Abstract
Osteoarthritis (OA) is an age-related disease with poorly understood pathogenesis. Recent studies have demonstrated that miRNA might play a key role in OA initiation and development. We reviewed recent publications and elucidated the connection between miRNA and OA cartilage anabolic and catabolic signals, including four signaling pathways: TGF-β/Smads and BMPs signaling, associated with cartilage anabolism; and MAPK and NF-KB signaling, associated with cartilage catabolism. We also explored the relationships with MMP, ADAMTS and NOS (NitricOxide Synthases) families, as well as with the catabolic cytokines IL-1 and TNF-α. The potential role of miRNAs in biological processes such as cartilage degeneration, chondrocyte proliferation, and differentiation is discussed. Collective evidence indicates that miRNAs play a critical role in cartilage degeneration. These findings will aid in understanding the molecular network that governs articular cartilage homeostasis and in to elucidate the role of miRNA in the pathogenesis of OA.
Keywords: miRNA, Cartilage, Homeostasis, Osteoarthritis, Chondrocytes.
INTRODUCTION
Osteoarthritis (OA) is the most common joint disorder in the elderly and places a significant financial burden on healthcare providers and governments [1-4]. Despite its high prevalence and substantial public health impact, the disease’s etiology is not fully understood. OA is a multifactorial disease with a strong genetic component and heritability estimates ranging from 40% to 65%, depending on the joint site [5].
Recent reports have demonstrated that microNAs (miRNAs) might play an important role in OA development. These small non-coding ~22 nucleotides (nt) long RNAs originate by a multistep process from miRNA genes localized in the genomic DNA. miRNAs are post-transcriptional regulators that bind to 3’-untranslated sequences on target messenger RNAs (mRNAs), usually resulting in translational repression or target degradation and gene silencing [6-8]. The human genome may encode thousands of miRNAs [9], which may target about one third of human protein-coding genes [10-13].
To date, more than 2588 mature miRNAs have been identified in humans (miRBase v21, available at http:// www.miRbase.org/cgi-bin/browse.pl?org=hsa). Translational inhibition with little or no influence on mRNA levels has been also reported [13]. Because each miRNA may suppress multiple mRNA targets (average ~200), and, at the same time, one mRNA can be targeted by many miRNAs, miRNAs constitute a complex network able to control a wide spectrum of cellular processes [13].
miRNAs participate in the regulation of almost every aspect of cell physiology [14-17]. Although the roles of miRNAs in cartilage development, bone regeneration, immune system, rheumatoid arthritis, and osteoarthritis have been described, there is no comprehensive and detailed review associated with the role of miRNA in the articular cartilage homeostasis. This review focuses on recent studies that shed light on the role of miRNAs in the maintenance of chondrocytes and the homeostasis of cartilage’s extra-cellular matrix (ECM) (Table 1). We reviewed recent publications and elucidated the connection between miRNA and OA cartilage anabolic and catabolic signals, including several signaling pathways: TGF-β/Smad, IGF, FGF and BMPs signaling, associated with cartilage anabolism; and MAPK and NF-KB signaling, associated with cartilage catabolism. We also explored the relationships with MMPs, ADAMTS (a disintegrin and metalloproteinase domain with thrombospondin motifs) and NOS (Nitric Oxide Synthases) families, as well as with the catabolic cytokines IL-1 and TNF-α [18].
Table 1.
Summary of miRNAs potentially involved in cartilage homeostasis.
| miRNAs | Target Gene(s) | Metabolic Type | Function | Specimens |
|---|---|---|---|---|
| miR-140 | SMAD3 | catabolism | Suppressing the Smad 2/3 pathway | Human chondrocytes [31] |
| MMP13 | anabolism | Inhibition of the matrix metalloprotease | Human cartilage C28/I2 cells [89] | |
| adamts5 | anabolism | Reduces cartilage matrix degradation | Mouse chondrocytes [39] | |
| dnpep | anabolism | Increases BMP signaling | Mouse chondrocytes [33] | |
| RALA | anabolism | Upregulates SOX9, ACAN, Col2a | Human mesenchymal stem cells [123] | |
| miR-146 | smad4 | catabolism | Reduces cellular responsiveness to TGF-β and increases apoptosis rate in chondrocytes | Primary chondrocytes from Sprague-Dawley rats [41] |
| IL-1β, COLL3a1 | anabolism | Inhibits (IL-1β)-induced signaling | Human knee synovial fibroblasts [106] | |
| miR-145 |
SMAD3 SOX9 |
catabolism | Down-regulates type II collagen and glycosaminoglycans concentration while up-regulating of MMP-13 expression | Human knee OA cartilage [45] |
| miR-29 | COL3a1, OSTEONECTIN | anabolism | Promotes osteogenesis, inhibits osteoblast differentiation | hMSCs [72-74] |
| miR-455 | SMAD2 ACVR2B | catabolism | Promotes a degradative chondrocyte response | Human hip articular cartilage [30] |
| miR-337 | tgf-β r2 | catabolism | Promotes anabolism, prevents cartilage degradation | Femoral head cartilage tissues from SD rats [59] |
| miR-483 | mmp13 bmp7 | anabolism | Promotes anabolic processes | Mouse knee OA cartilage [61] |
| miR-92a | noggin3 | catabolism | Has pro-catabolic and anti- anabolic activities | Zebrafish embryo cartilages [62] |
| miR-302 | BMP 2R | anabolism | Shows pro- anabolic activities | The hADSCs [66] |
| TGF-ΒR2 | catabolism | Shows pro- catabolic activities | The hADSCs [66] | |
| miR-199a | smad1 | anabolism | Shows pro- anabolic activities | C3H10T1/2 murine mesenchymal stem cells [55] |
| miR-26 | smad1 | anabolism | Shows pro- anabolic activities | Mouse C2C12 [54] |
| miR-135 | smad5 | anabolism | Shows pro- anabolic activities | Mouse premyogenic C2C12 cells [53] |
| miR-24 | p16INK4a (Cdkn2a) | catabolism | Reduces production of the two matrix remodeling enzymes, MMP1 and MMP13 | Mouse primary OA chondrocytes [112] |
| miR-155 | MMP1 MMP3 | catabolism | Inhibits production of MMP1 and MMP13 | Human joint synovial tissue [91] |
| miR-127 | MMP13 | catabolism | Surpresses production of MMP1 and MMP13 | Human knee OA cartilage and chondrocytes [111] |
| miR-148 | MMP13, COL10A1 ADAMTS5 | catabolism | Shows pro-anabolic and anti- catabolic activities | Human OA articular cartilage and chondrocytes [104] |
| miR-602 | SHH MMP13 | catabolism | Negatively regulattes the expression of SHH and MMP-13 | Human OA articular cartilage and chondrocytes [136] |
| miR-608 | SHH MMP13 | catabolism | Negatively regulates the expression of SHH and MMP-13 | Human OA articular cartilage and chondrocytes [136] |
| miR-125 |
ADAMTS4 MMP13 |
catabolism | Negatively regulates the expression of ADAMTS4 and MMP-13 | Human OA articular cartilage and chondrocytes [109, 110] |
| miR-27 | MMP13 | catabolism | Negatively regulates the expression of MMP-13 | Human OA articular cartilage and chondrocytes [40] |
| miRNAs | Target Gene(s) | Metabolic Type | Function | Specimens |
| miR-22 | PPARΑ, BMP7 | anabolism | Negatively regulates the expression of PPARA and BMP7, blocked inflammatory and catabolic changes | Human articular cartilage and chondrocytes [76] |
| miR-9 |
MMP13 (indirect) PRTG |
catabolism | Negatively regulates the expression of MMP-13 | Rabbit articular chondrocytes and Human knee OA cartilage [92] |
| miR-558 |
COX-2
MMP-1 MMP-13 |
catabolism | Negatively regulates the expression of MMP-13, MMP-1, COX-2 | Human knee OA cartilage and chondrocytes [43] |
| miR-488 | ZIP8 MMP-13 | catabolism | Reduces cartilage degradation | Human OA cartilage and chondrocytes [100] |
| miR-320 | ADAMTS5 | catabolism | Negatively regulates the expression of ADAMTS5 | Human knee OA cartilage and chondrocytes [99] |
| miR-18 | IGF-1 | anabolism | Inhibits proliferation of cartilage cells. | Deer antler tip cartilage And chondrocytes [130] |
| miR-203 | MMP-1 and IL-6 (indirect) | catabolism | Increases secretion of MMP-1 and IL-6 via the NF-kB pathway | Human articular synovial tissue ang cell [113] |
| miR-181 | MMP13 (indirect) | catabolism | Increases production of MMP13 | Human knee OA cartilage [95] |
| miR-193 |
COL2, AGGRECAN, AND SOX9 |
catabolism | Downregulates anabolic factors such as type 2 collagen, aggrecan, and SOX9 | Human knee OA cartilage and chondrocytes [99] |
miRNAs AND TRANSFORMING GROWTH FACTOR (TGF)-BETA SIGNALING
Transforming growth factor (TGF)-β is a secreted homodimeric pleiotropic protein and has three isoforms: TGF-β1, TGF-β2, and TGF-β3 [18, 19]. TGF-β plays a dual role
of regulating matrix catabolism by modifying gene expression in cartilage homeostasis [16]. Activated TGF-β binds to the TGF-β type I receptor ALK1, resulting in phosphorylation of Smad1, Smad5, or Smad8, which form a complex with the co-Smad Smad4 and translocate to the nucleus to promote cartilage matrix catabolism [16]. Meanwhile, the activated TGF-β type II receptor ALK5 results in phosphorylation of Smad2 or Smad3 which form a complex with the co-Smad Smad4 and translocate to the nucleus to promote cartilage matrix anabolism (Fig. 1) [16, 18, 20]. (Fig. 1) summarizes miRNAs that regulate the TGF-β signaling pathway.
