Abstract
Bone has multiple functions, both morphologically and physiologically, and it frequently features in the pathological condition, including fracture and osteoporosis. For bone regeneration therapy, the regulation of osteoblast differentiation is important. MicroRNA (miRNA)s are short noncoding RNA which regulate gene expression at the post-transcriptional level. MiRNAs play an important role not only in a variety of other cellular processes including differentiation, proliferation, and apoptosis but also in the pathogenesis of human diseases. Recently, miRNAs have been known to participate in osteoblast differentiation by regulating several signaling pathways including transcription factors. New insight into the mechanism during osteogenes is affected by miRNAs has been gained. Moreover, therapeutic trials for bone diseases including osteoporosis, fracture and bone defects targeting miRNAs have been examined in animal models. MiRNA therapy will enable development of a bone regeneration therapy.
Keywords: Bone, Osteoblast, microRNA, Transcription factor, Signaling pathway.
INTRODUCTION
Bone disorders including trauma, tumor metastasis and metabolic diseases are often seen in daily clinical situations, and bone regeneration has been one of the main topics in the field of regenerative medicine. The usual treatment for bone defects or non-union, has been autologous bone grafts. However, due to a limited volume available for harvesting, there is a need for alternative methods using a scaffold such as hydroxyapatite, which is sometimes combined with loading cells or growth factors [1-3]. A single dose of cells or growth factors is sometimes used for the acceleration of bone formation at the fracture site [4]. In our super-aging society, the solution to osteoporosis is a major problem, due to its predisposition to increased risk fracture [5]. Elucidation of the pathogenesis of osteoporosis and pharmaceutical development, have opened the door to various types of drugs being developed for osteoporosis [6]. However, a more efficient therapeutic strategy is needed. Indeed, to develop an effective bone regeneration therapy, deep and thorough knowledge about bone metabolism together with novel findings is required.
Bone has multiple functions morphologically and physiologically, and it is often enforced into the pathological condition such as fracture, metastasis, and osteoporosis. Bone tissue maintains its homeostasis by continuous tissue remodeling by balancing bone formation and resorption (Fig. 1). Multicellular activities especially involving osteoblasts andosteoclasts play a crucial role in bone remodeling, and detailed biology has been investigated in these cells. Osteoblasts function to synthesize the extracellular matrix (ECM) and regulate mineralization [7]. Conversely, osteoclasts play a role in bone resorption, and osteoblasts and osteoclasts also interact during bone remodeling [8]. There are two pathways in bone formation, endochondral or intramembranous ossification. During the endochondral ossification process, chondrocytes are differentiated from mesenchymal stem cells (MSC) and produce a cartilaginous matrix, after which mineralization occurs in this matrix. In contrast, intramembranous ossification is the process without chondrocyte differentiation. For bone formation, angiogenesis/vasculogenesis is also an important factor, which enables oxygen and some nutrients to reach the osteogenesis site [9]. Several growth factors such as VEGF (vascular endothelial growth factor) and bFGF (basic fibroblast growth factor) contribute to vascular formation during bone formation. Several drugs or cell transplantation, or the administration of drugs or growth factors such as bFGF for the fracture site to enhance osteogenesis and angiogenesis have been examined in bone regeneration medicine. These bioactive factors have been developed in tissue engineering of regeneration medicine. Besides growth factors, genetic materials including nucleic acids which enable gene expressions at the transcriptional and post-transcriptional level have emerged [10]. Based on RNA interference technology, gene expressions are regulated by stimulating gene expression for bone formation or inhibition of antagonistic gene expression using antisense oligonucleotides. As for drugs for bone regeneration, bisphosphonate and parathyroid hormone (PTH) have showed good effect on bone regeneration. Bisphosphonates prevent bone resorption, and have been widely used for osteoporosis clinically. There are several reports highlighting that the combination of bisphosphonate and scaffold could exhibit good bone formation [11, 12]. PTH have the potential to enhance bone formation, and have been also used for the treatment of osteoporosis, and its application for bone regeneration medicine recently has been attractive [13]. To deliver the biochemical signals from the bioactive factors to cells or tissues effectively, temporal and spatial control of these factors is essential for bone regeneration. Therefore, the technique of fabrication of the scaffold and the combination of bioactive factors have been developed [14].
Fig. (1).
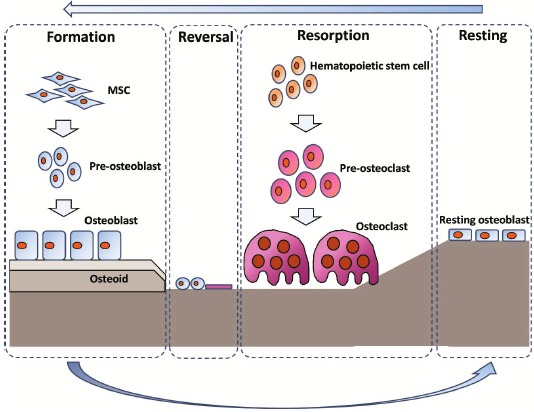
Bone remodeling. Bone remodeling consisted of formation, reversal, resorption and resting phase. Osteoblasts synthesize new bone, and osteoclasts resorb bone. After the bone resorption by osteoclast, pre-osteoblasts are prepared at the resorbed site in the reversal phase. Osteoblasts proliferate and synthesize the bone matrix. In resting phase, osteoblasts terminally differentiate into lining cells or osteocytes after the restoration of the bone surface.
The differentiation of osteoblasts is regulated by several signaling pathways including BMP (bone morphogenetic protein), Notch, and Wnt signaling pathways. These signaling pathways interact during the osteoblast differentiation with many transcriptional activators or repressors, and the transcriptional factor RUNX2, playing a crucial role in these dynamic interaction for osteoblast differentiation at all stages. Osteoblast maturation requires the additional transcriptional factor, Osterix. It has recently become clear that microRNA (miRNA)s contribute to the regulation of complicated interactions, such as transcription factors and signaling pathways during osteoblast differentiation. With this in mind, therapeutic trials for bone regeneration targeting miRNA in vivo have been conducted. This review will focus on the regulation of bone formation by miRNAs and it will also analyze an in-vivo study of bone regeneration using miRNA.
miRNAs
MiRNAs are short (around 22 nt) non-coding RNAs which regulate gene expression at post transcriptional level. MiRNAs are evolutionally conserved across phyla, and exhibit a tissue-specific or developmental stage–specific expression pattern [15, 16]. MiRNAs play an important role not only in a variety of other cellular processes including differentiation, proliferation, and apoptosis but also in the pathogenesis of human diseases [17-21].
