Abstract
Worldwide, lava caves host colorful microbial mats. However, little is known about the diversity of these microorganisms, or what role they may play in the subsurface ecosystem. White and yellow microbial mats were collected from four lava caves each on the Azorean island of Terceira and the Big Island of Hawai’i, to compare the bacterial diversity found in lava caves from two widely separated archipelagos in two different oceans at different latitudes. Scanning electron microscopy of mat samples showed striking similarities between Terceira and Hawai’ian microbial morphologies. 16S rRNA gene clone libraries were constructed to determine the diversity within these lava caves. Fifteen bacterial phyla were found across the samples, with more Actinobacteria clones in Hawai’ian communities and greater numbers of Acidobacteria clones in Terceira communities. Bacterial diversity in the subsurface was correlated with a set of factors. Geographical location was the major contributor to differences in community composition (at the OTU level), together with differences in the amounts of organic carbon, nitrogen and copper available in the lava rock that forms the cave. These results reveal, for the first time, the similarity among the extensive bacterial diversity found in lava caves in two geographically separate locations and contribute to the current debate on the nature of microbial biogeography.
Keywords: Azores, Hawai’i, lava cave, microbial communities
Introduction
Lava caves, which are caves formed by lava flows, develop microbial mats that cover their floors, walls and ceilings with remarkable colors and patterns (Figure 1). These mats are commonly found in moist lava caves; however very little is known about their diversity and the role they play in the biogeochemistry of this subsurface environment. Lava caves, like other caves, often have relatively stable temperatures and humidity throughout the year once into the dark zone (Culver and Pipan 2009; Palmer 2007; Poulson and White 1969), but are still considered extreme environments because of their low nutrient availability and productivity (Barton 2006; Moore and Sullivan 1997; Northup and Welbourn 1997). Similar to other caves, light is not available beyond the twilight zone in lava caves, forcing their inhabitants, including chemolithotrophic bacteria (Northup and Lavoie 2001), to rely on other energy sources. Furthermore, many of the factors that influence surface ecosystems are lacking in caves, including wide changes in temperature and humidity, or weathering effects such as strong wind and ultraviolet exposure (Northup et al. 2010; Snider et al. 2009a).
Fig. 1.
Macroscopic images of example white and yellow microbial mats and colonies from Hawai’ian (A, B, C) and Azorean (D, E) lava caves. Example yellow colonies are indicated with arrows; example white colonies are indicated with lines with an open circle on the end. ©Kenneth Ingham. Reproduced by permission of Kenneth Ingham. Permission to reuse must be obtained from the rightsholder.
The isolated nature of caves makes them a model ecosystem in which to study factors driving biological diversification (Culver and Pipan 2009). Cave environments are particularly isolated, thus limiting the ability of species to migrate; consequently, cave-adapted species tend to exhibit a high degree of endemism (Christman et al. 2005; Culver and Pipan, 2009; Elliott 2007; Sharratt et al. 2000). Moreover, discovery of volcanic activity and of lava caves on Mars increased the interest in their study on Earth, for lava caves may constitute a suitable environment to find evidence of fossil or extant life on Mars (Boston et al. 1992; Boston et al. 2001; Boston 2010).
Basaltic rock underlays large areas of the Martian surface, and recent evidence suggests that lava caves are present there (Léveillé and Datta 2010). It is thought that caves on Mars would retain liquid water longer than the surface, and the subsurface would offer protection from the harsh solar radiation, providing a possible refuge for life or the remnants of life forms (Boston 2010; Léveillé and Datta 2010; Northup et al. 2011; Popa et al. 2012). By studying lava caves on Earth, the groundwork is being laid for studies that will be relevant for life detection on Mars.
Despite these motivations, little is known about the diversity and establishment of microbial communities in lava caves. Particularly, the environmental factors, both inside and outside the cave that might contribute to microbial community composition, are poorly understood. The few published studies that exist on cave biogeochemistry focus on carbon cycling in karst caves (Simon et al. 2007) and to a lesser degree on nitrogen (Tetu et al. 2012). Notably, microbial mats in lava caves have received less attention than the epilithic microorganisms of limestone caves (de los Ríos et al. 2011; Hathaway et al. this issue; Northup and Welbourn 1997; Northup et al. 2008; Northup et al. 2011; Snider 2010; Garcia et al. 2009; Moya et al. 2009).
Culture-dependent techniques have been the common method applied in the few studies that have been done of the composition of these mats, often referred to as “slimes.” Stoner and Howarth (1981), the first to describe these mats in Hawai’ian lava caves, found fungi and aerobic bacteria and suggested that the white and brown slimes are important sites of nutrient cycling (e.g., nitrogen). Studies done in lava caves in Washington, USA, have found slime consisting of different species of bacteria, including Actinobacteria in the antibiotic producing genus Streptomyces (Staley and Crawford 1975). Recent studies of other types of volcanic terrain have also reported novel chemolithotrophic bacteria (Cockell et al. 2009; Gomez-Alvarez et al. 2007; King 2007; Popa et al. 2012; Stott et al. 2008). Acidobacteria, Alpha- and Gammaproteobacteria, Actinobacteria, and Cyanobacteria dominate bacterial communities on volcanic surface terrain in Hawai’i, where composition is controlled by local differences in the environment and nature of the volcanic deposits (Gomez-Alvarez et al. 2007).
The unique nature of caves makes them well suited for looking at questions regarding the biogeography of bacteria. Because bacteria are thought to disperse very easily, and to have a cosmopolitan distribution, the hypothesis “everything is everywhere, but the environment selects” (Baas-Becking 1934) often applies to microbial biogeography (see Hortal 2011; Fontaneto and Hortal 2012 for reviews) and we therefore might predict that bacteria in caves have lower endemism than that seen in cave animals. The “everything is everywhere” hypothesis has been repeatedly challenged however, and many studies report spatial variations in microbial diversity not related with environmental variability (see reviews in Fontaneto 2011). We do not know whether the microbial community composition of lava caves is similar worldwide, or if local influences in each lava cave result in unique microbial communities. In fact, it is predicted that microbial diversity follows the same biodiversity scaling rules of other organisms (Bell et al. 2005; Green and Bohannan 2006), which will imply some levels of endemism and spatial patterning in both alpha, beta and gamma diversities.
To investigate microbial biogeography in lava caves, we compared bacterial community composition in lava caves on the Big Island, Hawai’i, and on the Azorean Island of Terceira, which represent semi-arid to tropical, and temperate climate regimes respectively, in two widely separated oceanic archipelagos. These two islands offer a range of lava caves with different precipitation levels and elevations at which the lava caves are located (Table 1). Both locations have humid lava caves that are dominated by white and yellow-pigmented microbial mats. According to the hypothesis “everything is everywhere, but the environment selects” (e.g., Finlay 2002) microbial mats of similar pigmentation (i.e. yellow versus white) will have a common microbial composition across sites.
Table 1.
Abiotic characteristics of study caves
| Cave Name | Sample Names (Color) {Distance from Entrance} | No. High Quality Sequences Recovered (Number of OTUs) | Location | Cave Entrance Elevation (m) | Surface Precipitation (mm)c | Cave Ageb | Cave Temp. (°C) | Percent Humidity |
|---|---|---|---|---|---|---|---|---|
| Bird Park | BP087 (Yellow) {50 m} | 103 (59) | Hawai’i | 1219 | 1778 | 2000 | 14.3 ± 0 | NA |
| BP089 (White) {30 m} | 43 (25) | |||||||
| Epperson’s | EP912 (Yellow){115 m} | 33 (30) | Hawai’i | 304 | 4013 | 1000 | 17.7 ± 0.1 | >95 |
| EP097 (White) {115 m} | 55 (35) | |||||||
| Kaumana | KA613 (Yellow) {150 m} | 61 (38) | Hawai’i | 610 | 5003 | 121 | 17.5 ± 0.5 | >95 |
| Cave | KA619 (White) {150 m} | 31 (26) | ||||||
| Kula Kai | KK135 (Yellow) {100 m} | 39 (25) | Hawai’i | 415 | 1016 | 1500 | 19 ± 0.4 | >95 |
| Caverns | KK138 (White) {100 m} | 26 (26) | ||||||
| Gruta da | AS252 (Yellow) {18 m} | 92 (31) | Azores | 330 | 1635 | 2000 | 14.9 ± 0.3 | >95 |
| Achada | AS253 (White) {18 m} | 49 (45) | ||||||
| Gruta da | GBL17 (Yellow) {75 m}a | 188 (100) | Azores | 422 | 1967 | >7130 | 16 ± 0.6 | >95 |
| Balcões | GBL128 (White) {10 m}a | 99 (55) | ||||||
| Gruta da | GBO52 (Yellow) {45 m} | 124 (55) | Azores | 255 | 1400 | >7130 | 15.0 ± 0.1 | 95 |
| Branca Opala | GBO654 (White) {50 m} | 74 (40) | ||||||
| Gruta dos | GP283 (Yellow) {200 m} | 181 (95) | Azores | 346 | 1728 | >7130 | 15.4 ± 0.3 | >95 |
| Principiantes | GP276 (White) {100 m} | 172 (92) |
Distnce is from closest skylight, not entrance. Distance to entrance of cave is 200 m for GBL17 and 135 m for GBL128.
Age of cave is an estimate for the flow in the region.