Fig. (1).
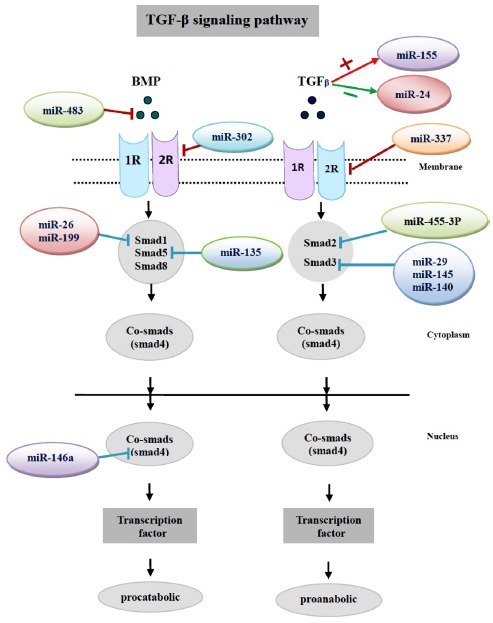
Cartilage matrix homeostasis is regulated dynamically by the TGF-β signaling pathway. The diagram shows the effect of miRNAs on the TGF-β signaling pathway. miR-199, and miR-26 target smad1 and miR-135 target smad5, and further promote the TGF-β Smad 2/3 pathway by suppressing the Smad 1/5/8 pathway from promoting an anabolic chondrocyte response. Similarly, miR-140, 145, 29 target Smad3 and then promote the TGF-β Smad 1/5/8 pathway by suppressing the Smad 2/3 pathway from promoting a degradative chondrocyte response. miR-146a regulates cartilage matrix homeostasis by targeting smad4. miR-483, and miR-337 influence cartilage matrix homeostasis via their effects on BMP7 and TGF-β2R, respectively. Conversely, miR-155 and miR-24 are negatively and positively regulated by TGF-β, respectively.
During OA pathogenesis, chondrocytes become metabolically active and disrupt the equilibrium between anabolism and catabolism, such that synthesis of aggrecan and type II collagen is reduced and production of matrix-degrading enzyme matrix metalloproteinase 13 (MMP-13) is enhanced [21]. In the multiple molecular events involved in OA cartilage destruction TGF-β signaling not only acts as a principal inducer of cartilage ECM synthesis [22, 23] but also counteracts catabolic effectors [24]. Disruption of TGF-β signaling in chondrocytes apparently leads to the degradation of cartilage ECM due to changing smad3 and elevated mmp-13 [25-29]. These data indicate an essential role of smad3 in maintaining the balance between cartilage matrix synthesis and degradation.
miRNAs play an integral part in regulating the expression of several TGF-β signaling molecules and, in turn, Smad proteins play a role in the regulation of miRNA expression and activity [20] miR-455-3p directly targets translation of smad2 and the activin type-2B receptor gene (ACVR2B), which signals through the Smad 2/3 pathway [30]. Studies by the same group [31] had previously shown that miR-140 targets smad3 expression and suggest that the increased miR-455 and -140 expression promotes the TGF-β Smad 1/5/8 pathway by suppressing the Smad 2/3 pathway, thereby promoting a degradative chondrocyte response (Fig. 1) [30]. miR-145 is up-regulated in response to TGF-β1 in rat mesenchymal stem cells [32]. The finding implied that miR-145 is probably involved in TGF-β signaling. Dnpep is a target of miR140. As expected, Dnpep expression was increased in miR140-null chondrocytes. Dnpep overexpression had a mild antagonistic effect on bone morphogenetic protein (BMP) signaling at a position downstream of smad activation. miR-140 null chondrocytes showed lower-than- normal basal BMP signaling, which was reversed by Dnpep knockdown [33].
miRNAs ARE ASSOCIATED WITH THE BALANCE BETWEEN TGF-BETA AND IL-1β
It is well known that interleukin-1 beta (IL-1β) contributes to the progression of OA [18, 34-38]. Interestingly, the balance between TGF-β signaling and IL-1β is also crucial for chondrocyte homeostasis. Recent studies demonstrated that miRNAs are implicated in the processes of OA cartilage breakdown triggered by IL-1β, including miR-140 [39], miR-27b [40], miR-146a [41], miR-9, miR-98 [42] and miR-558 [43].
For example, TGF-β counteracts IL-1β up-regulation of MMP-13 and down-regulation of ECM-related genes [24]. On the other hand, TGF-β-stimulated expression of type II collagen and aggrecan was abrogated by IL-1β [44]. miR-145 expression is significantly up-regulated in OA chondrocytes and also in response to IL-1β stimulation. These findings implied that miR-145 is possibly involved in OA pathogenesis [45]. Modulation of miR-145 efficiently affected the IL-1β-induced ECM degradation in OA chondrocytes. Over-expression of miR-145 aggravated IL-1β-induced down-regulation of aggrecan and type II collagen, and further decreased IL-1β-induced production of MMP-13. Conversely, inhibition of miR-145 reversed the catabolic effect resulting from IL-1β treatment. Moreover, SMAD3, an essential factor for chondrocyte homeostasis, was directly regulated by miR-145 [45] (Fig. 1). Smad3 is regulated by miR-145 via binding one of the predicted seed sites, which results in the suppression of smad3 expression at both mRNA and protein levels (Fig. 1). Additionally, researchers observed an inverse correlation between miR-145 and smad3 expression in OA chondrocytes stimulated with IL-1β [45].
Several miRNAs have been identified as participating in the processes of disrupted cartilage homeostasis in response to IL-1β. Treatment of human chondrocytes with IL-1β suppresses expression of miR-140 [39]. Two Smad3 target genes associated to OA cartilage degradation, PAI-1 and TIMP-3, can be regulated by miR-145 in OA chondrocytes stimulated with IL-1β. Both PAI-1 and TIMP-3 are important cartilage endogenous inhibitors of catabolic proteases, including plasmin, MMPs and ADAMs. Moreover, the effects of a miR-145 inhibitor in preventing IL-1β induced down-regulation of COL2A1, PAI-1, TIMP-3 and up-regulation of MMP-13 mRNA expression were abrogated by the addition of SMAD3 siRNA [46, 47].
miRNAs AND SMADS
A recent study [48] revealed that a selective group of miRNAs are directly regulated by signal transducers of the TGF-β/BMP pathway, the Smads. The majority of miRNAs in this group contain a consensus sequence (R-SBE) in the stem region of their primary transcripts that is similar to the Smad binding element (SBE). miR-140 was identified as a miRNA containing a putative R-SBE. Receptor-regulated Smads (R-Smads) bind directly to R-SBE in pri-miRNAs in response to TGF-β or BMP-4 stimulation to provide a platform for the recruitment of miRNA-processing enzymes, Drosha and DGCR8, thus facilitating pri-miRNA cleavage by Drosha. miR-140 can also suppress the TGF-β pathway through repressing the expression of smad3 at the protein level (Fig. 1) [31, 49]. Interestingly, a recent study identifies Smad3 as a repressor that regulates the expression of miR-140 by binding to this miRNA in OA chondrocytes [49]. These results indicate that the interaction between miR-140 and smad3 may form a negative feedback loop in OA chondrocytes.