MiRNAs are firstly transcribed into primary miRNAs (pri-miRNA)s by RNA polymerase. Pri-miRNAs harboring a hairpin structure are subsequently processed into 70-100 nt as precursor miRNA (pre-miRNA)s by the RNase Ⅱ type molecule Drosha and its cofactor DGCR8 within the nucleus. Then, pre-miRNAs are exported to the cytoplasm by exportin 5 and subsequently processed by the RNase Ⅲ type protein Dicer into a double-stranded RNA. Double-stranded miRNAs are unwound into single-stranded miRNA by a helicase, and one-strand miRNA as mature miRNA forms the RNA-induced silencing complex (RISC)-miRNA complex. The mature miRNA selectively binds mainly to the complementary 3’ untranslated region of its target mRNA including the seed sequence which comprises around 8 nt at 5 end of the mature miRNA. Then, miRNAs negatively regulated gene expression through mRNA degradation or inhibition of translation [22, 23]. Aberrant expression of miRNAs causes the loss of regulation of their target gene expression, subsequently leading to diseases such as cancer, heart failure, and autoimmune diseases.
MiRNAs AND BONE FORMATION
MiRNAs in Osteoblasts
Osteoblasts play a crucial role in bone formation. Differentiation of mature osteoblasts from MSCs regulates many factors such as several signaling pathways, transcription factors, and cytokines. Several studies have revealed that miRNAs regulate osteoblast differentiation, proliferation, mineralization and extracellular matrix synthesis. Some miRNAs promote osteoblast differentiation. MiRNA (MiR)-15b promotes osteoblast differentiation of human MSC (hMSC)s through inhibiting the BMP inhibitor, BMPER (BMP binding endothelial regulator) [24]. MiR-20a promotes osteoblast differentiation from hMSC by targeting PPARγ (Peroxisome Proliferator-Activated Receptor γ), Bambi, and Crim1, which up-regulate BMPs and Runx2 [25]. MiR-30c and miR-130b also promote osteoblast differentiation of hMSCs [24]. MiR-96 promotes osteoblast differentiation of hMSCs with increasing FABP4 (fatty acid binding protein4) expression [26]. MiR-199a also promotes osteoblast differentiation by decreasing SOX9 expression [26]. MiR-29b promotes osteoblast differentiation through targeting HDAC4 (Histone deacetylase 4), TGF (transforming growth factor)-β3, ACVR2A (activating A receptor type 2A), CTNNBIP (catenin β-interacting protein), and DUSP2 (dual specificity phosphatase 2) in mouse MC3T3 cells [27]. MiR-210 promotes osteoblast differentiation of ST2 stromal cell through inhibition of ACVR1B in mouse ST2 cells [28]. MiR-1228 promotes 1,25-dihydroxy-vitamin D-induced osteoclast differentiation of primary human osteoblasts through the inhibition of BMP2K (BMP-2 kinase) [29]. These miRNAs enhance osteoblast differentiation to inhibit the negative regulation of osteoblasts.
MiRNAs also work as negative regulators in osteoblast differentiation. MiR-23a~27a~24-2 inhibits osteoblast differentiation in mouse MC3T3-E1 cells by targeting SATB-2 (special AT-rich sequence binding protein 2) which is the positive regulator of bone formation by Runx2 synthesis, resulted in Runx2 suppression [30]. MiR-34b/c inhibits osteoblast differentiation and proliferation through decreasing SATB-2 and cyclin D1, CDK (cyclin-dependent kinase)4 and CDK6 in mouse primary osteoblast [31]. MiR-100 inhibits osteoblast differentiation of human adipose stem cell (hADSC)s by inhibiting BMPR2 (BMP receptor type Ⅱ) [32]. MiR-125b is recognized as a negative regulator of osteoblast differentiation. Over-expression of miR-125b decreases ALP (alkalin phosphatase) activity and down regulation increases ALP activity [33]. However, a possible target gene has not been confirmed. The expression of miR-138 was down-regulated during osteoblast differentiation of hMSCs [34]. Over-expression of miR-138 inhibits osteoblast differentiation of hMSC through the targeting of focal adhesion kinase, which promotes osteoblast differentiation. MiR-141 and miR-200a inhibit osteoblast differentiation by targeting Dlx5 (distal-less homeobox 5) in mouse MC3T3-E1 cells [35]. MiR-146a inhibits osteoblast differentiation from hADSC by targeting IRAK1(interleukin receptor-associated kinase 1) [36]. MiR-182 impedes osteoblast differentiation and proliferation through targeting FoxO1 (forkhead box protein O1) in mouse MC3T3-E1 cells and C3H10T1/2 cells [37]. MiR-370 inhibits BMP2 induced osteoblast differentiation by targeting ETS1 or BMP2 in mouse primary osteoblast and MC3T3-E1 cells [38]. MiR-378 inhibits osteoblast differentiation by targeting GaINT7 (UDP-N-acetyl-alpha-D-galactosamine:polupeptide N-acetylgalactosaminyl-transferase 7) in MC3T3-E1 cells [39].
MiRNA and Transcriptional Factors in Osteogenesis
Runx2 (Core-binding factor alpha, Cbfa1) is the key transcription factor in osteoblast differentiation and mineralization via the regulation of downstream genes for the osteoblast phenotype [40-42]. Runx2 binds to the promoter region of osteoblast specific genes and regulates their expression level, such as ALP, OP (osteopontin), BSP (bone sialoprotein), type 1 collagen and osteocalcin in the nucleus. The expression of Runx2 increased during the early mineralization stage of osteoblast differentiation. Runx2 is essential for the determination of osteoblasts. MSCs aredifferentiated to osteoblast by the up-regulation of Runx2, while down-regulation of Runx2 induces MSCs to chondrogenesis. The expression of Runx2 relating to osteogenesis is affected by miRNAs. Several miRNAs directly suppress Runx2. It has been reported that miR-23a, miR-27a, miR-24-2, miR-30cd, miR-31, miR-34c, miR-106a, miR-133a, miR-135a, miR-137, miR-155, miR-204, miR-205, miR-211, miR-217, miR-218, miR-222, miR-338, and miR-433 inhibit osteoblast differentiation through the targeting of RUNX2 [24, 30, 31, 43-48]. MiR-764-5p promotes osteoblast differentiation by inhibiting Runx2 degradation [49]. MiR-3960 expression was upregulated by Runx2, and miR-3960 can promote osteoblast differentiation via targeting HOXA2 (homeobox A2) which is Runx2 inhibitor [50].