Precipitation data from http://rainfall.geography.hawaii.edu/ (Giambelluca et al. 20013) for Hawai’i and http://www.climaat.angra.uac.pt/ for Terceira.
We alternatively predict that the composition of bacterial communities will differ widely between environmentally similar Hawai’ian and Azorean lava caves, due to recent challenges to the “everything is everywhere” hypothesis. Furthermore, we hypothesize that, as a result of colonization and evolution on remote archipelagoes under unique selective regimes in isolated cave habitats, novel organisms, those sharing less than 97% identity with known bacteria, are going to be found in the microbial communities of lava caves in the Azores and Hawai’i. Analysis of the community composition of these lava caves will help us to further understand factors controlling microbial diversity and community assemblages that may extend to other habitats, and perhaps even to other planets.
Methods
Microbial Mat and Site Description, and Sample Collection: Hawai’i and Terceira
The study caves in both Hawai’i and Terceira contained both white and yellow microbial mats that vary in relative predominance across the caves and may occur separately or with differently colored colonies intermixed (Figure 1). Both colors of microorganisms occur as distinct colonies (e.g., Figure 1A) or as microbial mats (Figure 1C) covering large portions of walls or ceilings. Yellow mats varied in color from a lemon yellow to a gold. Many of the microbial mats or colonies exhibit hydrophobicity during wetter times of the year. The mats can be thin with distinct microbial colonies, or 1–3 mm thick. Colonies sometimes grow on an organic ooze that coats some walls (Figure 1D), but an effort was made to collect our samples from sites with no organic ooze underlying the colonies.
Four lava caves, Kula Kai Caverns in the Kipuka Kanohina Cave Preserve, Kaumana, Epperson’s, and Bird Park, a.k.a. Kipuka Puaulu, were selected on the Big Island of Hawai’i, in the Pacific Ocean located at 19° 43′ N 155° 5′ W. The caves represent a variety of abiotic factors, such as temperature, elevation, and yearly precipitation. At each site, entrance elevation was recorded. Cave temperature and humidity (wet bulb/dry bulb) were taken every 50 m within the cave with an IMC Digital Thermometer probe and averaged. Average area rainfall is presented in Table 1.
Small samples of wall rock covered with yellow or white microbial mats were collected aseptically from the four Hawai’ian Island lava caves, under a National Park Service collecting permit or permission of land-owners. Two samples from each cave, one yellow and one white, were selected for collection based on uniformity of color. Samples were covered with sucrose lysis buffer (Giovannoni et al. 1990) to preserve the DNA, and transported to the laboratory where they were stored at −80°C until DNA extraction. Samples of lava rock for chemical analysis from each cave were selected based on the absence of biological matter on the rock. Samples were collected from loose rock on the cave floor that had obviously fallen off the cave wall and placed in a sterile Whirl Pak bag for transportation to the laboratory.
Terceira is located in the Atlantic Ocean, in the center of the Azores island chain at 38° 44′ N, 27° 17′ W, approximately 1,500 km west of the coast of mainland Portugal. White and yellow samples from Terceira were collected from Gruta dos Principiantes, Gruta da Achada, Gruta da Branca Opala and Gruta dos Balcões using the same protocol as in Hawai’i. These caves were selected based on both similar and contrasting abiotic factors as compared to the Hawai’ian caves (Table 1).
Scanning Electron Microscopy
Samples were examined on a JEOL 5800 SEM equipped with an energy dispersive X-ray analyzer (EDX), at high vacuum with an accelerating voltage of 15KeV with a beam current between 0.1 to 0.01 ηA. Rock chips with microbial mats were mounted directly on SEM sample stubs in the field, air dried, and coated with Au-Pd metal for imaging in the laboratory.
Chemical Analysis of Lava Rock
Single rock samples from each cave were crushed and then pulverized using a SPEX Shatter box (SPEX, Metuchen, NJ). The samples were then sieved through a 200 mesh sieve. About 0.1 to 0.2 g of each sample were digested in solutions of nitric acid and hydrofluoric acid for analysis of iron, magnesium, phosphorous, sulfur, copper, lead, and zinc content. The samples were analyzed using a Perkin Elmer Optima 5300 DV ICP-AES, following a method equivalent to US EPA 2007.
A subsample of the same pulverized rock sample was used to determine percentage of nitrogen and organic carbon by high-temperature combustion. The samples were dissolved in 6 N HCl prior to analysis to remove carbonate (Harris et al. 2001). The gases resulting from the high temperature combustion were eluted on a gas chromatography column and detected by thermal conductivity and integrated to yield organic carbon and nitrogen content (Pella 1990a; 1990b). Analyses were performed on a ThermoQuest CE instruments NC2100 Elemental Analyzer.
DNA Extraction, Amplification, and Sequencing
DNA was extracted and purified using the MoBio PowerSoil™ DNA Isolation Kit using the manufacturer’s protocol (MoBio, Carlsbad, CA), with the exception of the substitution of bead beating for 1.5 min (Biospec Products, Bartlesville, OK, USA) instead of vortexing for cell lysis. 16S rDNA sequences were amplified with universal bacterial primers 46 forward (5′-GCYTAAYACATGCAAGTCG-3′) and 1409 reverse (5′-GTGACGGGCRGTGTGTRCAA-3′) (Northup et al. 2010).
Reactions were carried out in a 25-μL volume with 1X PCR buffer with 1.5 mM Mg2+, 0.4 μM of each primer, 0.25 mM of each dNTPs, 5 μg of 50 mg/mL BSA (Ambion, Austin, TX, USA) and 1U AmpliTaq LD (Applied Biosystems, Foster City, CA, USA). Amplification was carried out under the following thermocyling conditions on an Eppendforf Mastercycler 5333 (Eppendorf, Hauppauge, NY, USA): 94°C for 5 min, followed by 31 cycles of 94°C for 30 sec, 50°C for 30 sec, 72°C for 1.5 min, and a final extension at 72°C for 7 min. Amplicons were cleaned and purified using the Qiagen PCR cleanup kit (Qiagen, Germantown, Maryland) and cloned using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA). Sequencing was carried out at the Washington University Genome Sequencing Facility with a total of 192 clones per sample initially sequenced.
Nucleotide Sequence Analysis for Phylogenetic and Community Analyses
Sequences were checked for quality, edited and assembled with Sequencher 4.8 (Gene Codes Corporation, Ann Arbor, MI, USA). Sequence orientation was checked with OrientationChecker (www.cardiff.ac.uk/biosi/research/biosoft). Chimeras were detected using the Mallard/Pintail software (http://www.bioinformatics-toolkit.org). Initial alignment was completed with Greengenes (greengenes.lbl.gov; DeSantis et al. 2006) and manually corrected using the BioEdit editor (www.mbio.ncsu.edu/BioEdit/bioedit.html), guided by 16S rRNA secondary structure considerations.
Sequences were then classified at the phylum level using the Ribosomal Database Project Classifier (RDP) (rdp.cme.msu.edu; Maidak et al. 2001). Alignments were then imported into the software ARB using a reference tree of either ~9000 full length sequences from RDP or 236,469 full-length sequences from Greengenes to determine close relatives from other volcanic habitats (green-genes.lbl.gov/Downloads/Sequence Data; Hugenholtz 2002; Ludwig et al. 2004). The software package mothur was used to determine OTUs at the 97% identity level, which was used throughout the study (Schloss et al. 2009).
Rarefaction curves and nonparametric richness estimates were generated with mothur to evaluate sampling efforts (Chao 1984; Schloss et al. 2009). Sequences were compared with the GenBank database using basic local alignment search tool (BLAST) to determine closest relatives (Altschul et al. 1997). Samples were also compared using the software FastUniFrac to determine if geography and color were important factors in determining community profiles using an unweighted Principal Coordinate Analysis (PCoA), with the category mapping feature (Hamady et al 2010). Sequences have been submitted to the NCBI Gen-Bank database (http://www.ncbi.nlm.nih.gov/genbank/) and assigned Accession numbers HM063010-HM063029, HM444833-HM445589 and HM545239-HM545243.
Sequences from this study were compared to sequences from terrestrial volcanic environments in Hawai’i (Gomez-Alvarez et al. 2007), the Canary Islands (Portillo and Gonzalez 2008), and Iceland (Cockell et al. 2009; Kelly et al. 2010). Sequences from this study also were compared to sequences from other cave environments available in GenBank. Comparison caves were selected if a sequence from that appeared as a near neighbor in the tree created by ARB. All sequences from that cave were then searched for in GenBank and used for comparison. Caves from which comparison sequences were derived included Lower Kane Cave, Wyoming (Engel et al. 2003, 2004), Mammoth Cave, Kentucky (Fowler, unpublished), Frasassi Cave, Italy (Macalady et al. 2007; Macalady et al. 2008; Vlasceanu et al. 2000), Altamira Cave, Spain (Portillo et al. 2008), Oregon Cave, Oregon (Fowler and Wade, unpublished), Nullarbor, Australia (Holmes et al. 2001), and Lechuguilla Cave, New Mexico (Northup et al. 2003; Spilde et al. 2005). Sequences from this study were trimmed to match the length of the sequences from the comparison studies, and very short sequences from the other studies were removed from the analysis. OTUs were defined at the 97% level and shared OTUs were determined using mothur.