Several target genes of miR-140 have been identified recently, including histone deacetylase 4 [50], platelet-derived growth factor receptor [51], CXC group of chemokine ligand 12 [52], and smad3. Other Smad proteins were also shown to be regulated by miRNAs during osteogenic differentiation. Smad5 is targeted by miR-135, which itself is down-regulated by bone morphogenic protein 2 [53], and smad1 is targeted by miR-26 during late osteoblast differentiation [54] (Fig. 1). miR-146a targets Smad4 through both mRNA degradation and translational repression. And miR-146a regulates chondrocytes and OA pathogenesis by inhibiting Smad4, a pivotal mediator of the TGF-β signaling pathway [41]. miR-199a significantly inhibited early chondrogenesis, as revealed by the reduced expression of early marker genes for chondrogenesis such as type II collagen and Sox9, whereas anti-miR-199a increased the expression of these chondrogenic marker genes [55]. miR-199a also inhibits Smad1/Smad4- mediated transactivation of target genes, and the over-expression of smad1 completely corrects miR-199a-mediated repression of early chondrogenesis [55]. Taking these findings together, miR-199a is the first BMP2 responsive miRNA found to adversely regulate early chondrocyte differentiation via direct targeting of the smad1 transcription factor [55].
miR-24 was repressed by TGF-β and it was demonstrated that this repression was Smad3-dependent during skeletal muscle differentiation [56] (Fig. 1). miR-155 is required for TGF-β-induced epithelial-mesenchymal transition, and is upregulated by TGF-β [57]. Two miRNA clusters (miR-106b-25 and miR-17-92) were also shown to modulate TGF-β signaling in different tumors [58] (Fig. 1). miR-337 significantly down-regulated the accumulation of TGF-βR2 protein as a target of miR-337 [59] (Fig. 1). Augmented miR-337 activity can promote anabolism of cartilaginous tissues. Furthermore, miR-337 can inhibit the activity of mmp3 to prevent cartilage degradation [59].
MicroRNAs 221 and 483-5p respond to the loss of chondrocyte matrix interaction by stimulating proliferation (by suppression of inhibitors of cell division) and suppression of matrix production (possibly by release of inhibition of the MAPK pathway), respectively [60]. The expression of miR-483 was negatively correlated with the expression of (mRNA) BMP7 and TGFβ and positively correlated with MMP13, while miR-483* was positively correlated with IL-1β. Surprisingly, there was no correlation between the expression of either miR-483 or miR-483* and IGF2. miR-483 expression is correlated with both anabolic (BMP7) and catabolic factors (MMP13) [61] (Fig. 1).
miR-92a is a component of the miR-17-92 cluster. Inactivation of miR-92a activity results in loss of cartilage due to a significant reduction in cell proliferation, differentiation, and survival of chondrogenic progenitors [62]. The BMP antagonist gene noggin3 (nog3) is a direct target of miR-92a (Fig. 1). Knockdown of miR-92a reduces p-Smad1/5/8 levels, but increases nog3 mRNA levels in the cartilage, resulting in severe cartilage loss due to impaired proliferation and differentiation as well as apoptosis of chondrogenic progenitors [62]. miR-302 targets type II BMP receptor and leads to the down-regulation of BMP signaling [63] (Fig. 1). Recent studies have shown that miR-302 targets epigenetic regulators (AOF1/2, MECP1- p66, MECP2 and MBD2) [64], cell-cycle regulators (Cyclin D1/D2, CDK2, BMI-1 and PTEN) [65], TGF-β regulators (Lefty1/2 and TGF-ΒR2) [66], BMP inhibitors (DAZAP2, SLAIN1, and TOB2) [65] and NR2F2 [67]. miR-302 increases the proliferation of hADSCs and inhibits their oxidant-induced cell death, which may be mediated by targeting CDKN1A and CCL5 [68].
miR-29 members (miR-29a, -b, -c) contribute to osteogenic differentiation by targeting fibrosis-associated markers (collagens I, III and osteonectin), histone deacetylase 4 (HDAC4), and TGF-β3 [69-71]. Up-regulation of miR-29b and -29c also plays important roles by inhibiting the expression of hdac4 and smad3 [72-74] (Fig. 1). Furthermore, FOXO3A was identified as another direct target of miR-29a. miR-29a can suppress chondrogenic differentiation of hMSCs likely by repressing FoxO3a and its downstream signaling [75]. miR-22 regulated the expressions of PPARα and BMP7 as its direct target genes, subsequently inhibiting the inflammatory and catabolic changes in OA chondrocytes [76].
miRNAs AND COL2 /AGGREACAN
miR-675 was shown to up-regulate the essential cartilage matrix component COL2A1 [77]. Decreased miR-221 induced an increase in collagen type II (Col2A1) [78]. miR-18a could significantly repress the expression of Col2a1 and aggrecan by gene transfection in HCS-2/8 cells [79]. miR-34a can induce up-regulation of Col2a1 and iNOS mediated by IL-1β in chondrocytes, and conversely, silencing miRNA-34a can prevent chondrocyte apoptosis [80]. miR-210 can promote collagen type 2 production from meniscus cells through upregulation of VEGF and FGF2 in synovial cells [81]. miR-21 represses the expression of GDF-5 (as the direct target of miR-21) by inducing GDF-5 mRNA decay during the regulation of chondrogenesis [82]. GDF-5 is closely related to the bone morphogenetic protein family and is a member of the TGF-β superfamily. Overexpression of GDF-5 increases chondrocyte cell growth and differentiation in chondrogenesis, while down-regulation of GDF-5 attenuates these processes [83, 84]. miR-21 promotes high proliferation and matrix synthesis (Col2a1 and aggrecan) of chondrocytes embedded in atelocollagen gel [85].
miR-1 plays an important role in the regulation of chondrocyte phenotype during growth plate development via direct targeting of histone deacetylase 4 (HDAC4). HDAC4 negatively regulates chondrocyte hypertrophy by inhibiting Runx2, a critical transcription factor for chondrocyte hypertrophy [86]. miR-1 also plays a role during the late stage of the differentiation process, maintaining the integrity of cartilage tissue [87].
miRNA AND CARTILAGE DEGRADING ENZYMES
miR-146a is significantly upregulated in human peripheral knee OA joint tissues [88]. Transfection of synthetic miR-146a significantly suppresses extracellular matrix-associated proteins (e.g., Aggrecan, MMP-13, ADAMTS-5, collagen II) in human knee joint chondrocytes and regulates inflammatory cytokines in synovial cells from human knee joints [88]. Exogenous supplementation of synthetic miR-146a significantly modulates inflammatory cytokines and pain-related molecules (e.g., TNFα, COX-2, iNOS, IL-6, IL8, RANTS and ion channel, TRPV1) [88]. miR-146a controls knee joint homeostasis and OA-associated algesia by balancing inflammatory responses in cartilage [88]. Expression of MMP-13 is inhibited by miRNA-140 in C28/I2 cells [89]. miR-140-/- mice showed age-related OA-like changes, characterized by proteoglycan loss and fibrillation of articular cartilage [90]. miR-140 plays dual roles in both cartilage development and homeostasis, in part via by regulating ADAMTS5 in OA [90].
Enforced expression of miR-155 in RASFs was found to repress the levels of MMP-3 [91]. Levels of miR-203 did not change upon stimulation with IL-1β, TNFα, or LPS. However, DNA demethylation with 5-AZAC increased the expression of miR-203. Enforced expression of miR-203 led to significantly increased levels of MMP-1 and IL-6. miR-9 negatively regulates the expression of MMP-13 [92] (Fig. 2), while another group observed that miR-22 blocks cartilage degradation by indirectly inhibiting MMP-13 expression via regulation of BMP-7 production [76]. Studies have shown that BMP-7 accelerates cartilage degradation via induction of MMP-13 expression in chondrocytes; therefore, inhibition of BMP-7 may be chondroprotective [93, 94].
Fig. (2).
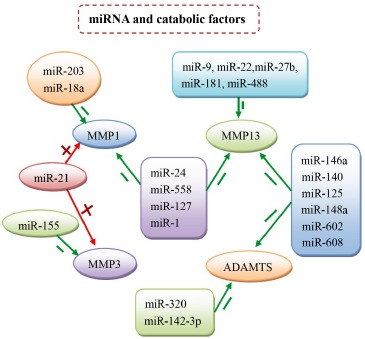
miRNAs and cartilage degenerating enzymes (MMP1, MMP3, MMP13 and ADAMTS). miR-9, 22, 27b, 181, 488 suppress cartilage matrix catabolism mainly by their effect on MMP13. miR-320 and 142-3p target ADAMTS; miR-203, miR-155, and miR-18a suppress the activation of MMP1. miR-146a, 140, 125, 148a, 602, 608 can regulate cartilage matrix homeostasis by acting on MMP13 and ADAMTS. Similarly, miR-24, 558, 127, 1 can suppress both MMP1 and MMP3. In addition, miR-21 increases the expression of MMP1 and MMP3.
miR-181b was significantly up-regulated in chondrocytes isolated from the cartilage of OA patients [95]. The use of a mimic or an inhibitor to alter miR-181b levels in chondroblasts and articular chondrocytes showed that attenuation of miR-181b reduced MMP-13 expression while inducing type II collagen expression. Furthermore, over-expression of anti-miR-181b significantly reduced the cartilage destruction caused by DMM surgery in mice [95].