Osterix is also an important transcription factor in osteoblast differentiation, which occurs through the prevention of osteoblast proliferation and the canonical Wnt signaling pathway, by inhibiting the expression of β-catenin [51, 52]. MiR-637 directly targets Osterix expression, and expression of miR-637 is decreased during osteoblast differentiation. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. MiR-637 also inhibits osteoblast differentiation, targeting COL4A1 and Osterix [53]. Osterix is also significant in mineralization. MiR-93 directly targets Osterix and over-expression of miR-93 inhibits osteoblast mineralization [54]. MiR-145 suppresses Osterix and klf4 (Kruppel-like factor 4), and klf4 inhibits miR-143 expression. Decreasing the expression of miR-143 promotes miR-145 expression, miR-143, miR-145, Osterix and klf4 constitute a feedback loop in osteoblast differentiation [55].
MiRNA AND SIGNALING PATHWAY IN OSTEOGENESIS
BMP/TGF β Pathway
BMP/TGFβ pathways play a major role in osteogenesis [56, 57]. A signal from BMP/ TGFβ is transmitted via canonical pathways, including Smad-dependent pathways and non-canonical pathways, including Smad-independent signaling pathways. Canonical pathways have eight types of Smad proteins, and activated Smads interact with a DNA promoter region to participate in transcription. Among Smad proteins, inhibitory Smad (Smad 6 and 7) negatively regulate a BMP/TGFβ signaling cascade. Non-canonical pathways include several signaling pathways, such as MAPK (Mitogen-Activated Protein Kinase)cascades and play an important role in osteoblast differentiation, proliferation and mineralization.
The BMP signaling pathway in osteoblast differentiation is also regulated by miRNAs (Fig. 2). miR-106a, miR-148a and miR-424 inhibit osteoblast differentiation of hMSCs by targeting BMPs [24]. The expression of miR-133 and miR-135 isdown-regulated, during BMP2- induced osteoblast differentiation from C2C12 cells, and over-expression of miR-133 and miR-135 impedes expression of osteogenic marker genes, through RUNX2 being targeted by miR-133, and Smad5 being targeted by miR-135 [58]. MiR-206 is also down-regulated during BMP2- induced osteoblast differentiation from C2C12 cells. Over-expression of miR-206 inhibits osteoblast differentiation, via the targeting of Connexin 43, which is necessary for osteoblast differentiation and function [54]. MiR-206 is expressed in osteoblasts and its expression decreases during osteoblast differentiation. MiR-206 expressing transgenic mice reveal low bone mass. MiR-208 decreases during BMP-induced osteogenesis, at which time it acts as an osteogenic inhibitor through binding 3’ UTR of Ets1, which stimulates osteopontin and Runx2. SMAD1 is an important factor in BMP signaling, and its increased expression augments osteoblast differentiation [60, 61]. MiR-26a decreases SMAD1 expression by binding its 3’ UTR, and down-regulation of miR-26a increases osteogenic marker genes, such as osteopontin, osteocalcin and COL1A1 [62]. MiR-199a-3p also inhibits osteoblast differentiation by targeting SMAD1, and miR-135 suppresses SMAD5 to inhibit osteoblast differentiation [59, 63]. MiR-20a is reported to be a positive regulator of the BMP signaling pathway, through the targeting of Bambi and Crim1, which are inhibitors of the BMP pathway. Furthermore, miR-20a can promote bone formation to up-regulate BMP2, BMP4, Runx2, Osterix/Sp7, Osteocalcin and Osteopontin.MiR-20a and miR-3960 could induce BMP2, while miR-370 down-regulated BMP2. Down-regulation of miR-370 expression induces an increase to its target gene expression, BMP2 and Ets1 [38, 47, 64]. MiR-221 and miR-1274a inhibit osteoblast differentiation by inhibiting the TGF-β signaling pathway [65].
Fig. (2).
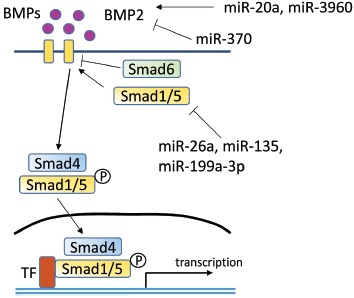
BMP signaling pathway including microRNA regulation.
