Abstract
Reversible protein phosphorylation is a crucial regulatory mechanism that controls many biological processes in eukaryotes. In plants, phosphorylation events primarily occur on serine (Ser) and threonine (Thr) residues, while in certain cases, it was also discovered on tyrosine (Tyr) residues. In contrary to plants, extensive reports on Tyr phosphorylation regulating a large numbers of biological processes exist in animals. Despite of such prodigious function in animals, Tyr phosphorylation is a least studied mechanism of protein regulation in plants. Recently, various chemical analytical procedures have strengthened the view that Tyr phosphorylation is equally prevalent in plants as in animals. However, regardless of Tyr phosphorylation events occuring in plants, no evidence could be found for the existence of gene encoding for Tyr phosphorylation i.e. the typical Tyr kinases. Various methodologies have suggested that plant responses to stress signals and developmental processes involved modifications in protein Tyr phosphorylation. Correspondingly, various reports have established the role of PTPs (Protein Tyrosine Phosphatases) in the dephosphorylation and inactivation of mitogen activated protein kinases (MAPKs) hence, in the regulation of MAPK signaling cascade. Besides this, many dual specificity protein phosphatases (DSPs) are also known to bind starch and regulate starch metabolism through reversible phosphorylation. Here, we are emphasizing the significant progress on protein Tyr phosphatases to understand the role of these enzymes in the regulation of post-translational modification in plant physiology and development.
Keywords: Carbohydrate binding module, Laforin, Kinases, Protein tyrosine phosphatase, Tyrosine phosphorylation, Signal transduction, Stress.
INTRODUCTION
Protein phosphorylation is a crucial post-translational event, first reported almost 50 years ago by Edwin Krebs and Edmond Fischer [1, 2]. Protein phosphorylation is known to alter the three dimensional structure, activity, cellular localization, and the stability of a protein, which in turn acts as an ‘on and off’ switch in numerous pathways regulating growth, differentiation, and oncogenesis [3-5]. Almost more than 70% of all the known proteins of eukaryotes are reversibly phosphorylated and hence, the importance of reversible protein phosphorylation to cellular regulation cannot be overstated [6]. Phosphorylation usually takes place at nine amino acids (Ser, Thr, Tyr, Cys, Arg, Lys, Asp, Glu and His) but most extensively phosphorylated amino acids are Ser, Thr and Tyr. Phosphoserines are the most abundant group of phosphoproteins followed by phosphothreonine and phosphotyrosine. In eukaryotes, phosphorylation on Ser, Thr, and Tyr is nearly 86.4%, 11.8% and 1.8%, respectively [2]. Although, the percentage of Tyr phosphorylation is relatively less as compared to the Ser and Thr residues, it is still considered as a crucial event in cells and is known to play significant role in various signaling and regulatory processes ineukaryotes (especially animals). Tyr phosphorylation is a fundamental mechanism for numerous important aspects of eukaryotic physiology [7]. However, it is true that the balance between protein tyrosine kinase (PTK) and protein tyrosine phosphatase (PTP) activity is the key for successful regulation of Tyr phosphorylation, as the numbers of PTKs are more than PTPs in the cell [8]. This is evident from the fact that, first PTP was identified and cloned ten years after the identification of first PTK [9].
PROTEIN TYROSINE PHOSPHATASES: STRUCTURAL FEATURES AND CLASSIFICATION
Although, different types of protein phosphatases have been discovered in plants [10, 11] and the role of many such phosphatases in various pathways are also known but our understanding of the protein Tyr phosphatase structure and function has largely been elucidated from animal and yeast systems where these enzymes were first discovered [12, 13]. PTPs are a family of enzymes whose structural diversity and complexity rival those of the PTKs. The PTKs are well known to share sequence similarity with protein serine/threonine kinases; however, the enzymes belonging to PTPs share no sequence similarity with protein serine/threonine phosphatases [14]. PTPs are characterized by the presence of active site signature motif (H/V)C (X)5R(S/T) in the conserved catalytic domain. This domain harbors the cysteine residue involved in the formation of phosphoenzyme reaction intermediate [15]. Structural domains like src homology 2 (SH2), PSD-95/Dlg/ZO-1 (PDZ), extracellular domains, etc. are also present in the PTP [16]. The three dimensional structure of over a dozen PTPs of animal origin has been determined, while not much work has yet been done in plants. The first crystal structure elucidation of protein phosphatase was that of PTP isolated from human placenta [17]. The crystal structure not only provides insight into the structural basis for catalysis but also provides insight into the substrate specificity. The PTPs so far characterized are known to possess both α helices and β sheets. The structure showed the presence of β sheet flanked by α helices on both sides [18]. Both the classes of PTPs (Tyr specific and dual-specificity) despite of having great difference in the amino acid sequences and the substrate specificity, showed high similarity in their crystal structure [19]. The active site of the PTP is located inside the crevices on the protein’s surface. The depth of active site cleft is more for Tyr specific phosphatases as compared to dual specificity phosphatases. Study on crystal structure of PTPs revealed that much deeper active site cleft in the Tyr-specific phosphatases selects exclusively pTyr-containing substrates, while the more shallow, active site cleft for the DSPs may contain both pTyr and pSer/pThr residues [20-23]. In addition, from structural study, we now recognised that the structure of the P-loop, including different residues (Cys and Arg, Ser/Thr, and Asp residues) are conserved in the PTPs, DSPs and LMWPTPs and thus, utilize a common strategy for phosphate monoester hydrolysis. In catalysis mechanism, Cys residue act as nucleophile and attack on phosphate ester to form a thiophosphate intermediate. The formation of thiophosphate is supported by a conserved aspartic acid (Asp181), which acts as a general acid for stabilization of the negative charge on the leaving thiolate group [19]. For the hydrolysis of intermediate product, Asp residue now serves as a general base by recruiting water molecule. Furthermore, Arg residue (Arg221) functions to facilitate the breakdown of the thiophosphate intermediate [24].
Earlier PTPs were categorized into three broad classes namely receptor-like PTPs, intracellular PTPs, and dual-specificity PTPs. These enzymes are unique among the PTPs in their capability to utilize phosphoserine and phosphothreonine as substrates in addition to phosphotyrosine [25]. While, on the basis of amino acid residues in their catalytic domains, PTPs are grouped into four sub-families i.e., type 1, 2, and 3 Cys-based PTPs, and the Asp-based phosphatases [26, 27]. The type-1 sub-family is the largest sub-family. This sub-family contains Cys in their catalytic domain. This is further subdivided into two sub-groups, based on the substrate specificity i.e., tyrosine specific classical PTPs and dual specificity phosphatases. The type-2 PTPs are low molecular weight PTPs and are tyrosine specific. Type-2 PTPs are frequently found in prokaryotes [26, 27]. The third sub-group of PTPs includes cell cycle regulators and show specificity towards both Tyr and Thr residues. Type-3 PTPs are known to dephosphorylate cyclin-dependent kinases (CDKs) causing inhibition of cell cycle progression [28]. Fourth sub-group of phosphatases are aspartic acid based PTPs, which comprises a heterogeneous group of phosphatases, that can be either Ser or Tyr specific, for example, EYA (Eyes Absents) phosphatase and HAD (Haloacid Dehalogenase) family of phosphatases in humans [29-31]. A widely used way to classify Tyr phosphatases is to categorize them into two main classes, one being Tyr specific PTPs and the other as dual specificity phosphatases (DSP) [32]. Both of these classes of Tyr phosphatases are well characterized in animals and microbes but not many of them are studied in plants.
Similarly, PTKs are an important family of regulatory enzymes in higher eukaryotes. The protein Ser/Thr kinases and protein Tyr kinases (PTKs) act on Ser/Thr and Tyr residues respectively, and are distinct from dual-specificity PTKs (DsPTKs). DsPTKs phosphorylates Ser, Thr, and Tyr. PTKs and DsPTKs share a common catalytic domain of around 250 amino acids containing 11 conserved subdomains (I to XI) [33]. At the N-terminal extremity of catalytic domain, subdomain II has a conserved lysine residue, known to be involved in ATP binding. The subdomain VII, located in the middle of the catalytic region contain aspartic acid residue, which is crucial for the catalytic activity of the kinase. Subdomain VI and VIII confers Ser/Thr and Tyr specificity, respectively. Consensus DL(R/A)A(A/R)N is specific for Tyr kinases whereas, the consensus DLKXXN is Ser/Thr-specific [7]. So far, a typical PTK could not be reported in plants; however, several DsPTKs have been identified through bioinformatics analysis [34].
IDENTIFICATION OF PROTEIN TYOROSINE PHOSPHATASES
The role of PTP in various cellular processes has recently been elucidated in organisms like Homo sapiens, Strongylocentrotus purpuratus, Plasmodium falciparum and others [24, 35]. Even though, several lines of evidences proved the presence of PTPs in various plant species, the complete PTPome has only been explored in Arabidopsis and rice [10, 11]. Tyr phosphorylation in plants was first described by Torruella et al. (1986), where they observed Tyr kinase activity in etiolated pea plants [36]. Since then, wide ranges of experimental tools have been employed to explore the presence of Tyr phosphorylation in plants [37-39]. Radioactive labeling has been extensively used for the detection of Tyr phosphorylation. The initial detection of phosphor-Tyr was based on radioactive labeling of 32P-ortho-phosphate of proteins present in the cells, followed by electrophoresis to separate the proteins and then partial acid hydrolysis. It was succeeded by thin layer chromatography and autoradiography of the hydrolysates, which led to the detection of phosphor-Ser, Phosphor-Thr, and phosphor-Tyr [40]. Due to limited occurrence of Phosphor-Tyr in comparison to phosphor-Ser and phosphor-Thr in the cell, its detection was quite difficult using conventional electrophoretic techniques [41]. An alternative approach for the detection of Tyr-phosphorylation makes use of monoclonal antibodies [42]. Phosphor-Tyr specific monoclonal antibodies are used for detecting phosphorylated-Tyr residues by using western blot technique. In this technique, antibodies are raised against immunizing rabbits with synthetic phosphopeptides representing the amino acid sequences, adjacent to the phosphorylation site of the target protein. This antibody largely recognized proteins containing phosphotyrosine. Afterwards, by using western blot technique multiple phosphoproteins could be resolved by their molecular weight. The presence of Tyr-phosphorylation in Pisum sativum was detected using this technique [35]. This technique has also been used to detect the presence of Tyr phosphorylated proteins in other plants such as Arachis hypogaea and Cocos nucifera [34, 41, 42].