Over-expression of miR-21 could attenuate the process of chondrogenesis by regulating chondrocyte proliferation and the expression of type II collagen and aggrecan. Furthermore, miR-21 significantly increased the levels of MMP1, MMP2, MMP3 and MMP9 [82] (Fig. 2). The differential expression of miR-142-3p alters ADAM9 expression and is partially responsible for the limb-type specific effects induced by TGF-β3 [96] (Fig. 2). Various studies have examined the influence of specific miRNAs on cartilage homeostasis and OA pathogenesis [76, 97]. Several reports have identified miRNA–gene target pairs that appear to be involved in cartilage homeostasis. These include miR-140-ADAMTS5, miR-483-ACAN, miR-509- SOX9, and miR-223-GDF5 [42, 76, 97, 98]. HOXA10, which is regulated by miR-320c, may regulate MMP13 through RUNX2 [99]. miR-199a-3p and miR- 193b may be involved in chondrocyte aging by regulating aggrecan, type 2 collagen, and SOX9 [99]. miRNA-320c may be involved in the juvenile properties of chondrocytes by regulating ADAMTS5 [99].
miR-488 acts as a positive role for chondrocyte differentiation/ cartilage development by inhibiting MMP-13 activity through targeting ZIP-8 [100] (Fig. 2). miR-365 is the first identified mechanically responsive miRNA that regulates chondrocyte differentiation via directly targeting HDAC4 [101]. Over-expression of HDAC4 reverses miR-365 stimulation of chondrocyte differentiation markers including Ihh, Col X, and Runx2. Moreover, inhibition of miR-365 abolishes mechanical stimulation of chondrocyte differentiation [101]. In addition, two other targets of miR-365, IL-6 and Bcl-2, have been identified [102, 103]. Overexpression of hsa-miR-148a increased COL2A1 and decreased COL10A1, MMP13 and ADAMTS5 gene expression [104].
miRNAs AND IL-1 IN CARTILAGE DEGENERATION
miR-146a functions in an anti-catabolic manner in articular cartilage by antagonizing the IL-1β-induced expression of cartilage degrading enzymes MMP13 [105] and ADAMTS5 [106, 107] (Fig. 2). miR-146 might regulate cytokine signaling by a negative feedback regulation loop involving down-regulation of IL-1 receptor associated kinase 1 (IRAK1) and TNF receptor associated factor 6 (TRAF6) [108]. Reduced miR-140 expression was observed in human OA cartilage [39, 76]. In vitro treatment of chondrocytes with IL-1β suppressed miR-140 expression. Transfection of chondrocytes with ds-miR-140 down-regulated IL-1β induced ADAMTS5 expression and rescued the IL-1β-dependent repression of AGGRECAN gene expression [39] (Fig. 2). Overexpression of miR-27b led to a significant reduction in IL-1- induced MMP-13 production, while inhibition of miR-27b in IL-1β stimulated chondrocytes increased MMP-13 protein expression in the culture supernatants [40] (Fig. 2).
miR-125 suppressed ADAMTS-4 mRNA expression by 72% and protein production by 62% following IL-1β stimulation [109]. In addition, miR-125b also suppressed MMP-13 [110] (Fig. 2). The expression of miR-127-5p was significantly reduced in OA cartilage [111]. miRNA- 127-5p suppressed IL-1–induced MMP-13 production as well as the activity of a reporter construct containing the 3-UTR of human MMP-13 mRNA. Interestingly, the IL-1 induced activation of JNK, p38, and NF-kB and expression of MMP-1 and cyclooxygenase 2 were significantly inhibited by miR-127-5p [111] (Fig. 2). When overexpressed in chondrocytes, p16INK4a induces the production of MMP1 and MMP13. miR-24 has been identified as a negative regulator of p16INK4a. p16INK4a expression was increased when miR-24 level was repressed upon IL-1βeta addition in OA cartilage [112] (Fig. 2).
Normal human articular cartilage expressed miR-558, and its expression was significantly lower in OA cartilage. Stimulation with IL-1β led to a significant reduction in miR-558 expression in normal and OA chondrocytes. IL-1β-induced activation of MAP kinase (MAPK) and nuclear factor-kB (NF-kB) decreased miR-558 expression and induced COX-2 expression in chondrocytes. Interestingly, IL-1β-induced activation of NF-kB and expression of matrix metalloproteinase (MMP)-1 and MMP-13 was significantly inhibited by miR-558 overexpression. miR-558 directly targets COX-2 and regulates IL-1β-stimulated catabolic effects in human chondrocytes [43] (Fig. 2). Elevated levels of miR-203 lead to increased secretion of MMP-1 and IL-6, indicating that miR-203 might be a proinflammatory and joint-destructive factor in RA [113] (Fig. 2). Over-expression of miR-203 resulted in increased secretion of NO, which was one of the major pro-inflammatory factors in OA via targeting TRPV4 in MCCs [114]. miR-221 is a negative modulator of chondrogenesis in chick limb mesenchymal cells and this occurs via Mdm2 down-regulation and subsequent inhibition of slug degradation [115]. The slug overexpression-induced apoptosis could be reversed by co-treatment with the miR-221 inhibitor, which decreased slug levels and enhanced cell migration [115]. miR-488 was significantly decreased in OA chondrocytes [100]. Exposure of IL-1β suppressed whereas exposure of TGF-β3 induced miR-488 in human articular chondrocytes isolated from biopsy samples of normal cartilages [100].
miRNAs AND SOX9 IN CARTILAGE HOMEOSTASIS
SOX9 inhibits the expression of its own repressor, miR-1247 in human chondrocytes, thus representing a negative feedback loop form of regulation [116]. The expression of SOX9 is tightly regulated during joint development [117] and is thought to be altered in joint disease [118]. SOX9 regulates the expression of several key genes required for normal cartilage function and maintenance [119, 120] and is also necessary for regulation of genes associated with cartilage hypertrophy [121, 122]. In addition to targeting cartilage matrix genes, SOX9 has more recently been shown to regulate expression of two miRNAs important for cartilage homeostasis, namely miR-140 [90, 123] and miR-675 [77]. Furthermore, SOX9 expression can be regulated by other miRNAs in different tissues; for example, brain-enriched miR-124 targets SOX9 during adult neurogenesis [124], and miR-145 also targets SOX9 in cartilage [125]. Transcription of miR-29a was negatively regulated by Sox9 and subsequent analyses demonstrated that overexpression of miR-29a inhibited the differentiation of hMSCs towards chondrocytes both in vitro and in vivo [75]. Over-expression of miR-675 can rescue COL2A1 levels in SOX9-depleted cells [77].
Sox9 is a master regulator for chondrocytes metabolism and it is a target gene of miR-145 [125, 126]. Overexpression of SOX9 could restore the expression levels of proteoglycans and type II collagen in OA articular cartilage [127]. It is perfectly reasonable to assume that the positive effect of miR-145 on IL-1β-induced catabolic events should be mediated by Sox9 [45]. miR-145 regulation of ACAN expression is not mediated by Smad3, but rather by Sox9 [46, 47]. miR-199a significantly inhibited early chondrogenesis, as revealed by the reduced expression of early marker genes for chondrogenesis such as type II collagen and SOX9, whereas anti-miR-199a increased the expression of these chondrogenic marker genes [55]. Decreased miR-221 induced increases in Sox9 and TRPS1 [78]. Sox5 and Sox6 activate COL2A1 and aggrecan genes in coordination with SOX9 [10, 33]. miR-194 suppresses the chondrogenic differentiation of ASCs by directly targeting Sox5 [128].
miRNAs AND NF-kB
Taganov et al. [108] reported that miR-146a is induced in response to lipopolysaccharide (LPS) and proinflammatory mediators in THP-1 cells by NF-kB. miR-18a is involved in the upregulation of TNFα-induced secretion of MMP-1, inflammatory cytokines, and chemokines [129] (Fig. 2). Moreover, using reporter gene assays, researchers identified TNFAIP-3 (tumor necrosis factor, alpha-induced protein 3) as a novel direct target of miR-18a, and they demonstrated that by repressing TNFAIP-3 expression, miR-18a enhances NF-kB signaling in RASFs [129]. miR-19b targets not only TNFAIP-3, but also other negative regulators of NF-kB signaling such as CYLD. miR-18a targets an inhibitor of STAT-3 signaling, the protein inhibitor of activated STAT-3 (PIAS-3) [130]. Interestingly, it has been shown that knockout of TNFAIP-3 in myeloid cells triggers the development of an erosive polyarthritis [131]; conversely, adenoviral delivery of TNFAIP-3 was shown to improve inflammation and bone destruction in collagen-induced arthritis [132]. These data show that the NF-kB signaling pathway is a crucial mediator of arthritis, and they imply that controlling NF-kB activity (directly via TNFAIP-3 or indirectly through miR-18a) in the different cell types involved in the pathogenesis of RA may be a promising therapeutic approach. Induction of IL-6 by miR-203 was inhibited by blocking of the NF-kB pathway. Basal expression levels of IL-6 correlated with basal expression levels of miR- 203 [114].