The Wnt/β-cateninsignaling Pathway
Wnt/β-catenin signaling plays a crucial role in bone remodeling, especially multiple- stage osteoblast differentiation and proliferation [66, 67]. Wnt signaling pathways include the non-β-catenin PCP pathway, calcium ion signaling, RHO family GTPase pathways, and the JNK pathways. Wnt up-regulates β-catenin, subsequently activating the transcription factors, TCF7 (transcription factor 7) and LEF1 (Lymphoid enhancer-binding factor 1), which target osteoprotegerin and RUNX2. MiRNAs also participate in the regulation of osteoblast differentiation by Wnt/β-catenin signaling (Fig. 3). Let-7f, miR-29, miR-218, miR-27, niR-142-3p and miR-335-5p are able to enhance the WNT/β-catenin signaling pathway and promote osteoblast differentiation. The expression of miR-29a and miR-29b is induced by canonical Wnt signaling which is a critical regulator of osteoblast differentiation. MiR-29a also activates canonical Wnt signaling, which forms a positive feedback loop to promote osteoblast differentiation. The target genes of miR-29 were validated as Dkk-1 (dikkopf-1), Kremen2 (Kringle domain, containing transmembrane protein), and SFRP2 (secreted frizzled related protein 2), which function as inhibitors of Wnt signaling [68]. A positive feedback loop of Wnt signaling, mediated by miR-29, promotes osteoblast differentiation through the inhibition of Dkk-1, Kremen2 and SFRP2. MiR-29 modulates many collagens and extracellular matrix proteins during bone formation. During osteoblast differentiation, the expression of miR-29 is low in its early phase but its expression increases in its later phase. The other validated target genes of miR-29a are COL1A1, COL3A1 and osteonectin which is a critical for the formation of collagen fibril [69]. A low level of miR-29 allows matrix deposition in the early phase of osteoblastogenesis. In the later phase, miR-29 up-regulates to suppress collagen synthesis during the subsequent mineral deposition. MiR-218 promotes osteoblast differentiation of MSCs by inhibition of the SOST (sclerostin), DKK2 (dickkopf 2), and SFRP2 (secreted frizzled related protein 2), which are Wnt signaling inhibitors [70]. Let-7f play an important role in the regulation of Wnt/β-catenin activity, subsequently inducing osteoblast differentiation by TIMP-1(tissue inhibitor of metalloproteinase-1) [71]. Let-7a promotes osteoblast differentiation of human unrestricted somatic stem cells via inhibition of NLK (nemo-like kinase) expression which is an inhibitor of the Wnt signaling pathway [65]. APC (adenomatous polyposis coli) prevent β-catenin translocation into the nucleus when canonical Wnt signaling is absent, miR-27 suppresses the expression of APC as its target gene, and miR-27 acts as a positive regulator for osteoblast differentiation through the activation of Wnt signaling [72]. DKK-1 maintains the skeletal homeostasis through inhibiting osteoblast differentiation by down-regulating Wnt signaling. MiR-335-5p has a role in down-regulating DKK-1 by binding 3’UTR of DKK-1, subsequently enhancing Wnt signaling [73]. MiR-221 and miR-1274a impede osteoblast differentiation by inhibiting Wnt signaling [65]. MiR-142-3p also promotes osteoblast differentiation by modulating Wnt signaling by targeting APC in human FOB1.19 cells [74].
Fig. (3).
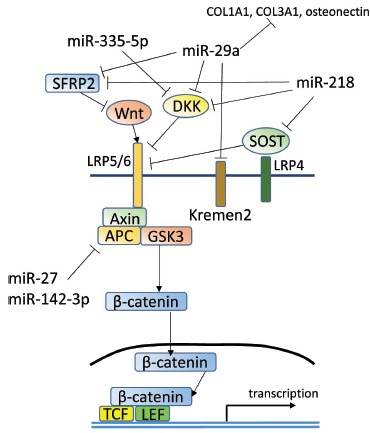
Wnt signaling pathway including microRNA regulation.
Notch Signaling
Notch signaling helps to promote differentiation of pre-osteoblasts into osteoblasts, while it suppresses differentiation of MSCs into pre-osteoblastic cells [75, 76]. MiR-34c down-regulatesnotch signaling including Notch1, Notch2, and Jag1, to maintain bone homeostasis [77].
MiRNA in Pathological Conditions Related to Bone
MiRNAs play a pivotal role in bone diseases, including osteoporosis and tumor metastasis. Osteoporosis is characterized by a low bone mineral density with increasing bone fragility. There are some subtypes in primary osteoporosis, relating to estrogen deficiency in postmenopausal women, and age-related osteoporosis in men and women. In primary osteoporosis, osteoclast activity increases, while osteoblast activity decreases, subsequently leading to loss of bone mineral density.
MiR-2861 is reported to be crucial in the pathogenesis of osteoporosis. MiR-2861 is intensely expressed in osteoblasts,its expression induced by BMP2 and it can up-regulate Runx2 through targeting HDAC5, subsequently causing osteoblast differentiation. However, down-regulation of miR-2861 inhibits bone formation [78].
MiR-133 is thought of as a potential biomarker for osteoporosis. The expression level of miR-133a in circulating monocytes of osteoporotic menopausal women is significantly higher than that of non-osteoporotic menopausal women. MiR-133 was also recognized as a negative regulator for bone formation through the suppression of inhibitory factors of osteoclastogenesis including CXCL11, CXCR3, and SLC39A1 [79]. MiR-214 is also reported to reduce bone formation through the targeting of ATF4 (activating transcription factor 4), resulting in decreasing ALP and osteocalcin expression [80]. MiR-214 shows higher expression in aged patients with multiple fractures, which indicates that miR-214 may play a role in the fracture of osteoporotic bone.MiR-21 is down-regulated in estrogen-deficient osteoporosis and its expression during osteogenesis of MSCs is suppressed by TNF (tumor necrosis factor)-α [81]. MiR-21 promotes the osteoblast differentiation of MSCs by inhibiting Spry1 (sprouty homolog 1), which is a negative regulator of osteoblast differentiation. Increasing expression of miR-21 can enhance bone formation in ovarectomized mice by down-regulation of Spry1.MiR-378 plays a role in osteoporosis and bone fracture by hyperglycemia in diabetes mellitus. High glucose conditions down-regulate miR-378 expression, and miR-378 expression up-regulates osteoblastic markers through the targeting of CASP3 (caspase 3) and activates the PI3K/Akt signaling pathway [82]. Eight miRNAs: miR-21, miR-23a, miR-24, miR-93, miR-100, miR-122a, miR-124a and miR-125b show higher expression in the serum of patients with osteoporosis compared to non-osteoporosis patients. MiR-21, miR-23a, miR-24, miR-25, miR-100 and miR-125b are up-regulated in the bone tissue of osteoporotic patients [83]. MiR-103a plays a role in osteoblast mechanotransduction. MiR-103a expression was down-regulated during cyclic mechanical stretch-induced osteoblast differentiation. Over-expression of miR-103a inhibits Runx2 through the binding of 3’ UTR of Runx2 mRNA. Inhibition of miR-103a in the hindlimb of unloading mice rescued mechanical unloading osteoporosis [84]. Osteoporosis is induced by glucocorticoids which cause osteoblasts to up-regulate the RANKL expression. Dexamethasone-induced RANKL expression in osteoblasts is down-regulated by miR-17 and miR-20a, which suggests that miR-17 and miR-20a play a role in glucocorticoid- induced osteoclast differentiation through the targeting of RANKL (Receptor activator of nuclear factor kappa-B ligand) expression in osteoblast cells [85]. MiR-34a is recognized as a suppressor of osteoclastogenesis. MiR-34a expression is down-regulated during osteoclast differentiation, and miR-34a nanoparticle treatment can effectively cause the condition of ovarectomy-induced osteoporosis mice to attenuate through the targeting of Tgif2 (transforming growth factor-β-induced factor 2) [86]. MiR-422a is up-regulated in circulatory monocytes of postmenopausal osteoporosis patients and it is thought to target CBL, CD266, IGF1, PAG1 and TOB2 [87]. Many miRNAs contribute in the bone metabolism and diseases (Table 1 and 2).