The specific components of the signal transduction pathways can be deciphered through loss-of-function of one of these elements. This can be done using pharmacological approaches, which can inactivate single component in intact cells of the organisms. This approach uses PTKs and PTPs inhibitors such as genistein and lavendustin, respectively [45, 46]. Because of the advancement of tools and techniques, PTPs in plants could also be identified after ten years of the discovery of PTP in animals. The first plant PTP, AtPTP1 was characterized in Arabidopsis followed by dual specificity phosphatase by Luan and co-workers [13, 46, 48]. Subsequently, Singh et al. (2010) classified PTPs as well as DSPs gene families for the first time in crop plant rice (Oryza sativa) [11]. Presence of Tyr phosphatase like gene has also been identified in other plants like tomato and Pinus sylvestris [49, 50]. Recent advances in the analytical techniques such as mass spectrometry and affinity purification have led to discovery of several tyrosine phosphatases in plants.
PROTEIN TYROSINE PHOSPHATASE GENE FAMILY IN PLANTS
As mentioned earlier, Try phosphatases have been widely studied in animals and they play critical roles in regulating a large number of cellular pathways. Luan and co-workers have identified the first plant Tyr phosphatase, AtPTP1 in Arabidopsis [13]. Later, orthologues of AtPTP1 were also reported in other plants species [51]. The first dual specificity phosphatase AtDsPTP1 was identified in Arabidopsis in 1998 [47]. AtDsPTP1 resembles animal MKPs and it can dephosphorylate both phospho-tyrosine and phospho-serine or phospho-threonine. AtPTP1 and AtDsPTP1 shared homology within their respective signature motifs in the catalytic core. The overall protein sequence of Tyr-specific PTP and DsPTP share little homology but these phosphatases contain the CX5R motif in their catalytic domain: (V/I)HCXAGXGR(S/T) [47]. Similarly, in animals, the role of DsPTPs in the inactivation and dephosphorylation of Tyr kinases and MAPKs has been widely reported [52, 53]. Subsequently, whole genome sequencing of Arabidopsis facilitated identification of many more genes, encoding proteins having catalytic core motif of PTPs [10]. Out of around 24 members of putative PTPs from Arabidopsis, one encodes a Tyr specific PTP, 22 genes encode dual-specificity protein Tyr phosphatases (DSPs or DsPTPs) and a single gene of low molecular weight PTP (LMWPTP) [10]. Furthermore, by using complete human genome as reference, some novel PTPs have been identified from the genomes of Arabidopsis thaliana, Chlamydomonas reinhardtii, Ostreococcus tauri, Oryza sativa, and Populus trichocarpa [54]. From this study a single member of PTP was identified from all the four species. Also, a single member of low molecular weight PTP (LMWPTP) was identified in A. thaliana, O. sativa, and C. reinhardtii. Moreover, 2 LMWPTP were present in P. trichocarpa [54]. Later on, genome-wide study from rice revealed that rice comprised of 23 DSPs, a single member of PTP and LMWPTP [11, 55]. Additionally, 29 PTPs have been identified from maize on the basis of sequence similarity of protein phosphatases from rice and Arabidopsis [56]. Interestingly, genomes of Arabidopsis, rice, and maize contain much lesser PTPs than the human genome, comprised of more than 100 PTPs including 60 DSPs. On the other hand, the number of protein kinases in Arabidopsis is twice the number found in human. This observation leads us to a conclusion that either the protein Tyr-specific phosphatases (PTPs) and DSPs are directed at more sites or the protein Tyr component is limited in plant phosphoproteoms.
FUNCTIONAL ROLE OF PROTEIN TYR PHOSPHORYLATION AND DSP IN PLANTS
Protein Tyr phosphorylation plays a major role in many signaling pathways. In animal cells, many physiological roles of Tyr phosphorylation such as regulation of growth, differentiation, or oncogenesis have been studied in detail [5]. On the other hand, Tyr phosphorylated proteins have been detected in higher plants and evidence for regulation by Tyr phosphorylation in plant physiological processes is beginning to emerge. This section of the review provides an overview of Tyr phosphorylation in signaling processes and their role in physiological responses in plant cells. These include regulation at the level of expression, localization, substrate specificity, and activity of these enzymes.
In Response to Abiotic Stress
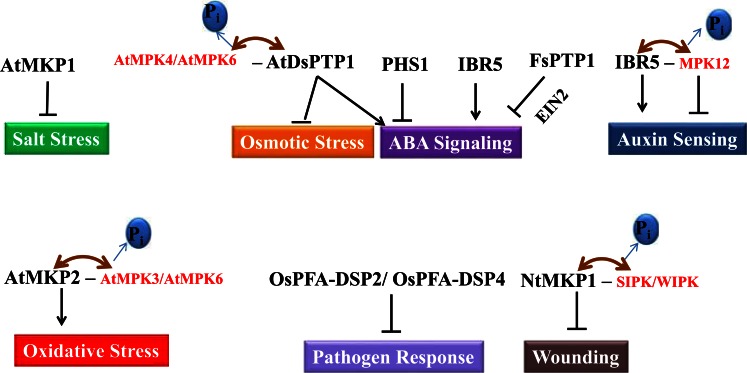
The growth and development of land plants is greatly affected by a variety of abiotic factors like dehydration, low temperature, salinity, heat, wounding, and pathogen infection. Consequently, plants exposed to diverse stress conditions, activate a complex set of distinct signaling pathways from perception of stress signal to amplification, transmission, and finally generation of responses, which helps in stress adaptation [57-59]. Protein phosphorylation and dephosphorylation, catalyzed by protein kinases and phosphatases play an important role in the co-ordination of various signal transduction pathways [5, 60]. One of the immediate and important responses under a particular condition is change in the gene expression that might be linked to regulation of that particular response. In Arabidopsis, the expression of AtPTP1, a protein Tyr phosphatase, was found to be manifold increased under salinity stress while reduced significantly in cold stress [13]. Recently, it was found that AtDsPTP1 (Dual-specificity protein phosphatase 1) mutant grew better than wild type (WT) under osmotic stress and hence acts as a negative regulator of osmotic stress (Fig. 1). Also, AtDsPTP1 regulated the expression of many genes associated with ABA biosynthesis and ABA-catabolism related genes such as NCED3 and CYP707A4 and hence positively regulates ABA accumulation [61]. Another report in chestnut showed CsDSP4 to be highly induced in the stems of chestnut seedlings exposed to low temperatures [62]. OsPFA-DSP1 (fungi atypical dual-specificity phosphatase from rice) is known to function as a negative regulator of drought stress in rice (Fig. 1). Overexpression of this DSP protein results into increased sensitivity of plants towards drought stress [63]. Arabidopsis, MAPK phosphatase 1 (MKP1) overexpression line is tolerant to salt stress. It is also proven that mutants lacking AtMPK1 are hypersensitive to salinity [64]. Furthermore, AtMKP1, is essential for UV resistance in Arabidopsis, as disruption of this gene showed hypersensitive phenotype on exposure to UV rays. Additionally, MKP1 is also known to be a key regulator of MAP kinases, MPK3 and MPK6 in response to UV stress [65, 66]. This has been known that AtMKP2 dephosphorylates AtMPK3 and AtMPK6; both of these proteins are positive regulator of oxidative stress signaling pathways and AtMKP2-silenced plants exhibit enhanced sensitivity to ozone stress (Fig. 1) [67]. Activation and deactivation occur rapidly and this transient signaling is controlled by MAPKs and MKPs. Both PTPs and DSPs play a vital role in deactivation of MAPKs and hence counteract the responses mediated by MAPKs.
Fig. (1).
Overview of responses regulated by plant Tyr phosphatases. Several genes encoding plant Tyr phosphatase are responsible for the regulation of different biological processes such as osmotic, oxidative, salt stresses, ABA signaling, auxin sensing, wounding, and pathogen response. Mitogen-activated protein kinase (MAPK) dephosphorylation by phosphatase is represented in red color while phosphatase is presented in black. The positive (↓) and negative (↓) regulatory role reflect the physiological function of the phosphatase. Brown double-headed arrow represents the phosphorylation and dephosphorylation event.
The phytohormone abscisic acid (ABA) regulates plant development and responses to many environmental constraints, shoot and root growth, stomatal closure, storage protein synthesis, and seed dormancy [68, 69]. The foremost components of the ABA signaling cascade includes calcium, G-proteins, kinases, phosphatases, phospholipase C and D, and redox signals produced by NADPH oxidase [70, 71]. The significance of the phosphorylation events in ABA signal transduction came into picture with the discovery of mutants defective in ABA responses. For example, ABI1 and ABI2 are type-2C protein phosphatases acting as negative regulators in several ABA responses [72, 73]. Under water deficit condition, ABA induces stomatal closure by reducing the turgor pressure of guard cells. It has been demonstrated that the regulation of stomatal closure is achieved by the role of PTPs. This phenomenon of stomatal closure is induced by H2O2, darkness, Ca2+ and ABA. All these factors may lead to alterations in intracellular calcium homeostasis, which then changes the K+ channel activity in plasma membrane as well as on tonoplast [74, 75]. The knockdown mutation in AtPHS1 gene, a DsPTP, leads to hypersensitivity to ABA during early development and stomatal closure, which suggest PHS1 as a negative regulator of ABA signaling pathway (Fig. 1) [76]. It was shown that several ABA-dependent responses are inhibited by phenylarsine oxide (PAO), a specific inhibitor of PTP [77]. Inhibitors of PTKs and PTPs activity interferes with the expression of ABA responsive genes such as RAB18 (responsive to ABA18) and it blocks the ABA regulated phenomenon such as stomatal closure [78]. The other effect of PAO has been reported in the inhibition of ABA-induced RAB16 gene expression in barley (Hordeum vulgare) aleurone protoplasts [79]. Moreover, PAO prevents stomatal closure in Commelina communis by reopening them, which might get closed either by the induction of ABA, high external Ca2+, darkness, and hydrogen peroxide [77]. Furthermore, PAO enhances the inhibition of germination in Arabidopsis suggesting PTP function as a negative regulator of the ABA signaling [80]. In conclusion, the above-mentioned aspects provides important insights related to Tyr phosphorylation signaling during abiotic stress conditions.