miRNAs AND HEDGEHOG (HH)
Activated hedgehog (HH) signaling can up-regulate the expression of MMP-13 and ADAMTS-5 in OA chondrocytes via transcription factor RUNX-2 [133-135] (Fig. 2). Activation of NF-kB or MAPK signaling negatively regulates the expression of SHH in OA chondrocytes. miR-602 and miR-608 bind the target sites present in the coding region of SHH mRNA and inhibit its expression in human chondrocytes in vitro [136]. These data provide an explanation for recent findings suggesting that expression of miR-602 and miR-608 is low and expression of SHH is high in damaged cartilage, while expression of miR-602 and miR-608 is high and expression of SHH is low in smooth cartilage [136].
miRNA AND OTHER METABOLIC CYTOKINES
Protogenin (PRTG) inhibits cell proliferation and survival in chondrogenic progenitors and articular chondrocytes. The abbrogation of miR-9 induction, which results in increased PRTG levels in OA pathogenesis, may be responsible for chondrocyte apoptosis [92]. Jones et al. suggested the involvement of miR-9 in OA bone and cartilage by mediating the IL-1β-induced production of TNF-α [42]. Over-expression of miR-203 resulted in increased secretion of NO, which was one of the major pro-inflammatory factors in OA via targeting TRPV4 in MCCs [114]. miRNA-23b and the small molecule PKA inhibitor H-89 induced differentiation into chondrocyte of hMSCs through down-regulation of protein kinase A (PKA) signaling [137]. H-89 induces the expression of aggrecan and miR-23b and inhibits MMP-2 and MMP-9 [138]. Thus, inhibiting PKA signaling using H-89 and/or miRNA-23b may be a useful tool for developing treatments for patients with degenerative arthritis [137]. miR-149 is down-regulated in OA chondrocytes, and this decrease may correlate with an increased expression of pro-inflammatory cytokines such as TNFa, IL-1β and IL-6 [139].
Controversial results and discrepancies have been found in studies of miRNA expression in human OA cartilage [140-142]. They may be due to the subpopulation of OA patients or the different stage of OA. Furthermore, there are several methods used to study miRNA profiles in the field, such as the Taqman MicroRNA array (Applied Biosystems, Thermo Fisher Scientific, Grand Island, NY) and µParaflo® Microarray (LC Sciences, Houston, TX). The correlation between the results obtained with different methods has not been fully explored.
SUMMARY
OA is a common disease with unclear pathogenesis. miRNAs are emerging as important modulators in human diseases. Recent findings indicate that miRNAs may also play a critical role in maintaining normal and OA cartilage homeostasis.
ACKNOWLEDGEMENTS
The project was supported by Grant R01AR059142 from NIH/NIAMS and an Institutional Development Award (IDeA) P20GM104937 from NIH/NIGMS, NSFC 81071495, 81171676 and 31271033, SXNSF 2011011042, SXISTCP 2013081050. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health. The authors gratefully acknowledge Ericka M. Bueno, Ph.D. for help with the manuscript preparation and editorial services.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Arden N., Nevitt M.C. Osteoarthritis: epidemiology. Best Pract. Res. Clin. Rheumatol. 2006;20(1):3–25. doi: 10.1016/j.berh.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence R.C., Felson D.T., Helmick C.G., Arnold L.M., Choi H., Deyo R.A., Gabriel S., Hirsch R., Hochberg M.C., Hunder G.G., Jordan J.M., Katz J.N., Kremers H.M., Wolfe F., National Arthritis Data Workgroup Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 2008;58(1):26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Prevalence of doctor-diagnosed arthritis and arthritis-attributable activity limitation --- United States, 2007-2009. MMWR Morb. Mortal. Wkly. Rep. 2010;59(39):1261–1265. [PubMed] [Google Scholar]
- 4.Tsezou A. Osteoarthritis year in review 2014: genetics and genomics. Osteoarthritis Cartilage. 2014;22(12):2017–2024. doi: 10.1016/j.joca.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Kraus V.B., Jordan J.M., Doherty M., Wilson A.G., Moskowitz R., Hochberg M., Loeser R., Hooper M., Renner J.B., Crane M.M., Hastie P., Sundseth S., Atif U. The Genetics of Generalized Osteoarthritis (GOGO) study: study design and evaluation of osteoarthritis phenotypes. Osteoarthritis Cartilage. 2007;15(2):120–127. doi: 10.1016/j.joca.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Nakasa T., Nagata Y., Yamasaki K., Ochi M. A mini-review: microRNA in arthritis. Physiol. Genomics. 2011;43(10):566–570. doi: 10.1152/physiolgenomics.00142.2010. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Sharon E., Spector Y., Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 10.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomankova T., Petrek M., Gallo J., Kriegova E. MicroRNAs: emerging regulators of immune-mediated diseases. Scand. J. Immunol. 2012;75(2):129–141. doi: 10.1111/j.1365-3083.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Stricker H.M., Gou D., Liu L. MicroRNA: past and present. Front. Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 15.Sonkoly E., Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J. Cell. Mol. Med. 2009;13(1):24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Wei X., Zhou J., Wei L. The age-related changes in cartilage and osteoarthritis. 2013 doi: 10.1155/2013/916530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirzamohammadi F., Papaioannou G., Kobayashi T. MicroRNAs in cartilage development, homeostasis, and disease. Curr. Osteoporos. Rep. 2014;12(4):410–419. doi: 10.1007/s11914-014-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto M., Nakasa T., Hikata T., Asahara H. Molecular network of cartilage homeostasis and osteoarthritis. Med. Res. Rev. 2008;28(3):464–481. doi: 10.1002/med.20113. [DOI] [PubMed] [Google Scholar]
- 19.Clark D.A., Coker R. Transforming growth factor-beta (TGF-beta). Int. J. Biochem. Cell Biol. 1998;30(3):293–298. doi: 10.1016/S1357-2725(97)00128-3. [DOI] [PubMed] [Google Scholar]
- 20.Gibson G., Asahara H. microRNAs and cartilage. J. Orthop. Res. 2013;31(9):1333–1344. doi: 10.1002/jor.22397. [DOI] [PubMed] [Google Scholar]
- 21.Sandell L.J., Aigner T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 2001;3(2):107–113. doi: 10.1186/ar148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redini F., Galera P., Mauviel A., Loyau G., Pujol J.P. Transforming growth factor beta stimulates collagen and glycosaminoglycan biosynthesis in cultured rabbit articular chondrocytes. FEBS Lett. 1988;234(1):172–176. doi: 10.1016/0014-5793(88)81327-9. [DOI] [PubMed] [Google Scholar]
- 23.van Beuningen H.M., van der Kraan P.M., Arntz O.J., van den Berg W.B. Transforming growth factor-beta 1 stimulates articular chondrocyte proteoglycan synthesis and induces osteophyte formation in the murine knee joint. Lab. Invest. 1994;71(2):279–290. [PubMed] [Google Scholar]
- 24.Takahashi N., Rieneck K., van der Kraan P.M., van Beuningen H.M., Vitters E.L., Bendtzen K., van den Berg W.B. Elucidation of IL-1/TGF-beta interactions in mouse chondrocyte cell line by genome-wide gene expression. Osteoarthritis Cartilage. 2005;13(5):426–438. doi: 10.1016/j.joca.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Serra R., Johnson M., Filvaroff E.H., LaBorde J., Sheehan D.M., Derynck R., Moses H.L. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. J. Cell Biol. 1997;139(2):541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., Chen L., Xu X., Li C., Huang C., Deng C.X. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell Biol. 2001;153(1):35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.G., Thuillier D., Chin E.N., Alliston T. Chondrocyte-intrinsic Smad3 represses Runx2-inducible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum. 2012;64(10):3278–3289. doi: 10.1002/art.34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q., Kim K.O., Sampson E.R., Chen D., Awad H., O’Brien T., Puzas J.E., Drissi H., Schwarz E.M., O’Keefe R.J., Zuscik M.J., Rosier R.N. Induction of an osteoarthritis-like phenotype and degradation of phosphorylated Smad3 by Smurf2 in transgenic mice. Arthritis Rheum. 2008;58(10):3132–3144. doi: 10.1002/art.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valdes A.M., Spector T.D., Tamm A., Kisand K., Doherty S.A., Dennison E.M., Mangino M., Tamm A., Kerna I., Hart D.J., Wheeler M., Cooper C., Lories R.J., Arden N.K., Doherty M. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 2010;62(8):2347–2352. doi: 10.1002/art.27530. [DOI] [PubMed] [Google Scholar]
