication or telomerase holoenzyme assembly, the induced defect is selectively in C-strand synthesis at chromosome termini. Overexpressed p19 might compete with telomerase holoenzyme for binding Polα–primase or uncouple Polα–primase regulation from telomerase.
On the basis of data described here and in previous studies, we propose that 7–4–1 is a telomerase-tethered CST. Tetrahymena CST association with the telomerase catalytic core improves the activity of reconstituted telomerase6, probably by stabilizing p50 folding. In addition, association of Tetrahymena CST with the telomerase catalytic core would target CST function to elongating telomeres and specificity would be enhanced by the reduced affinity of direct ssDNA binding of Tetrahymena CST compared with that of Saccharomyces cerevisiae Cdc13 or human CST19. Overexpressed p45 or p19 appeared to cause a general telomere replication defect, perhaps owing to aberrant binding of G-strand DNA. However, given the low physiological expression levels of telomerase subunits3,5, CST’s function may be to initiate telomere C-strand fill-in by Polα–primase in coordination with G-strand synthesis by telomerase. In Euplotes crassus undergoing chromosome fragmentation and new telomere addition, large telomerase complexes contain at least one subunit of primase20, thus suggesting that under conditions of particularly high demand for telomere synthesis, both CST and Polα–primase may be preassembled with the ciliate telomerase catalytic core. In summary, we suggest that stable assembly of Tetrahymena CST (7–4–1) into telomerase holoenzyme enables efficient, precise coordination of telomerase with CST-recruited Polα–primase and thereby enables coordination of telomere G-strand and C-strand synthesis.
ONLINE METHODS
Protein expression and purification
Coding sequence–optimized T. thermophila p19 (residues 2–163) was expressed in Escherichia coli strain BL21(DE3) from a modified pET-28a vector that contained a SUMO protein fused after the N-terminal His6 tag. Selenomethionine labeling of p19 was achieved by expression in E. coli strain B834(DE3) supplemented with (+)-l-selenomethionine in synthetic selenomethionine expression medium (Molecular Dimensions). Cells were harvested after induction for 16 h with IPTG (0.1 mM for native p19 and 0.3 mM for SeMet-substituted p19) at 23 °C. Cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 10% glycerol, 3 mM imidazole, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 5 mM benzamidine, and 4 mM β-mercaptoethanol); this was followed by lysis via sonication, and the cell debris was removed by ultracentrifugation (180,000g) for 30 min. The supernatant was mixed with Ni-NTA agarose beads (Qiagen) and rocked for 2 h at 4 °C before washing with 20 bead volumes of lysis buffer. The p19 protein was released to lysis buffer by overnight on-bead Ulp1 protease digestion. This was followed by further purification via gel-filtration chromatography on a HiLoad Superdex 75 column (GE Healthcare) equilibrated with buffer TN (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.5 mM TCEP). Finally, p19 was concentrated to 80 mg/ml and stored at −80 °C. The SeMet-labeled p19 was purified similarly and concentrated to 60 mg/ml with 2 mM TCEP.
Coding sequence–optimized T. thermophila p45N (residues 2–129) fused with an N-terminal His6-SUMO tag and p19 (residues 7–162) fused with an N-terminal GST tag were coexpressed in BL21(DE3) with 0.4 mM IPTG in TB medium at 23 °C. Similarly to the purification of p19, the p45N–p19 complex was mixed with Ni-NTA agarose beads and rocked for 2 h at 4 °C before elution with 250 mM imidazole. After the His6-SUMO tag was removed by Ulp1 protease treatment for 2 h, the protein complex was bound with glutathione-Sepharose (GE Healthcare) for 2 h at 4 °C. After washing with 20 bead volumes of lysis buffer, the p45N–p19 complex was released into lysis buffer by overnight on-bead PreScission protease (GE Healthcare) digestion. The complex was further purified by gel-filtration chromatography on a HiLoad Superdex 200 column (GE Healthcare) equilibrated with buffer (25 mM Tris-HCl, pH 8.0, and 150 mM NaCl). Finally, the p45N–p19 complex was concentrated to 30 mg/ml and stored at −80 °C. The SeMet-substituted complex was purified similarly.
Coding sequence–optimized T. thermophila p45C (residues 170–373) fused with an N-terminal His6-SUMO tag was expressed in BL21(DE3) with 0.1 mM IPTG in LB medium at 23 °C. Similarly to the purification of p19, p45C was mixed with Ni-NTA agarose beads and rocked for 2 h at 4 °C before washing with 20 bead volumes of lysis buffer. The p45C protein was released to lysis buffer by overnight on-bead Ulp1 protease digestion. This was followed by further purification via gel-filtration chromatography on a HiLoad Superdex 75 column (GE Healthcare) equilibrated with buffer (25 mM Tris-HCl, pH 8.0, and 150 mM NaCl). Finally, p45C was concentrated to 30 mg/ml and stored at −80 °C. The SeMet-labeled p45C was purified similarly.