In Response to Hormone and Development
Plant hormones are well-known to assist responses to stress and adaptation. Phytohormones have a diverse effect on plant growth and development. They influence nearly whole plant lifecycle starting from seed germination to maturation. It is well-known fact that ABA and gibberellins play critical roles in seed dormancy and germination [81, 82]. Tyr dephosphorylation is known as a key regulatory mechanism in post-germination arrest of seeds, mediated by ABA. By using an inhibitor of Tyr phosphatases, PAO, it was realized that Tyr dephosphorylation might function as a negative regulator of MAPK during the post-germination arrest of development mediated by ABA [7, 78, 80]. Constitutive expression of FsPTP1 (a tyrosine-specific phosphatase from Fagus sylvatica) in Arabidopsis caused plants to be insensitive towards ABA with simultaneous induction of EIN2 (ethylene insensitive2), a key gene of ethylene signaling. Additionally, the expression of ABA marker genes (RAB18 and RD29) was reduced and expression of the EIN2 gene was higher in FsPTP1 overexpression lines. Further, it has been suggested that negative effect of FsPTP1 in ABA signaling might be regulated by the modulation of ethylene signaling (Fig. 1) [83]. Additionally, Yoo et al. (2008) have reported the role of MKK9-MPK3/MPK6 cascade in the regulation of ethylene-insensitive3 (EIN3)-mediated transcription in ethylene signaling and shown to act downstream of the ethylene receptors [84].
The phytohormone auxin is considered as one of the key regulator of plant growth and development. It was observed that phytohormone auxin (synthetic auxin; 2,4-D) and cytokinin (kinetin) caused a higher degree of Tyr phosphorylation in Arabidopsis hypocotyls [85]. On the basis of sequence similarity, IBR5, encoding a putative DSP like protein in Arabidopsis was found as a positive regulator of both auxin and ABA [86]. ibr5 null mutants were less sensitive to auxin and auxin transport inhibitors. Similarly, seedlings of these mutants were comparable in phenotype with other auxin-response mutants, with long root and short hypocotyl in light, fewer lateral roots, increased leaf serration, defective vascular patterning, and reduced accumulation of an auxin-inducible reporter (DR5-GUS) [86, 87]. The double mutant of ibr5 with an auxin receptor mutant, tir1, improves auxin resistance compared to that of either parent [87]. All these facts clearly suggest that IBR5 promotes auxin responses independently and downstream of TIR1-mediated degradation of Aux/IAA-proteins. Also, it has been found that IBR5 interacted in vitro and in vivo with MPK12 and IBR5 dephosphorylated MPK12 [88]. On the basis of plant genetic studies, it was speculated that MPK12 act as a negative regulator in IBR5-regulated auxin signaling, as suppression of MPK12 in transgenic plants results in the upregulation of auxin-responsive genes and exhibited auxin-hypersensitive root growth phenotype (Fig. 1) [88]. Recently, it was revealed that IBR5 alternatively spliced to generate two isoforms i.e. IBR5.1 and IBR5.3, which may have diverse as well as overlapping functions in growth and development of plant [89].
Brassinosteroid (BR) is a plant steroid hormone, involved in growth regulation. It stimulates cell elongation and its deficiency causes plants to grow as dwarfs in light and have a light-grown phenotype in the dark. Most of the BR mutants showed reduced fertility, delayed senescence and altered vascular development [90]. It was observed that many brassinosteroid increased the level of phosphorylation on Tyr residues. Western blotting with monoclonal antibodies in Pisum sativum showed that brassinolide (BL) induced the phosphorylation of leaf proteins on Tyr residues [91]. It is a well-known fact that BRI1 (BRASSINOSTEROID INSENSITIVE 1), which is the receptor of BRs, is a leucine-rich repeat kinase, located in the plasma membrane and interacts with the cytosolic protein BAK1 (BRI1-ASSOCIATED RECEPTOR KINASE 1) in BR signaling [92, 93]. Recent studies revealed that both BRI1 and BAK1 can autophosphorylate on Tyr, therefore also designated as dual specificity protein Tyr kinases (DsPTKs) [94, 95]. The autophosphorylation of BRI1 and BAK1 are necessary for root growth inhibition induced by BRs [94]. Furthermore, it was seen that autophosphorylation/dephosphorylation of the GSK3-like kinase, BRASSINOSTEROID INSENSITIVE2 (BIN2) on Tyr 200 is a critical switch in downstream regulation of BR signaling [96]. Moreover, upon BR perception, BRI1 phosphorylates the BRI1 KINASE INHIBITOR 1 (BKI1) on Tyr-211, and release BKI1 into the cytosol thus facilitating the formation of an active signaling complex [97]. All these results clearly suggest the involvement of Tyr phosphorylation/dephosphorylation in the plant developmental processes controlled by auxins and BRs.
Leaf senescence is the final stage of leaf development and regulated by numerous environmental and endogenous signals, such as age, developmental cues, and plant growth regulators [98, 99]. AtMKP2 mutant plants displayed an early senescence compared to wild type and AtMKP2 overexpressing plants. However, overexpression of AtMKP2 did not prolong the plant life span [100]. Also, a MAPK cascade that involved MKK9 and MPK6, was shown to play an important role in regulation of leaf senescence in Arabidopsis [101]. Loss-of-function of MKK9 exhibited delayed leaf senescence, while overexpression of MKK9 causes premature senescence in leaves and in whole Arabidopsis plants. Additionally, in vitro phosphorylation revealed that MPK6 is a direct and downstream target of MKK9 [101]. Loss-of-function mutant of MPK6 also showed delayed leaf senescence. These evidences suggested that PTPs also play an important role in regulation of leaf senescence.
In higher plants embryo development and seed germination are also controlled by Tyr phosphorylation. Associations between protein Tyr phosphorylation and plant embryogenesis have also been shown in Daucus carota and Coco nucifera [102, 103]. A subset of well-known PTK inhibitors, including genistein, prevented the establishment of zygotic polarity in Fucus, by inhibition of germination [104]. Rudrabhatla et al. (2006) revealed that many genes of DsPTKs family were expressed particularly during seed germination and early seedling stages in Arabidopsis [35]. Tyr phosphorylation also regulates the dynamics and organization of microtubules in the plant cell. Enhanced Tyr phosphorylation after treatment with sodium orthovanadate (PTP inhibitor) induced intense root growth and led to shortened elongation zone [105]. Similar targets of Tyr phosphorylation, other than MAPKs are profilin and actin [106, 107]. The kinases that phosphorylate profilin and actin may be Tyr kinase or dual specificity kinase and this question is still remains to be answered.
In animals, loss-of-function mutations in PTEN causes tumor, hence termed as a tumor suppressor gene [108]. PTEN having a PTP catalytic core motif is a DsPTP that show significant similarity with the cytoskeleton-interacting protein tensin [109, 110]. AtPTEN1 is an Arabidopsis DsPTP similar to PTEN (Phosphatase and tensin homolog, a tumor suppressing phosphatase in human) and is required for pollen maturation following cell division [111, 112]. AtPTEN1 expresses exclusively in pollen grains and is necessary for pollen development. RNA interference mediated silencing of AtPTEN1 caused pollen cell death after mitosis [111]. Presence of different Tyr-phosphorylated proteins at different embryo developmental stages, revealed the strong correlation between protein Tyr phosphorylation and plant development
Expression Profile of Rice DSPs in Abiotic Stress, Nutrition Deficiency and Hormone Treatment
From literature survey of plant DSPs, it was evident that most of the work has been carried out on dicot model plant Arabidopsis, but knowledge related to these phosphatases is miniscule in crop plant rice. Several reports proposed the involvement of these phosphatases in regulation of stress signaling and tolerance, and development in Arabidopsis. It is quite obvious that these phosphatases might also be involved in related functions in rice. So, we carried out expression analysis of these putative DSPs under abiotic stress, nutrition deficiency and hormone treatment in plants. The expression analysis of rice DSPs hints toward the possible functional role of this family under these conditions.