- 30.Swingler T.E., Wheeler G., Carmont V., Elliott H.R., Barter M.J., Abu-Elmagd M., Donell S.T., Boot-Handford R.P., Hajihosseini M.K., Münsterberg A., Dalmay T., Young D.A., Clark I.M. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64(6):1909–1919. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- 31.Pais H., Nicolas F.E., Soond S.M., Swingler T.E., Clark I.M., Chantry A., Moulton V., Dalmay T. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA. 2010;16(3):489–494. doi: 10.1261/rna.1701210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayorga M.E., Penn M.S. miR-145 is differentially regulated by TGF-β1 and ischaemia and targets Disabled-2 expression and wnt/β-catenin activity. J. Cell. Mol. Med. 2012;16(5):1106–1113. doi: 10.1111/j.1582-4934.2011.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura Y., Inloes J.B., Katagiri T., Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol. Cell. Biol. 2011;31(14):3019–3028. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldring M.B., Marcu K.B. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res. Ther. 2009;11(3):224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westacott C.I., Sharif M. Cytokines in osteoarthritis: mediators or markers of joint destruction? Semin. Arthritis Rheum. 1996;25(4):254–272. doi: 10.1016/S0049-0172(96)80036-9. [DOI] [PubMed] [Google Scholar]
- 36.Daheshia M., Yao J.Q. The interleukin 1beta pathway in the pathogenesis of osteoarthritis. J. Rheumatol. 2008;35(12):2306–2312. doi: 10.3899/jrheum.080346. [DOI] [PubMed] [Google Scholar]
- 37.Goldring M.B. Osteoarthritis and cartilage: the role of cytokines. Curr. Rheumatol. Rep. 2000;2(6):459–465. doi: 10.1007/s11926-000-0021-y. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi M., Squires G.R., Mousa A., Tanzer M., Zukor D.J., Antoniou J., Feige U., Poole A.R. Role of interleukin-1 and tumor necrosis factor alpha in matrix degradation of human osteoarthritic cartilage. Arthritis Rheum. 2005;52(1):128–135. doi: 10.1002/art.20776. [DOI] [PubMed] [Google Scholar]
- 39.Miyaki S., Nakasa T., Otsuki S., Grogan S.P., Higashiyama R., Inoue A., Kato Y., Sato T., Lotz M.K., Asahara H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60(9):2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhtar N., Rasheed Z., Ramamurthy S., Anbazhagan A.N., Voss F.R., Haqqi T.M. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62(5):1361–1371. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Huang J., Dai L., Yu D., Chen Q., Zhang X., Dai K. miR-146a, an IL-1β responsive miRNA, induces vascular endothelial growth factor and chondrocyte apoptosis by targeting Smad4. Arthritis Res. Ther. 2012;14(2):R75. doi: 10.1186/ar3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S.W., Watkins G., Le Good N., Roberts S., Murphy C.L., Brockbank S.M., Needham M.R., Read S.J., Newham P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009;17(4):464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Park S.J., Cheon E.J., Kim H.A. MicroRNA-558 regulates the expression of cyclooxygenase-2 and IL-1β-induced catabolic effects in human articular chondrocytes. Osteoarthritis Cartilage. 2013;21(7):981–989. doi: 10.1016/j.joca.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Roman-Blas J.A., Stokes D.G., Jimenez S.A. Modulation of TGF-beta signaling by proinflammatory cytokines in articular chondrocytes. Osteoarthritis Cartilage. 2007;15(12):1367–1377. doi: 10.1016/j.joca.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B., Kang X., Xing Y., Dou C., Kang F., Li J., Quan Y., Dong S. Effect of microRNA-145 on IL-1β-induced cartilage degradation in human chondrocytes. FEBS Lett. 2014;588(14):2344–2352. doi: 10.1016/j.febslet.2014.05.033. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto G., Aoki T., Nakamura H., Tanzawa K., Okada Y. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4). FEBS Lett. 2001;494(3):192–195. doi: 10.1016/S0014-5793(01)02323-7. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Lee S.R., Arai K., Lee S.R., Tsuji K., Rebeck G.W., Lo E.H. Lipoprotein receptor-mediated induction of matrix metalloproteinase by tissue plasminogen activator. Nat. Med. 2003;9(10):1313–1317. doi: 10.1038/nm926. [DOI] [PubMed] [Google Scholar]
- 48.Davis B.N., Hilyard A.C., Nguyen P.H., Lagna G., Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell. 2010;39(3):373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tardif G., Pelletier J.P., Fahmi H., Hum D., Zhang Y., Kapoor M., Martel-Pelletier J. NFAT3 and TGF-β/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res. Ther. 2013;15(6):R197. doi: 10.1186/ar4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M.K., Clark I., Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 51.Eberhart J.K., He X., Swartz M.E., Yan Y.L., Song H., Boling T.C., Kunerth A.K., Walker M.B., Kimmel C.B., Postlethwait J.H. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat. Genet. 2008;40(3):290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicolas F.E., Pais H., Schwach F., Lindow M., Kauppinen S., Moulton V., Dalmay T. Experimental identification of microRNA-140 targets by silencing and overexpressing miR-140. RNA. 2008;14(12):2513–2520. doi: 10.1261/rna.1221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z., Hassan M.Q., Volinia S., van Wijnen A.J., Stein J.L., Croce C.M., Lian J.B., Stein G.S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. USA. 2008;105(37):13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luzi E., Marini F., Sala S.C., Tognarini I., Galli G., Brandi M.L. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J. Bone Miner. Res. 2008;23(2):287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 55.Lin E.A., Kong L., Bai X.H., Luan Y., Liu C.J. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J. Biol. Chem. 2009;284(17):11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M., Shao N., Yang X. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36(8):2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong W., Yang H., He L., Zhao J.J., Coppola D., Dalton W.S., Cheng J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008;28(22):6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrocca F., Vecchione A., Croce C.M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68(20):8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 59.Zhong N., Sun J., Min Z., Zhao W., Zhang R., Wang W., Tian J., Tian L., Ma J., Li D., Han Y., Lu S. MicroRNA-337 is associated with chondrogenesis through regulating TGFBR2 expression. Osteoarthritis Cartilage. 2012;20(6):593–602. doi: 10.1016/j.joca.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Yang M., Zhang L., Gibson G.J. Chondrocyte miRNAs 221 and 483-5p respond to loss of matrix interaction by modulating proliferation and matrix synthesis. Connect. Tissue Res. 2015;56(3):236–243. doi: 10.3109/03008207.2015.1018384. [DOI] [PubMed] [Google Scholar]
- 61.Qi Y., Ma N., Yan F., Yu Z., Wu G., Qiao Y., Han D., Xiang Y., Li F., Wang W., Gao X. The expression of intronic miRNAs, miR-483 and miR-483*, and their host gene, Igf2, in murine osteoarthritis cartilage. Int. J. Biol. Macromol. 2013;61:43–49. doi: 10.1016/j.ijbiomac.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Ning G., Liu X., Dai M., Meng A., Wang Q. MicroRNA-92a upholds Bmp signaling by targeting noggin3 during pharyngeal cartilage formation. Dev. Cell. 2013;24(3):283–295. doi: 10.1016/j.devcel.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 63.Kang H., Louie J., Weisman A., Sheu-Gruttadauria J., Davis-Dusenbery B.N., Lagna G., Hata A. Inhibition of microRNA-302 (miR-302) by bone morphogenetic protein 4 (BMP4) facilitates the BMP signaling pathway. J. Biol. Chem. 2012;287(46):38656–38664. doi: 10.1074/jbc.M112.390898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin S.L., Chang D.C., Lin C.H., Ying S.Y., Leu D., Wu D.T. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39(3):1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lipchina I., Elkabetz Y., Hafner M., Sheridan R., Mihailovic A., Tuschl T., Sander C., Studer L., Betel D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25(20):2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R., Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu S., Wilson K.D., Ghosh Z., Han L., Wang Y., Lan F., Ransohoff K.J., Burridge P., Wu J.C. MicroRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cells. 2013;31(2):259–268. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J.Y., Shin K.K., Lee A.L., Kim Y.S., Park H.J., Park Y.K., Bae Y.C., Jung J.S. MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 2014;5:e1385. doi: 10.1038/cddis.2014.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z., Hassan M.Q., Jafferji M., Aqeilan R.I., Garzon R., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284(23):15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapinas K., Kessler C., Ricks T., Gronowicz G., Delany A.M. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 2010;285(33):25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kapinas K., Kessler C.B., Delany A.M. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 2009;108(1):216–224. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7(3):e33766. doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J. Biol. Chem. 2012;287(30):25255–25265. doi: 10.1074/jbc.M112.357053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winbanks C.E., Wang B., Beyer C., Koh P., White L., Kantharidis P., Gregorevic P. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 2011;286(16):13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guérit D., Brondello J.M., Chuchana P., Philipot D., Toupet K., Bony C., Jorgensen C., Noël D. FOXO3A regulation by miRNA-29a Controls chondrogenic differentiation of mesenchymal stem cells and cartilage formation. Stem Cells Dev. 2014;23(11):1195–1205. doi: 10.1089/scd.2013.0463. [DOI] [PubMed] [Google Scholar]