Table 1.
Summary of microRNAs which stimulate bone formation. *; putative target gene.
| Osteoblast Differentiation | microRNA | Cell | Target Gene | Reference |
|---|---|---|---|---|
| Stimulation | Let-7a | Human USSC | NLK | [65] |
| Let-7f | Human MSC | axin2 | [71] | |
| miRNA-15b | Human MSC | BMPR2* | [24] | |
| miR-20a | Human MSC | PPARγ, Bambi, Crim1 | [25] | |
| miR-21 | Human and mouse MSC | Spry1 | [81] | |
| miR-27 | Human FOB1.19 cells | APC | [72] | |
| miR-29a/b/c | Mouse primary osteoblast, MC3T3 cells | osteonectin, ACVR2A, CTNNBIP, DUSP2, HDAC4, TGFβ3 | [27, 68, 69] | |
| miR-30c | Human MSC | CAMTA1*, CXCL12*, ITGB1*, FLT1* | [24] | |
| miR-96 | Human MSC | FABP4 | [26] | |
| miR-130b | Human MSC | CAMTA1*, CD44*, GDF6*, PDGFRA*, COL9A3* | [24] | |
| miR-142-3p | Human FOB1.19 cells | APC | [74] | |
| miR-199a | Human MSC | SOX9 | [26] | |
| miR-210 | Mouse ST2 cells | ACVR1B | [28] | |
| miR-218 | Mouse bone marrow stromal cells | DKK2, SFRP2, SOST | [70] | |
| miR-335-5p | Mouse MC3T3-E1 cells, MLO-A5 cells | DKK1 | [73] | |
| miR-1228 | Human primary osteoblast | BMP2K | [29] | |
| miR-2861 | Mouse ST2 cells | HDAC4 | [64] | |
| miR-3960 | Mouse osteoblast | HOXA2 | [64] |
Table 2.
Summary of microRNAs which inhibit bone formation. *; putative target gene.
| Osteoblast Differentiation | microRNA | Cell | Target Gene | Reference |
|---|---|---|---|---|
| Inhibition | miRNA-23a/27a/24-2 | Mouse MC3T3-E1 cells | SATB2, RUNX2 | [30] |
| miR-26a | Human ADSC | SMAD1 | [62] | |
| miR-31 | Human MSCs | RUNX2, BMPR2 | [24] | |
| miR-34b/c/d | Mouse primary osteoblast, MC3T3-E1 cells | SATB2, RUNX2, Cyclin D1, CDK4, CDK6 | [31, 77] | |
| miR-93 | Mouse primary osteoblast | Sp7 | [54] | |
| miR-100 | Human ADSC | BMPR2 | [32] | |
| miR-103a | Human FOB1.19 cells, human MSC | RUNX2 | [84] | |
| miR-106a | Human MSC | CBFB* | [24] | |
| miR-125b | Mouse ST2 cells | ErbB2 | [33] | |
| miR-133a | Mouse C2C12 cells, mouse MC3T3-E1 cells | RUNX2 | [58] | |
| miR-135a | Mouse C2C12 cells, mouse MC3T3-E1 cells | RUNX2, SMAD5 | [58] | |
| miR-137 | Mouse MC3T3-E1 cells | RUNX2 | [43] | |
| miR-138 | Human MSC | PTK2 | [34] | |
| miR-141 | Mouse MC3T3-E1 cells | DLX5 | [35] | |
| miR-143 | Mouse odontoblast | osterix, klf4 | [55] | |
| miR-145 | Mouse odontoblast | osterix, klf4 | [55] | |
| miR-146a | Human ADSC | IRAK1 | [36] | |
| miR-148a | Human MSC | RUX2*, BMP3*, BMP8A*, BMP8B*, BMMPR1B*, BMPR2* | [24] | |
| miR-155 | Human ligament fibroblast | SOCS1 | [44] | |
| miR-182 | Mouse MC3T3-E1 cells, mouse C3H10T1/2 cells | FoxO1 | [37] | |
| miR-199a-3p | U2-OS cell | SMAD1 | [63] | |
| miR-200a | Mouse MC3T3-E1 cells | DLX5 | [35] | |
| miR-204 | Mouse MC3T3-E1 cell, mouse C3H10T1/2 cells, mouse ST2 cells, mouse C2C12 cells | RUNX2 | [43, 48] | |
| miR-205 | MC3T3-E1 cells | RUNX2 | [43, 47] | |
| miR-206 | Mouse C2C12 cells | Cx43 | [59] | |
| miR-208 | Mouse primary osteoblast, MC3T3-E1 cells | ETS1 | [60, 61] | |
| miR-211 | Mouse C3H10T1/2 cells, mouse ST2 cells, mouse C2C12 cells | RUNX2 | [48] | |
| miR-214 | Mouse MC3T3-E1 cells, human FOB1.19 cells | ATF4 | [80] | |
| miR-217 | Mouse MC3T3-E1 cells | RUNX2 | [43, 47] | |
| miR-218 | Mouse MC3T3-E1 cells, mouse MSC | SOST, DKK2, SFRP2 | [70] | |
| miR-221 | Human USSCs | Wnt and TGFβ signaling pathway* | [65] | |
| miR-222 | Human ligament fibroblast | RUNX2* | [44] | |
| mR-338 | MC3T3-E1 cells | RUNX2 | [43] | |
| miR-370 | Mouse primary osteoblast/MC3T3-E1 cells | ETS1 | [38] | |
| miR-378 | MC3T3-E1 cells | GaINT7 | [39, 82] | |
| miR-424 | Human MSC | BMP8A*, BMPR2*, BMPR1A* | [24] | |
| miR-433 | Mouse C3H10T1/2 cells | RUNX2 | [46] | |
| miR-637 | Human primary osteoblast/MSC | COL4A1, Osterix | [53] | |
| miR-1274a | Human USSCs | Wnt and TGFβ signaling pathway* | [65] |
Bone Regenerative Medicine Using miRNA
Several therapeutic trials have focused on miRNAs to investigate bone disease (Table 3). As for osteoporosis, the role of miRNAs in pathogenesis has been gradually elucidated. Systemic administration of antagonic miR-214 from the tail vein in ovarectomized mice has been shown to increase BMD [80].