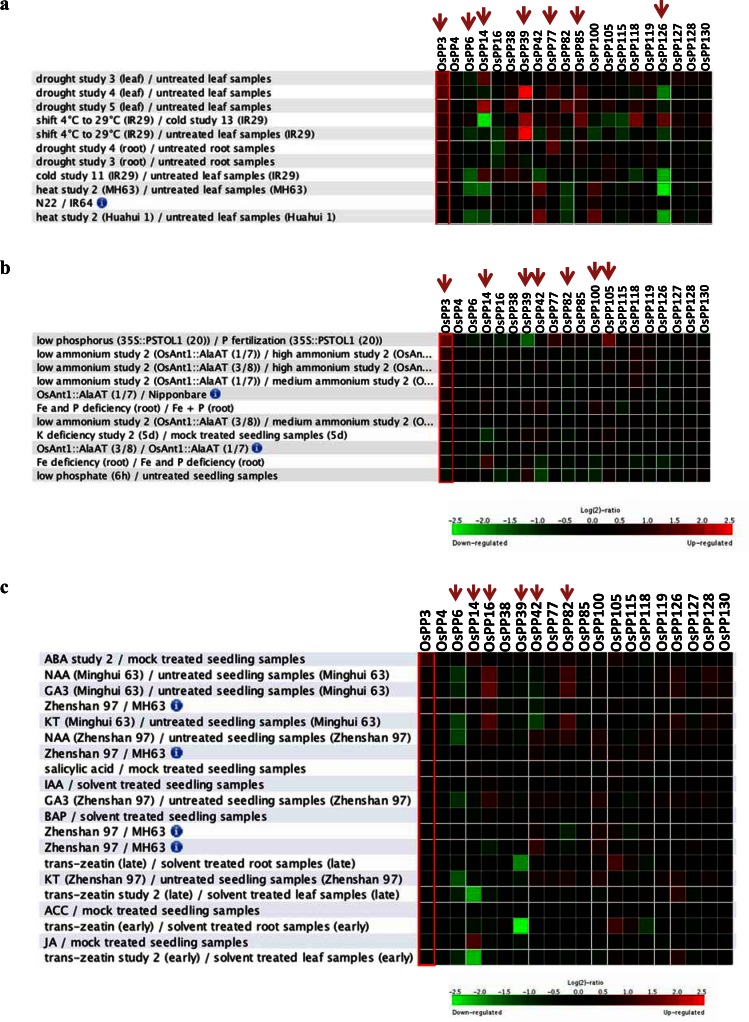
By using Genevestigator database, we have analyzed the whole DSP gene family, and found that some of the rice DSPs were differentially expressed during different stress conditions in rice. Expression of OsPP3 (LOC_ Os01g20940), which encodes rice PHS1, was significantly upregulated under drought and cold stresses in leaf and root tissue and no expression detected under heat stress. OsPP6 (LOC_Os01g29469) and OsPP126 (LOC_Os12g05660) exhibited downregulation under all the three conditions. OsPP14 (LOC_Os01g53710) showed high expression in drought and low expression in cold and heat stresses. OsPP39 (LOC_Os02g53160), OsPP77 (LOC_Os05g44910) and OsPP85 (LOC_Os06g20340) exhibited high expression under cold and drought stresses (Fig. 2a). Moreover, Arabidopsis STARCH EXCESS4 (AtSEX4), a close ortholog of rice OsPP42, was significantly regulated under cold, salt, and heat stress treatments [113]. Also, microarray expression data from Solanum tuberosum, exhibited that a dual specificity protein tyrosine phosphatase 1 was downregulated under salinity stress [113].
Fig. (2).
Expression profile of rice DSPs under abiotic stress (a), nutrition deficiency (b), and hormones (c). On the left side tissue types, conditions and treatment controls are mentioned. Gene names are listed on the top of heat map. Green color indicates the down regulation and red color is for up regulation of the transcript level of individual DSP genes. Expression profile of selected genes is shown on the basis of differential expression. Red arrow above the heat map indicates the genes, which are differentially expressed.
One of the major constrains for plant growth and development is mineral nutrient deficiency, which affects agricultural yield worldwide [115]. From microarray analysis for K+ and Ca2+ deficiency conditions in rice, several of the rice DSPs were not found to be affected under K+ and Ca2+ mineral deficiency [115, 116]. However, OsPP3 (LOC_ Os01g20940) was found to be upregulated under low phosphorus and low ammonium conditions, whereas downregulated in low potassium, low iron and low phosphate conditions. OsPP14 (LOC_Os01g53710) showed low expression under K+ deficiency while, exhibited high expression under Fe2+ deficiency (Fig. 2b). OsPP39 (LOC_Os02g53160) showed downregulation and OsPP105 (LOC_Os09g05020) showed high expression under low phosphorus. OsPP42 (LOC_Os03g01750), OsPP82 (LOC_Os06g05870), OsPP100 (LOC_Os08g29160) genes showed downregulation under low Fe2+ and low phosphate conditions (Fig. 2b). Rice DSP OsPP6 (LOC_Os01g29469) was significantly downregulated after the treatment with phytohormone NAA (naphthaleneacetic acid), GA3 (gibberellic acid), KT (kinetin), and trans-zeatin. The regulation of OsPP14 (LOC_Os01g53710) and OsPP39 (LOC_Os02g53160) genes was highly reduced upon treatment with trans-zeatin. OsPP3 (LOC_Os01g20940) and OsPP82 (LOC_Os06g05870) showed up-regulation after treatment with NAA and GA3 (Fig. 2c).
It is widely known that plants can sense, process, respond to environmental stimuli and activate related-gene expression to adapt different stress conditions. The expression analysis, of rice DSP genes revealed differential expression pattern of some of the DSPs like OsPP3, OsPP6, OsPP14, OsPP3, OsPP126, etc. under stress conditions. The biological function of this group of PTPs in plants, especially in rice, has not yet been elucidated. Thus, it can be speculated that these genes might contribute towards stress tolerance in plants and may be an important regulator for plant adaptation under abiotic stress, nutrition deficiency and hormone signaling. Moreover, this expression analysis provides a platform towards the in planta functional characterization of genes.
In Response to Biotic Stress
In the environment, plants encounter a variety of biotic stresses including bacteria, viruses, fungi, parasitic worms and insects, which strongly affect the plant growth and development and hence the productivity [117]. To counteract the biotic stresses, plants have developed active defense mechanism to protect themselves and one of the mechanisms, they adopt is transcriptional activation of a large number of genes to induce defense responses [118]. Reactive oxygen species (ROS) play an important role in controlling many biological processes, such as gene expression, activation of transcription factors, redox balance, programmed cell death (PCD) and regulation of the mitogen-activated protein kinase (MAPK) pathways [119, 120]. H2O2 is an important component of ROS and prevent the growth, penetration and proliferation of pathogen by stimulating program cell death (PCD) [121, 122]. Studies have shown that two orthologous genes from rice and Arabidopsis, OsPFA-DSP2 and AtPFA-DSP4 act as negative regulator of the pathogen responses in transgenic plants through H2O2-mediated pathway, respectively (Fig. 1) [123]. The OsPFA-DSP2 overexpressioning lines showed increased sensitivity to Magnaporthe grisea, and inhibited the accumulation of H2O2 as well as suppressed the expression of pathogenesis-related (PR) genes after fungal infection. Also, Arabidopsis plants overexpressing AtPFA-DSP4, exhibited sensitivity to Pseudomonas syringae, and decreased accumulation of H2O2.
Presently, MAPKs have been characterized in higher plants and the importance of Tyr phosphorylation in plant cells cannot be ignored. MAPKs play a vital role in the signal transduction pathways in higher plants [124]. It is well known that PTPs plays a major role in regulation of MAPK cascade and their expression is often induced by various stress signals [124]. For example, Arabidopsis MPK6 is activated by various pathogenic stimuli and abiotic stress signals [126, 127]. Also, the activation of MAPK is associated with Tyr phosphorylation [128]. For instance, AtMPK4 in Arabidopsis phosphorylated at Tyr residue in order to get activated. If AtPTP1 dephosphorylates this Tyr residue then it leads to complete loss of kinase activity [13]. The role of MAPK cascades in plant defense is well established but very little is known about plant Tyr phosphatases. MKP1 is associated with pathogenic responses in plants. Nicotiana tabaccum MAPK phosphatase1 (NtMKP1) over-expression showed abolished salicylic acid-induced protein kinase (SIPK) mediated cell death. SIPK is known as the ortholog of Arabidopsis MPK6. Moreover, induction of NtMKP1 inhibits wound induced MAPK (WIPK), an ortholog of AtMPK3 [129, 130]. A fungal elicitor induced Tyr phosphorylation of the 47-kD myelin basic protein kinase (MBP) was verified by immunoblots and immunoprecipitation analysis with anti-phospho-Tyr antibody in tobacco cells [131]. Additionally, Arabidopsis thaliana Columbia ecotype introgressed mpk1 mutant plants exhibited constitutive defense responses such as inhibited growth, enhanced expression of defense response markers such as PR genes, accumulation of salicylic acid and resistance to the bacterial pathogen Pseudomonas syringae [132]. Mutations in the SNC1 (SUPPRESSOR OF npr1-1 CONSTITUTIVE1) also reduced the growth defects and PR gene accumulation in mkp1 mutants. Recent studies have revealed that AtMKP1 negatively regulates MPK6-mediated pathogen-associated molecular patterns (PAMP) responses and resistance against bacteria, as mkp1/mpk6 double mutants showed suppressed defense responses [133].
Apart from its role in BR signaling, BAK1 has been shown to play an important role in plant defense mechanisms. The phosphorylation of BAK1 at Tyr-610 residue affects the expression of several defense-related genes [96]. Mass spectrometry (MS) analysis of BIK1 autophosphorylation and transphosphorylation by BAK1 identified three BIK1 tyrosine phosphorylation sites Y150, Y234, and Y250 [134]. Transgenic complementation assays suggested that, this Tyr phosphorylation was important in BIK1-mediated plant innate immunity because mutation of Tyr to phenylalanine (Phe) failed to complement the bik1 mutant deficiency in immunity [134]. BAK1/SERK3 binds to FLS2, the flagellin receptor, and positively regulates FLS2-mediated innate immunity in Arabidopsis [135]. The above-mentioned data clearly indicate the involvement of Tyr phosphorylation in regulation of defense responses mediated by MPKs and PTPs.
In Starch Metabolism
Starch belongs to a group of most abundant water-insoluble biomolecules on earth. Starch in plants and glycogen in animals are the major carbohydrates reserve for most of the organisms. Glucose residues of both, starch and glycogen are linked via α-1,4-glycosidic bonds and branched via α-1,6-glycosidic linkages. Amylopectin is a major component of starch (containing 70% or more of starch) and branching in amylopectin allows formation of insoluble semicrystalline nature of starch granules [136, 137]. In plants, starch synthesis occurs during daytime as a result of photosynthesis along with sucrose. Since, plants cannot transport starch in its native form, these are first converted to sucrose at night by combination of enzymes [136]. In most of the plants, sucrose is an essential carbon source for energy and growth.