- 76.Iliopoulos D., Malizos K.N., Oikonomou P., Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3(11):e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudek K.A., Lafont J.E., Martinez-Sanchez A., Murphy C.L. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J. Biol. Chem. 2010;285(32):24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lolli A., Lambertini E., Penolazzi L., Angelozzi M., Morganti C., Franceschetti T., Pelucchi S., Gambari R., Piva R. Pro-chondrogenic effect of miR-221 and slug depletion in human MSCs. Stem Cell Rev. 2014;10(6):841–855. doi: 10.1007/s12015-014-9532-1. [DOI] [PubMed] [Google Scholar]
- 79.Ohgawara T., Kubota S., Kawaki H., Kondo S., Eguchi T., Kurio N., Aoyama E., Sasaki A., Takigawa M. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583(6):1006–1010. doi: 10.1016/j.febslet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 80.Abouheif M.M., Nakasa T., Shibuya H., Niimoto T., Kongcharoensombat W., Ochi M. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 2010;49(11):2054–2060. doi: 10.1093/rheumatology/keq247. [DOI] [PubMed] [Google Scholar]
- 81.Kawanishi Y., Nakasa T., Shoji T., Hamanishi M., Shimizu R., Kamei N., Usman M.A., Ochi M. Intra-articular injection of synthetic microRNA-210 accelerates avascular meniscal healing in rat medial meniscal injured model. Arthritis Res. Ther. 2014;16(6):488. doi: 10.1186/s13075-014-0488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., Jia J., Yang S., Liu X., Ye S., Tian H. MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes. Exp. Mol. Med. 2014;46:e79. doi: 10.1038/emm.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yeh L.C., Mallein-Gerin F., Lee J.C. Differential effects of osteogenic protein-1 (BMP-7) on gene expression of BMP and GDF family members during differentiation of the mouse MC615 chondrocyte cells. J. Cell. Physiol. 2002;191(3):298–309. doi: 10.1002/jcp.10094. [DOI] [PubMed] [Google Scholar]
- 84.Shimaoka H., Dohi Y., Ohgushi H., Ikeuchi M., Okamoto M., Kudo A., Kirita T., Yonemasu K. Recombinant growth/differentiation factor-5 (GDF-5) stimulates osteogenic differentiation of marrow mesenchymal stem cells in porous hydroxyapatite ceramic. J. Biomed. Mater. Res. A. 2004;68(1):168–176. doi: 10.1002/jbm.a.20059. [DOI] [PubMed] [Google Scholar]
- 85.Kongcharoensombat W., Nakasa T., Ishikawa M., Nakamae A., Deie M., Adachi N., Mohamed A., Ochi M. The effect of microRNA-21 on proliferation and matrix synthesis of chondrocytes embedded in atelocollagen gel. Knee Surg. Sports Traumatol. Arthrosc. 2010;18(12):1679–1684. doi: 10.1007/s00167-010-1111-7. [DOI] [PubMed] [Google Scholar]
- 86.Li P., Wei X., Guan Y., Chen Q., Zhao T., Sun C., Wei L. MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development. FASEB J. 2014;28(9):3930–3941. doi: 10.1096/fj.13-249318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sumiyoshi K., Kubota S., Ohgawara T., Kawata K., Nishida T., Shimo T., Yamashiro T., Takigawa M. Identification of miR-1 as a micro RNA that supports late-stage differentiation of growth cartilage cells. Biochem. Biophys. Res. Commun. 2010;402(2):286–290. doi: 10.1016/j.bbrc.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 88.Li X., Gibson G., Kim J.S., Kroin J., Xu S., van Wijnen A.J., Im H.J. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 2011;480(1-2):34–41. doi: 10.1016/j.gene.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang Z.J., Zhuang H., Wang G.X., Li Z., Zhang H.T., Yu T.Q., Zhang B.D. MiRNA-140 is a negative feedback regulator of MMP-13 in IL-1β-stimulated human articular chondrocyte C28/I2 cells. Inflamm. Res. 2012;61(5):503–509. doi: 10.1007/s00011-012-0438-6. [DOI] [PubMed] [Google Scholar]
- 90.Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S., Takada S., Lotz M.K., Ueno-Kudo H., Asahara H. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24(11):1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stanczyk J., Pedrioli D.M., Brentano F., Sanchez-Pernaute O., Kolling C., Gay R.E., Detmar M., Gay S., Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 92.Song J., Kim D., Chun C.H., Jin E.J. MicroRNA-9 regulates survival of chondroblasts and cartilage integrity by targeting protogenin. Cell Commun. Signal. 2013;11:66. doi: 10.1186/1478-811X-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Im H.J., Muddasani P., Natarajan V., Schmid T.M., Block J.A., Davis F., van Wijnen A.J., Loeser R.F. Basic fibroblast growth factor stimulates matrix metalloproteinase-13 via the molecular cross-talk between the mitogen-activated protein kinases and protein kinase Cdelta pathways in human adult articular chondrocytes. J. Biol. Chem. 2007;282(15):11110–11121. doi: 10.1074/jbc.M609040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muddasani P., Norman J.C., Ellman M., van Wijnen A.J., Im H.J. Basic fibroblast growth factor activates the MAPK and NFkappaB pathways that converge on Elk-1 to control production of matrix metalloproteinase-13 by human adult articular chondrocytes. J. Biol. Chem. 2007;282(43):31409–31421. doi: 10.1074/jbc.M706508200. [DOI] [PubMed] [Google Scholar]
- 95.Song J., Lee M., Kim D., Han J., Chun C.H., Jin E.J. MicroRNA-181b regulates articular chondrocytes differentiation and cartilage integrity. Biochem. Biophys. Res. Commun. 2013;431(2):210–214. doi: 10.1016/j.bbrc.2012.12.133. [DOI] [PubMed] [Google Scholar]
- 96.Kim D., Song J., Kim S., Kang S.S., Jin E.J. MicroRNA-142-3p regulates TGF-β3-mediated region-dependent chondrogenesis by regulating ADAM9. Biochem. Biophys. Res. Commun. 2011;414(4):653–659. doi: 10.1016/j.bbrc.2011.09.104. [DOI] [PubMed] [Google Scholar]
- 97.Murata K., Yoshitomi H., Tanida S., Ishikawa M., Nishitani K., Ito H., Nakamura T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 2010;12(3):R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen Y.H., Hung M.C., Li L.Y. EZH2: a pivotal regulator in controlling cell differentiation. Am. J. Transl. Res. 2012;4(4):364–375. [PMC free article] [PubMed] [Google Scholar]
- 99.Ukai T., Sato M., Akutsu H., Umezawa A., Mochida J. MicroRNA-199a-3p, microRNA-193b, and microRNA-320c are correlated to aging and regulate human cartilage metabolism. J. Orthop. Res. 2012;30(12):1915–1922. doi: 10.1002/jor.22157. [DOI] [PubMed] [Google Scholar]
- 100.Song J., Kim D., Lee C.H., Lee M.S., Chun C.H., Jin E.J. MicroRNA-488 regulates zinc transporter SLC39A8/ZIP8 during pathogenesis of osteoarthritis. J. Biomed. Sci. 2013;20:31. doi: 10.1186/1423-0127-20-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guan Y.J., Yang X., Wei L., Chen Q. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011;25(12):4457–4466. doi: 10.1096/fj.11-185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qin B., Xiao B., Liang D., Xia J., Li Y., Yang H. MicroRNAs expression in ox-LDL treated HUVECs: MiR-365 modulates apoptosis and Bcl-2 expression. Biochem. Biophys. Res. Commun. 2011;410(1):127–133. doi: 10.1016/j.bbrc.2011.05.118. [DOI] [PubMed] [Google Scholar]
- 103.Xu Z., Xiao S.B., Xu P., Xie Q., Cao L., Wang D., Luo R., Zhong Y., Chen H.C., Fang L.R. miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-kappaB. J. Biol. Chem. 2011;286(24):21401–21412. doi: 10.1074/jbc.M110.198630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vonk L.A., Kragten A.H., Dhert W.J., Saris D.B., Creemers L.B. Overexpression of hsa-miR-148a promotes cartilage production and inhibits cartilage degradation by osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2014;22(1):145–153. doi: 10.1016/j.joca.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 105.Fosang A.J., Last K., Knäuper V., Murphy G., Neame P.J. Degradation of cartilage aggrecan by collagenase-3 (MMP-13). FEBS Lett. 1996;380(1-2):17–20. doi: 10.1016/0014-5793(95)01539-6. [DOI] [PubMed] [Google Scholar]
