Abstract
Background
The addition of sub therapeutic doses of antibiotics to cattle feed for growth promotion is a contributory factor to antibiotic resistance, thus an alternative to antibiotics is needed in animal feed additives.
Objective
To determine the antimicrobial activity of cow's intestinal Lactic acid bacteria (LAB) against enteric commensals.
Method
Escherichia coli, Klebsiella species (spp) and LAB were isolated from thirty different cow faecal samples and the LAB identified by partial sequencing of 16S rRNA. The antimicrobial activity of the LAB was determined against the test Escherichia coli and Klebsiella spp.
Results
Five species of LAB were isolated from thirty cow faecal samples and identified as Enterococcus hirae (8), Enterococcus durans (6), Enterococcus faecium (1), Enterococcus faecalis (1) and Weissella confusa (1). Viable cells and cell free supernatant (CFS) of the LAB were able to inhibit the growth of the test organisms with the largest zone of inhibition by the viable cells being 26mm against Escherichia coli CB6 produced by Enterococcus hirae CO6A while Weissella confusa CO29M and Enterococcus hirae CO2A produced the largest zones of inhibition (26mm) against Klebsiella CB2.
Conclusion
This study shows that LAB from cow faeces possess considerable antimicrobial activity against resistant Escherichia coli from the same environment.
Keywords: Antibacterial activities, lactic acid bacteria, cow faeces, enteric pathogens
Introduction
Livestock sustain the livelihood of millions of people in the world in both developing and developed countries1. The meat from cattle, goat, sheep, pig and poultry including the offals are the main sources of daily per capita consumption of animal protein2 while beef is the major source of animal protein supply among most Nigerian rural households and cities. The demand for animal protein far exceeds the supply. In order to meet the increasing demand for animal protein, management of cattle usually involves the addition of antibiotics to the cattle feeds and water for growth promotion, prophylaxis, metaphylaxis and therapy. Since the discovery and development of the first antibiotics prior to the second world war, these drugs have played an important role in curing disease in humans and animals. In 1946 experiments showed that low, sub therapeutic levels of antibiotics could increase feed efficiency and growth in food animals, and the addition of various antibiotics to feed for livestock was initiated3. The prevention of disease transmission and enhancement of growth and feed efficiency are critical in modern animal husbandry, and there has been widespread incorporation of antibiotics into animal feeds in many countries4. The use of antibiotics in such practices is the principal contributing factor to the emergence and dissemination of antimicrobial resistance among bacterial pathogens and commensals that have food animal reservoirs5.
Currently, the potential for agricultural antibiotics to contribute to the development of antibiotic-resistant bacteria of human concern is the subject of intense debate and research6,7. Antibiotics are now forbidden in animal feeds in the European Union countries and it is expected that this may be extended to Nigeria and other developing countries in the nearest future, hence there is an urgent need for an effective alternative to the use of antibiotics as animal feed additives.
Probiotics are live microorganisms which when administered in adequate amounts confer a health benefit on the host8. Bacteria proposed for probiotic uses are usually categorised as lactic acid bacteria, commonly used bacteria include various species of Lactobacillus, Bifidobacterium and Streptococcus as well as some Enterococcus species9. There is now increasing evidence that selected probiotic strains can provide health benefits to their human and animal hosts and it is noteworthy that the Food and Agriculture Organization of the United Nations and the World Health Organization have stated that there is adequate scientific evidence to indicate that there is potential for probiotics to provide health benefits. An alternative and effective approach to antibiotic administration to livestock is the use of probiotics, which can help to improve gut microbial balance and therefore the natural defence of the animal against pathogenic bacteria10.
Therefore the objective of this study was to isolate, characterize and identify lactic acid bacteria from cow intestines as well as test their antimicrobial activity against potential pathogens from the same habitat, these potential probiotics organisms may serve as biotherapeutic agents and a useful alternative to the use of antibiotics in animal feed supplements.
Materials and methods
Bacterial isolation
Thirty cow faecal samples were randomly collected from apparently healthy cows in Ibadan, Oyo state Nigeria over a period of three months (between May and July, 2012) using sterile universal bottles. Microbiological analysis was carried out within one hour of sample collection. One gram of the cow faeces was added to 9ml of sterile peptone water serving as the diluent and homogenized with a vortex mixer. Ten fold dilution was carried out, 1 ml each from dilutions of × 10−6 of the homogenized mixture of each sample was inoculated into:
MRS agar (Oxoid, UK) by pour plate method and incubated at 37°C for 48h under microaerophilic condition using CampyGen™ (Oxoid, UK).
MacConkey agar by pour plate method and incubated at 37°C under aerobic condition, for the determination of Escherichia coli and Klebsiella spp.
For the determination of lactic acid bacteria, colonies were picked according to differences in their morphology on MRS agar plates, all strains under examination were tested for Grams staining reaction and catalase production. Only Gram positive and catalase-negative isolates were selected. Pure cultures presumed to be lactic acid bacteria were kept for long term storage in eppendorf tubes containing MRS broth with 50% glycerol at −20°C.
For the isolation of the test E.coli and Klebsiella spp used as indicator for the determination of the antimicrobial activity of the lactic acid bacteria, colonies with characteristic pink colour on MacConkey agar after 24hrs of incubation at 37°C were further cultured in Eosin methylene blue agar, only colonies with characteristic metallic sheen that were indole positive, methyl red positive, Voges-proskauer negative and citrate negative were selected as E. coli while organisms that were indole negative, methyl red negative and citrate positive were selected as Klebsiella spp.
The antibiotic susceptibility of the E. coli was determined (result not shown), the test strain was found to be resistant to cefotaxime, ceftazidine and ceftriaxone in addition to erythromycin and clindamycin but sensitive to amoxicillin/clavulanic acid, cefoxitin, amoxicillin, aztreonam and imipenem.
Viable count of lactic acid bacteria.
After incubation under microaerophilic condition at 37°C for 48h, colonies were counted based on differences in their morphology on MRS agar plates to determine the colony forming unit of lactic acid bacteria per gram of the cow faeces.
Identification of the LAB isolates by sequencing of 16S rRNA gene.
Genetic characterization of the LAB isolates was performed by Polymerase Chain Reaction targeted to the 16S rRNA gene using the universal set of primers, forward primer: BSF8N 5′-AGAGTTTGATCMTGGCTCAG-′3 and reverse primer: BSR534 5′-ATTACCGCGGCTGCTGGC-′3. The amplification of 16S rRNA region of isolates was performed using the following conditions, 10 min of initial denaturation at 95°C, 45 cycles of annealing at 95°C for 15s, 30s at 55°C, 30s at 72°C, a single 7-minute extension at 72°C and finally set on hold at 4 °C. The PCR products were analysed on 1% agarose gel in TAE buffer containing ethidium bromide and visualized under UV light.
The PCR products were purified and sequenced for the identification of the isolates with the primer BSF8N 5′-AGAGTTTGATCMTGGCTCAG-′3 using the Sanger single read sequencing with Applied Biosystems 3730xl DNA sequencer at GATC biotech Germany. The sequences were compared with others present in GenBank database using the Ez Taxon Server version 2.1.11.
Determination of the antimicrobial activity of the lactic acid bacteria.
The agar overlay method of 12 as modified by 13 was employed to determine the ability of the viable lactic acid bacteria strains to inhibit the growth of the indicator pathogens, E. coli CB6 and Klebsiella CB2 isolated from the same habitat as the lactic acid bacteria. A loopful of LAB in MRS broth was inoculated on MRS agar plate as a thick line of about 2mm and about 30mm long at a good distance away from the edge of the plates and incubated under microaerophilic condition at 37°C for 24h. After incubation, the MRS agar plates were overlaid with approximately 0.2ml × 107 CFU/ml of an overnight broth culture of the test pathogens inoculated in 10ml of Mueller Hinton soft agar (with 0.7% agar-agar).The overlay was allowed to set, and incubated at 37°C under aerobic condition. The plates were then examined for clear zone of inhibition around the line of the LAB and the clear zones were measured.
Determination of the production of bacteriocin-like inhibitory substance by the lactic acid bacteria strains.
The lactic acid bacteria that produced clear zones of inhibition were grown in MRS broth overnight under microaerophilic condition at 37°C for 24h to determine their ability to produce bacteriocin-like inhibitory substances. The LAB cultures were centrifuged at 12,000 X g for 10 mins at 4°C and the supernatant was decanted into sterile test tubes. The antimicrobial activity of the cell free supernatant was determined twice (i.e before and after neutralization to pH 6.5 with 1M NaOH) using the agar well diffusion assay. 50µl of the supernatant was placed in 6mm wells cut with a cork borer into cooled Mueller Hinton agar plates inoculated with 0.2ml ×107 CFU/ml of the test pathogens, the supernatant was allowed to diffuse by leaving the plates at room temperature for 1h before incubation at 37°C for 24h.
Results
Seventeen lactic acid bacteria were isolated from thirty faecal samples collected from different healthy cows, for the purpose of determination of their antagonistic activities against Escherichia coli and Klebsiella spp isolated from the same environment.
The colony forming unit per gram of the presumed LAB was carried out on the faecal samples with the level of lactic acid bacteria in cow faeces ranging from 3.8 ×107 to 1.0 × 108 cfu/g has shown in table 1.
Table 1.
Colony forming unit/gram of lactic acid bacteria in cow faecal samples.
| Sample code | CFU/gram |
| Sample 1M | 6.2 × 107 |
| Sample 2A | 5.5 × 107 |
| Sample 4M | 4.4 × 107 |
| Sample 6A | 8.0 × 107 |
| Sample 10M | 4.2 × 107 |
| Sample 12A | 5.0 × 107 |
| Sample 13M | 4.5 × 107 |
| Sample 17A | 6.0 × 107 |
| Sample 18A | 2.5 × 107 |
| Sample 19M | 1.0 × 108 |
| Sample 20A | 4.1 × 107 |
| Sample 21M | 6.9 × 107 |
| Sample 26A | 6.3 × 107 |
| Sample 27M | 9.6 × 107 |
| Sample 28M | 7.1 × 107 |
| Sample 29M | 5.0 ×107 |
| Sample 30A | 3.8 ×107 |
The isolates were presumptively identified as lactic acid bacteria by morphological characteristic, Grams reaction and catalase test. Only isolates with small circular morphology, cream to off white colours, raised, smooth with entire margin colonies, which were also positive for Grams reaction and were catalase negative (table not shown) were selected for further investigation. The isolates were identified molecularly by sequencing the 16S rRNA genes. Only 17 isolates had their 16SrRNA genes amplified as confirmed by agarose gel electrophoresis after polymerase chain reaction.
Five species of lactic acid bacteria were isolated and identified as Enterococcus hirae, Enterococcus durans, Enterococcus faecium, Enterococcus faecalis and Weissella confusa with their percentage homology as shown in table 2.
Table 2.
Identification of the isolated LAB by 16S rRNA genes showing percentage similarities
| Lactic acid bacteria | % Accuracy |
| Enterococcus hirae CO1M | 99.797 |
| Enterococcus hirae CO2A | 100 |
| Enterococcus hirae CO4M | 100 |
| Enterococcus hirae CO6A | 99.770 |
| Enterococcus durans CO10M | 99.751 |
| Enterococcus durans CO12A | 99.748 |
| Enterococcus hirae CO13M | 100 |
| Enterococcus durans CO17A | 99.574 |
| Enterococcus durans CO18A | 97.546 |
| Enterococcus hirae CO19M | 99.309 |
| Enterococcus faecium CO20A | 97.026 |
| Enterococcus faecalis CO21M | 100 |
| Enterococcus hirae CO26A | 100 |
| Enterococcus hirae CO27M | 93.469 |
| Enterococcus durans CO28M | 99.638 |
| Weissella confusa CO29M | 100 |
| Enterococcus durans CO30A | 100 |
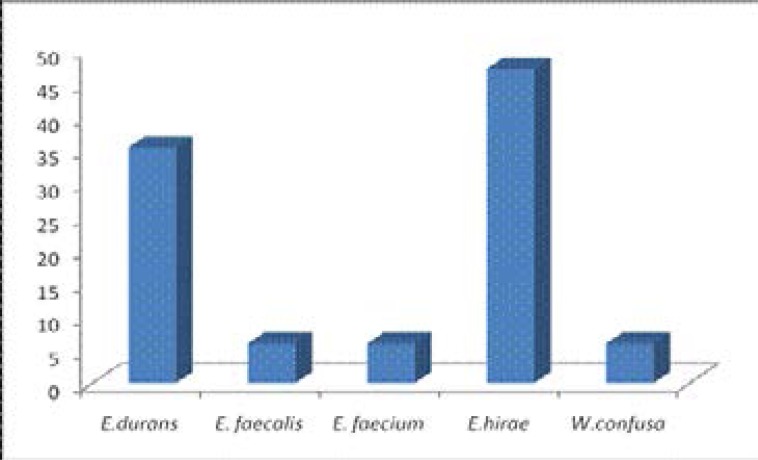
Sixteen (16) isolates (94.12%) belong to the genus Enterococcus while one (1) isolate (5.88%) belong to Weissella. The organism with the highest frequency of occurrence among the Enterococcus species is Enterococcus hirae with 50% occurrence and 47.06% among the total isolates, Enterococcus faecalis and Enterococcus faecium both had the lowest frequency of occurrence of 6.25% each among the Enterococcus spp and 5.88% among the total isolates, the same for Weissella confusa. Enterococcus durans had 37.5% occurrence among the Enterococcus spp and 35.29% among the total isolates as seen in fig 1.
Fig 1.
Percentage occurrence of lactic acid bacteria isolated from cow faeces
The viable LAB isolates were tested for their antimicrobial activity against the indicator pathogens, E. coli CB6 and Klebsiella CB2 using the agar overlay method. The diameter of the zones of inhibition (table 3) showed that all of the isolates have varying antibacterial activity against the test pathogens.
Table 3.
Inhibition of cow enteric E.coli and Klebsiella spp by viable LAB isolated from cow faeces. (Radius of zone of inhibition measured in mm)
| Isolated Lactic Acid Bacteria | E.coli CB6 | Klebsiella CB2 |
| Enterococcus hirae CO1M | 10 | 12 |
| Enterococcus hirae CO2A | 11 | 13 |
| Enterococcus hirae CO4M | 12 | 9 |
| Enterococcus hirae CO6A | 13 | 10 |
| Enterococcus durans CO10M | 10 | 11 |
| Enterococcus durans CO12A | 11 | 9 |
| Enterococcus hirae CO13M | 10 | 8 |
| Enterococcus durans CO17A | 12 | 9 |
| Enterococcus durans CO18A | 10 | 7 |
| Enterococcus hirae CO19M | 8 | 6 |
| Enterococcus faecium CO20A | 9 | 9 |
| Enterococcus faecalis CO21M | 10 | 12 |
| Enterococcus hirae CO26A | 9 | 8 |
| Enterococcus hirae CO27M | 12 |
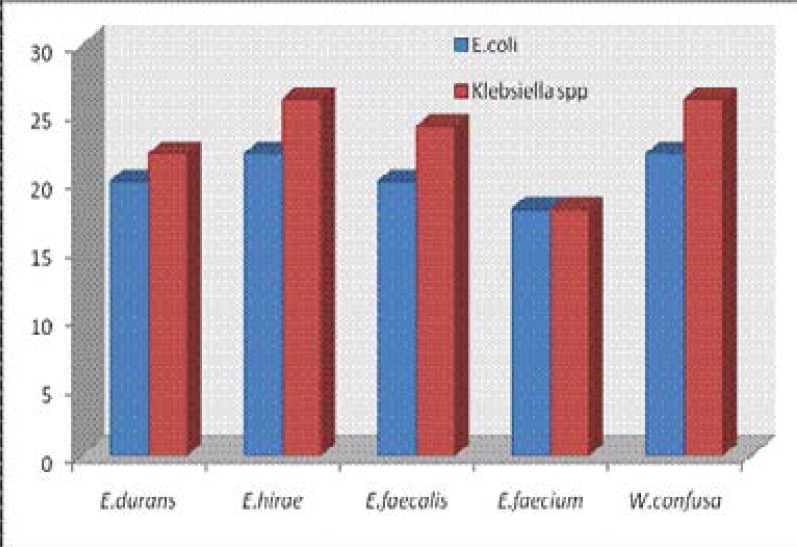
Enterococcus hirae CO6M showed the highest activity (26 mm) against E. coli CB6 while Weisella confusa CO29M and E.hirae CO2M both showed the highest activity (26mm) against Klebsiella CB2. The highest zones of inhibition by LAB against the test organisms is shown in fig 2. The lowest antibacterial activity against E.coli CB6 (10mm) and Klebsiella CB2 (14mm) was shown by E. durans CO30A.
Fig 2.
Highest zones of inhibition by LAB against the test organisms
The cell free supernatant of the LAB were also tested against E.coli CB6 and Klebsiella CB2 isolated from cow intestine and Staphylococcus aureus ES5 of clinical origin, the result of which is shown in table 4. The cell free supernatant from all the isolates had antagonistic activity against the test E.coli. The cell free metabolites of E.durans CO2IA and E.hirae CO19M showed no activity against Klebsiella spp. Only E.hirae CO6A produced no activity against Staph. aureus used in the test. After the neutralization of the cell free supernatant, no obvious antimicrobial activity was observed against the test organisms.
Table 4.
Inhibition of the test pathogens by cell free supernatant of the isolated LAB strains. (Diameter of zone of inhibition measured in mm)
| Isolated Lactic Acid Bacteria | E.coli CB6 |
Klebsiella CB2 |
Staphylococcus aureus ES5 |
| Enterococcus hirae CO1M | 8mm | 7mm | 8mm |
| Enterococcus hirae CO2A | 10mm | 11mm | 10mm |
| Enterococcus hirae CO4M | 11mm | 9mm | 8mm |
| Enterococcus hirae CO6A | 10mm | 8mm | Nil |
| Enterococcus durans CO10M | 8mm | 9mm | 8mm |
| Enterococcus durans CO12A | 7mm | nil | 8mm |
| Enterococcus hirae CO13M | 9mm | 8mm | 7mm |
| Enterococcus durans CO17A | 9mm | 8mm | 7mm |
| Enterococcus durans CO18A | 8mm | 8mm | 7mm |
| Enterococcus hirae CO19M | Nil | nil | 8mm |
| Enterococcus faecium CO20A | 10 | 8mm | 10mm |
| Enterococcus faecalis CO21M | 9mm | 10mm | 8mm |
| Enterococcus hirae CO26A | 9mm | 10mm | 9mm |
| Enterococcus hirae CO27M | 11mm | 9mm | 10mm |
| Enterococcus durans CO28M | 10mm | 7mm | 8mm |
| Weissella confusa CO29M | 10mm | 8mm | 8mm |
| Enterococcus durans CO30A | 7mm | 8mm | 7mm |
Discussion
Lactic acid bacteria are represented among the members of the normal microflora and inhabit the digestive tract of many animal species including human beings, numerous species of lactic acid bacteria can be detected in the digestive tract but their prevalence and distribution varies according to the animal species with which they are associated14,13 isolated lactic acid bacteria with probiotics potentials from fermented Nigerian dairy foods, raw cow milk, whey and intestine of cows. In this study, lactic acid bacteria were isolated from cow faeces.
The isolated LAB were identified presumptively based on phenotypic properties and more accurately by molecular method. Although for identification of LAB, phenotypic methods have been most commonly15. More recently, genetic techniques, such as 16S rDNA sequencing have been developed which allows a more consistent and accurate identification of individual strains16. The identification of the isolated lactic acid bacteria in this study was done by the sequencing of the 16S rRNA genes of the isolates.
Five lactic acid bacteria species were isolated belonging to two genera; Enterococcus hirae (8), Enterococcus durans (6), Enterococcus faecium (1), Enterococcus faecalis (1) and Weissella confusa (1). This result is in agreement with the work of 17 where Enterococcus spp. was isolated from 65 (90.2%) of 72 cattles' faecal specimens in Denizli region of Turkey, the species isolated include Enterococcus durans, Enterococcus hirae, Enterococcus faecalis, Enterococcus faecium and Enterococcus raffinosus although the most commonly isolated species from their study was E. faecalis while the most isolated species in this study was Enterococcus hirae. Isolation of Weissella confusa from cow faeces is in agreement with the report of 13, where Weissella confusa with antimicrobial activity against uropathogens was isolated from cow intestines in Nigeria.
Lactic acid bacteria can produce antimicrobial agents that exert strong antagonistic activity against many microorganisms, including pathogenic and spoilage microorganisms. Metabolites such as organic acids (lactic and acetic acid), hydrogen peroxide, ethanol, diacetyl, acetaldehyde, acetoine, carbon dioxide, reuterin, reutericyclin and bacteriocins18, are examples of antimicrobial agents produced by LAB. Organic acid produced by LAB leads to a reduction in pH levels and increases the production of hydrogen peroxide19. These products exhibit antibacterial activity against various pathogenic microorganisms, including Gram-positive and Gram negative bacteria20. One of the important WHO/FAO criteria for the selection of organism for probiotic purpose is their ability to display antimicrobial activity against pathogenic bacteria.
Lactic acid bacteria isolated in this study displayed significant antagonistic activity against E.coli and Klebsiella spp isolated from the same habitat as the lactic acid bacteria. Many researchers have isolated Enterococcus spp with antimicrobial activities against pathogens including E.coli and Klebsiella spp, from various sources including canine feces, boiler meat samples, swine feces, wild waterfowl faeces, and human faeces21. Weisella confusa isolated from cow intestine has been demonstrated to inhibit E.coli implicated in urinary tract infection13. Among all the antagonistic substances, bacteriocin production is often proposed as a beneficial characteristic of probiotic lactic acid bacteria22, which may facilitate the establishment of a probiotic strain in the competitive environment of the gut23. Although production of bacteriocin was not detected in the studied lactic acid bacteria, they all still showed appreciable antimicrobial activity against the test pathogens, this suggests that their antibacterial activity was as a result of other antimicrobial substances other than bacteriocin.
It can be inferred from this study that the apparently healthy state of these animals habouring these potential pathogens suggests that the lactic acid bacteria present in the gastrointestinal tracts of these cows are responsible for controlling the pathogen carriage and colonisation of the gut by harmful or pathogenic bacteria through bacterial antagonism and competitive exclusion of pathogens from the intestines.
Conclusion
Multidrug resistant bacteria may arise as a result of selection pressure in cattle and other food animals, as a result of use of sub therapeutic doses of antibiotics in their feeds. These organisms may find their way to the human community through the food chain, contamination of meat processing equipments, faecal waste runoff to water bodies used for drinking and other domestic purposes etc. Lactic acid bacteria have been demonstrated in this study to produce antimicrobial activity against resistant strain of Escherichia coli used in this study, interestingly the lactic acid bacteria, Klebsiella spp and E. coli strain were isolated from the same habitat. This suggests that lactic acid bacteria from cow intestine can be used in cow feed additives in lieu of antibiotics to achieve the same purpose with the advantage of being cost effective and a safe alternative without the fear of encouraging microbial antibiotics resistance that is transferable to the human population. However, further research needs to be done to ascertain that the isolated and identified antimicrobial producing lactic acid bacteria isolated from this study fulfill other criteria required by the WHO and FAO for the selection of probiotic strains in animals.
References
- 1.Morgavi DP, Forano E, Martin C, Newbold CJ. Microbial Ecosystem and methanogenesis in ruminants. Animal. 2010;4(7):1024–1036. doi: 10.1017/S1751731110000546. [DOI] [PubMed] [Google Scholar]
- 2.Muhammad-Lawal A, Balogun GS. Animal protein consumption among rural households in Kwara State, Nigeria. African Journal of General Agriculture. 2007;3(1):21–27. [Google Scholar]
- 3.Taylor DJ. A realistic assessment of the risks of antimicrobial use in animals and its effects on food safety. Pig J. 1997;40:46–59. [Google Scholar]
- 4.Khachatourians GG. Agricultural use of antibiotics and the evolution and transfer of antibiotics-resistant bacteria. Can Med Assoc J. 1998;159:1129–1136. [PMC free article] [PubMed] [Google Scholar]
- 5.Food and Agriculture Organization / World Health Organization, author. Expert Workshop on Non-Human Antimicrobial Usage and Antimicrobial Resistance: Scientific assessment Geneva. 2003. Dec 1–5, [Google Scholar]
- 6.Aarestrup FM. Occurrence, selection and spread of resistance to antimicrobial agents used for growth pro-motion for food animals in Denmark. APMIS. 2000;108:5–48. [PubMed] [Google Scholar]
- 7.Wegener HC, Aarestrup FM, Jensen LB, Hammerum AM, Bager F. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerging Inf Dis. 1999;5(3):329–335. doi: 10.3201/eid0503.990303. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Agriculture Organization / World Health Organization, author. Guidelines for the evaluation of probiotics in food Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Ontario, Canada: 2002. April 30, May 1. [Google Scholar]
- 9.Morrow LE, Gogineni V, Malesker MA. Probiotics in the intensive care unit. Nutr Clin Prac. 2012;27(2):235–241. doi: 10.1177/0884533612440290. [DOI] [PubMed] [Google Scholar]
- 10.Patterson JA, Burkholder KM. Application of prebiotics and probiotics in poultry production. Poultry Sci. 2003;82:627–631. doi: 10.1093/ps/82.4.627. [DOI] [PubMed] [Google Scholar]
- 11.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim Y W. Ez Taxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;5:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 12.Magnusson J, Schnurer J. Lactobacillus coryniformis subsp. Coryniformis strain Si3 produces a broad spectrum proteinaceous antifungal compound. Appl Environ Microbiol. 2001;67:1–5. doi: 10.1128/AEM.67.1.1-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayeni FA, Adeniyi BA, Ogunbanwo ST, Tabasco Raquel, Paarup Torsten, Pelaez Carmen, Requen Teresa. Inhibition of uropathogens by lactic acid bacteria isolated from dairy foods and cow's intestine in western Nigeria. Arch Microbiol. 2009;191:639–648. doi: 10.1007/s00203-009-0492-9. [DOI] [PubMed] [Google Scholar]
- 14.Tannock GW. The microecology of Lactobacilli inhabiting the gastrointestinal tract. Adv Microb Ecol. 1990;11:147–171. [Google Scholar]
- 15.Corsetti A, Lavermicocca P, Morea M, Baruzzi F, Tosti N, Gobbetti M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int J Food Microbiol. 2001;64:95–104. doi: 10.1016/s0168-1605(00)00447-5. [DOI] [PubMed] [Google Scholar]
- 16.Buddhiman T, Jyoti PT, Ulrich S, Charles MAPF, Michael G, Wilhelm HH. Phenotypic and genotypic identification of lactic acid bacteria isolated from ethnic fermented bamboo tender shoots of North East India. International Journal of Food Microbiology. 2008;121:35–40. doi: 10.1016/j.ijfoodmicro.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Mete E, Kaleli I. Species distribution and antibiotic resistance of Enterococci isolated from cattle farmers and cattles. Mikrobiyol Bul. 2006;4:75–80. [PubMed] [Google Scholar]
- 18.Jagoda Suskovic, Kos Blazenka, Beganovic Jasna, Pavunc Andreja Lebos, Habjanic Ksenija, Matosic Srecko. Antimicrobial Activity of Lactic Acid Bacteria. Food Technol Biotechnol. 2010;48(3):296–307. [Google Scholar]
- 19.Ponce AG, Moreira MR, Valle CE, Roura SI. Preliminary characterization of bacteriocinlike substances from lactic acid bacteria isolated from organic leafy vegetables. Food Science and Technology. 2008;41(3):432–441. [Google Scholar]
- 20.Maragkoudakis PA, Mountzouris KC, Psyrras D, Cremonese S, Fischer J, Canter MD, Tsakalidou E. Functional properties of novel protective lactic acid bacteria and application in raw chicken meat against Listeria monocytogenes and Salmonella enteridis. International Journal of Food Microbiology. 2009;130(3):219–226. doi: 10.1016/j.ijfoodmicro.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 21.Theppangna W, Murase T, Tokumaru N, Chikumi H, Shimizu E, Otsuki K. Screening of the enterocin genes and antimicrobial activity against pathogenic bacteria in Enterococcus strains obtained from different origins. J Vet Med Sci. 2007;69(12):1235–1239. doi: 10.1292/jvms.69.1235. [DOI] [PubMed] [Google Scholar]
- 22.Klaenhammer TR, Kullen MJ. Selection and design of probiotics. Int J Food Microbiol. 1999;50:45–57. doi: 10.1016/s0168-1605(99)00076-8. 1999. [DOI] [PubMed] [Google Scholar]
- 23.Fooks LJ, Gibson GR. In vitro investigations of the effect of probiotics and prebiotics on selected human intestinal pathogens. FEMS Microbiol Ecol. 2002;39:67–75. doi: 10.1111/j.1574-6941.2002.tb00907.x. 2002. [DOI] [PubMed] [Google Scholar]