Results
Comparison of Hawai’ian and Azorean Yellow and White Microbial Communities: Scanning Electron Microscopy Results
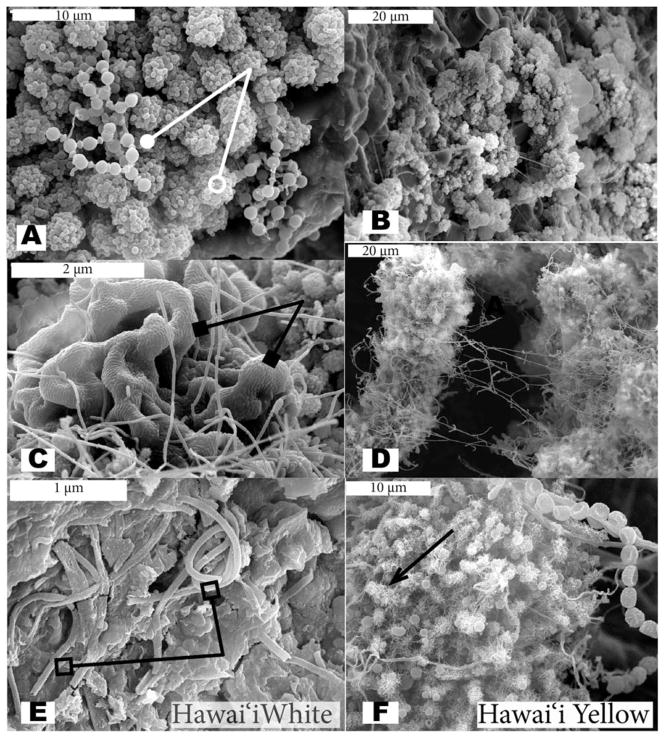
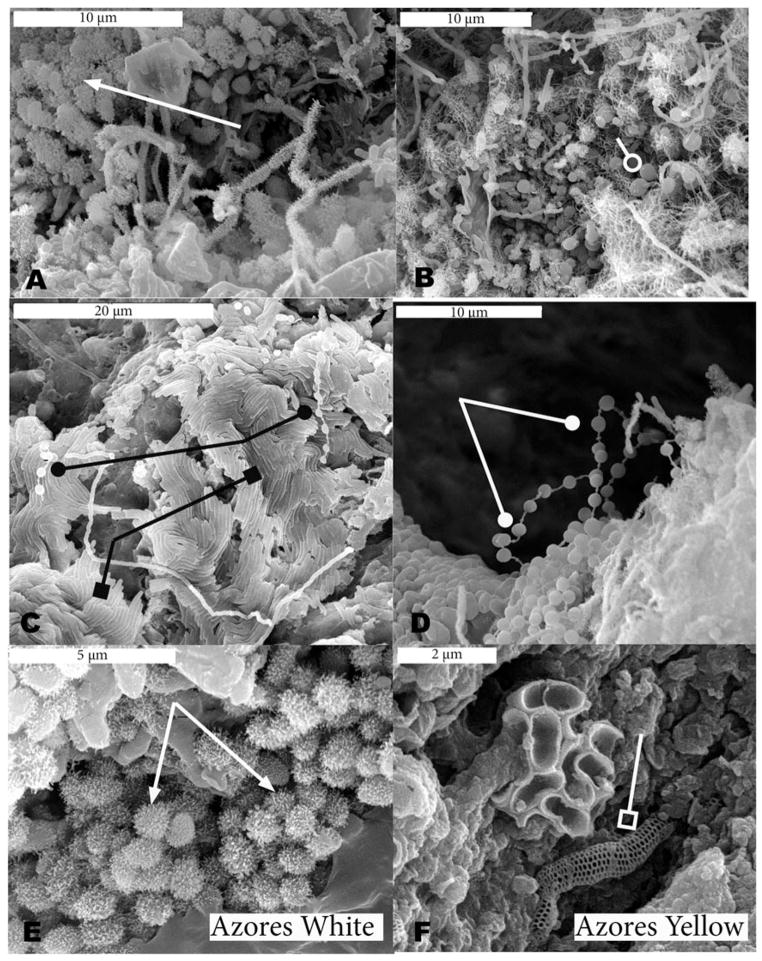
Scanning electron microscopy revealed many similarities among the microbial communities in both archipelagoes and several of the morphologies observed are very distinctive in appearance (Figures 2 and 3). Coccoidal morphologies with and without abundant thread-like or knobbed appendages (putative fimbrae or pili) and elongate structures with many thread-like appendages were seen in samples from both islands (Figures 2A, B, D, F, and 3B and E). The putative pili are much more common in the yellow microbial mats, while the cocci with knobby appendages are more common in the white microbial mats of the Azores (e.g., Figures 3A and 3E). Reticulated filaments (Melim et al. 2008) were detected in a white sample from Kula Kai Caverns (Hawai’i) (Figure 2E) and in a yellow sample from Gruta dos Balcões (Azores) (Figure 3F), but were not as commonly observed as other morphologies.
Fig. 2.
Scanning Electron Micrographs (SEM) of white (left column) and yellow (right column) microbial mats from Hawai’ian lava caves. Beads-on-a-string morphology is indicated by a line ending in a solid circle (A), while example cocci are indicated by a line ending in an open circle (A). The reticulated filaments are indicated by a line ending in an open square (E); example thread-like appendages are indicated by an arrow (F); and ordered rows of rods are indicated by a line ending in solid square (C).
Fig. 3.
Scanning Electron Micrographs (SEM) of white (left column) and yellow (right column) microbial mats from Azorean lava caves. Beads-on-a-string morphology is indicated by a line ending in a solid circle (D), while example cocci are indicated by a line ending in an open circle (B). The reticulated filaments are indicated by a line ending in an open square (F); example knob-like appendages are indicated by an arrow (A, E); and ordered rows of rods are indicated by a line ending in solid square (C).
In both Hawai’ian (2C) and Azorean (3C) white samples, biofilms with ordered rows of rods were observed. A common morphology observed in several samples was the beads-on-a-string morphology (Figures 2A and 3C and D), although the morphology was more commonly observed in white microbial mat samples. Segmented filaments were only observed in white microbial mats, while chains of cocci were only observed in one yellow Hawai’ian sample. All of the structures described above are presumed to be bacterial or archaeal due to their size (approximately 1 micron or less in diameter). Putative protozoa and algae were observed rarely in samples from both islands.
Chemical Analysis of Lava Rock
The analyses of lava rock from each cave showed higher amounts of sulfur, organic carbon and nitrogen associated with the basalt substrates of the lava cave walls from Terceira as a whole, although none of these elements were statistically significantly different between the two locations (Table 2). This may be due to the size of the data set. There were higher levels of magnesium and copper in Hawai’i, with copper being the only element statistically significantly different between Hawai’i and Terceira (p = 0.0012).
Table 2.
Rock chemistry
| Cave | MgOa | FeOa | MnOa | CaOa | P2O5a | SO4a | Cob | Cub | Pbb | Znb | % N | % OC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bird Park | 8.02 | 9.90 | 0.21 | 9.86 | 0.074 | 0.05 | 81.25 | 77.08 | BDL | 165.63 | BDL | 0.138 |
| Eppersons | 1.46 | 14.07 | 0.12 | 5.10 | 0.141 | 0.04 | 52.27 | 67.05 | BDL | 125.00 | BDL | 0.098 |
| Kaumana | 1.44 | 5.63 | 0.10 | 5.19 | 0.363 | 0.06 | 64.50 | 100.00 | 136 | 151.91 | BDL | 0.089 |
| Kuli Kai | 1.96 | 6.74 | 0.10 | 4.67 | 0.099 | 0.05 | 44.66 | 94.86 | BDL | 79.60 | 0.003 | 0.124 |
| Achada | 2.71 | 8.36 | 0.13 | 6.54 | 0.146 | 0.04 | 44.05 | BDLc | BDL | 104.76 | 0.015 | 0.369 |
| Balcones | 0.13 | 5.45 | 0.08 | 2.77 | 0.122 | 0.07 | 37.80 | 27.60 | 3.51 | 69.40 | 0.006 | 0.143 |
| Branca Opala | 2.05 | 7.33 | 0.10 | 6.74 | 0.121 | 0.06 | 50.00 | 1.14 | BDL | 98.86 | 0.059 | 1.303 |
| Principiantes | 1.87 | 7.64 | 0.10 | 6.64 | 0.087 | 0.07 | 52.08 | 35.42 | BDL | 129.17 | 0.011 | 0.278 |
All elements are accurate to ±0.01%.
Data reported in percent weight of oxides.
Data reported in ppm.
Below detection limit. Detection limit for Cu is 0.54 ppm, limit for Pb is 0.420 ppm, limit for %N is 0.01%.