- 106.Glasson S.S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H.L., Flannery C.R., Peluso D., Kanki K., Yang Z., Majumdar M.K., Morris E.A. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434(7033):644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 107.Stanton H., Rogerson F.M., East C.J., Golub S.B., Lawlor K.E., Meeker C.T., Little C.B., Last K., Farmer P.J., Campbell I.K., Fourie A.M., Fosang A.J. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434(7033):648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 108.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matsukawa T., Sakai T., Yonezawa T., Hiraiwa H., Hamada T., Nakashima M., Ono Y., Ishizuka S., Nakahara H., Lotz M.K., Asahara H., Ishiguro N. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res. Ther. 2013;15(1):R28. doi: 10.1186/ar4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu N., Zhang L., Meisgen F., Harada M., Heilborn J., Homey B., Grandér D., Ståhle M., Sonkoly E., Pivarcsi A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012;287(35):29899–29908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park S.J., Cheon E.J., Lee M.H., Kim H.A. MicroRNA-127-5p regulates matrix metalloproteinase 13 expression and interleukin-1β-induced catabolic effects in human chondrocytes. Arthritis Rheum. 2013;65(12):3141–3152. doi: 10.1002/art.38188. [DOI] [PubMed] [Google Scholar]
- 112.Philipot D., Guérit D., Platano D., Chuchana P., Olivotto E., Espinoza F., Dorandeu A., Pers Y.M., Piette J., Borzi R.M., Jorgensen C., Noel D., Brondello J.M. p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res. Ther. 2014;16(1):R58. doi: 10.1186/ar4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stanczyk J., Ospelt C., Karouzakis E., Filer A., Raza K., Kolling C., Gay R., Buckley C.D., Tak P.P., Gay S., Kyburz D. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63(2):373–381. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hu F., Zhu W., Wang L. MicroRNA-203 up-regulates nitric oxide expression in temporomandibular joint chondrocytes via targeting TRPV4. Arch. Oral Biol. 2013;58(2):192–199. doi: 10.1016/j.archoralbio.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 115.Kim D., Song J., Jin E.J. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J. Biol. Chem. 2010;285(35):26900–26907. doi: 10.1074/jbc.M110.115105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Martinez-Sanchez A., Murphy C.L. miR-1247 functions by targeting cartilage transcription factor SOX9. J. Biol. Chem. 2013;288(43):30802–30814. doi: 10.1074/jbc.M113.496729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee J.S., Im G.I. SOX trio decrease in the articular cartilage with the advancement of osteoarthritis. Connect. Tissue Res. 2011;52(6):496–502. doi: 10.3109/03008207.2011.585409. [DOI] [PubMed] [Google Scholar]
- 119.Lefebvre V., Huang W., Harley V.R., Goodfellow P.N., de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell. Biol. 1997;17(4):2336–2346. doi: 10.1128/MCB.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Crombrugghe B., Lefebvre V., Behringer R.R., Bi W., Murakami S., Huang W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000;19(5):389–394. doi: 10.1016/S0945-053X(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 121.Dy P., Wang W., Bhattaram P., Wang Q., Wang L., Ballock R.T., Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell. 2012;22(3):597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ikegami D., Akiyama H., Suzuki A., Nakamura T., Nakano T., Yoshikawa H., Tsumaki N. Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development. 2011;138(8):1507–1519. doi: 10.1242/dev.057802. [DOI] [PubMed] [Google Scholar]
- 123.Yang J., Qin S., Yi C., Ma G., Zhu H., Zhou W., Xiong Y., Zhu X., Wang Y., He L., Guo X. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011;585(19):2992–2997. doi: 10.1016/j.febslet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 124.Cheng L.C., Pastrana E., Tavazoie M., Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martinez-Sanchez A., Dudek K.A., Murphy C.L. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J. Biol. Chem. 2012;287(2):916–924. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang B., Guo H., Zhang Y., Chen L., Ying D., Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011;6(7):e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cucchiarini M., Thurn T., Weimer A., Kohn D., Terwilliger E.F., Madry H. Restoration of the extracellular matrix in human osteoarthritic articular cartilage by overexpression of the transcription factor SOX9. Arthritis Rheum. 2007;56(1):158–167. doi: 10.1002/art.22299. [DOI] [PubMed] [Google Scholar]
- 128.Xu J., Kang Y., Liao W.M., Yu L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One. 2012;7(3):e31861. doi: 10.1371/journal.pone.0031861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Trenkmann M., Brock M., Gay R.E., Michel B.A., Gay S., Huber L.C. Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheum. 2013;65(4):916–927. doi: 10.1002/art.37834. [DOI] [PubMed] [Google Scholar]
- 130.Brock M., Trenkmann M., Gay R.E., Gay S., Speich R., Huber L.C. MicroRNA-18a enhances the interleukin-6-mediated production of the acute-phase proteins fibrinogen and haptoglobin in human hepatocytes. J. Biol. Chem. 2011;286(46):40142–40150. doi: 10.1074/jbc.M111.251793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Matmati M., Jacques P., Maelfait J., Verheugen E., Kool M., Sze M., Geboes L., Louagie E., Mc Guire C., Vereecke L., Chu Y., Boon L., Staelens S., Matthys P., Lambrecht B.N., Schmidt-Supprian M., Pasparakis M., Elewaut D., Beyaert R., van Loo G. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 2011;43(9):908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 132.Hah Y.S., Lee Y.R., Jun J.S., Lim H.S., Kim H.O., Jeong Y.G., Hur G.M., Lee S.Y., Chung M.J., Park J.W., Lee S.I., Park B.H. A20 suppresses inflammatory responses and bone destruction in human fibroblast-like synoviocytes and in mice with collagen-induced arthritis. Arthritis Rheum. 2010;62(8):2313–2321. doi: 10.1002/art.27545. [DOI] [PubMed] [Google Scholar]
- 133.Tetsunaga T., Nishida K., Furumatsu T., Naruse K., Hirohata S., Yoshida A., Saito T., Ozaki T. Regulation of mechanical stress-induced MMP-13 and ADAMTS-5 expression by RUNX-2 transcriptional factor in SW1353 chondrocyte-like cells. Osteoarthritis Cartilage. 2011;19(2):222–232. doi: 10.1016/j.joca.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 134.Lin A.C., Seeto B.L., Bartoszko J.M., Khoury M.A., Whetstone H., Ho L., Hsu C., Ali S.A., Alman B.A. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat. Med. 2009;15(12):1421–1425. doi: 10.1038/nm.2055. [DOI] [PubMed] [Google Scholar]
- 135.Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J.P., Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011;7(1):33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 136.Akhtar N., Makki M.S., Haqqi T.M. MicroRNA-602 and microRNA-608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis Rheumatol. 2015;67(2):423–434. doi: 10.1002/art.38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ham O., Lee C.Y., Song B.W., Lee S.Y., Kim R., Park J.H., Lee J., Seo H.H., Lee C.Y., Chung Y.A., Maeng L.S., Lee M.Y., Kim J., Hwang J., Woo D.K., Chang W. Upregulation of miR-23b enhances the autologous therapeutic potential for degenerative arthritis by targeting PRKACB in synovial fluid-derived mesenchymal stem cells from patients. Mol. Cells. 2014;37(6):449–456. doi: 10.14348/molcells.2014.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ham O., Song B.W., Lee S.Y., Choi E., Cha M.J., Lee C.Y., Park J.H., Kim I.K., Chang W., Lim S., Lee C.H., Kim S., Jang Y., Hwang K.C. The role of microRNA-23b in the differentiation of MSC into chondrocyte by targeting protein kinase A signaling. Biomaterials. 2012;33(18):4500–4507. doi: 10.1016/j.biomaterials.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 139.Santini P., Politi L., Vedova P.D., Scandurra R., Scotto d’Abusco A. The inflammatory circuitry of miR-149 as a pathological mechanism in osteoarthritis. Rheumatol. Int. 2014;34(5):711–716. doi: 10.1007/s00296-013-2754-8. [DOI] [PubMed] [Google Scholar]
- 140.Iliopoulos D., Malizos K.N., Oikonomou P., Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3(11):e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jones S.W., Watkins G., Le Good N., Roberts S., Murphy C.L., Brockbank S.M., Needham M.R., Read S.J., Newham P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009;17(4):464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 142.Yamasaki K., Nakasa T., Miyaki S., Ishikawa M., Deie M., Adachi N., Yasunaga Y., Asahara H., Ochi M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60(4):1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]