Table 3.
In vivo study using miRNA for bone regeneration.
| Disease | Model | microRNA | Administration Method | Animal | Reference | |
|---|---|---|---|---|---|---|
| 1 | Osteoporosis | Ovariectomy | antimir-214 | Systemic injection | Mouse | [80] |
| 2 | Fracture | Femoral fracture | antimir-92a | Systemic or local injection | Mouse | [88] |
| 3 | Bone defect | 8mm diameter bony defect on calvarium | BMSC transfected antimir-31 on poly glycerol sebacute scafffold | Implantation | Rat | [89] |
| 4 | Bone defect | 5mm diameter bony defect on calvarium | ADSC transfected with antimir-31 on β-TCP | Implantation | Rat | [90] |
| 5 | Bone defect | 5mm diameter bony defect on calvarium | MSC transfected with antimir-26a on hydrogel | Implantation | Mouse | [91] |
| 6 | Normal | Subcutanous | Freeze dried allograft bone wraped with BMSC sheet transfected with antimir-138 | Implantation | Mouse | [93] |
| 7 | Bone defect | 10mm diameter orbital wall bony defect | BMSC transfected with antimir-31 on β-TCP | Implantation | Canin | [94] |
| 8 | Bone defect | 4mm diameter bony defect on calvarium | ADSC transfected with photoactivated miR-148b mimic -SNP conjugate on polycaprolactom | Implantation | Mouse | [95] |
Fractures are often seen in the clinical setting, and a long time is needed to achieve bone union. MiR-92a expression in the serum of patients with fractures was down-regulated and it recovered on day 21. Systemic or local administration of anti-miR-92a has been shown to enhance fracture healing with increasing callus formation and neovascularization in mice with a femoral fracture [88].
For repair ofa critical bone defect, an autologous bone graft is not sufficient, so the combination of an artificial bone graft and stem cells or growth factors to repair large bone defects has been examined. To this end, miRNAs have been recently studied,considered to be a promising new player.
Down-regulation of miR-31 in bone marrow MSCs enhances osteogenesis. Therefore, the effect of MSCs, with the miR-31 knock-down on critical-size bone defects,has been investigated. MSCs, expressed by anti-miR-31 and seeded on polyglycerol sebacate (PGS) scaffolds, were implanted into critical-sized calvarial defects in rats. PGS scaffolds with anti-miR-31-expressing MSCs were found to effectively repair a bone defect with good biocompatibility [89].
The effect of ADSC, modified with anti-miR-31 for a critical-sized bone defect, was also examined. Anti-miR-31 modified ADSCs, loaded onto β-TCP (β-tricalcium phosphate), was implanted to repair the critical-sized calvarial defects in rats. As a result, increasing bone volume, bone mineral density and decreasing scaffold residue brought about the dramatic repair of bone defects [90].
For bone formation, not only osteogenesis but also angiogenesis are important. MiR-26a plays a role in both osteogenesis and angiogenesis. Bone marrow MSCs transfected with anti-miR-26a, combined with hydrogel,were implanted into a critical-sized calvarial bone defect, revealing good bone repair with osteogenesis and angiogenesis [91].
For bone repair, polymeric nanofibers, which mimic natural extracellular matrix morphology,comprise one of the best scaffolds. MiR-29a inhibits ECM synthesis through the targeting of osteonectin. Therefore, down-regulation of miR-29a may lead to an increase in ECM production. Anti-miR-29a-loaded gelatin nanofibers, combined with osteoblastic murine MC3T3-E1 cells, exhibited abundant osteonectin with increasing IGF-1 (insulin growth factor-1) and TGF-β1. The combination of an ECM-mimicking nanostructured scaffold and osteogenic oligonucleotide holds much promise for the purpose ofbone repair [92].
The combination of cell sheet technology with miRNAs for regenerative medicine has been investigated. Bone marrow MSCs sheets, delivered with anti-miR-138, show the enhancement osteogenesis of cell sheets. Down-regulation of endogenous miR-138 in the cell sheets are able to enhance Runx2 expression. Anti-miR-138-delivered cell sheets also have the potential to be a novel therapeutic strategy for large bone defects [93].
Bone marrow MSCs, transfected with anti-miR-31, were seeded onto β-TCP scaffolds, then implanted into bone defects in canines. Anti-miR-31 modified MSCs, with a β-TCP scaffold,were found to efficiently repair bone defects, with increasing bone mineral density and new bone volume [94].
Conjugate miR-146b mimic and silver nanoparticles were added into hADSCs, and loaded to matrigel or polycaprolacton scaffolds. It was discovered that they could heal the critical-size bone defect in nude mice [95].