Coding sequence–optimized genes of full-length p75 and p19 were cloned into Bac-to-Bac vectors containing an N-terminal MBP tag and N-terminal His-tag, respectively, preceding the multiple cloning sites (Invitrogen). p45 was cloned into a Bac-to-Bac vector with no tag. For protein expression, Sf9 cells were infected at ~1.5 × 106 cells per milliliter with a multiplicity of infection of 1 plaque-forming unit per milliliter of recombinant baculovirus. The cells were harvested after 72 h by centrifugation and stored at −80 °C. Cells were resuspended in 50 ml of lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM 2-mercaptoethanol, 5 mM benzamidine, and 1 mM PMSF) and incubated on ice for 15 min. The cells were lysed by sonication, and cell debris was removed by centrifugation. The supernatant was applied to Ni-NTA beads (Qiagen) and eluted with 250 mM imidazole and then applied to amylose beads (NEB) and eluted with 20 mM maltose. MBP tags and His-tags were removed by 3C protease. Proteins were further purified by gel-filtration chromatography (Superdex 200) with buffer (25 mM Tris-HCl, pH 8.0, and 150 mM NaCl). Proteins were concentrated to 15 mg/ml by Centricon 10 centrifugal filters (Millipore) and stored at −80 °C.
Crystallization, data collection, and structure determination
Crystals of native p19 were grown by sitting-drop vapor diffusion at 4 °C. The precipitant well solution consisted of 4.0 M sodium formate, pH 7.0, and 2% (v/v) PEG 400. Crystals were gradually transferred into a harvesting solution containing 5 M sodium formate, pH 7.0; this was followed by flash-freezing in liquid nitrogen for storage. Crystals of SeMet-labeled p19 were grown in solution consisting of 3.9 M sodium formate and 0.3 M NDSB-195. Data sets were collected under cryogenic conditions (100 K) at the Advanced Photon Source (APS) beamlines 21-ID-D and 21-ID-F. A 1.9-Å single-wavelength anomalous dispersion (Se-SAD) data set of p19 was collected at the Se-peak wavelength (0.97983 Å) and was processed by HKL2000 (ref. 21). One selenium atom was located and refined, and the single-wavelength anomalous diffraction data phases were calculated with Phenix22. The initial SAD map was substantially improved by solvent flattening. An initial model was automatically built into the modified experimental electron density. The model was then refined with a native data set with a 1.7-Å resolution (wavelength 0.97983 Å) with manual building in Coot23. In the final Ramachandran plot, the favored and allowed residues were 98.0% and 100.0%, respectively.
Crystals of SeMet-labeled p45N–p19 complex were grown by sitting-drop vapor diffusion. The precipitant well solution consisted of 4.1 M LiCl, 50 mM HEPES sodium, pH 7.0, 10 mM MgCl2, and Silver Bullets Additive A10 (Hampton Research). Crystals were protected by a harvesting solution (5 M LiCl, 50 mM HEPES sodium, pH 7.1, and 10 mM MgCl2). Data sets were collected under cryogenic conditions (100 K) at the Advanced Photon Source (APS) beamlines 21-ID-D and 21-ID-F. A 2.2-Å Se-SAD data set of p45N–p19 was collected at the Se-peak wavelength (0.97776 Å) and was processed by HKL2000 (ref. 21). Three selenium atoms were located and refined, and the single-wavelength anomalous diffraction data phases were calculated with Phenix22. The model was then refined with manual building in Coot23. In the final Ramachandran plot, the favored and allowed residues were 95.7% and 100.0%, respectively.
Crystals of native p45C were grown by sitting-drop vapor diffusion at 4 °C. The precipitant well solution consisted of 0.08 M magnesium acetate tetrahydrate, 0.05 M sodium cacodylate trihydrate, pH 6.5, and 15% (v/v) PEG 400. Crystals were gradually transferred into a harvesting solution containing 5 M sodium formate, 0.05 M sodium cacodylate trihydrate, pH 6.5, and 15% (v/v) PEG 400; this was followed by flash-freezing in liquid nitrogen for storage. Crystals of SeMet-labeled p45C were grown in similar conditions. Data sets were collected under cryogenic conditions (100 K) at the Shanghai Synchrotron Radiation Facility (SSRF) beamlines BL18U1 and BL19U1. A 2.8-Å SeMet-SAD data set of p45C was collected at the Se-peak wavelength (0.97876 Å) and was processed by HKL2000 (ref. 21). Two selenium atoms were located and refined, and the single-wavelength anomalous diffraction data phases were calculated with Phenix22. The initial SAD map was substantially improved by solvent flattening. The model was then refined with a native data set with a 2.3-Å resolution (wavelength 0.97876 Å) in Phenix, together with manual building in Coot23. In the final Ramachandran plot, the favored and allowed residues were 98.8% and 100.0%, respectively. All the structural figures were generated with PyMOL (http://www.pymol.org/).