Molecular Phylogenetics and Community Composition
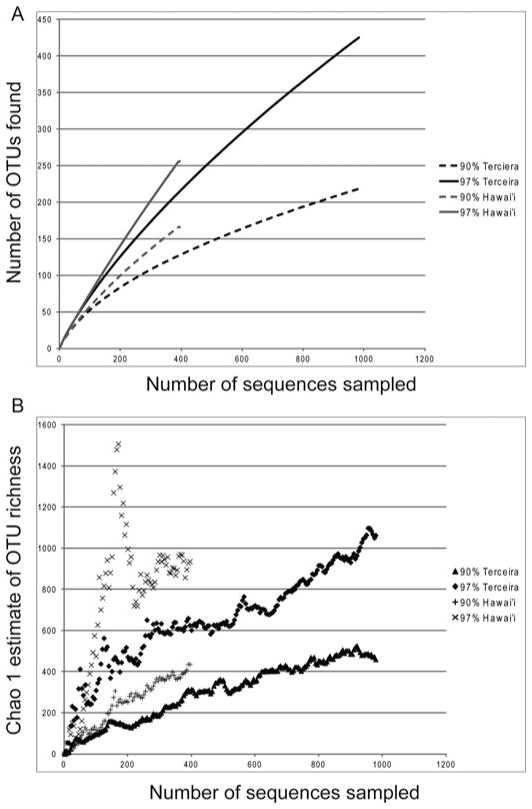
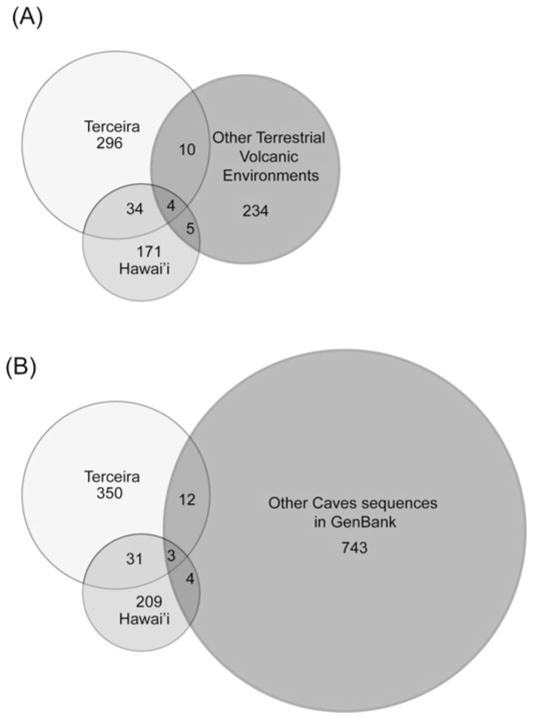
A total of 1370 full-length nonchimeric sequences were obtained from the 16 clone libraries. The number of sequences and OTUs found in each sample are reported in Table 1. The rarefaction curves showed that the full extent of the diversity has not been sampled (Figure 4). Such results are common in bacterial diversity studies (Bent and Forney 2008; Hughes et al. 2001), and deep coverage of microbial communities is beyond the scope of this research. Operational taxonomic units (OTU) were defined at 97% sequence similarity, and the sequences sampled cluster into 609 OTUs. There were only 34 (5%) shared OTUs out of all OTUs defined between the two islands.
Fig. 4.
Rarefaction and Chao1 Estimate of Richness for Hawai’i and Terceira. Collection curves estimated at 97% and 90% sequence similarity. (A) Rarefaction curves show that sampling at both taxonomic levels do not captures all diversity present. (B) Chao1 estimate indicate that Hawai’i has higher diversity than Terceira.
Thirty percent of the sequences were more closely related to other sequences in the same cave, whereas 44% were most closely related to a sequence in another cave on the same island. Only 26% of the sequences were most closely related to a sequence from the other archipelago. Chao1 estimates of taxonomic richness at the 97% identity level showed that the microbial communities of the sampled lava caves of Hawai’i are estimated to be more diverse than those sampled on Terceira Island (Figure 4).
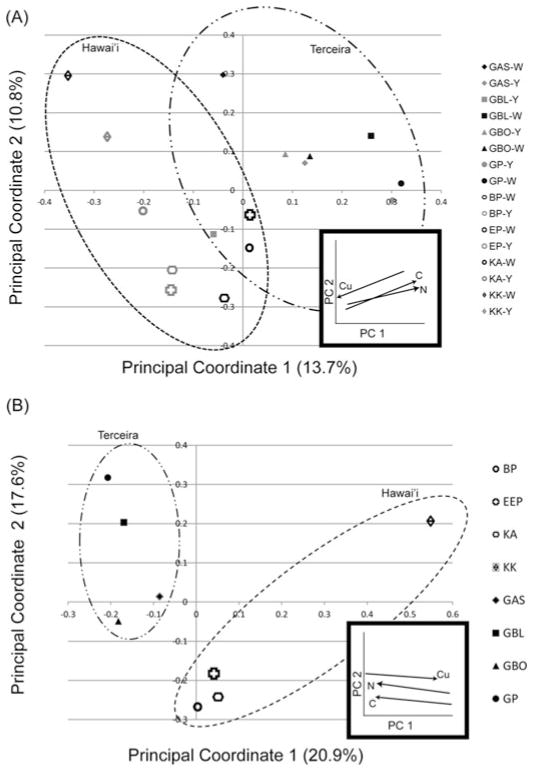
The PCoA (Figure 5A) showed a clustering along Principal Coordinate 1, which corresponds to geographical location as well as the levels of some elements in the rock composition (e.g., nitrogen, organic carbon, copper) and accounts for 13.7% of the variation. The amounts of nitrogen and organic carbon increase from left to right, that is the Terceira samples have more of these elements than Hawai’ian samples, while copper increases from right to left, as the Hawai’ian samples had more copper than those from Terceira (Figure 5A inset).
Fig. 5.
Principal Coordinate Analysis of all samples performed with the software UniFrac. Clone libraries from Azores are indicated by solid symbols, and Hawai’ian ones by open symbols. Yellow samples are indicated by dark grey, and white samples by light grey. Names of clone libraries are listed on the right. Number in parenthesis indicated percent variance explained by principal component. Insets shows general trend in elements that appear to influence the PCoA, as determined with Fast Unifrac, with the amount of the element increasing toward the arrowhead. BP: Bird Park, EP: Epperson’s Cave, KAU: Kaumana Cave, KK: Kula Kai, GBO: Gruta da Branca Opala, GAS: Gruta da Achada, GP: Gruta dos Principiantes, GBL: Gruta dos Balcões. (A) PCoA by individual samples. (B) PCoA with samples grouped by cave.
In the Hawai’ian samples Principal Coordinate 2 appears to correspond to precipitation, and accounts for 10.8% of the variation. The trend in precipitation is not as apparent for Principal Coordinate 2 in the Terceira samples. None of the other factors tested, mat color, elevation, temperature or other chemicals composition, showed clear grouping patterns. The PCoA performed on sequences grouped by cave instead of by sample showed an even greater influence of geography with clustering along Principal Coordinate 1 accounting for 20.9% of the variation. When the sequences from Hawai’i and Terceira were grouped by archipelago location, the communities were found to be statistically different using the Libshuff function of mothur (p < 0.001; Singleton et al. 2001).
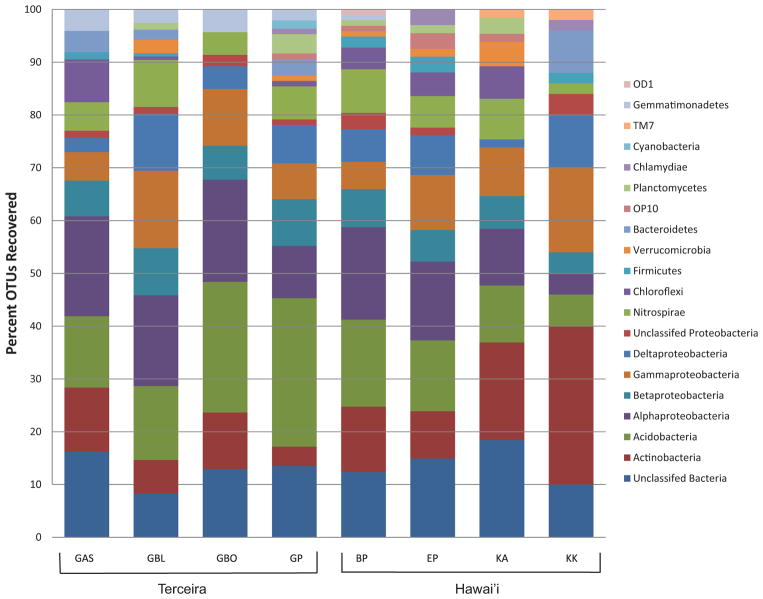
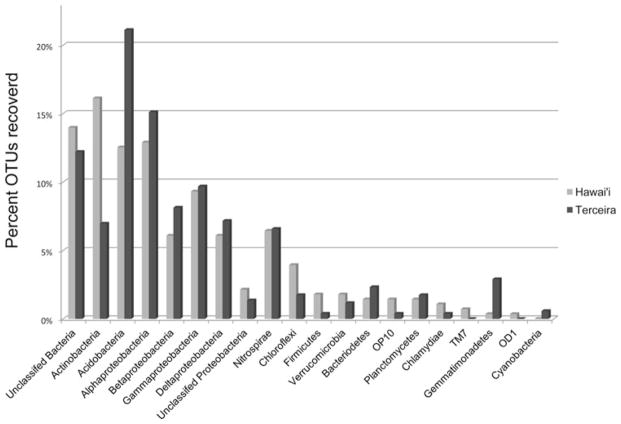
Fifteen phyla were identified across 16 clone libraries using the Ribosomal Database Project (RDP) analysis tools after OTUs were defined (Figures 6 and 7). The largest percentage of OTUs from the Hawai’ian communities were identified as Actinobacteria (16%), yet Acidobacteria were the most numerous in the Terceira communities (21%) (Figure 7). Alphapro-teobacteria represented approximately 13% of the Hawai’ian OTUs and 15% of the Azorean OTUs (Figure 7). Fourteen percent of Hawai’ian sequences and 12% of the Terceira OTUs could not be assigned to a known phylum.
Fig. 6.
Bar chart of phyla found in each lava cave based on OTUs (Operational Taxonomic Units) defined at 97% identity. BP: Bird Park, EP: Epperson’s Cave, KAU: Kaumana Cave, KK: Kula Kai, GBO: Gruta da Branca Opala, GAS: Gruta da Achada, GP: Gruta dos Principiantes, GBL: Gruta dos Balcões.
Fig. 7.
Phyla recovered from each location as a percent of the total OTUs recovered.
Other taxa comprising a substantial portion of the community from both locations include Nitrospirae, Gammaproteobacteria, Betaproteobacteria and Deltaproteobacteria (Figures 6 and 7). Phyla with less than 3% of the recovered OTUs included Cyanobacteria, Planctomycetes, Chlamydiae, Verru-comicrobia, Gemmatimonadetes, Chloroflexi, Firmicutes, Bacteroidetes, TM7, OD1, and OP10 (Figure 7).
When sequences from this study were compared to the ARB database, nearest neighbors from other caves also were found, including those from Lower Kane Cave, Wyoming (Engel et al. 2003, 2004), Mammoth Cave, Kentucky (Fowler, unpublished), Frasassi Cave, Italy (Macalady et al. 2007; Macalady et al. 2008; Vlasceanu et al. 2000), Altamira Cave, Spain (Portillo et al. 2008), Oregon Caves, Oregon (Fowler and Wade unpublished), Nullarbor, Australia (Holmes et al. 2001), and Lechuguilla Cave, New Mexico (Northup et al. 2003; Spilde et al. 2005). The sequences from this study were compared with 1495 unique sequences from the above listed caves available from GenBank to determine the overlap in diversity with other cave environments. When compared at the 97% similarity level, only three OTUs (0.22%) overlapped in all three data sets (Hawai’i, Terceira and other caves), and 19 OTUs (1.4%) overlapped between either Hawai’i or Terceira and the other caves, as compared to the 34 OTUs shared between the Terceira and Hawai’i (Figure 8B).
Fig. 8.
Venn Diagrams. (A) Comparison at 97% similarity level of sequences from terrestrial volcanic environments. (B) Comparison at 97% similarity level of sequences from this study as compared to other full length cave sequences available in Gen-bank. The total number of OTUs found in Hawai’i and Terceira is different in each case based on the length of the fragment analyzed.
Approximately seven percent of the defined OTUs had close matches in GenBank from other volcanic environments when compared using BLAST. The sequences from this study were compared to 414 sequence fragments approximately 390 bp long, from terrestrial volcanic environments in Hawai’i (Gomez-Alvarez et al. 2007), the Canary Islands (Portillo and Gonzalez 2008), and Iceland (Cockell et al. 2009; Kelly et al. 2010). There were only four OTUs (0.7%) shared by all three data sets, while 14 OTUs (2.7%) were shared between the terrestrial environments and Terceira and nine (1.7%) were shared between Hawai’i and the terrestrial environments (Figure 8A).
Discussion
Biodiversity and Similarities between Archipelagoes
It is worth noting that this study represents one of the few culture-independent studies of the diversity associated with microbial mats in lava caves (but see de los Rios et al. 2011; Garcia et al. 2009; Moya et al. 2009; Snider et al. 2009b), and one of the first to compare lava cave microbial communities that encompasses widely isolated locations and different climatic regimes (but see Northup et al. 2011; Porca et al. 2012).
Our phylogenetic survey revealed at least 15 named bacterial phyla, supporting earlier findings that lava caves, like other types of subterranean habitats, contain considerable microbial diversity (Jones et al. 2008; Northup et al. 2010; Shabarova and Pernthaler 2010). There was notable variation among caves in the phyla found, especially in the rarer phyla (Figure 6). Based on the traditional view of the way microorganisms globally distribute in space, i.e., the “everything is everywhere” hypothesis, one might expect to find similar microbial communities in caves of Hawai’i and Terceira (see Green and Bohannan 2006). At the phylum level, many of the same phyla were found in both locations, with a high percentage of recovered sequences belonging to the Actinobacteria and the Acidobacteria.
However, at the OTU level, there was only 5% overlap between the two archipelagoes. The shared OTUs belonged to Proteobacteria, Acidobacteria, Actinobacteria, Nitrosiprae and unclassified phyla, which are many of the most common phyla found in the lava caves. Moreover, sequences were more likely to be most closely related to a sequence from either the same cave or the same island than between islands. Additionally there were 254 and 179 singletons in Terceira and Hawai’i, respectively. If microbial distribution is cosmopolitan, as the “everything is everywhere” hypotheses suggests (Baas-Becking 1934), we would expect a higher percent of shared OTUs between the two archipelagoes. Although we acknowledge that our sample size for the rock chemistry is small, the lack of statistically significant differences in the chemical analysis of the lava rock between the two island leads us to consider these lava caves to be similar environments for the colonization of bacteria. This in turn suggests that we should reject the claim that it is the environment that is most determining the diversity in the lava caves. Instead, we believe that the distribution of lava cave organisms is not cosmopolitan, as we are seeing a substantial difference in the diversity in geographically distant areas of similar environments. This conclusion is partially supported by the results of Porca et al. (2012) who compared our yellow microbial mat sequences with their study of limestone caves, concluding that the microbial mat libraries in over half of their study caves were significantly different from our caves.
The limitations on using morphology as a classifying tool, along with our incomplete knowledge of the taxonomy of bacterial species are two of the biggest hindrances to determining if “everything is everywhere” (Hortal 2011). The SEM results from this study can highlight one of the challenges with the identification and classification of microorganisms. The SEM results suggest that many similar looking bacteria are found in the two remote islands. However, the DNA phylogeny shows that these similar looking bacteria are most likely different “species.”
Hortal (2011) suggests that as we learn more about the taxonomy and phylogeny, and refine the tree of life, the number of bacterial species that are considered to be cosmopolitan will decline and will support biogeographical patterns in microorganisms. The high level of novel diversity in both the Hawai’ian and Terceira lava caves, as well as the global distribution of this habitat, make lava caves a particularly good environment in which to fill in part of the phylogenetic tree with novel species, as well as to study patterns of biogeography.
Biodiversity Comparisons Between Lava Caves and Other Volcanic and Cave Environments
Out of 253 OTUs from four studies of volcanic surface environments encompassing the surface flows in Hawai’i (Gomez-Alvarez et al. 2007) and the Canary Islands (Portillo and Gonzalez 2008), and volcanic glass from Iceland (Cockell et al. 2009; Kelly et al. 2010), only 19 OTUs were shared with the 609 OTUs found in this study, of which only four were found in both Hawai’i and the Azores (Figure 8). The diversity of sequences recovered was also very different between surface and cave environments. Cyanobacteria were commonly found in the surface environments (Gomez-Alverez et al. 2007), which would be expected given their primary use of sunlight as an energy source. Cyanobacteria were only rarely found in the lava caves of Terceira, and never in Hawai’i. Cyanobacterial DNA has been found in other caves, but only rarely, and those found are assumed to be using an alternative energy pathway since there is no light in the caves (Dichosa 2008), or to be remnant DNA from recently transported surface Cyanobateria.
Firmicutes were also more commonly found in surface environments, comprising from 0.6% to 10% of the OTUs recovered (Cockell et al. 2009; Gomez-Alvarez et al. 2007; Kelly et al. 2010; Portillo and Gonzalez 2008), while less than 1% of the OTUs recovered from Hawai’i and Terceira lava caves belonged to the Firmicutes. The volcanic soil from Hawai’i had the smallest amount of recovered Firmicutes, with only 0.6% of sequences recovered belonging to the phylum, which is comparable to the amounts found within the caves in Hawai’i. None of the above studies on volcanic surface environments found the phylum Nitrospirae, which comprised 6% and 7% of the Terceira and Hawai’ian OTUs respectively. Although 7% had a closest known relative from a volcanic surface environment, the similarity at the OTU level was much smaller (Figure 7).
Patterns of Species Richness and Endemism
The Chao1 estimates of taxonomic richness show that the Hawai’ian lava caves have higher richness than the Terceira lava caves. The same trend has been reported by previous studies on snails and plants on the two archipelagos that found that Hawai’i had considerably more diversity in both groups (Borges et al. 2009; Price 2009; Whittaker et al. 2008). Hawai’i and the Azores have already been designated as a “hotspot” for bacterial/archaeal, as well as eukaryotic, diversity (Donachie et al. 2004 Myers et al. 2000).
Although macrobiotic diversity is not necessarily an indicator of microbial diversity, there are other studies that show that biodiversity patterns for microorganisms loosely follow those for macroorganisms (Fontaneto 2011; Green and Bohannan 2006 (review); Horner-Devine et al. 2007). The basis for this diversity in microorganisms; however, needs to be put in the context of environmental factors important to bacteria, such as rock chemistry and other abiotic and historical factors.
Caves have been shown to harbor high levels of endemism (Culver et al. 2000; Elliott 2007; Sharratt et al. 2000). Culver et al. (2000) found that approximately 30% of troglobionts and stygobionts in U.S. caves were endemic to a single cave. In the Azores, a majority of the troglobionts occurred in one or a few caves, and most species have single island endemism (Borges et al. 2012; Reboleira et al. 2011). The results in this study indicate that this trend can be extended to cave bacterial diversity as well. Although we acknowledge that the total diversity of bacteria has not been sampled within these caves, less than 5% of the OTUs found in lava caves occur in other caves or in other volcanic environments.
The Role of Environmental Factors
Environmental heterogeneity is currently considered as a major driver of patterns of spatial distribution of microorganisms (Green and Bohannan 2006). To address the question of what drives diversity in lava caves, we considered environmental and abiotic factors such as color, precipitation, elevation, temperature, lava rock geochemistry and geographic location (latitude/longitude).
The caves on Terceira have a temperature range of 14°–18°C, depending on the season, among the caves, while the lava caves of Hawai’i have a similar range of 14°–19°C. The temperature range within each cave can vary depending on the structural characteristics of the cave, including the presence of skylights and multiple entrances, as well as how far into the cave the temperature is measured. Abiotic factors, such as relative humidity, have been shown to influence the invertebrate communities of lava caves, with drier caves being particularly poor in diversity for instance (Howarth 1982; Martín and Oromí 1988; Sharratt et al. 2000). Cave length has also been shown to influence the number of troglobiont species, with long caves having more species (Borges et al. 2012). The role of environmental factors in our study is less pronounced.
The UniFrac PCoA showed that location, and to a lesser extent the amount of nitrogen, organic carbon, and copper were the driving component of Principal Coordinate 1, accounting for the largest part of the variability within the data (Figure 5A). The lack of statistical difference in the rock chemistry indicates that while the amounts of carbon, nitrogen, and copper are involved in determining community composition, they are not the major force at work. There is a trend along Principal Coordinate 2 in the Hawai’ian samples to correlate with precipitation. The cave with the lowest precipitation in Hawai’i, Kula Kai, grouped separately from the others with higher precipitation. The trend can be seen in both the samples individually (Figure 5A), and when the samples are grouped by cave (Figure 5B). This trend does not translate as well to the samples from Terceira. The trend could indicate that there is a cutoff in precipitation, in which a certain minimum amount of rainfall will allow for different bacteria to flourish.
None of the other factors tested produced clear groupings suggesting that geography is the largest predictor of community composition. Geography influences many environmental factors in the lava caves, notably the lava chemistry as well as the pool of potential colonizers. We acknowledge the strong influence of geography may be due to the limited number of clones sequenced per library, and the limited number of microbial and rock samples analyzed (low sampling spatial intensity). The lack of influence of essential nutrients such as phosphorous and sulfur, may suggest that there are sufficient quantities of these elements in the environment to foster diversity, but not an over abundance, which could drive alternative energy pathways.
It also may suggest that a larger data set is needed to see differences in the amounts of nutrients available in the rock. Future studies will investigate the variety of possible energy pathways that bacteria may be using in the lava caves, as well as include larger data sets in order to strengthen any statistical significance. The slightly larger, but not statistically significant, amounts of carbon and nitrogen in the lava caves of Terceira is likely due to the agricultural use of the land above the caves, one of the main drivers of biotic homogenization in the Azores (Borges et al. 2008).
The results of the PCoA also highlight the strong influence of microenvironments on the community structure. Samples from within the same cave did not necessarily have similar community structure (Figure 5A), suggesting that small differences in the chemistry of the lava or other microenvironmental changes, may strongly control the diversity found. We assumed that the lava within a cave would be similar in composition throughout the cave, but there may be subtle but important variations throughout each cave which were not detected with our analyses. Furthermore other factors such as distance from entrances may influence the community structure more strongly than the over all environmental factors shared by the cave such as precipitation or elevation.
When community composition is investigated by cave rather than by sample, the influence of location is more pronounced. It is likely that the unique habitat characteristics of each lava cave, and areas within each cave, determines the community composition, in addition to dispersal constraints. In Hawai’ian terrestrial lava flows, soil parameters and trace gas profiles of the lava unique to each site have been shown to influence the bacterial community composition (Gomez-Alvarez et al. 2007). Future studies should try to include additional analyses of the geochemical/mineralogical characteristics of the cave microbial mat habitat, as well as other abiotic factors of the lava caves. Specifically, overlying soil development and age of the original lava flow may give a better understanding of the environmental parameters that are driving community assemblage and diversity in the lava caves.
The pool of potential colonizers also would play a role in the resulting community structure in lava caves. Researchers hypothesize that bacteria may be washed into shallow lava caves, or be introduced by plant roots, which can often be seen in lava caves (Pereira et al. in press; Snider et al. 2009b; Stoner and Howarth 1981). Snider’s (2010) results suggest that the roots may be a conduit for nutrients and microorganisms from the surface. Although comparisons among soil samples above the lava caves to determine if there is substantial overlap between the community structures of the two environments was beyond the scope of this study, this inquiry should be tested in the future.
The findings of this study suggest that although the surface may be a source for continual immigration of colonizers, many immigrants die off as they are unable to adapt to the unique environments in the lava caves. The presence of Cyanobacteria in the caves from Terceira may be an indication of this, as our study only showed the presence of these bacteria, not whether they are currently alive in the caves. Other studies underway show that there is an approximately 10% overlap in OTUs in the lava caves of Lave Beds National Monument and OTUs from the overlying surface (Northup, personal communication, January 2012). Those that are able to adapt to the new environment have time to evolve as the caves offer a presumed isolation from the surface, allowing for adaptation leading to the evolution of novel taxa as suggested by the lack of close relatives for many of the cave clones. Our analysis of the overlap between Terceira and Hawai’ian lava caves OTUs with other cave bacterial sequences showed high alpha-diversity with very little overlap, meaning that bacterial communities found in each cave are different.
The lack of support for our hypothesis that color would be a predictive factor of community composition has come as somewhat of a surprise (Figure 5). Color is a very visible distinguishing factor in microbial mat appearance, but many microbial mats, especially white mats, look indistinguishable among lava caves. EDX analysis of the SEM samples was used to look for elemental differences within microbial mats of different colors. Most areas of the samples were heavily covered in microbial mats, which revealed only carbon and sometimes oxygen when analyzed, but some areas with less coverage of microbial mat were analyzed for elemental composition.
EDX analysis of these areas revealed the presence of aluminum, magnesium, and/or silica (possible clays), calcium carbonate based on the height of the calcium, carbon, and oxygen peaks, silica and oxygen, suggesting some opal, iron oxides, and rarely, titanium. Significant differences in these findings were not observed between yellow and white mats. Our prior work has shown that yellow coloration is not dependent on the presence of iron-oxides and is most likely due to pigmentation differences (unpublished data). There are several possible reasons why color does not appear to be a main predictive factor in community composition. First, none of our clone libraries were sampled in-depth enough to capture all of the biodiversity. It may be that with more sequencing, a clearer pattern may emerge.
Second, color may also be the result of metabolic processes, and differences in metabolic processes were not investigated by our analyses of phyla. Finally, color may be due to the presence of other microorganisms that were not included in this study, such as Fungi. Soil fungi such as Fusarium spp. are known to produce pigments, and occur in other cave systems (Northup et al. 1994). Although there is only minimal evidence of fungi in our SEM studies, it is likely that fungal species are present based on their occurrence in caves in general (Nieves-Rivera 2003). Additionally, Stoner and Howarth (1981) reported fungi in lava caves in Hawai’i, including Bird Park Lava Cave, one of our study caves, and preliminary results from Kaumana Cave, another of our Hawai’ian study caves (Perry, unpublished results), indicate the presence of fungi in the lava caves. Future studies will further examine the significance of mat color.
Conclusions
This study provides the first in depth comparison of microbial communities of lava caves from different archipelagos and climatic regimes and provides insight into what factors may control microbial species community composition. These results indicate that geographic location may be important in determining the composition of the bacterial communities, and that Hawai’i is more diverse than Terceira. The results also lend support to conservation of these lava caves, as novel bacteria were found throughout. The study also highlights the need for further exploration of these ecosystems as both sources of novel bacteria and as natural laboratories in which to answer larger ecological questions.
Acknowledgments
We would like to thank Dr. P. Boston, D. Medville, H. Medville, E. Davis, M. Warner, L. Fleming, and D. and B. Coons for assistance with field work in Hawai’i and A. Dapkevicius, A.R. Varela, F. Pereira, P. Cardoso, C. Gaspar and A.F. Rodrigues with field work in Terceira. For permits and access to the lava caves we thank the Cave Conservancy of Hawai’i, Hawai’i Volcanoes National Park, Os Montanheiros, and the landowners. We thank Frank Haworth, Joaquín Hortal, Andrea Porras-Alfaro, Cristina Riquelme Gabriel and Robert Sinsabaugh for conversations and comments on previous drafts that greatly improved this manuscript.
Funding
This work would not have been possible without the generous funding provided by Fundação para a Ciência e Tecnologia (FCT, Portugal), under the project PTDC/AMB/70801/2006 (Maria de Lurdes N. E. Dapkevicius, P.I.) and the Undergraduate Opportunities Program, Museum of Southwestern Biology (NSF-DEB 0731350, Joseph Cook, P.I.). This work was also supported by the Cave Conservancy of the Virginias, the Graduate Research Allocation Committee at UNM Biology, UNM Biology Grove Scholarship, the Student Research Allocation Committee at UNM, the National Speleological Society, the New Mexico Space Grant Consortium, the New Mexico Alliance for Minority Participation Program, the New Mexico Geological Society, and Kenneth Ingham Consulting. We acknowledge support from the UNM Molecular Biology Facility, which is supported by NIH grant number P20GM103452.
Footnotes
Color versions of one or more of the figures in the article can be found online at www.tandfonline.com/ugmb.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas-Becking LGM. Geologie of Inleiding Tot de Milieukunde. The Hague: Stockum and Zoon; 1934. [Google Scholar]
- Barton HA. Introduction to cave microbiology: A review for the non-specialist. J Cave Karst Stud. 2006;68:43–54. [Google Scholar]
- Bell T, Ager D, Song J-I, Newman JA, Thompson IP, Lilley AK, van der Gast CJ. Larger islands house more bacterial taxa. Science. 2005;308:1884. doi: 10.1126/science.1111318. [DOI] [PubMed] [Google Scholar]
- Bent SJ, Forney LJ. The tradedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2008;2:689–695. doi: 10.1038/ismej.2008.44. [DOI] [PubMed] [Google Scholar]
- Borges PAV, Cardoso P, Amorim IR, Pereira F, Constância JP, Nunes JC, Barcelos P, Costa A, Gabriel R, Dapkevicius ML. Volcanic caves: priorities for conserving the Azorean endemic troglobiont species. Int J Speleol. 2012;41:101–112. [Google Scholar]
- Borges PAV, Amorim IR, Cunha R, Gabriel R, Martins AF, Silva L, Costa A, Vieira V. Azores. In: Gillespie R, Clagu D, editors. Encyclopedia of Islands. Berkeley, California: University of California Press; 2009. pp. 70–75. [Google Scholar]
- Borges PAV, Ugland KI, Dinis FO, Gaspar C. Insect and spider rarity in an oceanic island (Terceira, Azores): true rare and pseudo-rare species. In: Fattorini S, editor. Insect Ecology and Conservation. Kerala, India: Research Signpost; 2008. pp. 47–70. [Google Scholar]
- Boston PJ. Location, location, location! Lava caves on Mars for habitat, resources, and the search for life. J Cosmol. 2010;12:3957–3979. [Google Scholar]
- Boston PJ, Ivanov MV, McKay CP. On the possibility of chemosynthetic ecosystems in subsurface habitats on Mars. Icarus. 1992;95:300–308. doi: 10.1016/0019-1035(92)90045-9. [DOI] [PubMed] [Google Scholar]
- Boston PJ, Spilde MN, Northup DE, Melim LA, Soroka DS, Kleina LG, Lavoie KH, Hose LD, Mallory LM, Dahm CN, Crossey LJ, Scelble RT. Cave biosignature suites: Microbes, minerals, and Mars. Astrobiology. 2001;1:25–55. doi: 10.1089/153110701750137413. [DOI] [PubMed] [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- Christman MC, Culver DC, Madden MK, White D. Patterns of endemism of the eastern North American cave fauna. J Biogeogr. 2005;32:1441–1452. [Google Scholar]
- Cockell CS, Olsson-Francis K, Herrera A, Meunier A. Alteration textures in terrestrial volcanic glass and the associated bacterial community. Geobiology. 2009;7:50–65. doi: 10.1111/j.1472-4669.2008.00184.x. [DOI] [PubMed] [Google Scholar]
- Culver DC, Master LL, Christman MC, Hobbs HH. Obligate Cave Fauna of the 48 contiguous United States. Conserv Biol. 2000;14(2):386–401. [Google Scholar]
- Culver DC, Pipan T. The Biology of Caves and other Subterranean Habitats. Oxford, UK: Oxford University Press; 2009. [Google Scholar]
- de los Ríos A, Bustillo MA, Ascaso C, Carvalho MR. Bioconstructions in ochreous speleothems from lava tubes on Terceira Island (Azores) Sediment Geol. 2011;236:117–128. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucl Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichosa A. PhD dissertation. University of New Mexico; Albuquerque, NM: 2008. Biogenicity and Microbial Community Composition of Desert Varnish and Cave Ferromanganese Deposits. [Google Scholar]
- Donachie SP, Hou S, Lee KS, Riley CW, Pikina A, Belisle C, Kempe S, Gregory TS, Bossuyt A, Boerema J, Liu J, Freitas TA, Malahoff A, Alam M. The Hawai’ian Archipelago: A microbial diversity hotspot. Microb Ecol. 2004;48:509–520. doi: 10.1007/s00248-004-0217-1. [DOI] [PubMed] [Google Scholar]
- Elliott WR. Zoogeography and biodiversity of Missouri caves and karst. J Cave Karst Stud. 2007;69:135–162. [Google Scholar]
- Engel AS, Lee N, Porter ML, Stern LA, Bennett PC, Wagner M. Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl Environ Microbiol. 2003;69:5503–5511. doi: 10.1128/AEM.69.9.5503-5511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AS, Porter ML, Stern LA, Quinlan S, Bennett PC. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria. FEMS Microbiol Ecol. 2004;51:31–53. doi: 10.1016/j.femsec.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Fontaneto D. Biogeography of microorganisms. Is everything small everywhere? Cambridge UK: Cambridge University Press; 2011. [Google Scholar]
- Fontaneto D, Hortal J. Microbial biogeography: is everything small everywhere? In: Ogilvie LA, Hirschl PR, editors. Microbial Ecological Theory: Current Perspectives. Norwich, UK: Horizon Scientific Press; 2012. p. 8798. [Google Scholar]
- Garcia MG, Moya M, Spilde MN, Stone FD, Northup DE. Discovering new diversity in Hawai’ian lava tube microbial mats. Proc Int Congr Speleol. 2009;1:364–369. [Google Scholar]
- Giambelluca TW, Chen Q, Frazier AG, Price JP, Chen Y-L, Chu P-S, Eischeid JK, Delparte DM. Online rainfall atlas of Hawai’i. Bull Amer Meteor Soc. 2012;94:313–316. [Google Scholar]
- Giovannoni SJ, DeLong EF, Schmidt TM, Pace NR. Tangential flow filtration and preliminary phylogenetic analysis of marine pi-coplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V, King GM, Nusslein K. Comparative bacterial diversity in recent Hawai’ian volcanic deposits of different ages. FEMS Microbiol Ecol. 2007;60:60–73. doi: 10.1111/j.1574-6941.2006.00253.x. [DOI] [PubMed] [Google Scholar]
- Green J, Bohannan BJM. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitation high-throughput phylogenetic analysis of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D, Horwath WR, van Kessell C. Acid fumigation of soils to remove carbonates prior to total organic carbon or carbon-13 isotopic analysis. Soil Sci Soc Am J. 2001;65:1853–1856. [Google Scholar]
- Holmes AJ, Tujula NA, Holley M, Contos A, James JM, Rogers P. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ Microbiol. 2001;3:256–264. doi: 10.1046/j.1462-2920.2001.00187.x. [DOI] [PubMed] [Google Scholar]
- Horner-Divine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA, Green JL, Kuske CR, Martiny JBH, Muyzer G, Overas L, Reysenbach AL, Smith VH. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology. 2007;88:1345–1353. doi: 10.1890/06-0286. [DOI] [PubMed] [Google Scholar]
- Hortal J. Geographical variation in the diversity of microbial communities: Research directions and prospects for experimental biogeography. In: Fontaneto D, editor. Biogeography of Microscopic Organisms; Is Everything Small Everywhere? Cambridge, UK: Systematics Association and Cambridge University Press; 2011. pp. 335–358. [Google Scholar]
- Howarth FG. Bioclimatic and geologic factors governing the evolution and distribution of Hawai’ian cave insects. Entomol Gen. 1982;8:17–26. [Google Scholar]
- Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:0003.0001–0003.0008. doi: 10.1186/gb-2002-3-2-reviews0003. reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. Counting the Uncountable: Statistical approaches to estimating microbial diversity. Appl Environ Micro. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DS, Lyon EH, Macalady JL. Geomicrobiology of biover-miculations from the Frasassi cave system, Italy. J Cave Karst Stud. 2008;70:78–93. [Google Scholar]
- Kelly LC, Cockell CS, Piceno YM, Andersen GL, Thorsteinsson T, Marteinsson V. Bacterial diversity of weathered terrestrial Icelandic volcanic glass. Microbiol Ecol. 2010;60:740–752. doi: 10.1007/s00248-010-9684-8. [DOI] [PubMed] [Google Scholar]
- King GM. Chemolithotrohic bacteria: Distributions, functions and significance in volcanic environments. Microbes Environ. 2007;22:309–319. [Google Scholar]
- Léveillé RJ, Datta S. Lava tubes and basaltic caves as astrobiological targets on Earth and Mars: a review. Planet Space Sci. 2010;58:592–598. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: A software environment for sequence data. Nucl Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalady JL, Dattagupta S, Scaperdoth I, Jones DS, Druschel GK, Eastman D. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2008;2008:590–601. doi: 10.1038/ismej.2008.25. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Jones DS, Lyon EH. Extremely acidic, pendulous cave wall biofilms from the Frasassi cave system, Italy. Environ Microbiol. 2007;9:1402–1414. doi: 10.1111/j.1462-2920.2007.01256.x. [DOI] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Farris RJ, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucl Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín JL, Oromí P. Two new species of Anataelia Bol. Dermaptera Pygidicranidae from caves and recent lava flows of Hierro and La Palma Canary Islands Spain. Mem Biospeologie. 1988;15:49–60. [Google Scholar]
- Melim LA, Northup DE, Spilde MN, Jones B, Boston PJ, Bixby RJ. Reticulated filaments in cave pool speleothems: Microbe or mineral? J Cave Karst Stud. 2008;70:135–141. [Google Scholar]
- Moore GW, Sullivan N. Speleology: Caves and the Cave Environment. St. Louis, MO: Cave Books; 1997. [Google Scholar]
- Moya M, Garcia MG, Spilde MN, Northup DE. Composition of Bacterial mats in El Malpias, National Monument, New Mexico, USA: comparison and contrasts with Bacterial communities in Hawai’i lava tubes. Proc Int Congr Speleol. 2009;2:709–713. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kents J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:856–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nieves-Rivera AM. Mycological survey of Río Camuy Caves Park, Puerto Rico. J Cave Karst Stud. 2003;65:23–28. [Google Scholar]
- Northup DE, Barns SM, Yu LE, Spilde MN, Schelble RT, Dano KE, Crossey LJ, Connolly CA, Boston PJ, Natvig DO, Dahm CN. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ Microbiol. 2003;5:1071–1086. doi: 10.1046/j.1462-2920.2003.00500.x. [DOI] [PubMed] [Google Scholar]
- Northup DE, Carr DL, Crocker MT, Cunningham KI, Hawkins LK, Leonard P, Welbourn WC. Biological investigations in Lechuguilla Cave. NSS Bull. 1994;56:54–63. [Google Scholar]
- Northup DE, Connolly CA, Trent A, Peck VM, Spilde MN, Welbourn WC, Natvig DO. The nature of Bacterial communities in Four Windows Cave, El Malpais National Monument, New Mexico, USA. AMCS Bull. 2008;19:119–125. [Google Scholar]
- Northup DE, Lavoie KH. Geomicrobiology of caves: a review. Geomicrobiol J. 2001;18:199–222. [Google Scholar]
- Northup DE, Melim LA, Spilde MN, Hathaway JJM, Garcia MG, Moya M, Stone FD, Boston PJ, Dapkevicius MLNE, Riquelme C. Lava Cave microbial communities within mats and secondary mineral deposits: Implications for life detection on other planets. Astrobiology. 2011;12(7):601–618. doi: 10.1089/ast.2010.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northup DE, Snider JR, Spilde MN, Porter ML, Van de Kamp JL, Boston PJ, Nyberg AM, Bargar JR. Diversity of rock varnish Bacterial communities from Black Canyon, New, Mexico. J Geophys Res. 2010;115:G02007. doi: 10.1029/2009JG001107. [DOI] [Google Scholar]
- Northup DE, Welbourn WC. Life in the twilight zone: Lava tube ecology. New Mex Bur Mine Miner Resour Bull. 1997;156:69–82. [Google Scholar]
- Palmer AN. Cave Geology. Dayton, OH: Cave Books; 2007. [Google Scholar]
- Pella E. Elemental organic analysis. Part 1. Historical developments. Am Lab. 1990a;22:116–125. [Google Scholar]
- Pella E. Elemental organic analysis. Part 2. State of the Art. Am Lab. 1990b;22:28–32. [Google Scholar]
- Pereira F, Borges PAV, Costa MP, Constância JP, Nunes JC, Barcelos P, Braga T, Gabriel R, Amorim IR. Catálogo das cavidades vulcânicas dos Açores (grutas lávicas, algares e grutas de erosão marinha) Direcção Regional do Ambiente, Horta. :286. in press. [Google Scholar]
- Popa R, Smith AR, Popa R, Boone H, Fisk M. Olivine-Respiring Bacteria Isolated from the Rock-Ice Interface in a Lava-Tube Cave, a Mars Analog Environment. Astrobiology. 2012;12:9–18. doi: 10.1089/ast.2011.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porca E, Jurado V, ŹGur-Bertok D, Saiz-Jimenez C, Pašić L. Comparative analysis of yellow microbial communities growing on the walls of geographically distinct caves indicates a common core of microorganisms involved in their formation. FEMS Microbiol Ecol. 2012;81:255–266. doi: 10.1111/j.1574-6941.2012.01383.x. [DOI] [PubMed] [Google Scholar]
- Portillo MC, Gonzalez JM, Saiz-Jimenez C. Metabolically active microbial communities of yellow and grey colonizations on the walls of Altamira Cave, Spain. J Appl Microbiol. 2008;104:681–691. doi: 10.1111/j.1365-2672.2007.03594.x. [DOI] [PubMed] [Google Scholar]
- Portillo MC, Gonzalez JM. Microbial communites and immigration in volcanic environments of Canary Islands (Spain) Naturwis-senschaften. 2008;95:307–315. doi: 10.1007/s00114-007-0330-3. [DOI] [PubMed] [Google Scholar]
- Poulson TL, White WB. The cave environment. Science. 1969;165:971–981. doi: 10.1126/science.165.3897.971. [DOI] [PubMed] [Google Scholar]
- Price J. Hawai’ian Islands, Biology. In: Gillespie R, Clagu D, editors. Encyclopedia of Islands. Berkeley, California: University of California Press; 2009. pp. 397–404. [Google Scholar]
- Reboleira ASPS, Borges PAV, Gonçalves F, Serrano ARM, Oromí P. The subterranean fauna of a biodiversity hotspot region—Portugal: An overview and its conservation. Int J Speleol. 2011;40:23–37. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: Open Source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabarova T, Pernthaler J. Karst pools in subsurface environments: collectors of microbial diversity or temporary residence between habitat types. Environ Microbiol. 2010;12:1061–1074. doi: 10.1111/j.1462-2920.2009.02151.x. [DOI] [PubMed] [Google Scholar]
- Sharratt NJ, Picker MD, Samways MJ. The invertebrate fauna of the sandstone caves of the Cape Peninsula (South Africa): patterns of endemism and conservation priorities. Biodivers Conserv. 2000;9:107–143. [Google Scholar]
- Simon KS, Pipan T, Culver DC. Conceptual model of the flow and distribution of organic carcon in caves. J Karst Stud. 2007;69:279–284. [Google Scholar]
- Singleton DR, Furlong MA, Rathbun SL, Whitman WB. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol. 2001;67:4374–4376. doi: 10.1128/AEM.67.9.4374-4376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider JR. Master’s Thesis. University of New Mexico; Albuquerque, NM: 2010. Comparison of Microbial Communities on Roots, Ceilings and Floors of Two Lava Tubes Caves in New Mexico. [Google Scholar]
- Snider JR, Goin C, Miller RV, Boston PJ, Northup DE. Ultraviolet radiation sensitivity in cave bacteria: Evidence of adaptation to the subsurface? Int J Speleol. 2009a;38:1–12. [Google Scholar]
- Snider JR, Moya M, Garcia MG, Spilde MN, Northup DE. Identification of the microbial communities associated with roots in lava tubes in New Mexico and Hawai’i. Proc Int Congr Speleol. 2009b;2:718–723. [Google Scholar]
- Spilde MN, Northup DE, Boston PJ, Schelble RT, Dano KE, Crossey LJ, Dahm CN. Geomicrobiology of cave ferromanganese deposits: A field and laboratory investigation. Geomicrobiol J. 2005;22:99–116. [Google Scholar]
- Staley JT, Crawford R. The biologist’s chamber: Lava tube slime. Cascade Caver. 1975;14:20–21. [Google Scholar]
- Stoner MF, Howarth FG. Community structure and niche differentiation in Hawai’ian lava tubes. In: Muller Dombois D, Bridges KW, Carson HL, editors. Island Ecosystems: Biological organization in selected Hawai’ian communities. Stroudsburg, Pennsylvania: Hutchinson Ross Publ. Co; 1981. pp. 318–336. [Google Scholar]
- Stott MB, Crowe MA, Mountain BW, Smirnova AV, Hou SB, Alam M, Dunfield PF. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ Microbiol. 2008;10:2030–2041. doi: 10.1111/j.1462-2920.2008.01621.x. [DOI] [PubMed] [Google Scholar]
- Tetu SG, Breakwell K, Elbourne LDH, Holmes AJ, Gillings MR, Paulsen IT. Life in the dark: metagenomic evidence that a microbial slime community is driven by inorganic nitrogen metabolism. ISME J. 2013;7:1227–1236. doi: 10.1038/ismej.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasceanu L, Sarbu SM, Engel AS, Kinkle BK. Acidic cave-wall biofilms located in the Frasassi Gorge, Italy. Geomicrobiol J. 2000;17:125–139. [Google Scholar]
- Whittaker RJ, Triantis KA, Ladle RJ. A general dynamic theory of oceanic island biogeography. J Biogeogr. 2008;35:977–994. [Google Scholar]