FUTURE PERSPECTIVES
Recent evidence shows that targeting miRNAs is effective as a novel therapeutic strategy in animal models. Actually, miRNA therapy, in clinical use for patients with chronic HCV (hepatitis C virus) infection, has already been conducted [96]. It was discovered that locked nucleic acid-modified DNA phosphorothioate antisense oligonucleotide, which sequesters mature miR-122 (Miravirsen), could reduce HCV RNA levels by subcutaneous injection. For clinical applicationin bone regeneration, the precise effects, including adverse effects, distribution and metabolism of administered miRNA mimic or antisense, should be examined in detail. As for adverse effects, the possibility that given miRNAs reinforce osteoblast phenotype in a detrimental environment should be considered. Theoretically, one miRNA has many target genes by computational analysis and simultaneously represses its target genes. It is possible that administered miRNA represses simultaneously not only one of the target genes which function against bone formation but also genetically important target genes. The cluster of miRNAs have been recognized to play an important role in the entire process of differentiation from stem cells [97]. Regulatory network by miRNA cluster during a characterizing a given cell phenotype is constituted by the regulation of repressors, co-activators or associated proteins, and these miRNAs ensure time related expression of genes [98, 99]. Administration of miRNA may affect these regulatory systems. In addition, the use of more effective delivery tools of miRNA mimic or antisense should be considered. Building on the findings on bone regeneration therapy from previous reports, miRNA mimic or antisense was administered by local or systemic injection, or loaded into the scaffolds. Bone turn-over consisted with the balance physiologically coordinated bone formation and resorption. To create a bone physiological microenvironment, it is desirable to co-culture of osteoblast and osteoclast in a scaffold. Previous study demonstrated that three dimensional co-culture system could promote osteoblast and osteoclast differentiation and well organized bone matrix formation [100]. The combination of these co-culture system and regulation of miRNA will enhance to create the morphologically normal bone tissue. In the case of cell modification, some transfection reagents or viral transfection were applied. Exosome has attracted attention as a novel drug delivery system. MiRNAs circulating in body fluid play a role in cell-to-cell and tissue-to-tissue communications. MiRNAs are packed in microvesicles, including exosomes, and are secreted from cells. Exosome-formed miRNAs are easily up-taken into the cells, and these features are applied to miRNA therapy. When synthetic miRNA are introduced into cells, they are reported to be enveloped in exosome, thus becoming exosome-formed miRNAs, which can be efficiently delivered into cells and can function with the same effect as when the lipofection method is used [101]. As other option to deliver of synthetic miRNA using exosome, artificial exosome which is artificially constructed mimetic structure of exosome is proposed [102]. These studies confirm that miRNA therapy for bone regeneration will be realized in the near future, thanks to the development of cell or drug delivery systems and novel scaffolds.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Kneser U., Schaefer D.J., Polykandriotis E., Horch R.E. Tissue engineering of bone: the reconstructive surgeon’s point of view. J. Cell. Mol. Med. 2006;10(1):7–19. doi: 10.1111/j.1582-4934.2006.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui L., Liu B., Liu G., Zhang W., Cen L., Sun J., Yin S., Liu W., Cao Y. Repair of cranial bone defects with adipose derived stem cells and coral scaffold in a canine model. Biomaterials. 2007;28(36):5477–5486. doi: 10.1016/j.biomaterials.2007.08.042. [DOI] [PubMed] [Google Scholar]
- 3.Bueno E.M., Glowacki J. Cell-free and cell-based approaches for bone regeneration. Nat. Rev. Rheumatol. 2009;5(12):685–697. doi: 10.1038/nrrheum.2009.228. [DOI] [PubMed] [Google Scholar]
- 4.Kawaguchi H., Oka H., Jingushi S., Izumi T., Fukunaga M., Sato K., Matsushita T., Nakamura K. TESK Group. A local application of recombinant human fibroblast growth factor 2 for tibial shaft fractures: A randomized, placebo-controlled trial. J. Bone Miner. Res. 2010;25(12):2735–2743. doi: 10.1002/jbmr.146. [DOI] [PubMed] [Google Scholar]
- 5.Bliuc D., Nguyen N.D., Nguyen T.V., Eisman J.A., Center J.R. Compound risk of high mortality following osteoporotic fracture and refracture in elderly women and men. J. Bone Miner. Res. 2013;28(11):2317–2324. doi: 10.1002/jbmr.1968. [DOI] [PubMed] [Google Scholar]
- 6.Bliuc D., Nguyen N.D., Milch V.E., Nguyen T.V., Eisman J.A., Center J.R. Mortality risk associated with low-trauma osteoporotic fracture and subsequent fracture in men and women. JAMA. 2009;301(5):513–521. doi: 10.1001/jama.2009.50. [DOI] [PubMed] [Google Scholar]
- 7.Shegarfi H., Reikeras O. Review article: bone transplantation and immune response. J. Orthop. Surg. (Hong Kong) 2009;17(2):206–211. doi: 10.1177/230949900901700218. [DOI] [PubMed] [Google Scholar]
- 8.Jin Z., Li X., Wan Y. Nuclear receptor regulation of osteoclast and bone remodeling. Mol. Endocrinol. 2014;30:m2014316. doi: 10.1210/me.2014-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saran U., Gemini Piperni S., Chatterjee S. Role of angiogenesis in bone repair. Arch. Biochem. Biophys. 2014;561:109–117. doi: 10.1016/j.abb.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Evans C.H., Ghivizzani S.C., Robbins P.D. Orthopedic gene therapy--lost in translation? J. Cell. Physiol. 2012;227(2):416–420. doi: 10.1002/jcp.23031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukozawa A., Ueki K., Marukawa K., Okabe K., Moroi A., Nakagawa K. Bone healing of critical-sized nasal defects in rabbits by statins in two different carriers. Clin. Oral Implants Res. 2011;22(11):1327–1335. doi: 10.1111/j.1600-0501.2010.02135.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim S.E., Suh D.H., Yun Y.P., Lee J.Y., Park K., Chung J.Y., Lee D.W. Local delivery of alendronate eluting chitosan scaffold can effectively increase osteoblast functions and inhibit osteoclast differentiation. J. Mater. Sci. Mater. Med. 2012;23(11):2739–2749. doi: 10.1007/s10856-012-4729-9. [DOI] [PubMed] [Google Scholar]
- 13.Esbrit P., Alcaraz M.J. Current perspectives on parathyroid hormone (PTH) and PTH-related protein (PTHrP) as bone anabolic therapies. Biochem. Pharmacol. 2013;85(10):1417–1423. doi: 10.1016/j.bcp.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Samorezov J.E., Alsberg E. Spatial regulation of controlled bioactive factor delivery for bone tissue engineering. Adv. Drug Deliv. Rev. 2015;84:45–67. doi: 10.1016/j.addr.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 17.De Santis G., Ferracin M., Biondani A., Caniatti L., Rosaria Tola M., Castellazzi M., Zagatti B., Battistini L., Borsellino G., Fainardi E., Gavioli R., Negrini M., Furlan R., Granieri E. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J. Neuroimmunol. 2010;226(1-2):165–171. doi: 10.1016/j.jneuroim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Townley-Tilson W.H., Callis T.E., Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int. J. Biochem. Cell Biol. 2010;42(8):1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai K.W., Hu L.Y., Wu C.W., Li S.C., Lai C.H., Kao H.W., Fang W.L., Lin W.C. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;49(11):969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 20.Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakasa T., Miyaki S., Okubo A., Hashimoto M., Nishida K., Ochi M., Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 24.Gao J., Yang T., Han J., Yan K., Qiu X., Zhou Y., Fan Q., Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell. Biochem. 2011;112(7):1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J.F., Fu W.M., He M.L., Xie W.D., Lv Q., Wan G., Li G., Wang H., Lu G., Hu X., Jiang S., Li J.N., Lin M.C., Zhang Y.O., Kung H.F. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;8(5):829–838. doi: 10.4161/rna.8.5.16043. [DOI] [PubMed] [Google Scholar]
- 26.Laine S.K., Alm J.J., Virtanen S.P., Aro H.T., Laitala-Leinonen T.K. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J. Cell. Biochem. 2012;113(8):2687–2695. doi: 10.1002/jcb.24144. [DOI] [PubMed] [Google Scholar]
- 27.Li Z., Hassan M.Q., Jafferji M., Aqeilan R.I., Garzon R., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284(23):15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno Y., Tokuzawa Y., Ninomiya Y., Yagi K., Yatsuka-Kanesaki Y., Suda T., Fukuda T., Katagiri T., Kondoh Y., Amemiya T., Tashiro H., Okazaki Y. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583(13):2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Lisse T.S., Chun R.F., Rieger S., Adams J.S., Hewison M. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J. Bone Miner. Res. 2013;28(6):1478–1488. doi: 10.1002/jbmr.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan M.Q., Gordon J.A., Beloti M.M., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA. 2010;107(46):19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei J., Shi Y., Zheng L., Zhou B., Inose H., Wang J., Guo X.E., Grosschedl R., Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol. 2012;197(4):509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng Y., Qu X., Li H., Huang S., Wang S., Xu Q., Lin R., Han Q., Li J., Zhao R.C. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 2012;586(16):2375–2381. doi: 10.1016/j.febslet.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 33.Mizuno Y., Yagi K., Tokuzawa Y., Kanesaki-Yatsuka Y., Suda T., Katagiri T., Fukuda T., Maruyama M., Okuda A., Amemiya T., Kondoh Y., Tashiro H., Okazaki Y. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 2008;368(2):267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 34.Eskildsen T., Taipaleenmäki H., Stenvang J., Abdallah B.M., Ditzel N., Nossent A.Y., Bak M., Kauppinen S., Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA. 2011;108(15):6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh T., Nozawa Y., Akao Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J. Biol. Chem. 2009;284(29):19272–19279. doi: 10.1074/jbc.M109.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho H.H., Shin K.K., Kim Y.J., Song J.S., Kim J.M., Bae Y.C., Kim C.D., Jung J.S. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J. Cell. Physiol. 2010;223(1):168–177. doi: 10.1002/jcp.22024. [DOI] [PubMed] [Google Scholar]
- 37.Kim K.M., Park S.J., Jung S.H., Kim E.J., Jogeswar G., Ajita J., Rhee Y., Kim C.H., Lim S.K. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J. Bone Miner. Res. 2012;27(8):1669–1679. doi: 10.1002/jbmr.1604. [DOI] [PubMed] [Google Scholar]
- 38.Itoh T., Ando M., Tsukamasa Y., Akao Y. Expression of BMP-2 and Ets1 in BMP-2-stimulated mouse pre-osteoblast differentiation is regulated by microRNA-370. FEBS Lett. 2012;586(12):1693–1701. doi: 10.1016/j.febslet.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Kahai S., Lee S.C., Lee D.Y., Yang J., Li M., Wang C.H., Jiang Z., Zhang Y., Peng C., Yang B.B. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4(10):e7535. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada S., Rodan G.A. Control of osteoblast function and regulation of bone mass. Nature. 2003;423(6937):349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- 41.Komori T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006;99(5):1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 42.Lian J.B., Stein G.S., Javed A., van Wijnen A.J., Stein J.L., Montecino M., Hassan M.Q., Gaur T., Lengner C.J., Young D.W. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 2006;7(1-2):1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Xie R.L., Croce C.M., Stein J.L., Lian J.B., van Wijnen A.J., Stein G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. USA. 2011;108(24):9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu F., Cui Y., Zhou X., Zhang X., Han J. Osteogenic differentiation of human ligament fibroblasts induced by conditioned medium of osteoclast-like cells. Biosci. Trends. 2011;5(2):46–51. doi: 10.5582/bst.2011.v5.2.46. [DOI] [PubMed] [Google Scholar]
- 45.Wu T., Zhou H., Hong Y., Li J., Jiang X., Huang H. miR-30 family members negatively regulate osteoblast differentiation. J. Biol. Chem. 2012;287(10):7503–7511. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim E.J., Kang I.H., Lee J.W., Jang W.G., Koh J.T. MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013;92(10):562–568. doi: 10.1016/j.lfs.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Xie R.L., Gordon J., LeBlanc K., Stein J.L., Lian J.B., van Wijnen A.J., Stein G.S. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J. Biol. Chem. 2012;287(26):21926–21935. doi: 10.1074/jbc.M112.340398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J., Zhao L., Xing L., Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28(2):357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo J., Ren F., Wang Y., Li S., Gao Z., Wang X., Ning H., Wu J., Li Y., Wang Z., Chim S.M., Xu J., Chang Z. miR-764-5p promotes osteoblast differentiation through inhibition of CHIP/STUB1 expression. J. Bone Miner. Res. 2012;27(7):1607–1618. doi: 10.1002/jbmr.1597. [DOI] [PubMed] [Google Scholar]
- 50.Hu R., Liu W., Li H., Yang L., Chen C., Xia Z.Y., Guo L.J., Xie H., Zhou H.D., Wu X.P., Luo X.H. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J. Biol. Chem. 2011;286(14):12328–12339. doi: 10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 52.Jensen E.D., Gopalakrishnan R., Westendorf J.J. Regulation of gene expression in osteoblasts. Biofactors. 2010;36(1):25–32. doi: 10.1002/biof.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J.F., Fu W.M., He M.L., Wang H., Wang W.M., Yu S.C., Bian X.W., Zhou J., Lin M.C., Lu G., Poon W.S., Kung H.F. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol. Biol. Cell. 2011;22(21):3955–3961. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Cheng P., Chen C., He H.B., