Abstract
Automated tracking of living cells in microscopy image sequences is an important and challenging problem. With this application in mind, we propose a global track linking algorithm, which links cell outlines generated by a segmentation algorithm into tracks. The algorithm adds tracks to the image sequence one at a time, in a way which uses information from the complete image sequence in every linking decision. This is achieved by finding the tracks which give the largest possible increases to a probabilistically motivated scoring function, using the Viterbi algorithm. We also present a novel way to alter previously created tracks when new tracks are created, thus mitigating the effects of error propagation. The algorithm can handle mitosis, apoptosis, and migration in and out of the imaged area, and can also deal with false positives, missed detections, and clusters of jointly segmented cells. The algorithm performance is demonstrated on two challenging datasets acquired using bright-field microscopy, but in principle, the algorithm can be used with any cell type and any imaging technique, presuming there is a suitable segmentation algorithm.
Index Terms: Cell tracking, data association, dynamic programming, multiple target tracking, track linking, Viterbi algorithm
I. Introduction
In cell and developmental biology, different cell types are studied either in cell cultures (in vitro), in live organisms or embryos (in vivo), or in live tissue (in situ). The biological questions can be extremely different from one another, but time-lapse microscopy is often a very important and powerful technique to study and analyze the cells [1]. Time-lapse microscopy can be used to characterize and quantify many different aspects of cell behavior, such as proliferation [2], mitosis (cell division) [3], [4], apoptosis (cell death) [5], migration [6], and morphology [7], that are important in the study of cancer [8], [9], embryogenesis [10], [11], stem cells [12]–[14], and many other topics in the fields of cell and developmental biology. In early works like [5], [10] cells were observed using transmission microscopy, and the images were sketched by hand at appropriate time intervals, or recorded on video tape in cases where all cells of interest were in the same focal plane. Nowadays, a large number of microscopy techniques are available, the cells or their nuclei can be labeled using fluorescent proteins or dyes, and sequences of 2-D images or 3-D z-stacks can be recorded by digital cameras. The analysis of the time-lapse sequences is however still performed manually in most cases, and is often very demanding [1]. Manual analysis is generally very time consuming, hard to reproduce, and can sometimes be subjective or biased by the expectations of the person performing the analysis. For these reasons, there is a large demand for automated or semi automated methods to perform the analysis.
Automated cell tracking algorithms can reduce the manual labor required, and make the analysis more quantitative and reproducible. Frequently, automated analysis not only reduces manual labor, but also enables experiments that would have been too labor intensive to perform using manual techniques. A good example of this can be found in [3], where ~190 000 image sequences were analyzed automatically to search through the entire human genome for genes associated with mitosis, migration, and survival. Furthermore, automated algorithms can be used to analyze subtle changes that are hard for a human to observe or quantify, as done in [15], where the fates of neural progenitor cells were predicted by looking at their appearances and motions. The cell tracking problem is however very challenging, and more research is required, as existing automated methods are still considered too error-prone for most long-term imaging experiments [1].
Reviews of existing automated cell tracking methods can be found in [16]–[19]. The authors of [17] and [18] also review particle tracking, which is a related field where sub-cellular particles are tracked in fluorescence microscopy [20], [21]. In particle tracking, the tracked objects are usually small enough to be represented as point objects, and they usually interact less with each other than cells do, but the tracking problem is similar to cell tracking in many other aspects. Cell tracking algorithms are normally classified into model evolution algorithms, where mathematical models of the cells are propagated in time [22]–[24], and tracking by detection algorithms, where the tracking problem is separated into finding the outlines of the cells (segmentation) and linking the detected outlines into tracks (track linking, data association, or tracking) [2], [25]–[27].
Model evolution is fundamentally different from tracking by detection in that mathematical representations of the entire objects are tracked, instead of just the object locations. This makes model evolution well suited for studies of morphological changes of cells imaged in high magnification. Model evolution algorithms generally require a high imaging frequency, but can use temporal information to increase the segmentation accuracy in cases where, due to low image quality or cell-cell contact, it is hard to segment the cells based on information from a single image. Initialization of new cells that appear in the first image or that migrate into the imaged area is however problematic and often requires a separate segmentation algorithm which operates on a single image. Model evolution algorithms often evolve mathematical representations of the contours of the cells by minimizing an energy functional. This is normally done by solving a PDE, and that is typically very time consuming, making the algorithms slow compared to tracking by detection algorithms. Faster model evolution algorithms have however been presented in the last few years [28], [29]. In [28], 3-D contours of cells are represented using discrete meshes, so that fast algorithms and hardware normally used for computer graphics can be used for processing. In [29], the energy functional is minimized without solving a PDE, by applying the fast level set-like framework and graph cuts. Tracking by detection algorithms can get by with lower imaging frequencies and are well suited for studies of migration and lineages of cells imaged in low magnification. The algorithms can use temporal information to find out where the cells go, by doing advanced data association. Another advantage of tracking by detection is that it breaks the tracking problem into the separate problems of segmentation and track linking, which can be solved independently. This often makes it possible to apply a track linking algorithm to new tracking applications simply by replacing the segmentation algorithm.
In this paper, we focus on tracking by detection, and present an algorithm that can be used to solve the track linking problem. The main challenge of the track linking problem is to perform data association despite errors in the segmentation. The segmented outlines in a single image can often be ambiguous in the sense that it is hard or impossible to determine how many cells the outlines contain, and the ambiguities can often persist for a large number of images. This makes it desirable to use information from a large number of future images, or ideally the entire image sequence, when the track linking is performed. An algorithm which makes use of the entire image sequence is called a batch algorithm [30]. Examples of batch algorithms can be found in [27], [30]. In cell tracking applications, the image sequences are normally recorded ahead of time and analyzed later, so there is very little explicit demand for algorithms that process the image sequences sequentially and causally, like conventional multiple target tracking algorithms used in for example surveillance applications. Despite this, there are to date almost no prior batch algorithms for cell tracking.
Given the above, we propose a batch algorithm for track linking, which uses information from all images in the image sequence in a probabilistic manner to make individual track linking decisions. The algorithm incorporates mitosis, apoptosis, and other events into the same probabilistic framework without using heuristic postprocessing algorithms or separate detection algorithms that make hard detection decisions ahead of time. The algorithm can handle false positive detections (also referred to as spurious detections or clutter), missed detections, and clusters of cells that are segmented jointly.
Many existing track linking algorithms for cell tracking perform the linking image by image. The algorithms thereby create tracks sequentially in time and extend the tracks in one image to detections in the next image, often by solving integer programming problems [9], [25]. Sometimes, information from future images is incorporated into the track linking using heuristics for initiation and termination of tracks, or using heuristic postprocessing algorithms. The capabilities of these methods are however limited, and the number of future time-points that can be considered is usually small. There are examples of more sophisticated algorithms where information from all future images can be used to make tracking decisions [27], [31], but the use of information from future images is still limited. In [27], track fragments with a low level of ambiguity are created using image by image matching and then the fragments are linked into longer tracks using integer programming. In [31], cell outlines are grouped into spatiotemporal tubes and then the tubes are either merged or split to maximize a set of likelihoods. Both [27] and [31] optimize tracks over the entire image sequence, but [27] makes final choices for most track linking decisions when the track fragments are created, and [31] is limited in the types of operations that can be performed on the spatiotemporal tubes. The method that we propose performs global optimization over all track linking decisions of the entire image sequence, and can thereby make use of more information from future images than [27], [31].
In addition to performing global data association using information from all time-points, the proposed algorithm can handle joint segmentation of multiple cells in clusters. Joint segmentation of multiple cells has been identified as the most problematic segmentation error for cell tracking [2], and is seldom treated explicitly by track linking algorithms. An example of an algorithm that can handle joint segmentation is [31], where outlines can be split at branching points where two outlines in one image overlap with the same outline in the prior or following image. Another example can be found in [6], [14], where extreme points on cell boundaries in one image are matched to extreme points on cell boundaries in the following image, to break clusters of cells. The method that we propose incorporates jointly segmented cells into the global track linking algorithm and does not rely on having accurate cell outlines, or outlines that overlap in adjacent images.
The proposed track linking algorithm starts with the hypothesis that there are no cells in the image sequence and then adds one cell track at a time, in a greedy way, which maximizes a probabilistically motivated scoring function in each iteration. The algorithm stops when the scoring function can not be further increased by adding an additional track. Every track addition is however optimal in the sense that no other single track addition can increase the scoring function more. This optimality is ensured by solving a dynamic programming problem over a state space diagram using the Viterbi algorithm. We also introduce a novel way of altering preexisting tracks at the same time as a new track is created, by introducing a notion of swap operations.
The idea of using dynamic programming to create entire tracks that are optimal with respect to some scoring function has been used for tracking of a single biological particle in [32]. Furthermore, the authors of [32] emphasize the power of the global track linking and they also call for a dynamic programming algorithm that can be used to track multiple objects, thereby providing inspiration for the algorithm we now present.
Dynamic programming has also been used to track multiple objects in [33], [34], where people are tracked in video recordings, with good results. In [33], [34] tracks are added one at a time using dynamic programming, without editing of previously generated tracks. This works well for tracking of people, but it often gives rise to a lot of tracking errors in cell tracking applications where a large number of cells are tracked. In [34], the authors also present an alternative dynamic programming algorithm which allows some editing of previously generated tracks. That algorithm is however not well suited for tracking of dividing cells, as the editing mechanism would not enforce the constraint that a daughter cell of a mitotic event must have a mother cell and a sister cell. Furthermore, the algorithm of [34] does not allow multiple objects to occupy the same detection, which is important for tracking of cells that form clusters. The use of dynamic programming is also well established in tracking for defense applications. The states in the state space diagram can in these applications correspond to discrete target states [35], manoeuvres of a target [36], or as in our case, to measurements [30]. In [30], two different multiple target tracking algorithms are presented. The first algorithm is similar to [33] in that there is one state space trellis per target, but the tracks are generated in parallel. The trellises are updated in a random order and a detection is included in a trellis if it fits the motion model of the tracked target sufficiently well. When a detection is included in a trellis, it can not be used in other trellises. The other algorithm tracks all of the targets using a single trellis. In this case, the states of the trellis represent data associations between all targets and all measurements. Gating is used to reduce the computational requirements of the algorithm, but the algorithm is still too computationally expensive to be suitable for cell tracking where there is a large number of targets. Furthermore, the algorithm would need to be extended to handle an unknown number of targets and initialization and termination of tracks, and that would most likely come at a very high computational price. Finally, it is unclear if cell tracking specific challenges like mitosis and groups of cells occupying the same detection could be incorporated into the algorithm. In general, traditional multiple target tracking algorithms designed for defense and surveillance applications perform poorly on biological tracking problems [21]. This is because they assume that the tracked targets exhibit smooth and predictable trajectories, while the motion of cells and other biological objects is very erratic and unpredictable [21].
A precursor to the algorithm proposed herein was presented in [37]. The prior algorithm has now been extended to handle cells that enter and leave the field of view, missed detections, and situations where the daughter cells in a mitotic event are not segmented in the same outline. Furthermore, we present a version of the algorithm with reduced complexity, which allows image sequences with thousands of cells to be processed, without sacrificing performance. We participated, with two different precursors of the algorithm described here, in the ISBI 2012 Particle Tracking Challenge [38] and in the ISBI 2013 Cell Tracking Challenge [39].1 We also participated in the ISBI 2014 Cell Tracking Challenge [40] with the algorithm that we propose in this paper, but the results of that challenge have not yet been presented in a scientific publication. In the ISBI 2013 Cell Tracking Challenge, our full cell tracking system had outstanding performance, due primarily to the track linking algorithm, and in the ISBI 2014 Cell Tracking Challenge, an updated version of the system achieved better performance than all other systems on all of the challenge datasets [40]. Furthermore, a very early version of the proposed track linking algorithm was used in [13] to investigate how the rigidity of the culture substrate affects the stemness and viability of Muscle Stem Cells (MuSCs). These examples show that the track linking algorithm that we propose can be applied to a broad range of tracking problems and that it can be used to answer biologically relevant questions. The unpublished version of the algorithm being presented here is more versatile than its precursors and therefore has an even broader applicability.
In Section II we formulate the track linking problem in more precise terms, and in Section III we describe the scoring function used to score different solutions to the track linking problem. The track linking algorithm, which is the main contribution of this work, is described in Section IV. We then test the algorithm in Section V on two very challenging 2-D datasets with MuSCs and primary myoblasts imaged using bright-field microscopy and compare our tracking results to tracking results produced using CellProfiler [41]. We also briefly discuss suitable algorithms for segmentation and scoring of tracking hypotheses. We do however stress that the proposed track linking algorithm also can be used to track other cell types imaged using other imaging techniques in both 2-D and 3-D, as was done in [39], as long as a suitable segmentation algorithm is available. The scoring function can, just like the segmentation algorithm, be replaced by a different scoring function, as long as it satisfies certain Markov properties described in Section III-B, but the described scoring function is much less sensitive to variations in the data than the segmentation algorithm is.
II. Problem Statement
The proposed algorithm can be used to link a set of detected cell outlines, produced by an application specific cell segmentation algorithm, into tracks. We assume that the segmentation algorithm outputs a set of detections , where Dt,i is detection number i in image t, Nt is the number of detections in image t, and T is the length of the image sequence. Every detection is a representation of an object in the image. Round cells are often represented using centroid coordinates and radii [26], while cells with variable morphology are often represented using pixel or voxel regions in the image or z-stack [25], but other representations are also possible. In tracking by model evolution, cells are usually represented using contours described by either parametric functions or by the zero level set of a scalar function defined over the image or z-stack [16], but such representations are not normally used for tracking by detection.
Ideally, every detection should correspond to exactly one cell. In all nontrivial cell tracking problems, there are however errors in the segmentation, so that a detection can correspond to zero cells, a single cell, or multiple cells. Fig. 1 shows an example with detections generated using the segmentation algorithm described in Section V-C.
Fig. 1.
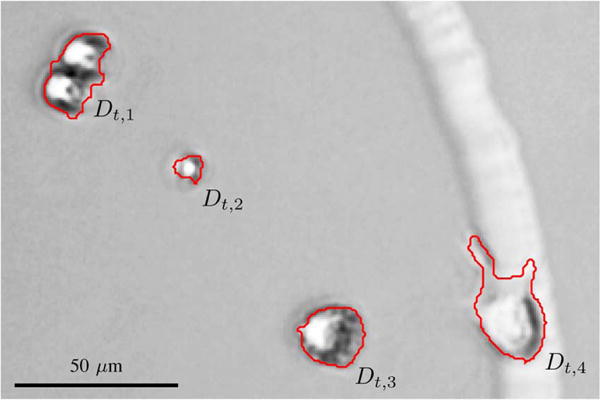
Detections of MuSCs imaged using bright-field microscopy, generated using the segmentation algorithm described in Section V-C. Detection Dt,1 contains two cells, detection Dt,2 is a false positive containing no cells, and detections Dt,3 and Dt,4 are both detections containing a single cell.
A. Representation of Tracks
Given the set of detections , we try to generate an accurate set of cell tracks. We represent a set of cell tracks and the mother–daughter relationships between them using an acyclic graph F with one or more connected components. Such a graph is usually referred to as a forest, or as a tree if it consists of a single connected component. The nodes of F represent individual cells at different time-points, and are labeled with the indexes of the detections that the cells occupy at those time-points. Furthermore, the last node of cells that undergo apoptosis are marked with a black cross, to differentiate them from cells that leave the imaged area. Cell migration is represented by links between pairs of nodes, and mitotic events are represented by two links from the last node of the mother cell, to the first nodes of the respective daughter cells. Fig. 2 shows an example of a forest graph F representing a set of cell tracks.
Fig. 2.
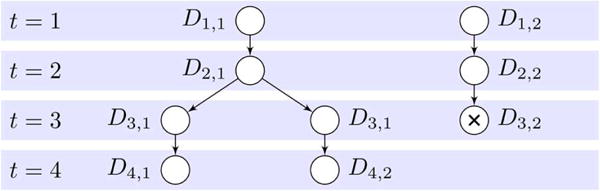
Forest graph F representing a set of four cell tracks in an image sequence with four images. In the first image, there are two cells that occupy detections D1, 1 and D1, 2 respectively. The cell that starts in D1, 2 migrates through detections D2, 2 and D3, 2 in images 2 and 3 and then undergoes apoptosis. The cell that starts in D1, 1 first migrates to detection D2, 1 in image 2 and then undergoes mitosis between image 2 and image 3. In image 3, the daughter cells are segmented jointly in D3, 1 and in image 4, they have migrated to D4, 1 and D4, 2, respectively.
III. Scoring Function
In order to score different solutions to the tracking problem, we introduce a set of event variables , that represent possible events that can take place in the image sequence. For every set of proposed tracks F, there is a unique set of assignments to the event variables . We then define a score g(F) for the set of tracks F, as the sum of the logarithmic probabilities of the assignments to the individual event variables, i.e.,
| (1) |
If all event variables are assumed to be a priori independent, the scoring function corresponds to the logarithm of the probability of the joint assignment to the set of event variables, required to produce the set of tracks F. In reality, there will surely be some dependence between the variables, but the forced independence assumption gives a scoring function which can be optimized effectively and which still has a strong probabilistic motivation.
A. Events
The set of event variables should be defined so that it contains events associated with cell counts in detections, migrations, and all mechanisms by which cells appear and disappear in the image sequence. In this paper, we include event variables associated with the number of cells in each detection (Ct,i), migrations inside the imaged area (Mt,i,j,τ), mitotic events (St,i), apoptotic events (At,i), migrations into the imaged area (It,i), and migrations out of the imaged area (Ot,i). The different variable types are explained further in Table I. The time index t goes between 1 and T for Ct,i, between 2 and T for It,i, and between 1 and T – 1 for all other event variables. Apoptotic events are not considered in the last image, as it is impossible to determine if a cell is dead one time step after the last time point. The detection indexes i and j go from 1 to Nt and from 1 to Nt+1 respectively. Missed detections are handled by letting a migration event occur between detections in images that are τ = 1,…,τmax time-points apart. A mitotic event that takes place between time t and time t + 1, where the mother cell occupies Dt,i and the two daughter cells occupy detections Dt+1,j and Dt+1,k, is represented by the mitotic event St,i and the two migration events Mt,i,j,1 and Mt,i,k,1. In the event variables associated with the forest F in Fig. 2, M1,1,1,1 = 1, as there is a cell migrating between D1,1 and D2,1, S2,1 = 1 and M2,1,1,1 = 1, as there is a mitotic event in D2,1 after which both daughter cells occupy D3,1, and M1,1,2,1, = 0, as there is no cell migrating between D1,1 and D2,2.
TABLE I.
Event Variables in the Scoring Function. If Multiple Cells Meet the Criteria of a Binary Event Variable, the Value is 1
| Variable | Event | Values |
|---|---|---|
|
| ||
| Ct,i | cell count | The number of cells in detection Dt,i. |
| Mt,i,j,τ | Migration | 1 if a cell migrates between Dt,i and Dt+τ,j, 0 otherwise. |
| St,i | Mitosis | 1 if a cell divides in Dt,i so that the daughter cells appear in image t+1,0 otherwise. |
| At,i | Apoptosis | 1 if a cell undergoes apoptosis (dies) in Dt,i, 0 otherwise. |
| It,i | migration into image | 1 if a cell is outside the imaged area at time t − 1 and migrates into Dt,i, 0 otherwise. |
| Ot,i | migration out of image | 1 if a cell is in Dt,i at time t and has left the imaged area at time t + 1, 0 otherwise. |
To optimize the performance of the track linking algorithm, it is judicious to include only the event types that occur in the data to be processed. If it is known a priori that none of the cells undergo mitosis, the mitotic events do not need to be included in ℰ, if no cells undergo apoptosis, the apoptotic events do not need to be included, if the cells are confined to some type of microwell the event variables for entering and leaving the field of view do not need to be included, and if there are no missed detections, τmax should be set to 1.
B. Scoring Function Requirements
All track linking algorithms have some kind of explicit or implicit scoring of tracks to choose between different linking options, and different track linking strategies impose different constraints on how the tracks can be scored. The proposed track linking algorithm, described in Section IV, requires a scoring function which is memoryless in the sense that the score associated with an event in a cell track does not depend on prior events in the track. This is equivalent to a Markov property saying that the event following the current time-point is independent of prior states of the cell, given the current state of the cell. For migration probabilities based on the locations of detections, the Markov property requires that the score associated with the migration Mt,i,j,τ must not depend on the location of the cell in images prior to image t, given that the location in image t is known. In many other tracking problems, this would be a severe limitation, as it effectively precludes the use of dynamic motion models. In cell tracking however, there is not a lot of performance to be gained from using a dynamic motion model, as cells frequently violate the assumption of smooth motion made by dynamic motion models. The Markov property also precludes the probability of mitosis, apoptosis, and other events from depending on the cells age, motility, or other properties that require data from multiple time-points.
The scoring function proposed in (1) is not the only possible scoring function satisfying the necessary Markov properties, but we have found that it is a good choice, as it defines an easily interpreted probabilistic framework and achieves high tracking performance. The event probabilities in the individual terms of the scoring function can be computed from the detections using either parametric probabilistic models for the different events, or classification algorithms, such as (multiclass) logistic regression and Gaussian discriminant analysis, trained on a manually generated ground truth dataset. The probabilities can also be replaced by heuristically chosen functions of detection features, as done in for example [9]. In Section V-D, we present suggested parametric models for the migration probabilities and for the probabilities that a cell enters or leaves the field of view in a particular detection. In Section V-E we present a way to model probabilities of cell counts in detections, mitosis, and apoptosis, using logistic regression. The models described in Section V have been shown to work well for tracking of adherent cell types imaged using bright-field microscopy, but they might require some modifications to handle other cell types and microscopy techniques.
IV. Track Linking Algorithm
Given a set of detections and the scoring function (1), the problem of generating an optimal set to tracks is a combinatorial optimization problem which can not be solved exactly, except for extremely small tracking problems. The track linking algorithm that we propose in this paper finds an approximate solution to this optimization problem by iteratively adding individual tracks to the image sequence in a greedy way. The following subsections explain the algorithm in detail.
A. Generating Tracks Using Operations
The addition of a track to F can be broken down into a sequence of operations, where each operation adds an additional node to F, links it to a preexisting node if necessary, and updates the appropriate event variables. If we assume that the track linking algorithm has created two of the three tracks in Fig. 2, so that there is one track passing through the detections D1,1, D2,1, D3,1, and D4,1, and one track passing though the detections D1,2, D2,2, and D3,2, the third track can be created using a sequence of two such operations. The first operation adds an additional node labeled D3,1, creates a link from the node labeled D2,1, to the new node, changes the value of the mitosis variable S2,1, from 0 to 1, indicating that a mitotic event occurs in detection D2,1, and increases the value of the count variable C3,1 from 1 to 2. The value of the migration variable M2,1,1,1 is already 1, so it is not changed. The second operation adds a node labeled D4,2, creates a link from the node added by the first operation to the new node, and increases the value of the migration variable M3,1,2,1 and the count variable C4,2 from 0 to 1. The change in the scoring function is induced by the changes to the values of the event variables. The set of allowed operations at a certain position in the sequence of operations is restricted based on the previous operation. For example, the second operation described above can not be replaced by an operation which adds a migration from D3,2 to D4,2, as the first operation created a cell in D3,1. These restrictions ensure the internal consistency of F after each addition of a track. The set of allowed operations and the constraints that the sequence of operations has to satisfy can be represented nicely using a state space diagram, as described in the next section.
B. State Space Diagram
The track linking algorithm that we propose starts from an empty set of tracks F and adds one track at a time until the scoring function can not be increased further by adding an additional track. In each image, an added track can either pass through one of the detections, not be present because the cell has yet to appear in the image sequence, or not be present because it has disappeared from the image sequence. These possibilities can be represented as states in a state space diagram, as shown in Fig. 3. A track also has the possibility to skip one or multiple images where the cell is missing from the segmentation, but that is represented using arcs spanning multiple layers of the state space diagram, as described later in this section. The state space diagram has one state associated with each detection Dt,i in the image sequence, a chain of “born later” states Yt for cells that have not yet appeared, a chain of “dead” states Xt for cells that have disappeared, a starting state A and an ending state B. The detection states have the same labels as the detections themselves. The arcs that leave a state represent the operations that can be performed on F, when a track under creation is in that state, so that every track that can be added to F corresponds to a path from A to B in the state space diagram.
Fig. 3.
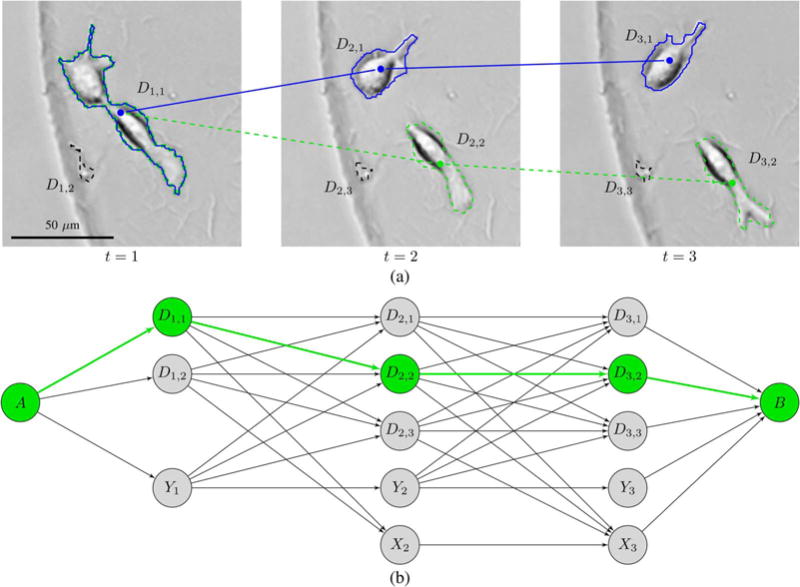
State space diagram (b) for the addition of a track to an image sequence with 3 images (a). There is a preexisting track shown in blue, and the state space diagram describes all the possible ways to add a second track. The roles of the nodes and the arcs between them are explained in Section IV-B. The arrows that go out of the “born later” states Y1, Y2, and Y3 represent both mitosis arcs and arcs for migration into the field of view. Arrows that go into the “dead” states X2 and X3 represent both apoptosis arcs and arcs for migration out of the field of view. Note that there is no X1 state, as we do not include cells in the tracking if they are dead in the first image. To avoid cluttering the state space diagram, we have set τmax to 1 so that only migrations between detections in adjacent images appear in the trellis. Even though this is a very small example with only three images and τmax = 1, there are 42 different paths through the state space diagram, assuming that only the maximum utility arcs between pairs of nodes are included in the trellis, so that each arrow represents a single arc. The path going through the three “born later” states represents the option of not adding a second track at all, but all of the other paths represent different ways in which a second track can be added. The correct way to add a second track, as sharing D1,1 with the blue track and then passing through D2,2 and D3,2 is shown as a green dashed track through the image sequence and a corresponding green path through the state space diagram.
The operations associated with the arcs can either start the creation of a new track in F, append an additional node to the end of the track under creation, or end the track under creation. Every such operation is associated with a change to the scoring function and that score change is placed as a utility on the arc in the state space diagram. The total change in the scoring function, that occurs when a particular track is added to F can therefore be computed as the sum of the utilities on the arcs in the corresponding path through the state space diagram.
An operation that initiates a track in detection i of the first image corresponds to an arc from state A to state D1,i, and an operation that initiates a track that enters the field of view in detection i of image t corresponds to an arc from state Yt−1 to state Dt,i. An operation that terminates a track in detection i of image t, through either apoptosis or migration out of the field of view, corresponds to an arc from state Dt,i to state Xt+1. An arc between two detection states Dt,i and Dt+τ,j represents an operation which appends detection Dt+τ,j after Dt,i in the track under creation. Mitotic events are introduced by creating an additional branch from a preexisting tree of nodes in F. If the mitotic event to be created occurs in detection Dt,i, which is already linked through migration to Dt+1,j, one of the daughter cells has to appear in Dt+1,j, and the other daughter cell can appear in any detection Dt+1,k of the next image. Operations which add such branches to F have arcs that go from Yt to Dt+1,k, and a utility which reflects the changes in St,i and Mt,i,k,1. Note here also that the introduction of a mitotic event in a preexisting track corresponding to a single cell creates a mother cell and two daughter cells in F: The mother cell and one of the daughter cells simply inherit the former and latter parts of the preexisting track, respectively, while the track of the second daughter cell is created by the remaining linking operations. Mitotic events can not be added at the end points of preexisting tracks. In Section IV-D, we define an additional type of arcs called swap arcs, which allow links in preexisting tracks to be changed during the creation of a new track. The main ideas of the algorithm are however easier to describe without swap arcs, so we save the description of them for later.
Given that the scoring function (1) has one term associated with every event variable, the utilities of the arcs in the state space diagram will involve only a small number of terms associated with the relevant event variables. For example, the utility associated with a migration arc between the detections Dt,i and Dt+τ,j corresponds to updating the values of the migration variable Mt,i,j,τ and the count variable Ct+τ,j, and can be written as
| (2) |
The first two rows of (2) correspond to changing Mt,i,j,τ from its current value to 1, and the last two rows correspond to adding 1 to Ct+τ,j. The score updates associated with other event types are a little more complicated, but follow the same pattern and can be found in Appendix A.
C. Finding the Highest Scoring Path
In the previous section, we showed that the change in the score of F, that occurs when a track is added, can be computed as the sum of the arc utilities along the corresponding path through the state space diagram. This means that finding the track which increases the score of F the most is equivalent to finding the path from A to B in the state space diagram, with the highest summed utility. This is equivalent to solving an instance of the well known shortest path problem. Given that the state space diagram has a trellis structure, we can solve the problem with linear complexity in T using a dynamic programming algorithm known as the Viterbi algorithm [42] in the communications literature. The value and computational efficiency of the Viterbi algorithm becomes evident when considering that the number of possible paths from A to B grows exponentially with T: A sequence with T = 100 images and 10 detections per image allows for more than 10100 different tracks in any iteration of the algorithm, even without considering swaps.
The Viterbi algorithm finds a solution to the optimization problem by using the fact that only the highest scoring path from A to an arbitrary state of image t can be the beginning of the highest scoring path from A to B. We let Et,i denote state number i at time-point t, and let Jt be the number of states at this time-point. The time index t goes from 0 to T+1, as there is a state A before the first image and a state B after the last image. There can be multiple arcs between a pair of states, but only the arc with the highest utility can be a part of the optimal path from A to B, so we define the arc set , containing the maximum utility arcs between all pairs of states. A path from A to an arbitrary state E corresponds to a set of operations that can be performed on F. We let gmax(E) denote the score of F after the operations corresponding to the maximum utility path from A to E have been performed. Furthermore we let the utility of an arc in , between the states Et,i and Et+τ,j, be denoted by Δg(Et,i, Et+τ,j). The Viterbi algorithm starts from the beginning of the state space diagram and finds the optimal paths to the states at each time-point. For t = 0, the optimal path consists of only the state A, and for subsequent time-points, the algorithm uses the fact that the optimal paths to states at earlier time-points are known. For each state Et,i at time t > 0, the algorithm determines the second to last state, denoted ϕ(Et,i), in the optimal path. The second to last state is found by comparing all candidates E, for which there is an arc (E, Et,i) in , and choosing the one for which gmax(E)+Δg(E, Et,i) is maximal. When the algorithm reaches B, the second to last state in the optimal path to every node is known, so then the algorithm can back-track from B to A to recreate the optimal path through the state space diagram, . Pseudo-code for the Viterbi algorithm can be found in Fig. 4. If gmax(B) > g(F), the sequence of operations corresponding to the optimal path are applied to F, the state space diagram is updated, and another iteration of the Viterbi algorithm is performed. Otherwise, g(F) can not be increased by adding an additional cell, will be {A, Y1,…,YT, B}, gmax(B), will be equal to g(F), and the track linking algorithm is terminated.
Fig. 4.

Description of the Viterbi algorithm, which is used to find the highest scoring path from A to B in the state space diagram.
While the states in the trellis are the same for each application of the Viterbi algorithm, the possible arcs and the arc utilities are changed in each iteration due to the change in F induced by the track added to F following the previous application of the Viterbi algorithm. It is also important to note that while the Viterbi algorithm solves the shortest path problem in a sequential manner from A to B (and then back to A in the backtracking step), this does not mean that the track linking itself is causal or sequential in time: The Viterbi algorithm achieves a globally and noncausally optimal track by propagating all candidates for the optimal track, and could have been implemented in the reverse order to find the optimal path from B to A, without altering the final solution.
D. Swaps
The state space diagram described in Section IV-B does not allow previously created tracks to be altered. This is problematic because track linking errors in preexisting tracks can not be corrected, and furthermore, incorrect links can hinder the creation of correct tracks in subsequent iterations, as illustrated in Fig. 5. In order to mitigate these problems, we introduce a swap operation, which modifies links in preexisting tracks during the creation of a new track. This is done by splitting a preexisting track in two parts and linking the second part of the preexisting track to the end of the track under creation. The same operation will also link a new state to the end of the first half of the preexisting track, so that the track linking algorithm can continue from there and generate a new continuation of the preexisting track. The scores at subsequent time-points are the same when a preexisting track is extended, as when a new track is extended, so the highest scoring way to modify F can still be found using the Viterbi algorithm, as described in Section IV-C. The only difference is that the added track can consist of multiple fragments linked to preexisting tracks at the time-points when swap operations are performed. In this way, the swap operation lets the track under creation take over a path which is already occupied by the second half of a preexisting track, and starts the process of finding a new continuation for that preexisting track.
Fig. 5.
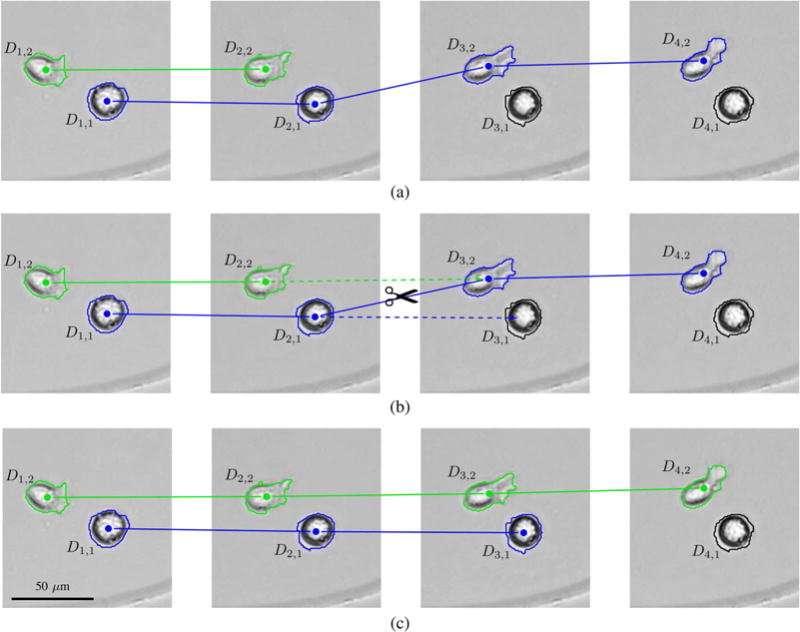
Illustration of a swap operation. First row (a) shows the tracks before the swap operation is performed. Detection D3,2 should be linked to the end of the green track under creation, but the incorrect preexisting blue track prevents this from being done using a migration arc in the state space diagram, as increasing the count variable C3,2 to 2 would lower the scoring function significantly. Instead, the shortest path through the state space diagram goes through a swap arc from D2,2 to D3,1. The swap operation associated with that arc links D3,2 to the end of the green track and changes the incorrect link between D2,1 and D3,2 in the blue track at the same time, as shown in (b). First the swap operation removes the incorrect link. Then it appends the second half of the preexisting blue track to the end of the green track under creation, and links D3,1 to the end of the first half of the preexisting blue track. Tracks after the swap operation are shown in (c). The next arc in the shortest path through the state space diagram determines how the blue track continues from D3,1, so to produce a correct tracking result, the next arc should be the migration arc from D3,1 to D4,1.
To formalize what a swap operation does, we consider a swap arc between the trellis states Dt,i and , which removes a link between detections and from a preexisting track. The procedure is exemplified in Fig. 5, for τ1 = τ2 = 1. In the general case, the track under creation, to which Dt,i was appended by the previous operation, takes over the second half of the preexisting track, starting with detection . Detection is appended after at the end of the first half of the preexisting track, which will be extended further or possibly terminated by the next operation.
The swap operation described above adds two new migration events and removes a preexisting migration event. The swap operations can however be generalized to allow other types of events to be added and removed. We do this by thinking of the beginnings of preexisting tracks as links from “born later” states to detection states in the state space diagram, and the terminations of tracks as links from detection states to “dead” states. This lets us treat the “born later” states and the “dead” states like detection states in the description above, so that swap events can change how and if tracks begin and end. As an example, the track under creation can take over the second half of a preexisting track and make the first half end in an apoptotic event. A more formal definition of generalized swap operations can be found in Appendix A.
E. Complexity and Implementation
If we assume that the length of the image sequence is T, that the maximum number of detections in an individual image is N, and that the number of tracks to be created is proportional to N, the worst case complexity of a naive implementation of the algorithm described above is . This is because the complexity of the Viterbi algorithm is linear in T, there can be N2 pairs of detections in images t and t+ τ, for τ = 1, …, τmax, and every such pair can have as many swap arcs between them as there are preexisting tracks. Since the number of tracks to be created is proportional to N, the average number of preexisting tracks will also be proportional to N, and the overall complexity of the algorithm will therefore be .
In practice, one can however restrict the number of migration and mitosis arcs entering and exiting detection states in the state space diagram. This can be done by discarding all but the L most likely migration events from and to every detection and the L most likely mitotic events for entering and leaving of any detection state. This reduces the number of swap arcs associated with a time-point in a preexisting track from to , and the overall complexity of the modified algorithm is therefore , where the constant L needs to be chosen so that correct events are not discarded when the set of arcs is pruned. In the experiments in Section V, and in many other cell tracking problems that we have worked on, L = 3 results in a very fast algorithm with the same performance as with larger values of L. With L = 3, the algorithm is fast enough to track thousands of objects in the same field of view.
Further, as the state space trellis changes from one iteration to the next, one can either generate a new trellis in each iteration (i.e., application of the Viterbi algorithm) or modify the trellis from the previous iteration, to reflect the changes that were made to F. The algorithm complexity is the same in both cases, but the run time is lowered significantly if the trellis is kept in memory and updated iteratively.
F. Postprocessing
When detections are assigned more than one track in the solution of the tracking problem, it is often necessary to split the detections so that each cell gets an outline of its own for the subsequent data analysis. Before the detections are split, the track links entering (or leaving) a detection with multiple cells can be interchanged without changing the assignments to the event variables, and consequently, by the structure of (1), without changing the score of F. After the splits, the different linking choices will however have different scores and it can therefore be necessary to change the links generated by the initial track linking in addition to just splitting the detections.
If the detections are represented using pixel or voxel regions, the problems of splitting detections and changing associations can be solved using the algorithm described in Section V-G, which splits detections using k-means clustering and changes the associations by solving an assignment problem. This chooses the best linking alternative for tracks through detections with multiple cells, based on the scoring function (1) and can also correct some tracking mistakes which can not be corrected by swap operations. The splitting of detections and the changes of assignments can however be performed using other algorithms. In [14], the problem of splitting outlines with multiple cells is solved by matching extreme points on the outline boundaries in adjacent images. The method in [14] has the potential to separate the cells more accurately than the method presented in Section V-G, but the algorithm requires that the number of cells in each detection is small and that the cell outlines have been segmented accurately. The algorithm in Section V-G does not suffer from these limitations and has the advantage that it can be used on both 2-D images and 3-D z-stacks.
V. Experiments and Real Life Performance
To test the proposed track linking algorithm, we implemented it in C++ and incorporated it into a cell tracking system that we have developed in MATLAB. The track linking algorithm is most easily implemented in an object oriented language and we chose C++ because of its efficiency. The cell tracking system is called the Baxter Algorithms, and we are currently working towards making the full software publicly available.
The track linking algorithm was tested on two datasets with MuSCs and myoblasts, respectively. The datasets are described in Section V-A, preprocessing of the images is described in Section V-B, the segmentation algorithm used to generate the detections is described in Section V-C, and the tracking performance is evaluated in Section V-H.
A. Data
For the performance evaluation, we use two bright-field microscopy datasets with MuSCs and myoblasts respectively. We chose these cell types because they are challenging to track due to their changing morphology, their motility, and the fact that the cells form clusters that are hard for segmentation algorithms to separate into individual cells. Bright-field microscopy was chosen because it gives less contrast than most other imaging techniques and therefore serves as a good benchmark. For display purposes, we have adjusted the brightness and contrast of the example images included in the paper to improve legibility. To someone who is used to looking at raw bright-field images, the problem of segmenting the cells may therefore seem easier than what it actually is.
1) MuSCs
In the first dataset, we use murine MuSCs, isolated and cultured according to protocols described in [13]. The MuSC data has previously been used for performance evaluation in [37] and it was analyzed from a biological perspective in [13], using an earlier version of the cell tracking system described here. In [13], the authors look at how the rigidity of the culture substrate affects the stemness and viability of MuSCs in culture, by culturing the cells on tissue culture plastic and a Poly Ethylene Glycol hydrogel respectively. The cells therefore come from two different culturing conditions, but that does not impact the performance evaluation.
The cells are confined in circular microwells, with a diameter of 600 μm, so that they can not leave the field of view. The dataset consists of 115 sequences of 1388 × 1040 pixel 8-bit images with cells imaged every 3 min for 33 h, using bright-field microscopy with 10× magnification. The image sequences start with 0–2 live cells and end with 0–14 live cells. When the fields of view to be imaged were chosen, they all contained at least one live cell, but a few of the cells died before the imaging started, and therefore there are a few image sequences without live cells. Image sequences where all cells die are truncated about 100 min after the last cell dies. Fig. 6 shows an image with 14 live cells and one dead cell at the end of the experiment.
Fig. 6.

Image from the end of the MuSC dataset.
The MuSC dataset is challenging to process because the cells move a lot and adhere to each other so that they can not be segmented into individual detections. In addition to this, there are a lot of background features and debris from the isolation procedure that can give rise to false positive detections. Furthermore, it is challenging to detect mitotic and apoptotic events as the events lack distinctive features in bright-field microscopy.
To estimate the parameters in the motion model described in Section V-D and to train the classifiers described in Section V-E, we use a separate training dataset with 52 image sequences of MuSCs isolated, cultured, and imaged in the same way as in the test dataset. The training dataset is 23 h long and has ground truth tracks generated through manual correction of tracks created using an earlier version of the algorithm described in this paper.
2) Myoblasts
To show that the algorithm can handle denser cultures and cells that enter and leave the field of view, we have also included a dataset with murine primary myoblasts, isolated and cultured according to the protocols described in [43]. The dataset consists of three sequences of 1384 × 1036 pixel 8-bit images with myoblasts imaged every 3 min for 24 h using bright-field microscopy with 10× magnification. The image sequences start with 25–32 live cells and end with 74–92 live cells. Fig. 7 shows an image with 92 cells at the end of one of the image sequences. A fourth training image sequence generated in the same way as the others was used to estimate parameters and train classifiers. This image sequence was first tracked using the parameters and the classifiers from the MuSC dataset and then corrected manually to produce a ground truth.
Fig. 7.
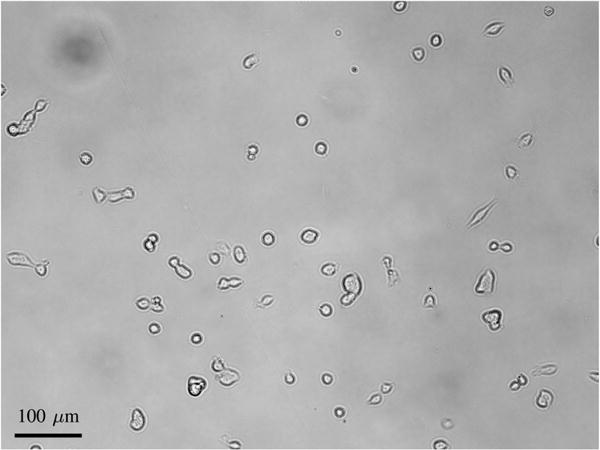
Image from the end of the myoblast dataset.
The myoblast dataset can be segmented with fewer false positive detections than the MuSC dataset, as there is no microwell and significantly less debris. Furthermore, the tracking is made easier by the fact that the number of apoptotic events is negligible. The high cell number in combination with rapid movement, severe clustering, and mitotic events that are hard to detect does however make the myoblast dataset slightly more challenging than the MuSC dataset. The proposed track linking algorithm does however perform well on this dataset as well.
B. Preprocessing
The MuSC dataset has circular microwells that need to be removed before the images can be segmented. To do this we first align the images in the image sequences using the Image Stabilizer plugin for ImageJ [44]. Then we detect the microwell border using a Hough transform for circular objects, applied to a gradient magnitude image computed using the Sobel operator, and crop the image to get a slightly smaller image with the microwell in the center. In the next step, we generate a background image by computing the median image through the time dimension of the image sequence. If Pt,i is pixel i in image t, the pixels of the static background image are given by
| (3) |
To speed up the processing, we only include every dth image, where d = max{1, floor(T/50)}, in the median computation. Finally, the background image is subtracted from every image in the sequence to produce a background subtracted image sequence. The illumination intensity in the images varies slightly over time, so to prevent this from causing problems in the background subtraction, the images are shifted using additive offsets so that they all have a mean intensity of 127.5.
C. Segmentation
To segment the images, in both datasets, we compute the standard deviation of the pixels in a small square region around every pixel and threshold the standard deviation image, as described in [45]. After that, we fill holes in the binary segmentation mask, apply morphological erosion [46] to compensate for the spread caused by the size of the square region, and remove small objects to reduce the number of false positive detections. This is a simple segmentation algorithm which performs reasonably well on most transmission microscopy images.
To separate clusters of cells into individual cells, we apply a watershed transform [46] to the standard deviation image. The watershed transform is confined to the foreground pixels of the binary segmentation mask and to avoid over-segmentation we apply some Gaussian smoothing and an h-minima transform [46] before the watershed transform is computed. To further reduce over-segmentation, we merge watershed regions without cells into adjacent watershed regions with cells, once the tracking has been performed.
D. Motion Model
There is no problem with missed detections in either of the datasets that we process in this section, and therefore we set τmax = 1, and consider only migrations between detections in adjacent images. To simplify the notation, we drop the last subscript on the migration variables and let Mt,i,j denote Mt,i,j,1.
In traditional multiple target tracking used in for example radar surveillance applications, it is very important to use an accurate dynamic motion model where the motion in one time-step depends on the motion in earlier time-steps. In cell tracking however, the objects to be tracked move very randomly and can make sudden large moves in unexpected directions, so there is not a lot to be gained from using a dynamic motion model.
We therefore model the motion of a cell as a Brownian random walk performed by its centroid. If xt is the centroid position of a cell in image t, the probability density of the centroid location in image t + 1 is assumed to follow a uniform Gaussian distribution with the mean xt and the covariance matrix σ2I2, where I2 is the 2 × 2 identity matrix and σ2 is the variance of the cell displacement, projected on one of the coordinate axes. The probability density at a candidate location xt+1 in image t + 1 is therefore given by
| (4) |
At the same location, the distribution of detections coming from other cells, debris, and background features is modeled as a uniform distribution over the image
| (5) |
where A is the area of the image.
If we assume that the prior probability for migration Mt,i,j to occur is , we can use Bayes’ rule to compute a posterior probability for the migration, given the distance between the centroids of Dt,i and Dt+1,j. If the centroids of Dt,i and Dt+1,j are denoted xt,i and xt+1,j, the posterior probability can be written
| (6) |
The prior probability is taken to be independent of t,i, and j, and is estimated from detection pairs in the training data.
Both datasets contain a lot of stationary or slowly moving debris, which fits the above motion model very well. To prevent such objects from giving rise to tracks, we truncate the posterior migration probabilities at 0.5, so that adding a migration (i.e., changing Mt,i,j from 0 to 1) can not increase the scoring function in and of itself. This prevents tracks through detections that look like debris based on the count classifier from being created due to positive migration arc utilities. The truncation of the migration score is however removed when the assignment problem described in Section IV-F is solved, as the algorithm will otherwise be unable to distinguish between different migrations with positive scores.
The probability that a cell in detection Dt,i migrates out of the field of view between time t and time t + 1 is computed as the product between a prior probability, , that there is a cell migrating out of Dt,i, and the probability that a cell migrating out of Dt,i ends up outside the field of view. The probability that the cell ends up outside the field of view is estimated as the fraction of the probability density, for the predicted centroid location in image t + 1, that is outside the imaged area. This gives the expression
| (7) |
where the prior probability is estimated from the data, and is taken to be independent of t and i. The probability Pr(It,i = 1), that a cell migrates from outside the imaged area into Dt,i is set equal to Pr(Ot,i = 1), as the cell culture is assumed to have the same properties on both sides of the image borders.
E. Classification
To estimate the probabilities of cell counts, apoptosis, and mitosis, we compute 74 different features for each detection and then use (multiclass) logistic regression [47] to compute posterior probabilities for different assignments to the event variables, given the features. The feature set is based on the feature set described in [7], but has some additional features described in Appendix B.
It is hard to train accurate cell count classifiers for detections with many cells, because the number of training examples is very small or even 0, and the features in different classes have similar values. To avoid these problems, we pool all of the training examples with K or more cells, so that the classifier returns the posterior probabilities Pr(Ct,i = 0), Pr(Ct,i = 1),…, Pr(Ct,i ≥ K). We then assume that the cell counts in the last class with probability Pr(Ct,i ≥ K) are given by a geometric distribution, so that
| (8) |
where
| (9) |
In the experiments presented herein we choose K = 2, and the parameter ρ is computed from the training dataset using maximum likelihood estimation. The use of the geometric distribution can be motivated by assuming that single cells attach to other cells or clusters of cells at a rate λ and that cells leave the clusters at a higher rate μ. This implies that the number of surplus cells in each detection follows a birth–death process, with a birth rate of λ and a death rate of μ, which results in a the geometric distribution (9), with ρ = λ/μ [48].
The apoptotic events are also hard to classify, because it is often hard to determine the exact time-point of apoptosis, and because the changes in the cell appearance are often subtle. Because of this, and the fact that the mitosis classifier can have false positives, the scoring function can sometimes be increased by adding a mitotic event followed by an apoptotic event killing one of the daughter cells one or a few time-steps later. To prevent this from happening, we truncate the classification probabilities for apoptosis at 0.5, just like we do with the probabilities for migration, so that the introduction of an apoptotic event (i.e., changing At,i from 0 to 1) can not increase the scoring function in and of itself. In a case where a cell actually undergoes apoptosis, the apoptotic event will be introduced by the algorithm anyway to avoid the decrease in the scoring function that would be associated with continuing the track through detections of other cells or detections that should not have any cells at all.
F. Track Linking
For the track linking, we used the algorithm described in Section IV. In the MuSC dataset, the cells were confined to microwells and therefore we did not include variables for migration into and out of the field of view in the set of event variables. In the myoblast dataset, there were only two apoptotic events, and therefore we did not include any apoptosis variables in the set of event variables. We use the generalized form of swap events, described in Section IV-D, that can change how preexisting tracks begin and end, but to simplify the implementation we do not allow swap operations to create new mitotic events. We do however allow swap operations to remove mitotic events and thereby change the origin of one of the daughter tracks and link the other daughter track to the end of the mother track using a migration event.
G. Postprocessing
To solve the postprocessing problem described in Section IV-F and give cells in detections with multiple cells separate outlines, we split the binary segmentation masks of the detections using k-means clustering [47] applied to the x- and y-coordinates of the mask pixels. The seeding of the clusters is done randomly with one seed per cell, and once the pixel clusters have been determined, they are assigned to the cells in the previous image by solving an assignment problem using the Hungarian algorithm [49]. Migrations in and out of the imaged region are handled by introducing dummy cells and dummy pixel clusters with matching-scores that correspond to migration in and out of the imaged region respectively. To keep the problem simple, we do not change the identities of the cells that undergo mitosis and apoptosis. The mother cells in mitotic events are however entered twice into the matching problem, so that the identities of the two daughter cells can be determined based on the migration distances from the mother cell, in accordance with how mitotic events are scored in Section III.
H. Algorithm Performance
The tracking results on representative image sequences of the MuSC dataset and the myoblast dataset are shown in Figs. 8 and 9, respectively. In both figures, the tracks and the outlines of the cells at the end of the image sequence are overlayed on the last image. For the MuSCs, the tracks of the entire image sequence are shown, but for the myoblasts, only the last 100 time-points of the tracks are shown, as the tracks would otherwise cover most of the image, making the figure hard to interpret. The tracking results shown in Figs. 8 and 9 can be viewed in their entirety in Supplemental Video 1 and Supplemental Video 2, respectively. The images and the videos provide some qualitative evidence that the proposed algorithm performs well on both cell types, but we will also provide quantitative performance measures to show that the method works well on all image sequences of the datasets, and provide some means to compare the algorithm performance to existing and future works. We also compare our tracking results with tracking results produced using CellProfiler [41]. In Section V-H-1 we describe how CellProfiler was used to track the cells, in Section V-H-2, we evaluate tracking performance, and in Section V-H-3 we evaluate run time.
Fig. 8.
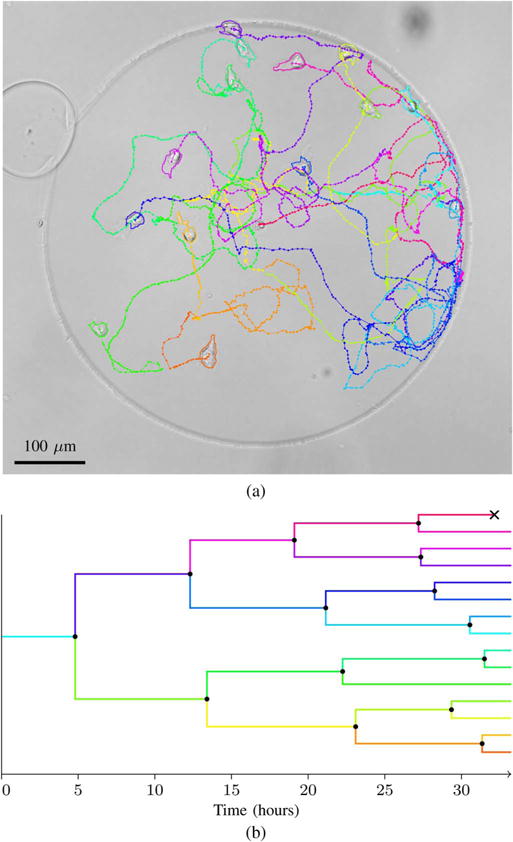
Representative tracking result from the MuSC dataset. Image (a) shows the tracks and the outlines at the end of the image sequence, overlayed on the last image of the sequence. The same image is shown without annotation in Fig. 6, and the entire image sequence can be viewed with annotation in Supplemental Video 1. Plot (b) shows the lineage tree corresponding to the tracks. Mitotic events are shown as black dots and apoptotic events are shown as black crosses.
Fig. 9.
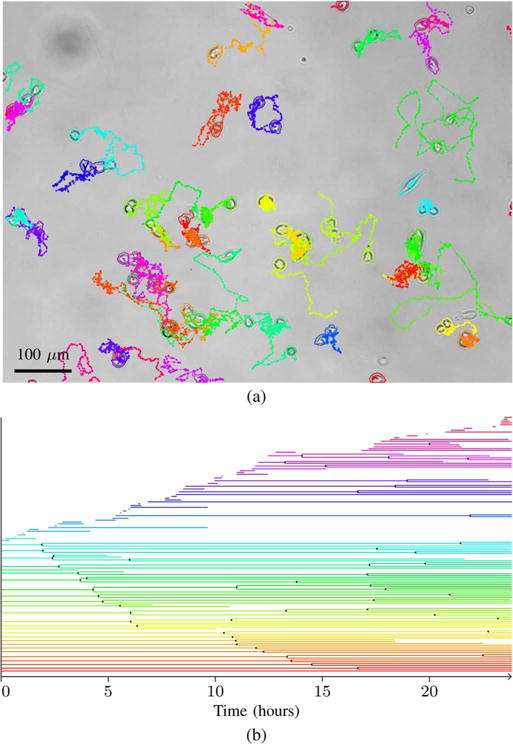
Representative tracking result from the myoblast dataset. Image (a) shows the last 100 time-points of the tracks, and the outlines at the end of the image sequence, overlayed on the last image of the sequence. The same image is shown without annotation in Fig. 7, and the entire image sequence can be viewed with annotation in Supplemental Video 2. Plot (b) shows the lineage trees corresponding to the tracks. Mitotic events are shown as black dots. Line segments that start after the first image, without a mitotic event, represent cells that migrate into the imaged area. Line segments that end before the last image represent cells that migrate out of the imaged area.
1) Tracking Using CellProfiler
In order to compare the proposed method to existing methods and software, we used the freely available software CellProfiler [41] to process our datasets. We saved the segmentation results produced by the algorithm described in Section V-C to images with object labels and loaded these into CellProfiler, so that we would be able to evaluate the tracking performance in isolation from the segmentation performance. For track linking, we used the LAP method in the TrackObjects module, which is an implementation of the track linking method presented in [20]. The algorithm is similar to [27] in that it first links detections into tracklets and then links the tracklets into longer tracks by solving a combinatorial optimization problem. In [20], the tracklets are created and linked into tracks by solving two different Linear Assignment Problems (LAPs). Like most other existing tracking systems, CellProfiler can not reason about the probability that a detection is a false positive. To increase the tracking performance, we therefore removed all objects with an area smaller than 250 pixels prior to tracking. We also introduced a minimum cell lifetime of 10 frames, so that short tracks caused by false positives would be excluded. Further, we disallowed merging of cells and adjusted parameters expressed in pixels to the scale of the the images. Once we had made these relatively natural problem specific adjustments, we spent several days testing different values for the remaining parameters, to maximize the performance measures described in Section V-H2. There were too many parameters to cover the entire parameter space in a systematic search, but nevertheless we are confident that all parameters were given sensible values. CellProfiler project files, containing all of the tracking parameters used for tracking of MuSCs and myoblasts are available to the reader as Supplemental File 1 and Supplemental File 2 respectively. Furthermore, tracking results for MuSCs and myoblasts, generated from the same image sequences as Supplemental Video 1 and Supplemental Video 2, can be viewed in Supplemental Video 3 and Supplemental Video 4, respectively.
2) Tracking Performance
To evaluate the tracking performance, we compare the computer generated tracks and the ground truth objects using a number of different performance measures. The overall accuracy of the tracks is measured using the track purity [50] and the object purity [50]. The accuracies with which the algorithms determine how many cells each detection contains is evaluated by looking at the confusion matrices for classification of detections into detections with 0, 1, and 2 or more cells. The accuracies by which mitotic and apoptotic events are detected are measured using the precision and the recall, with which the events were detected. The precision and the recall are defined as
| (10) |
and
| (11) |
where tp is the number of correctly detected events (true positives), fp is the number of falsely detected events (false positives), and fn is the number of events that were not detected (false negatives).
There is not yet any consensus, in the field of cell tracking, about what performance measures should be used to evaluate algorithm performance. We chose the parameters presented above because they are easy to compute and because they capture the most important aspects of the track linking performance. Another important reason for choosing these performance measures is that they make it possible to compare our results to the results presented in [37] and [27]. The performance measures track purity, object purity, and mitosis recall are used for performance evaluation in [27], but in [27], object purity is referred to as target effectiveness and the mitosis recall is referred to as mitosis branching correctness.
We think that it is important to strive for a consensus about which performance measures to use for evaluation of cell tracking algorithms. The Cell Tracking Challenges [39], [40] is a great initiative to achieve this goal. In [39], [40], many different types of tracking errors are combined into a single performance measure called TRA. We however chose not to use this measure in this paper, because we want to study different aspects of the tracking performance separately. Furthermore, the TRA measure is designed to indicate how much time it would take for a human to correct all tracking errors, and therefore the measure gives high weights to tracking errors where cells are missing, and lower weights to linking errors. The TRA measure is therefore an appropriate measure when entire systems for cell tracking are evaluated, but given that the focus of this paper is on track linking, we found the measures presented above to be more appropriate.
Track purity is a measure of the degree to which the computer generated tracks follow ground truth objects and object purity is a measure of the degree to which the ground truth objects are followed by computer generated tracks. The track purity of a single track is computed by finding the object that the track follows in most images and dividing the number of images where the object is followed by the length of the track in frames. Object purity is computed in the same way, but with the roles of the tracks and the objects reversed. This means that the object purity of a single object is the largest number of images that it is followed by the same track, divided by the length of the object trajectory in frames.
For simplicity, a track is said to be following an object in a frame if its outline overlaps with the outline of the object in at least one pixel in that frame. This corresponds to an F-measure coverage threshold of 0 in [50]. This allows cells to be misplaced inside clusters, as long as the outlines overlap in at least one pixel. The outlines of cells which are not in clusters are the same in the ground truth and in the tracking results, so they either overlap in all pixels or in no pixels. To formalize this, we let ni and nj denote the lengths (in frames) of track i and object trajectory j respectively. Further, we let nij denote the number of images in which track i follows object j, and let nji denote the number of images in which object j is followed by track i. Then the track purity of track i can be written as [50, Eq. (9)]
| (12) |
and the object purity of object j can be written as [50, Eq. (10)]
| (13) |
The track or object purity of a set of tracks or objects is computed by averaging the values of the individual tracks or objects in the set. We use a weighted average where the trajectories are weighted by their length to prevent short trajectories of cells that move back and forth across the image boundary from skewing the results. For two mitotic events to be considered matching, they are not allowed to be separated in time by more than 50 min, the mother cells have to match in the last image of the mother cell that divided first, and each daughter cell has to match the corresponding ground truth daughter cell, in the first image of the cell that appears last of the two cells to be matched. The threshold of 50 min is taken from [27], so that our results can be compared to the results in that publication. For two apoptotic events to be considered matching, they are not allowed to be separated in time by more than 50 min, and the cells have to match in the last image of the cell that dies first. For mitotic and apoptotic events, cells can be considered to match if their outlines overlap in at least one pixel, but an event in the computer generated tracks can only match one ground truth event and vice versa.
The performance measures achieved by the proposed algorithm and by CellProfiler on the MuSC dataset and the myoblast dataset are shown in Tables II and III, respectively.2 The corresponding confusion matrices for classification of cell counts in detections are shown in Tables IV and V. The track and object purities of the proposed algorithm are very high on both datasets considering the challenges of false positive detections and clustering of cells. CellProfiler achieves lower (worse) values on both measures in the MuSC dataset. On the myoblast dataset CellProfiler achieves a track purity comparable to the track purity of the proposed method at the cost of a significantly lower object purity. Both the precision and the recall of mitotic events are high on the MuSC dataset for the proposed method. The performance measures for mitosis are slightly lower on the myoblast dataset, but the performance is still good considering that the cell density is much higher and that there is more clustering of cells than in the MuSC dataset. When the cell culture is dense and has a lot of clusters, it becomes very challenging to detect mitotic events as the tracks in adjacent images provide less context and as the mitotic cells that are segmented jointly with other cells are very hard to identify using classification techniques. As noted above, the problem of detecting apoptotic events correctly is more challenging than detecting mitotic events, as the changes in appearance when the cell dies are often very subtle. Therefore, the proposed method achieves slightly lower precision and recall for apoptosis than for mitosis in the MuSC dataset. The performance is however still very good considering that classification of dead and live cells can be challenging even for a human observer. CellProfiler detects many of the mitotic events in both datasets and even has a higher mitosis recall than the proposed method in the myoblast dataset. CellProfiler does however introduce a lot of incorrect mitotic events as well, so the mitosis precision is very low on both datasets. Typically, the algorithm generates a lot of spurious tracks by introducing mitotic events and later the tracks disappear through incorrect apoptotic events. Like almost all other cell tracking algorithms, CellProfiler does not model apoptotic events explicitly, and therefore both the precision and the recall are very low for apoptosis. The mitotic events that were correctly detected by the proposed algorithm in the MuSC dataset had delays with an average and a standard deviation of 2.0 ± 9.1 min and the corresponding delays for apoptotic events were 0.35 ± 16 min. For CellProfiler, the same delays were 27 ± 13 min and 5.5 ± 26 min. On the myoblast dataset, the mitosis delays were −0.78 ± 12min and 5.7 ± 19 min for the proposed method and CellProfiler, respectively. From the delay values it is clear that the proposed method is a lot better at determining the exact time of the different events and that it is harder to determine the exact time of an apoptotic event than a mitotic event. The detection of mitotic events is very often delayed in CellProfiler as a result of the two daughter cells being segmented together in a number of images following the mitotic event. In the myoblast dataset, the amount of apoptosis was negligible, and it was therefore not meaningful to include any performance measures for apoptosis in Table III.
TABLE II.
Tracking Performance on the MuSC Dataset
| Parameter | proposed | CellProfiler |
|---|---|---|
| track purity | 0.92 | 0.68 |
| object purity | 0.83 | 0.69 |
| mitosis precision | 0.79 | 0.17 |
| mitosis recall | 0.80 | 0.55 |
| apoptosis precision | 0.72 | 0.01 |
| apoptosis recall | 0.66 | 0.10 |
TABLE III.
Tracking Performance on the Myoblast Dataset
| Parameter | proposed | CellProfiler |
|---|---|---|
| track purity | 0.85 | 0.86 |
| object purity | 0.81 | 0.54 |
| mitosis precision | 0.59 | 0.17 |
| mitosis recall | 0.60 | 0.67 |
TABLE IV.
Confusion Matrices for the Number of Cells in Each Detection in the MuSC Dataset
| true # | 0 | 64249 | 1009 | 252 | 39884 | 25626 | 0 |
| 1 | 9422 | 68986 | 253 | 4522 | 74139 | 0 | |
| ≥ 2 | 172 | 1167 | 4166 | 0 | 5505 | 0 | |
|
| |||||||
| 0 | 1 | ≥ 2 | 0 | 1 | ≥ 2 | ||
| estimated # proposed method |
estimated # CellProfiler |
||||||
TABLE V.
Confusion Matrices for the Number of Cells in Each Detection in the Myoblast Dataset
| true # | 0 | 3012 | 336 | 41 | 2071 | 1318 | 0 |
| 1 | 1716 | 39472 | 2809 | 1048 | 42949 | 0 | |
| ≥ 2 | 3 | 2189 | 11520 | 3 | 13709 | 0 | |
|
| |||||||
| 0 | 1 | ≥ 2 | 0 | 1 | ≥ 2 | ||
| estimated # proposed method |
estimated # CellProfiler |
||||||
Even though we optimized the settings in CellProfiler for our datasets, we recognize that the developers of the algorithms and the CellProfiler software were not using our datasets during the development phase of their cell tracking method. We do however still believe that the comparison supports our claim that the tracking results that we have produced can not be achieved using existing methods and software. To give the reader an additional indication about what the numbers in Tables II and III mean in terms of tracking performance, we note that the method presented in [27] achieves a track purity of 0.81, an object purity of 0.87, and a mitosis recall of 0.65. The method in [27] achieves very good tracking results on cells from the cell line C2C12 imaged using phase-contrast microscopy, and is shown to outperform the highly cited, earlier work [23]. Given that we have applied our algorithm to other datasets, we can not objectively compare our performance values to the performance values in [27]. The fact that the proposed method achieves similar and in some cases higher performance values than the method in [27] does however support our claim that the proposed method produces tracks of high quality. Furthermore, phase-contrast microscopy is normally preferred to bright-field microscopy for cell tracking applications as it gives higher image contrast and as mitotic events can be identified by an increased brightness. It is therefore noteworthy that our algorithm produces good tracking results, and especially that mitotic events are identified with both high precision and high recall. In [27], mitotic events are handled using a mitosis detection algorithm specifically designed for phase-contrast microscopy [4]. Compared to the method used in [27], the proposed algorithm for handling of mitotic events has the advantages that it is independent of the microscopy technique used, and that it treats mitotic events in a probabilistic manner without making hard classification decisions before the tracking is started.
For further comparisons with other track linking algorithms, we refer the reader to [39], where an earlier version of the proposed track linking algorithm is compared on equal terms to five other methods on eight different datasets of real and simulated fluorescence microscopy images. In [39], the different track linking algorithms were used together with different segmentation algorithms, so good tracking performance was somewhat dependent on having a good segmentation algorithm. The earlier version of our algorithm did however achieve the highest track linking performance of the six methods, on five of the eight datasets, in combination with a simple segmentation algorithm based on Gaussian bandpass filtering followed by thresholding and separation of cells in clusters using the watershed transform. A compiled version of the software that we used to process the datasets was made available when [39] was published, and can be downloaded from [40]. In the ISBI 2014 Cell Tracking Challenge, the algorithm that we propose herein was compared to 7 other methods on 14 different datasets generated using fluorescence microscopy, phase contrast microscopy, differential interference contrast (DIC) microscopy, and simulated fluorescence microscopy. The bandpass filtering algorithm described above was used to process most of the datasets, but a segmentation algorithm based on local variance (similar to the one described in Section V-C) was used on a phase contrast dataset and a segmentation algorithm based on ridge detection was used on a DIC dataset. Our cell tracking system achieved the highest performance of all the competing systems on all 14 datasets [40].
3) Run Time
To give the reader a rough idea about the run time of the proposed algorithm we measured the time it took to process the MuSC dataset and the myoblast dataset. The processing was done on a desktop computer with an Intel Core i7-3930K 3.2 GHz processor with six cores, and 64 GB of RAM memory, running MATLAB 2013b in Windows 7. It took 26 h and 48 min to process the MuSC dataset and 1 h and 6 min to process the myoblast dataset. This corresponds to 1.6 s per image for the MuSC dataset, and 2.8 s per image for the myoblast dataset. Table VI shows how this run time was distributed among different processing tasks. Most of the time was spent on image stabilization, segmentation, and feature computation. In the myoblast dataset, no image stabilization was performed, and the segmentation took less time than in the MuSC dataset as no background subtraction was performed, but the myoblast dataset still took almost twice as long to process per image, as the feature computation took much longer due to the large number of detections. In both datasets, the time consumed by the track linking algorithm was negligible compared to the other tasks, showing that it would be possible to process datasets where the number of cells per image is significantly higher, before the processing power becomes the limiting factor. The track linking algorithm itself processed 65 images per second in the myoblast dataset and images per second in the MuSC dataset.
TABLE VI.
Run Times of Different Processing Steps, in Seconds per Image
| Task | MuSCs | Myoblasts |
|---|---|---|
| stabilization | 0.47 | NA |
| cropping | 0.067 | NA |
| segmentation | 0.54 | 0.44 |
| feature computation | 0.48 | 1.7 |
| computation of scores | 0.0068 | 0.18 |
| track linking | 0.000082 | 0.015 |
| post processing | 0.0053 | 0.25 |
| other | 0.048 | 0.15 |
| all | 1.6 | 2.8 |
In the run time evaluation, we processed a single image sequence at a time, to simplify the evaluation and the interpretation of the results. By processing one image sequence on each processor core, the run time for the MuSC dataset was reduced to 7 h and 44 min and the run time for the myoblast dataset was reduced to 24 min, corresponding to speed-ups by factors of 3.5 and 2.8, respectively. For the MuSC dataset, the speed-up was much less than a factor of 6, indicating that the processor speed is not the limiting factor when all six cores are used in parallel, but the limited speedup is partly explained by the fact that the segmentation algorithm is implemented using the fast Fourier transform, which has built in parallelization in MATLAB. Cell-Profiler uses parallel processing by default, and required 35 h and 26 min to process the MuSC dataset and 44 min to process the myoblast dataset, even though the tasks of stabilization, cropping, and segmentation had been replaced by loading of label images. This shows that our cell tracking system as a whole is competitive when it comes to run time.
VI. Conclusion
We have presented a global track linking algorithm for cell tracking, which uses information from all images in an image sequence to make local linking decisions. The algorithm can handle false detections, missed detections, detections containing multiple cells, mitosis, apoptosis, and cells migrating in and out of the field of view. All of the different types of events are incorporated into the same probabilistic framework and there is no need for heuristic postprocessing algorithms or hard decisions made by detection algorithms, to handle for example mitosis or apoptosis.
The algorithm starts from an empty set of tracks and adds one track at a time in a greedy way which gives the maximum increase to a probabilistically motivated scoring function, until the scoring function can not be increased further by adding an additional track. During the addition of a new track, preexisting tracks can be modified, so that a large number of cells can be tracked without problems with incorrect tracks blocking the creation of correct tracks in subsequent iterations. In each iteration, the track to be added is found by solving an optimization problem using the Viterbi algorithm. The track linking algorithm can be made to have linear complexity in the number of images and quadratic complexity in the number of detections in each image, and it is therefore suitable for tracking in long image sequences with a large number of cells in each image. The track linking algorithm is independent of the method used for image segmentation and can therefore be applied to a broad spectrum or problems in both 2-D and 3-D. The algorithm has been shown to give good performance on two very challenging datasets with MuSCs and myoblasts imaged using bright-field microscopy.
Supplementary Material
Acknowledgments
The authors would like to thank the reviewer who alerted us to the prior work on Viterbi algorithms in tracking for defense applications, and the reviewer who suggested expanding the performance evaluation with information about how well the proposed algorithm determines how many cells each detection contains. We would also like to thank the reviewers who prompted the inclusion of the comparison with CellProfiler.
This work was supported in part by the Swedish Research Council (VR) under Grant 621-2011-5884, in part by the National Institutes of Health (NIH) under Grant 1R01 HL096113, and in part by the California Institute for Regenerative Medicine (CIRM) under Grant RT1-01001-1.
Appendix A: Scoring Function Updates
This appendix describes the utilities of the different arc types in the state space diagram described in Section IV-B. These utilities are equal to the change in the scoring function g(F), that occurs when the operation associated with the arc is performed on F.
An arc from A to Dt,i represents adding a new cell, which is present in the first image, to F, and has the utility
| (14) |
A migration arc from Dt,i to Dt+τ,j represents appending the detection Dt+τ,j to the end of the track under creation, which currently ends in detection Dt,i, and has the utility
| (15) |
A mitosis arc from Yt to Dt+1,j represents adding a new daughter cell starting in detection Dt+1,j, by introducing a mitotic event in a preexisting track. This breaks the preexisting track into two segments, where the first segment becomes the mother cell, and the second segment becomes the other daughter cell. This corresponds to adding an additional branch to one of the trees in F. If the preexisting track that will become the mother cell passes through detection Dt,i, the utility of the arc is given by
| (16) |
An apoptosis arc from Dt,i to Xt+1 represents terminating the cell under creation with an apoptotic event, and has the utility
| (17) |
An arc from Yt to Dt+1,j for migration into the image adds a new cell to F, and has the utility
| (18) |
An arc from Dt,i to Xt+1 for migration out of the image terminates the track under creation, and has the utility
| (19) |
A swap arc from Dt,i to , which breaks a migration link between detections and in a preexisting track, will append the later section of the preexisting track after Dt,i at the end of the track under creation, append at the end of the first section of the preexisting track, and let the now extended first section be the new track under creation. This arc has the utility
| (20) |
where F′ is a modified version of F, where the swap operation has been performed.
In a generalized swap, which can change if and how a preexisting track starts and ends, the three migrations can be replaced by migration into the field of view, mitosis followed by migration, migration out of the field of view, and apoptosis. In particular, the first migration can be replaced by It+1,n, followed by , Ot,i, or At,i, the second migration can be replaced by , followed by , , or , and the third migration can be replaced by , followed by , , or , where k1∈[1,…,Nt] and . To produce a valid swap operation, the third event must start in the same state of the state space diagram as the second event and end in the same state as the first event.
Appendix B: Features Used for Classification
For classification of cell counts, mitosis, and apoptosis, we use the feature set described in [7], combined with some additional features. The feature set in [7] has 18 shape-based features derived from the binary segmentation mask and 21 appearance-based features derived from the pixel intensities inside the segmented outline. Out of the 18 shape-based features, 13 are based on different image moments of the whole pixel region and five are based on the boundary of the region. The appearance-based features are the mean, standard deviation, and skew of the image itself, the gradient computed at three different scales, and the Laplacian computed at three different scales. The feature set in [7] was designed to classify morphologies of individually segmented cells and identify false positive detections. Therefore, we had to extend the feature set for detection of temporal events like mitosis and apoptosis, and for classification of detections containing multiple cells.
To increase the classification performance in cell tracking applications, we removed the cell centroid from the original feature set, for classification of myoblasts. For classification of MuSCs we instead replaced the centroid by the distance between the centroid and the center of the microwell. Furthermore, we extended the appearance-based features by using the minimum, maximum, mean, standard deviation, and skew of the image itself, the gradient and Laplacian images described above, the background image, the difference between the previous image and the current image, and the difference between the next image and the current image. In addition to these features, we also added the mean absolute difference from the mean intensity, of the image itself, and the two difference images.
We also added the two features and , which measure the amounts of image gradient that are perpendicular and parallel, to the boundary of detection Dt,i, respectively. To compute the features, we first compute the distance transform of the segmented pixel region, where the pixel values of the transform represent the distance to the closest background pixel. Then we compute the gradient components of the distance transform, and , and use them together with the gradient components of the original image, Gx(x,y) and Gy(x,y), to define the features
| (21) |
and
| (22) |
where (x, y) represents pixel coordinates. These two features can for example be used to identify detections with multiple cells, as the image gradients at the borders between the cells have large components that are parallel to the detection boundary. A similar idea is used in [26], where the amount of false positive detections is reduced by observing that the image intensity is close to constant along the boundaries of Hematopoietic Stem Cells imaged using phase-contrast microscopy.
Footnotes
This work was published in part at the 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona, Spain, 2012.
This paper has supplementary downloadable material available at http://ieeexplore.ieee.org, provided by the authors.
In [38] and [39], the focus is on datasets and performance measures that can be used to compare different algorithms for segmentation and tracking, and therefore the publications only provide high level descriptions of the precursors to the algorithm presented here.
If the track purities and the object purities of the individual tracks or objects are not weighted by the lengths of the trajectories, the average track purities of the proposed method would be 0.90 and 0.78, and the average object purities would be 0.84 and 0.90, for MuSCs and myoblasts respectively. For the MuSC dataset, the unweighted measures are very close to the weighted measures, as most tracks are of similar length. For the myoblast dataset however, the unweighted measures are slightly different, as equal emphasis is put on long tracks and short track fragment at the image boundary. We think that the weighted measures are more appropriate, but we present the unweighted measures as well, to allow comparisons with as many other works as possible.
Contributor Information
Klas E. G. Magnusson, Department of Signal Processing, ACCESS Linnaeus Centre, KTH Royal Institute of Technology, 100 44 Stockholm, Sweden.
Joakim Jaldén, Department of Signal Processing, ACCESS Linnaeus Centre, KTH Royal Institute of Technology, 100 44 Stockholm, Sweden.
Penney M. Gilbert, Institute of Biomaterials and Biomedical Engineering, University of Toronto, Toronto, ON, M5S 3G9 Canada, and with the Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, ON, M5S 3E1 Canada University Health Network Arthritis Program, Toronto, ON, M5G 2M9 Canada.
Helen M. Blau, Baxter Laboratory for Stem Cell Biology, Department of Microbiology and Immunology, and the Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA 94305 USA
References
- 1.Coutu DL, Schroeder T. Probing cellular processes by long-term live imaging—Historic problems and current solutions. J Cell Sci. 2013;126(17):3805–3815. doi: 10.1242/jcs.118349. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport DH, Becker T, Mamlouk AM, Schicktanz S, Kruse C. A novel validation algorithm allows for automated cell tracking and the extraction of biologically meaningful parameters. PLoS One. 2011;6(11):e27315. doi: 10.1371/journal.pone.0027315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464(7289):721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huh S, Eom S, Bise R, Yin Z, Kanade T. Mitosis detection for stem cell tracking in phase-contrast microscopy images. Proc IEEE Int Symp Biomed Imag From Nano to Macro. 2011:2121–2127. [Google Scholar]
- 5.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44(6):817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 6.Bise R, Kanade T, Yin Z, Huh S-I. Automatic cell tracking applied to analysis of cell migration in wound healing assay. Proc IEEE Annu Int Conf IEEE Eng Med Biol Soc. 2011:6174–6179. doi: 10.1109/IEMBS.2011.6091525. [DOI] [PubMed] [Google Scholar]
- 7.Theriault DH, Walker ML, Wong JY, Betke M. Cell morphology classification and clutter mitigation in phase-contrast microscopy images using machine learning. Mach Vis Appl. 2012;23(4):659–673. [Google Scholar]
- 8.Chen X, Zhou X, Wong ST. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans Biomed Eng. 2006 Apr;53(4):762–766. doi: 10.1109/TBME.2006.870201. [DOI] [PubMed] [Google Scholar]
- 9.Li F, Zhou X, Ma J, Wong S. Multiple nuclei tracking using integer programming for quantitative cancer cell cycle analysis. IEEE Trans Med Imag. 2010 Jan;29(1):96–105. doi: 10.1109/TMI.2009.2027813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulston JE, Schierenberg E, White JG, Thomson J. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100(1):64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 11.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322(5904):1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 12.Eilken HM, Nishikawa S-I, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457(7231):896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329(5995):1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bise R, Li K, Eom S, Kanade T. Reliably tracking partially overlapping neural stem cells in DIC microscopy image sequences. MICCAI Workshop OPTMHisE. 2009:67–77. [Google Scholar]
- 15.Cohen AR, Gomes FL, Roysam B, Cayouette M. Computational prediction of neural progenitor cell fates. Nat Methods. 2010;7(3):213–218. doi: 10.1038/nmeth.1424. [DOI] [PubMed] [Google Scholar]
- 16.Zimmer C, Zhang B, Dufour A, Thébaud A, Berlemont S, Meas-Yedid V, Marin J-C. On the digital trail of mobile cells. IEEE Signal Process Mag. 2006 May;23(3):54–62. [Google Scholar]
- 17.Meijering E, Dzyubachyk O, Smal I, van Cappellen WA. Tracking in cell and developmental biology. Semin Cell Dev Biol. 2009;20(8):894–902. doi: 10.1016/j.semcdb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Rohr K, Godinez WJ, Harder N, Wörz S, Mattes J, Tvaruskó W, Eils R. Tracking and quantitative analysis of dynamic movements of cells and particles. Cold Spring Harb Protoc. 2010 Jun;(6) doi: 10.1101/pdb.top80. [DOI] [PubMed] [Google Scholar]
- 19.Kanade T, Yin Z, Bise R, Huh S, Eom S, Sandbothe MF, Chen M. Cell image analysis: Algorithms, system and applications. IEEE Workshop Appl Comput Vis. 2011:374–381. [Google Scholar]
- 20.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenouard N, Bloch I, Olivo-Marin J-C. Multiple hypothesis tracking for cluttered biological image sequences. IEEE Trans Pattern Anal Mach Intell. 2013 Nov;35(11):2736–2750. doi: 10.1109/TPAMI.2013.97. [DOI] [PubMed] [Google Scholar]
- 22.Zimmer C, Labruyere E, Meas-Yedid V, Guillen N, Olivo-Marin J-C. Segmentation and tracking of migrating cells in videomicroscopy with parametric active contours: A tool for cell-based drug testing. IEEE Trans Med Imag. 2002 Oct;21(10):1212–1221. doi: 10.1109/TMI.2002.806292. [DOI] [PubMed] [Google Scholar]
- 23.Li K, Miller ED, Chen M, Kanade T, Weiss LE, Campbell PG. Cell population tracking and lineage construction with spatiotemporal context. Med Image Anal. 2008;12(5):546–566. doi: 10.1016/j.media.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dzyubachyk O, van Cappellen WA, Essers J, Niessen WJ, Meijering E. Advanced level-set-based cell tracking in time-lapse fluorescence microscopy. IEEE Trans Med Imag. 2010 Mar;29(3):852–867. doi: 10.1109/TMI.2009.2038693. [DOI] [PubMed] [Google Scholar]
- 25.Al-Kofahi O, Radke RJ, Goderie SK, Shen Q, Temple S, Roysam B. Automated cell lineage construction: A rapid method to analyze clonal development established with murine neural progenitor cells. Cell Cycle. 2006;5(3):327–335. doi: 10.4161/cc.5.3.2426. [DOI] [PubMed] [Google Scholar]
- 26.Kachouie NN, Fieguth P, Ramunas J, Jervis E. Probabilistic model-based cell tracking. Int J Biomed Imag. 2006;2006:1–10. doi: 10.1155/IJBI/2006/12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bise R, Yin Z, Kanade T. Reliable cell tracking by global data association. Proc IEEE Int Symp Biomed Imag, From Nano to Macro. 2011:1004–1010. [Google Scholar]
- 28.Dufour A, Thibeaux R, Labruyere E, Guillen N, Olivo-Marin J-C. 3-D active meshes: Fast discrete deformable models for cell tracking in 3-D time-lapse microscopy. IEEE Trans Image Process. 2011 Jul;20(7):1925–1937. doi: 10.1109/TIP.2010.2099125. [DOI] [PubMed] [Google Scholar]
- 29.Maška M, Daněk O, Garasa S, Rouzaut A, Munoz-Barrutia A, Ortiz-de Solorzano C. Segmentation and shape tracking of whole fluorescent cells based on the Chan-Vese model. IEEE Trans Med Imag. 2013 Jun;32(6):995–1006. doi: 10.1109/TMI.2013.2243463. [DOI] [PubMed] [Google Scholar]
- 30.Pulford G, La Scala B. Multihypothesis Viterbi data association: Algorithm development and assessment. IEEE Trans Aerosp Electron Syst. 2010 Apr;46(2):583–609. [Google Scholar]
- 31.Padfield D, Rittscher J, Roysam B. Spatio-temporal cell segmentation and tracking for automated screening. Proc IEEE Int Symp Biomed Imag, From Nano to Macro. 2008:376–379. [Google Scholar]
- 32.Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans Image Process. 2005 Sep;14(9):1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 33.Berclaz J, Fleuret F, Fua P. Robust people tracking with global trajectory optimization. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit Workshop. 2006;1:744–750. [Google Scholar]
- 34.Pirsiavash H, Ramanan D, Fowlkes CC. Globally-optimal greedy algorithms for tracking a variable number of objects. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit Workshop. 2011:1201–1208. [Google Scholar]
- 35.Barniv Y. Dynamic programming solution for detecting dim moving targets. IEEE Trans Aerosp Electron Syst. 1985 Jan;(1):144–156. [Google Scholar]
- 36.Pulford GW, La Scala BF. MAP estimation of target manoeuvre sequence with the expectation-maximization algorithm. IEEE Trans Aerosp Electron Syst. 2002;38(2):367–377. [Google Scholar]
- 37.Magnusson KEG, Jaldén J. A batch algorithm using iterative application of the Viterbi algorithm to track cells and construct cell lineages. Proc 9th IEEE Int Symp Biomed Imag. 2012:382–385. [Google Scholar]
- 38.Chenouard N, et al. Objective comparison of particle tracking methods. Nat Methods. 2014;11(3):281–289. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maška M, et al. A benchmark for comparison of cell tracking algorithms. Bioinformatics. 2014;30(11):1609–1617. doi: 10.1093/bioinformatics/btu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cell Tracking Challenge [Online]. Available: http://www.codes-olorzano.com/celltrackingchallenge/Cell_Tracking_Challenge/Welcome.html accessed: 2014-07-22
- 41.Carpenter AE, et al. Cellprofiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forney GD., Jr The Viterbi algorithm. Proc IEEE. 1973 Mar;61(3):268–278. [Google Scholar]
- 43.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125(6):1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li K. The Image Stabilizer Plugin for ImageJ [Online] 2008 Feb; Available: http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html.
- 45.Wu K, Gauthier D, Levine MD. Live cell image segmentation. IEEE Trans Biomed Eng. 1995 Jan;42(1):1–12. doi: 10.1109/10.362924. [DOI] [PubMed] [Google Scholar]
- 46.Soille P. Morphological Image Analysis: Principles and Applications. New York: Springer-Verlag; 2003. [Google Scholar]
- 47.Bishop CM, Nasrabadi NM. Pattern Recognition and Machine Learning. New York: Springer; 2006. [Google Scholar]
- 48.Gross D, Shortle JF, Thompson JM, Harris CM. Fundamentals of Queueing Theory. New York: Wiley; 2013. [Google Scholar]
- 49.Papadimitriou CH, Steiglitz K. Combinatorial Optimization: Algorithms and Complexity. Mineola, NY: Dover; 1998. [Google Scholar]
- 50.Smith K, Gatica-Perez D, Odobez J-M, Ba S. Evaluating multi-object tracking. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit Workshops. 2005:36–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