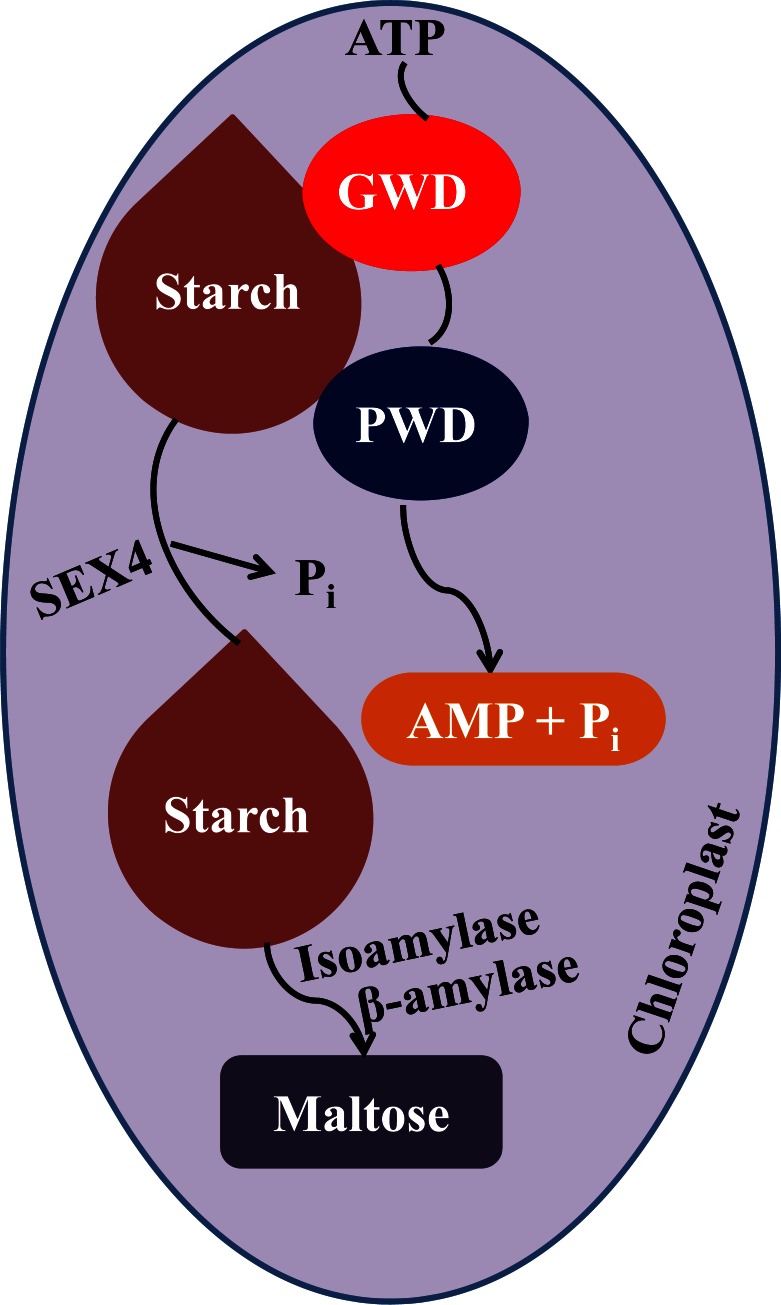
Plants have two glucan dikinases, which phosphorylate starch, i.e. glucan water dikinase (GWD) and phosphoglucan water dikinase (PWD) [138-140]. Both enzymes add beta-phosphate groups from ATP to glucose residues within starch polymers. The addition of charged and hydrophilic phosphate groups may interrupt the organization of the starch granule matrix by unwinding the amylopectin double helices and make it a better substrate for exoamylolytic attack. Glucan hydrolyzing enzymes, such as exoamylases, endoamylase, and debranching enzymes are involved in the subsequent degradation of starch [137]. For the complete degradation of starch, removal of phosphate group added by GWD/PWD is essential (Fig. 3) [138-140]. The products obtained from the degradation i.e., maltose and glucose are exported from the chloroplast to the cytosol for metabolism processes. GWD phosphorylates the C-6 positions of glucosyl residues, whereas PWD phosphorylates the C-3 positions [141]. Plants contain two glucan phosphatases for dephosphorylation of starch and promote degradation by amylases (Fig. 3) [142-144]. Glucan phosphatases are members of the PTP family. Mutation in a glucan phosphatase, STARCH EXCESS4 (SEX4), also known as PTPKIS (protein-tyrosine phosphatase kinase interaction sequence) and DSP4 (dual-specificity phosphatase 4) results in stunted plant growth and delayed flowering, due to accumulation of starch [142, 145]. Arabidopsis plants having disrupted DSP4 are impaired in starch degradation during the night-time and thus, increased accumulation of starch [146]. The starch-binding capacity of DSP4 is regulated by light through redox and pH [145]. SEX4 dephosphorylates the starch and as a result, allowed it to be further hydrolyzed by β-amylase3 (BAM3) and isoamylase3 (ISA3). Some of the recent studies have shown that SEX4 preferentially dephosphorylate the C6-position and another glucan phosphatases, LIKE SEX FOUR2 (LSF2 or DSP5) specifically dephosphorylates the C3-position of starch glucose [144, 147]. SEX4/DSP4 and LSF2/DSP5 proteins both contain a chloroplast targeting peptide, a DSP domain, and a unique C-terminal (CT) motif [142, 144, 148, 149]. Chloroplast targeting peptide facilitate the localization of proteins to the site of starch metabolism i.e. chloroplast. Among all the plant DSPs, CT motif was detected in SEX4, which is needed for protein stability and function [149]. Moreover, SEX4 contains a carbohydrate-binding module (CBM) that is common in starch-interacting enzymes and modify substrate surfaces to facilitate enzymatic hydrolysis [150, 151].
Fig. (3).
The proposed pathway of starch degradation in chloroplast.
With the help of bioinformatics analysis, it has been known that lafora DSP of humans, fungal DSP and DSPs of some plants share a wide spread ancient carbohydrate binding domain (CBD) [148]. These DSPs plays important role in glycogen metabolism in animals and starch metabolism in plants. Two of the DSPs of Arabidopsis At3g52180 (DSP4) and At3g01510 (DSP5) have been shown to display sequence and functional homology with the human laforin, which is a DSP containing carbohydrate binding motif and any mutation in this protein results in lafora disease (LD), a neurodegenerative disorder with epilepsy in humans [148]. Both in animals and plants, laforin (dual specificity proteins) regulate carbohydrate accumulation indicating that both of them play an essential role in storage form of carbohydrate (starch or glycogen) metabolism. Hence, in spite of the significant differences in the enzymes directly involved in starch metabolism in plants and glycogen metabolism in mammals, both SEX4 and Laforin plays a key role in regulation of glucan metabolism.
CONCLUSION AND FUTURE PERSPECTIVE
Despite of the identification of large number of protein Tyr kinases and phosphatases in the regulation of number of responses in animal system, the role of Tyr phosphorylation in plants is still under explored. The most likely reason for this is absence of a typical Tyr kinases as well as phosphatases in plants. However, plants possess few PTP and DSP like enzymes but so far no PTK type enzyme has been identified in plants. Recently, several genome-wide studies and large scale proteomic analysis have facilitated identification of phospho-Tyr proteins in plants. Protein Tyr phosphorylation plays important role in many plant processes and majorly found in the regulation of developmental, abiotic and biotic stress responses (as summerized in Fig. 1, 2). However, the impact of protein Tyr phosphorylation on the regulation of large number of physiological processes in plant is not completely understood and revelations are just beginning to emerge. Recent genetic, molecular, and biochemical studies have shed light on some aspect of their functions and the mode of action. Nonetheless, more plant Tyr-phosphorylated proteins still require to be identified. Improved analytic technologies such as mass spectrometry (MS) can be employed to identify more relevant phosphorylation sites in proteins. Genetic studies can be used to decipher the role of a specific enzyme by using knockdown and knockouts with the complementation substitution of kinases and phosphatases causing visible change in phenotype [152]. Similarly, the impact on the phosphorylation level of a particular protein by the overexpression of a phosphatase can be analyzed. Subsequently, various domain prediction tools could be used to study set of Tyr-directed protein kinases and linked to their substrate. With detail in planta functional role of these Tyr phosphatases, some of these can be utilized for development of future crop by genetic intervention to enhance the crop productivity and quality, especially the seed quality and enhancement of yield under stress conditions.
ACKNOWLEDGEMENTS
GKP is thankful to Department of Biotechnology (DBT) and Department of Science and Technology (DST), India for supporting research work. AS acknowledge Council for Scientific and Industrial Research (CSIR), India for research fellowship.
LIST OF ABBREVIATIONS
- ABA
= Abscisic acid
- DSP
= Dual specific phosphatase
- MAPK
= Mitogen-activated protein kinase
- PAO
= Phenyl arsine oxide
- PTK
= Protein tyrosine kinase
- PTP
= Protein tyrosine phosphatase
- ROS
= Reactive oxygen species
- SA
= Salicylic acid
- SEX4
= Starch excess 4
- SH2
= Src homology 2
- Ser
= Serine
- Thr
= Threonine
- Tyr
= Tyrosine
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Krebs E.G., Kent A.B., Fischer E.H. The muscle phosphorylase b kinase reaction. J. Biol. Chem. 1958;231(1):73–83. [PubMed] [Google Scholar]
- 2.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Boyle W.J. Growth factors and tyrosine kinase receptors during development and cancer. Curr. Opin. Oncol. 1992;4(1):156–162. doi: 10.1097/00001622-199202000-00021. [DOI] [PubMed] [Google Scholar]
- 4.Sun H., Tonks N.K. The coordinated action of protein tyrosine phosphatases and kinases in cell signaling. Trends Biochem. Sci. 1994;19(11):480–485. doi: 10.1016/0968-0004(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 5.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 2009;21(2):140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gnad F., Forner F., Zielinska D.F., Birney E., Gunawardena J., Mann M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol. Cell. Proteomics. 2010;9(12):2642–2653. doi: 10.1074/mcp.M110.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghelis T. Signal processing by protein tyrosine phosphorylation in plants. Plant Signal. Behav. 2011;6(7):942–951. doi: 10.4161/psb.6.7.15261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter T. The genesis of tyrosine phosphorylation. Cold Spring Harb. Perspect. Biol. 2014;6(5):a020644. doi: 10.1101/cshperspect.a020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson K.I., Brummer T., O’Brien P.M., Daly R.J. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem. J. 2009;418(3):475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 10.Kerk D., Bulgrien J., Smith D.W., Barsam B., Veretnik S., Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002;129(2):908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A., Giri J., Kapoor S., Tyagi A.K., Pandey G.K. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith R.D., Walker J.C. Plant Protein Phosphatases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:101–125. doi: 10.1146/annurev.arplant.47.1.101. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q., Fu H.H., Gupta R., Luan S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell. 1998;10(5):849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walton K.M., Dixon J.E. Protein tyrosine phosphatases. Annu. Rev. Biochem. 1993;62:101–120. doi: 10.1146/annurev.bi.62.070193.000533. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z.Y. Protein-tyrosine phosphatases: biological function, structural characteristics, and mechanism of catalysis. Crit. Rev. Biochem. Mol. Biol. 1998;33(1):1–52. doi: 10.1080/10409239891204161. [DOI] [PubMed] [Google Scholar]
- 16.Li L., Dixon J.E. Form, function, and regulation of protein tyrosine phosphatases and their involvement in human diseases. Semin. Immunol. 2000;12(1):75–84. doi: 10.1006/smim.2000.0209. [DOI] [PubMed] [Google Scholar]
- 17.Barford D., Flint A.J., Tonks N.K. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263(5152):1397–1404. [PubMed] [Google Scholar]
- 18.Zhang Z.Y. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu. Rev. Pharmacol. Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 19.Barford D. Colworth Medal Lecture. Structural studies of reversible protein phosphorylation and protein phosphatases. Biochem. Soc. Trans. 1999;27(6):751–766. doi: 10.1042/bst0270751. [DOI] [PubMed] [Google Scholar]
- 20.Jia Z., Barford D., Flint A.J., Tonks N.K. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. . Science . 1995;268(5218):1754–1758.. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 21.Dunn D., Chen L., Lawrence D.S., Zhang Z.Y. The active site specificity of the Yersinia protein-tyrosine phosphatase. J. Biol. Chem. 1996;271(1):168–173. doi: 10.1074/jbc.271.1.168. [DOI] [PubMed] [Google Scholar]
- 22.Yuvaniyama J., Denu J.M., Dixon J.E., Saper M.A. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272(5266):1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 23.Stewart A.E., Dowd S., Keyse S.M., McDonald N.Q. Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation. Nat. Struct. Biol. 1999;6(2):174–181. doi: 10.1038/5861. [DOI] [PubMed] [Google Scholar]
- 24.Stone R.L., Dixon J.E. Protein-tyrosine phosphatases. J. Biol. Chem. 1994;269(50):31323–31326. [PubMed] [Google Scholar]
- 25.Denu J.M., Stuckey J.A., Saper M.A., Dixon J.E. Form and function in protein dephosphorylation. Cell. 1996;87(3):361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 26.Neel B.G., Tonks N.K. Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 1997;9(2):193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 27.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Mustelin T. Protein Phosphatase Protocols. Totowa: Humana Press; 2006. A brief introduction to the protein phosphatase families. pp. 9–22. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson-Rosenthal C., Millar J.B. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16(6):285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Gohla A., Birkenfeld J., Bokoch G.M. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 2005;7(1):21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- 31.Rebay I., Silver S.J., Tootle T.L. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21(3):163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Moorhead G.B., Trinkle-Mulcahy L., Ulke-Lemée A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 2007;8(3):234–244. doi: 10.1038/nrm2126. [DOI] [PubMed] [Google Scholar]
- 33.Tonks N.K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 34.Hanks S.K., Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–596. [PubMed] [Google Scholar]
- 35.Rudrabhatla P., Reddy M.M., Rajasekharan R. Genome-wide analysis and experimentation of plant serine/ threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 2006;60(2):293–319. doi: 10.1007/s11103-005-4109-7. [DOI] [PubMed] [Google Scholar]
- 36.Byrum C.A., Walton K.D., Robertson A.J., Carbonneau S., Thomason R.T., Coffman J.A., McClay D.R. Protein tyrosine and serine-threonine phosphatases in the sea urchin, Strongylocentrotus purpuratus: identification and potential functions. Dev. Biol. 2006;300(1):194–218. doi: 10.1016/j.ydbio.2006.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torruella M., Casano L.M., Vallejos R.H. Evidence of the activity of tyrosine kinase(s) and of the presence of phosphotyrosine proteins in pea plantlets. J. Biol. Chem. 1986;261(15):6651–6653. [PubMed] [Google Scholar]
- 38.Barizza E., Lo Schiavo F., Terzi M., Filippini F. Evidence suggesting protein tyrosine phosphorylation in plants depends on the developmental conditions. FEBS Lett. 1999;447(2-3):191–194. doi: 10.1016/s0014-5793(99)00272-0. [DOI] [PubMed] [Google Scholar]
- 39.Carpi A., Di Maira G., Vedovato M., Rossi V., Naccari T., Floriduz M., Terzi M., Filippini F. Comparative proteome bioinformatics: identification of a whole complement of putative protein tyrosine kinases in the model flowering plant Arabidopsis thaliana. Proteomics. 2002;2(11):1494–1503. doi: 10.1002/1615-9861(200211)2:11<1494::AID-PROT1494>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Ndimba B.K., Chivasa S., Hamilton J.M., Simon W.J., Slabas A.R. Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics. 2003;3(6):1047–1059. doi: 10.1002/pmic.200300413. [DOI] [PubMed] [Google Scholar]
- 41.Peck S.C., Nühse T.S., Hess D., Iglesias A., Meins F., Boller T. Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell. 2001;13(6):1467–1475. doi: 10.1105/tpc.13.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter T. Life Sciences for the 21st Century. Hoboken, NJ: Wiley; 2004. Protein phosphorylation: what does the future hold? pp. 191–223. [Google Scholar]
- 43.Blaydes J.P., Vojtesek B., Bloomberg G.B., Hupp T.R. The development and use of phospho-specific antibodies to study protein phosphorylation. Methods Mol. Biol. 2000;99:177–189. doi: 10.1385/1-59259-054-3:177. [DOI] [PubMed] [Google Scholar]
- 44.Islas-Flores I., Oropeza C., Hernandez-Sotomayor S.M. Protein phosphorylation during coconut zygotic embryo development. Plant Physiol. 1998;118(1):257–263. doi: 10.1104/pp.118.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudrabhatla P., Rajasekharan R. Developmentally regulated dual-specificity kinase from peanut that is induced by abiotic stresses. Plant Physiol. 2002;130(1):380–390. doi: 10.1104/pp.005173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akiyama T., Ishida J., Nakagawa S., Ogawara H., Watanabe S., Itoh N., Shibuya M., Fukami Y. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 1987;262(12):5592–5595. [PubMed] [Google Scholar]
- 47.Onoda T., Iinuma H., Sasaki Y., Hamada M., Isshiki K., Naganawa H., Takeuchi T., Tatsuta K., Umezawa K. Isolation of a novel tyrosine kinase inhibitor, lavendustin A, from Streptomyces griseolavendus. J. Nat. Prod. 1989;52(6):1252–1257. doi: 10.1021/np50066a009. [DOI] [PubMed] [Google Scholar]
- 48.Gupta R., Huang Y., Kieber J., Luan S. Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 1998;16(5):581–589. doi: 10.1046/j.1365-313x.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- 49.Gupta R., Luan S. Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol. 2003;132(3):1149–1152. doi: 10.1104/pp.103.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fordham-Skelton A.P., Chilley P., Lumbreras V., Reignoux S., Fenton T.R., Dahm C.C., Pages M., Gatehouse J.A. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 2002;29(6):705–715. doi: 10.1046/j.1365-313x.2002.01250.x. [DOI] [PubMed] [Google Scholar]
- 51.Kovaleva V., Cramer R., Krynytskyy H., Gout I., Gout R. Analysis of tyrosine phosphorylation and phosphotyrosine-binding proteins in germinating seeds from Scots pine. Plant Physiol. Biochem. 2013;67:33–40. doi: 10.1016/j.plaphy.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 52.Fordham-Skelton A.P., Skipsey M., Eveans I.M., Edwards R., Gatehouse J.A. Higher plant tyrosine-specific protein phosphatases (PTPs) contain novel amino-terminal domains: expression during embryogenesis. Plant Mol. Biol. 1999;39(3):593–605. doi: 10.1023/a:1006170902271. [DOI] [PubMed] [Google Scholar]
- 53.Keyse S.M., Emslie E.A. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359(6396):644–647. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- 54.Keyse S.M. Protein phosphatases and the regulation of MAP kinase activity. Semin. Cell Dev. Biol. 1998;9(2):143–152. doi: 10.1006/scdb.1997.0219. [DOI] [PubMed] [Google Scholar]
- 55.Kerk D., Templeton G., Moorhead G.B. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008;146(2):351–367. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh A., Pandey G.K. Protein phosphatases: a genomic outlook to understand their function in plants. J. Plant Biochem. Biotechnol. 2012;21(1):100–107. [Google Scholar]
- 57.Wei K., Pan S. Maize protein phosphatase gene family: identification and molecular characterization. BMC Genomics. 2014;15:773. doi: 10.1186/1471-2164-15-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shinozaki K., Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007;58(2):221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 59.Sharma M., Singh A., Shankar A., Pandey A., Baranwal V., Kapoor S., Tyagi A.K., Pandey G.K. Comprehensive expression analysis of rice Armadillo gene family during abiotic stress and development. DNA Res. 2014;21(3):267–283. doi: 10.1093/dnares/dst056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pandey G.K., Kanwar P., Pandey A. Global Comparitive Analysis of CBL-CIPK Gene Families in Plants. SpringerBriefs in Plant Sciences; 2014. [Google Scholar]
- 61.Luan S. Tyrosine phosphorylation in plant cell signaling. Proc. Natl. Acad. Sci. USA. 2002;99(18):11567–11569. doi: 10.1073/pnas.182417599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu R., Liu Y., Ye N., Zhu G., Chen M., Jia L., Xia Y., Shi L., Jia W., Zhang J. AtDsPTP1 acts as a negative regulator in osmotic stress signalling during Arabidopsis seed germination and seedling establishment. J. Exp. Bot. 2015;66(5):1339–1353. doi: 10.1093/jxb/eru484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berrocal-Lobo M., Ibañez C., Acebo P., Ramos A., Perez-Solis E., Collada C., Casado R., Aragoncillo C., Allona I. Identification of a homolog of Arabidopsis DSP4 (SEX4) in chestnut: its induction and accumulation in stem amyloplasts during winter or in response to the cold. Plant Cell Environ. 2011;34(10):1693–1704. doi: 10.1111/j.1365-3040.2011.02365.x. [DOI] [PubMed] [Google Scholar]
- 64.Liu B., Fan J., Zhang Y., Mu P., Wang P., Su J., Lai H., Li S., Feng D., Wang J., Wang H. OsPFA-DSP1, a rice protein tyrosine phosphatase, negatively regulates drought stress responses in transgenic tobacco and rice plants. Plant Cell Rep. 2012;31(6):1021–1032. doi: 10.1007/s00299-011-1220-x. [DOI] [PubMed] [Google Scholar]
- 65.Ulm R., Ichimura K., Mizoguchi T., Peck S.C., Zhu T., Wang X., Shinozaki K., Paszkowski J. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 2002;21(23):6483–6493. doi: 10.1093/emboj/cdf646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ulm R., Revenkova E., di Sansebastiano G.P., Bechtold N., Paszkowski J. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 2001;15(6):699–709. doi: 10.1101/gad.192601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.González Besteiro M.A., Ulm R. Phosphorylation and stabilization of Arabidopsis MAP kinase phosphatase 1 in response to UV-B stress. J. Biol. Chem. 2013;288(1):480–486. doi: 10.1074/jbc.M112.434654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee J.S., Ellis B.E. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 2007;282(34):25020–25029. doi: 10.1074/jbc.M701888200. [DOI] [PubMed] [Google Scholar]
- 69.Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danquah A., de Zelicourt A., Colcombet J., Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014;32(1):40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Schroeder J.I., Kwak J.M., Allen G.J. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410(6826):327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- 72.Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl.):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gosti F., Beaudoin N., Serizet C., Webb A.A., Vartanian N., Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11(10):1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25(3):295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 75.Xiong L., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl.):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sreenivasulu N., Harshavardhan V.T., Govind G., Seiler C., Kohli A. Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene. 2012;506(2):265–273. doi: 10.1016/j.gene.2012.06.076. [DOI] [PubMed] [Google Scholar]
- 77.Quettier A.L., Bertrand C., Habricot Y., Miginiac E., Agnes C., Jeannette E., Maldiney R. The phs1-3 mutation in a putative dual-specificity protein tyrosine phosphatase gene provokes hypersensitive responses to abscisic acid in Arabidopsis thaliana. Plant J. 2006;47(5):711–719. doi: 10.1111/j.1365-313X.2006.02823.x. [DOI] [PubMed] [Google Scholar]
- 78.MacRobbie E.A. Evidence for a role for protein tyrosine phosphatase in the control of ion release from the guard cell vacuole in stomatal closure. Proc. Natl. Acad. Sci. USA. 2002;99(18):11963–11968. doi: 10.1073/pnas.172360399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghelis T., Bolbach G., Clodic G., Habricot Y., Miginiac E., Sotta B., Jeannette E. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008;148(3):1668–1680. doi: 10.1104/pp.108.124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knetsch M., Wang M., Snaar-Jagalska B.E., Heimovaara-Dijkstra S. Abscisic Acid Induces Mitogen-Activated Protein Kinase Activation in Barley Aleurone Protoplasts. Plant Cell. 1996;8(6):1061–1067. doi: 10.1105/tpc.8.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reyes D., Rodríguez D., Nicolás G., Nicolás C. Evidence of a role for tyrosine dephosphorylation in the control of postgermination arrest of development by abscisic acid in Arabidopsis thaliana L. Planta. 2006;223(2):381–385. doi: 10.1007/s00425-005-0135-6. [DOI] [PubMed] [Google Scholar]
- 82.Chen S.Y., Kuo S.R., Chien C.T. Roles of gibberellins and abscisic acid in dormancy and germination of red bayberry (Myrica rubra) seeds. Tree Physiol. 2008;28(9):1431–1439. doi: 10.1093/treephys/28.9.1431. [DOI] [PubMed] [Google Scholar]
- 83.Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., Liu C., Feng Y., Cao X., Xie Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013;9(6):e1003577. doi: 10.1371/journal.pgen.1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alonso-Ramírez A., Rodríguez D., Reyes D., Jiménez J.A., Nicolás G., Nicolás C. Functional analysis in Arabidopsis of FsPTP1, a tyrosine phosphatase from beechnuts, reveals its role as a negative regulator of ABA signaling and seed dormancy and suggests its involvement in ethylene signaling modulation. Planta. 2011;234(3):589–597. doi: 10.1007/s00425-011-1426-8. [DOI] [PubMed] [Google Scholar]
- 85.Yoo S.D., Sheen J. MAPK signaling in plant hormone ethylene signal transduction. Plant Signal. Behav. 2008;3(10):848–849. doi: 10.4161/psb.3.10.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang H.J., Lin Y.M., Huang D.D., Takahashi T., Sugiyama M. Protein tyrosine phosphorylation during phytohormone-stimulated cell proliferation in Arabidopsis Hypocotyls. Plant Cell Physiol. 2003;44(7):770–775. doi: 10.1093/pcp/pcg082. [DOI] [PubMed] [Google Scholar]
- 87.Monroe-Augustus M., Zolman B.K., Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15(12):2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strader L.C., Monroe-Augustus M., Bartel B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 2008;8:41. doi: 10.1186/1471-2229-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J.S., Wang S., Sritubtim S., Chen J.G., Ellis B.E. Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 2009;57(6):975–985. doi: 10.1111/j.1365-313X.2008.03741.x. [DOI] [PubMed] [Google Scholar]
- 90.Jayaweera T., Siriwardana C., Dharmasiri S., Quint M., Gray W.M., Dharmasiri N. Alternative splicing of Arabidopsis IBR5 pre-mRNA generates two IBR5 isoforms with distinct and overlapping functions. PLoS One. 2014;9(8):e102301. doi: 10.1371/journal.pone.0102301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh A.P., Savaldi-Goldstein S. Growth control: brassinosteroid activity gets context. J. Exp. Bot. 2015;66(4):1123–1132. doi: 10.1093/jxb/erv026. [DOI] [PubMed] [Google Scholar]
- 92.Fedina E.O., Karimova F.G., Tarchevsky I.A. Effect of brassinolide on tyrosine phosphorylation of pea leaf proteins. Biochemistry (Mosc.) 2006;71(4):423–429. doi: 10.1134/s0006297906040109. [DOI] [PubMed] [Google Scholar]
- 93.Gendron J.M., Wang Z.Y. Multiple mechanisms modulate brassinosteroid signaling. Curr. Opin. Plant Biol. 2007;10(5):436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clouse S.D. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23(4):1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oh M.H., Wang X., Kota U., Goshe M.B., Clouse S.D., Huber S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106(2):658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oh M.H., Wang X., Wu X., Zhao Y., Clouse S.D., Huber S.C. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. USA. 2010;107(41):17827–17832. doi: 10.1073/pnas.0915064107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009;11(10):1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaillais Y., Hothorn M., Belkhadir Y., Dabi T., Nimchuk Z.L., Meyerowitz E.M., Chory J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25(3):232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guo Y. Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 2013;82(6):519–528. doi: 10.1007/s11103-012-9974-2. [DOI] [PubMed] [Google Scholar]
- 100.Kim J., Chang C., Tucker M.L. To grow old: regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015;6:20. doi: 10.3389/fpls.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Z., Peng J., Wen X., Guo H. Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 2012;54(8):526–539. doi: 10.1111/j.1744-7909.2012.01136.x. [DOI] [PubMed] [Google Scholar]
- 102.Zhou C., Cai Z., Guo Y., Gan S. An arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009;150(1):167–177. doi: 10.1104/pp.108.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barizza E., Lo Schiavo F., Terzi M., Filippini F. Evidence suggesting protein tyrosine phosphorylation in plants depends on the developmental conditions. FEBS Lett. 1999;447(2-3):191–194. doi: 10.1016/s0014-5793(99)00272-0. [DOI] [PubMed] [Google Scholar]
- 104.Islas-Flores I., Oropeza C., Hernandez-Sotomayor S.M. Protein phosphorylation during coconut zygotic embryo development. Plant Physiol. 1998;118(1):257–263. doi: 10.1104/pp.118.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Corellou F., Potin P., Brownlee C., Kloareg B., Bouget F.Y. Inhibition of the establishment of zygotic polarity by protein tyrosine kinase inhibitors leads to an alteration of embryo pattern in Fucus. Dev. Biol. 2000;219(2):165–182. doi: 10.1006/dbio.1999.9603. [DOI] [PubMed] [Google Scholar]
- 106.Yemets A., Sheremet Y., Vissenberg K., Van Orden J., Verbelen J.P., Blume Y.B. Effects of tyrosine kinase and phosphatase inhibitors on microtubules in Arabidopsis root cells. Cell Biol. Int. 2008;32(6):630–637. doi: 10.1016/j.cellbi.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 107.Guillén G., Valdés-López V., Noguez R., Olivares J., Rodríguez-Zapata L.C., Pérez H., Vidali L., Villanueva M.A., Sánchez F. Profilin in Phaseolus vulgaris is encoded by two genes (only one expressed in root nodules) but multiple isoforms are generated in vivo by phosphorylation on tyrosine residues. Plant J. 1999;19(5):497–508. doi: 10.1046/j.1365-313x.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- 108.Kameyama K., Kishi Y., Yoshimura M., Kanzawa N., Sameshima M., Tsuchiya T. Tyrosine phosphorylation in plant bending. Nature. 2000;407(6800):37. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 109.Maehama T., Dixon J.E. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9(4):125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 110.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., Bigner S.H., Giovanella B.C., Ittmann M., Tycko B., Hibshoosh H., Wigler M.H., Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 111.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M., Eng C., Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16(1):64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 112.Gupta R., Ting J.T., Sokolov L.N., Johnson S.A., Luan S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell. 2002;14(10):2495–2507. doi: 10.1105/tpc.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y., Li S., Zhou L.Z., Fox E., Pao J., Sun W., Zhou C., McCormick S. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 2011;68(6):1081–1092. doi: 10.1111/j.1365-313X.2011.04761.x. [DOI] [PubMed] [Google Scholar]
- 114.Ma T.L., Wu W.H., Wang Y. Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol. 2012;12:161. doi: 10.1186/1471-2229-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou J., Wang X., Jiao Y., Qin Y., Liu X., He K., Chen C., Ma L., Wang J., Xiong L., Zhang Q., Fan L., Deng X.W. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol. Biol. 2007;63(5):591–608. doi: 10.1007/s11103-006-9111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shankar A., Singh A., Kanwar P., Srivastava A.K., Pandey A., Suprasanna P., Kapoor S., Pandey G.K. Gene expression analysis of rice seedling under potassium deprivation reveals major changes in metabolism and signaling components. PLoS One. 2013;8(7):e70321. doi: 10.1371/journal.pone.0070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shankar A., Srivastava A.K., Yadav A.K., Sharma M., Pandey A., Raut V.V., Das M.K., Suprasanna P., Pandey G.K. Whole genome transcriptome analysis of rice seedling reveals alterations in Ca(2+) ion signaling and homeostasis in response to Ca(2+) deficiency. Cell Calcium. 2014;55(3):155–165. doi: 10.1016/j.ceca.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 118.Ribot C., Hirsch J., Balzergue S., Tharreau D., Nottéghem J.L., Lebrun M.H., Morel J.B. Susceptibility of rice to the blast fungus, Magnaporthe grisea. J. Plant Physiol. 2008;165(1):114–124. doi: 10.1016/j.jplph.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 119.Agarwal P., Parida S.K., Mahto A., Das S., Mathew I.E., Malik N., Tyagi A.K. Expanding frontiers in plant transcriptomics in aid of functional genomics and molecular breeding. Biotechnol. J. 2014;9(12):1480–1492. doi: 10.1002/biot.201400063. [DOI] [PubMed] [Google Scholar]
- 120.Nanda A.K., Andrio E., Marino D., Pauly N., Dunand C. Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 2010;52(2):195–204. doi: 10.1111/j.1744-7909.2010.00933.x. [DOI] [PubMed] [Google Scholar]
- 121.Torres M.A. ROS in biotic interactions. Physiol. Plant. 2010;138(4):414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 122.Kotchoni S.O., Gachomo E.W. The reactive oxygen species network pathways:an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J. Biosci. 2006;31(3):389–404. doi: 10.1007/BF02704112. [DOI] [PubMed] [Google Scholar]
- 123.Qin G., Liu J., Cao B., Li B., Tian S. Hydrogen peroxide acts on sensitive mitochondrial proteins to induce death of a fungal pathogen revealed by proteomic analysis. PLoS One. 2011;6(7):e21945. doi: 10.1371/journal.pone.0021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.He H., Su J., Shu S., Zhang Y., Ao Y., Liu B., Feng D., Wang J., Wang H. Two homologous putative protein tyrosine phosphatases, OsPFA-DSP2 and AtPFA-DSP4, negatively regulate the pathogen response in transgenic plants. PLoS One. 2012;7(4):e34995. doi: 10.1371/journal.pone.0034995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu J., Zhang S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015;20(1):56–64. doi: 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 126.Keyse S.M., Emslie E.A. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359(6396):644–647. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- 127.Xu J., Chua N.H. Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 2012;31(8):1975–1984. doi: 10.1038/emboj.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Evrard A., Kumar M., Lecourieux D., Lucks J., von Koskull-Döring P., Hirt H. Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ. 2013;1:e59. doi: 10.7717/peerj.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Orton R.J., Sturm O.E., Vyshemirsky V., Calder M., Gilbert D.R., Kolch W. Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway. Biochem. J. 2005;392(Pt 2):249–261. doi: 10.1042/BJ20050908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Katou S., Karita E., Yamakawa H., Seo S., Mitsuhara I., Kuchitsu K., Ohashi Y. Catalytic activation of the plant MAPK phosphatase NtMKP1 by its physiological substrate salicylic acid-induced protein kinase but not by calmodulins. J. Biol. Chem. 2005;280(47):39569–39581. doi: 10.1074/jbc.M508115200. [DOI] [PubMed] [Google Scholar]
- 131.Seo S., Katou S., Seto H., Gomi K., Ohashi Y. The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 2007;49(5):899–909. doi: 10.1111/j.1365-313X.2006.03003.x. [DOI] [PubMed] [Google Scholar]
- 132.Suzuki K., Shinshi H. Transient Activation and Tyrosine Phosphorylation of a Protein Kinase in Tobacco Cells Treated with a Fungal Elicitor. Plant Cell. 1995;7(5):639–647. doi: 10.1105/tpc.7.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bartels S., Anderson J.C., González Besteiro M.A., Carreri A., Hirt H., Buchala A., Métraux J.P., Peck S.C., Ulm R. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21(9):2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anderson J.C., Bartels S., González Besteiro M.A., Shahollari B., Ulm R., Peck S.C. Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 2011;67(2):258–268. doi: 10.1111/j.1365-313X.2011.04588.x. [DOI] [PubMed] [Google Scholar]
- 135.Lin W., Li B., Lu D., Chen S., Zhu N., He P., Shan L. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA. 2014;111(9):3632–3637. doi: 10.1073/pnas.1318817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 137.Smith A.M., Zeeman S.C., Smith S.M. Starch degradation. Annu. Rev. Plant Biol. 2005;56:73–98. doi: 10.1146/annurev.arplant.56.032604.144257. [DOI] [PubMed] [Google Scholar]
- 138.Zeeman S.C., Smith S.M., Smith A.M. The diurnal metabolism of leaf starch. Biochem. J. 2007;401(1):13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- 139.Ritte G., Lloyd J.R., Eckermann N., Rottmann A., Kossmann J., Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc. Natl. Acad. Sci. USA. 2002;99(10):7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baunsgaard L., Lütken H., Mikkelsen R., Glaring M.A., Pham T.T., Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41(4):595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 141.Kötting O., Pusch K., Tiessen A., Geigenberger P., Steup M., Ritte G. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137(1):242–252. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ritte G., Heydenreich M., Mahlow S., Haebel S., Kötting O., Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580(20):4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 143.Niittylä T., Comparot-Moss S., Lue W.L., Messerli G., Trevisan M., Seymour M.D., Gatehouse J.A., Villadsen D., Smith S.M., Chen J., Zeeman S.C., Smith A.M. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J. Biol. Chem. 2006;281(17):11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 144.Kötting O., Santelia D., Edner C., Eicke S., Marthaler T., Gentry M.S., Comparot-Moss S., Chen J., Smith A.M., Steup M., Ritte G., Zeeman S.C. STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2009;21(1):334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Santelia D., Kötting O., Seung D., Schubert M., Thalmann M., Bischof S., Meekins D.A., Lutz A., Patron N., Gentry M.S., Allain F.H., Zeeman S.C. The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell. 2011;23(11):4096–4111. doi: 10.1105/tpc.111.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sokolov L.N., Dominguez-Solis J.R., Allary A.L., Buchanan B.B., Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. USA. 2006;103(25):9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zeeman S.C., Smith S.M., Smith A.M. The diurnal metabolism of leaf starch. Biochem. J. 2007;401(1):13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- 148.Hejazi M., Fettke J., Kötting O., Zeeman S.C., Steup M. The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 2010;152(2):711–722. doi: 10.1104/pp.109.149914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kerk D., Conley T.R., Rodriguez F.A., Tran H.T., Nimick M., Muench D.G., Moorhead G.B. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 2006;46(3):400–413. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- 150.Vander Kooi C.W., Taylor A.O., Pace R.M., Meekins D.A., Guo H.F., Kim Y., Gentry M.S. Structural basis for the glucan phosphatase activity of Starch Excess4. Proc. Natl. Acad. Sci. USA. 2010;107(35):15379–15384. doi: 10.1073/pnas.1009386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boraston A.B., Bolam D.N., Gilbert H.J., Davies G.J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 2004;382(Pt 3):769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Machovic M., Janecek S. Starch-binding domains in the post-genome era. Cell. Mol. Life Sci. 2006;63(23):2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]