Microscale thermophoresis measurements of the interaction between the 7–4–1 complex and ssDNAs
Microscale thermophoresis (MST) is a new immobilization- free technique for the analysis of biomolecular interactions24. MST measurements were performed with a NanoTemper Monolith NT.115 instrument (NanoTemper Technologies). In brief, 20 nM 5′-fluorophore–labeled wild-type or mutant telomeric ssDNAs were first incubated for 20 min on ice with different concentrations of the 7–4–1 complex in a solution of 25 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Then 5 μL of the samples was loaded into standard treated capillaries, and MST measurements were collected at 25 °C at 20% infrared-laser power and 50% light-emitting-diode power. The laser-on and laser-off intervals were 30 s and 5 s, respectively. NanoTemper Analysis 1.2.20 software was used to fit the data and to determine the apparent Kd values. All measurements were collected at least three times.
Yeast two-hybrid assay
The yeast two-hybrid assays were performed with L40 strains containing pBTM116 and pACT2 (Clontech) fusion plasmids. The colonies containing both plasmids were selected on –Leu –Trp plates. The β-galactosidase activities were measured by liquid assay25.
Tetrahymena strain construction and cell growth
Transgenes encoding p19-FZZ, p19(R38E)-FZZ, p19(Y145A)-FZZ, p19(M147W)-FZZ, p19(I161R)-FZZ, p45-FZZ, p45(A109R)-FZZ, p45(E112R)-FZZ, and p45(M113W)-FZZ were targeted for integration at the BTU1 locus. The BTU1 promoter and open reading frame were replaced with the open reading frame of interest under expression control by ~1 kb of the MTT1 promoter26. A synthetic open reading frame for the wild-type or mutant protein was followed by the F-tev-ZZ tag with the triple-FLAG peptide (F) and tandem Protein A domain (ZZ) modules separated by a tobacco etch virus protease (tev)–cleavage site. Transformation and taxol selection against the endogenous BTU1 locus in strain CU522 were performed as previously described1. Genotypes were verified by Southern blotting and western blotting for the transgene-encoded protein. Cells were grown in modified Neff medium (0.25% proteose peptone, 0.25% yeast extract, 0.5% dextrose, and 30 μM FeCl3) at 30 °C to mid-log phase (3.5 × 105 cells/ml). For affinity purifications, cells were then starved in 10 mM Tris, pH 8.0, for 16 h and refed with modified Neff medium containing 0.5 μg/ml cadmium for 4 h before harvesting. Extracts of cells grown in cadmium were used for two-step affinity purification of wild-type and mutant p19 and p45 proteins with a C-terminal F-tev-ZZ tag. Importantly, C-terminal F-tev-ZZ fusion to p19 or p45 does not disrupt protein function, as demonstrated by integration of the tag cassette after the protein open reading frame at the endogenous gene loci9. For telomere-length assays, a final concentration of 0.5 μg/ml cadmium was added directly to the mid-log-phase growth culture.
Subunit association and telomerase activity assays
Untagged proteins expressed in rabbit reticulocyte lysate were labeled by [35S]methionine incorporation and tested for interaction with FLAG-tagged partner as previously described5. Holoenzyme was purified from cell extract by tandem steps of IgG agarose and anti-FLAG resin binding, as previously described3. Activity assays were performed after anti-FLAG resin elution with Tetrahymena telomerase reaction buffer containing 50 mM Tris acetate, pH 8.0, 2 mM MgCl2, 10 mM spermidine, and 5 mM β-mercaptoethanol. Product-synthesis reactions additionally contained [α-32P]dGTP mixed with 300 nM unlabeled dGTP, 200 μM dTTP, and 200 nM DNA primer (GT2G3)3. Reactions were allowed to proceed for 3 min at room temperature. A 5′-end–radiolabeled oligonucleotide was added to telomerase products before precipitation as a recovery control. Products were resolved by denaturing gel electrophoresis and detected by phosphorimager analysis with a Typhoon Trio.
Genomic-DNA purification and hybridization
For genomic-DNA purification, cells from 1.5 ml of mid-log-phase culture were washed with 10 mM Tris, pH 8.0, and DNA was extracted as previously described3. Purified DNA was digested with MseI, recovered by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and ethanol precipitation, resolved with denaturing 6% acrylamide/7 M urea/0.6× TBE gel electrophoresis, transferred to Hybond-N+ membrane, and probed with (C4A2)3 or (T2G4)3 oligonucleotide 5′-radiolabeled with polynucleotide kinase. Blots were hybridized overnight in 4× Denhardt’s solution (0.08% bovine serum albumin, 0.08% Ficoll 400, and 0.08% polyvinylpyrrolidone) with 0.9 M NaCl, 90 mM Na3C6H5O7, and 0.1% SDS and then washed in buffer containing 30 mM NaCl, 3 mM Na3C6H5O7, and 0.1% SDS at 45 °C. Data were collected with a Typhoon Trio Imager. The 5′-end–radiolabeled DNA ladder (100-bp ladder, Invitrogen) migration positions should be considered length approximations because sequence could influence migration in denaturing gel-electrophoresis conditions.
Quantification of telomere length
Telomere DNA-strand and 3′-overhang lengths were determined with denaturing-gel Southern blotting to detect the length range of the G strand and C strand separately9. The intensity profiles of telomeric-repeat hybridization were quantified to determine the mean length of each telomere strand27. Mean telomere length was calculated as the weighted mean of the optical density (OD) as described previously27. Briefly, OD data at each position were recovered by densitometry analysis with ImageJ and adjusted for background. Lengths were determined by the following equation: mean telomere length = Σ(ODi)/Σ(ODi/Li), where ODi is the optical density at position i, and Li is the telomere length at position i. Values for telomere lengths in cells expressing p19 R38E were determined from a shorter exposure of the same blot to remain in the linear range for quantification.
Supplementary Material
Acknowledgments
We are grateful to the National Center for Protein Science Shanghai (Protein Expression and Purification system) for instrument support and technical assistance. We thank L. Wu, D. Yao, W. Qin and R. Zhang at Shanghai Synchrotron Radiation Facility for help with data collection and processing. This work was supported by grants from the Ministry of Science and Technology of China (2013CB910402 to M.L.), the National Natural Science Foundation of China (31330040 to M.L. and 31270787 to Z.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010201 to M.L.), the US National Science Foundation (Graduate Research Fellowship under grant DGE-1106400 to H.U.) and the US National Institutes of Health (GM54198 to K.C.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Footnotes
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5DOF (p19), 5DOK (p45C) and 5DOI (p45N–p19 complex).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
B.W. and T.T. were responsible for the structural determination and biochemical experiments; H.U. for Tetrahymena telomerase activity assay and telomere-length quantification; J.S. for p45C crystallization; Y.Z., S.L., Z.Z. and J.C. for some of the protein purification; and J.S.B. for data collection and processing. K.C., J.W. and M.L. contributed to overall design, interpretation of the studies and manuscript preparation.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Witkin KL, Collins K. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witkin KL, Prathapam R, Collins K. Mol Cell Biol. 2007;27:2074–2083. doi: 10.1128/MCB.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min B, Collins K. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Z, et al. Proc Natl Acad Sci USA. 2011;108:20357–20361. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Upton HE, Hong K, Collins K. Mol Cell Biol. 2014;34:4200–4212. doi: 10.1128/MCB.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J, et al. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong K, et al. Mol Cell Biol. 2013;33:3962–3971. doi: 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, et al. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan C, Rice C, Harkisheimer M, Schultz DC, Skordalakes E. PLoS ONE. 2013;8:e66756. doi: 10.1371/journal.pone.0066756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelinas AD, et al. Proc Natl Acad Sci USA. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart JA, Chaiken MF, Wang F, Price CM. Mutat Res. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen LY, Lingner J. Nucleus. 2013;4:277–282. doi: 10.4161/nucl.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, et al. Cell Res. 2011;21:258–274. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mer G, et al. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 15.Jacob NK, Skopp R, Price CM. EMBO J. 2001;20:4299–4308. doi: 10.1093/emboj/20.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob NK, Kirk KE, Price CM. Mol Cell. 2003;11:1021–1032. doi: 10.1016/s1097-2765(03)00131-x. [DOI] [PubMed] [Google Scholar]
- 17.Linger BR, Morin GB, Price CM. Mol Biol Cell. 2011;22:4161–4170. doi: 10.1091/mbc.E11-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob NK, Lescasse R, Linger BR, Price CM. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen LY, Redon S, Lingner J. Nature. 2012;488:540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 20.Ray S, Karamysheva Z, Wang L, Shippen DE, Price CM. Mol Cell Biol. 2002;22:5859–5868. doi: 10.1128/MCB.22.16.5859-5868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretti P, Freeman K, Coodly L, Shore D. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 26.Shang Y, et al. Proc Natl Acad Sci USA. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M, et al. Nat Protoc. 2010;5:1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.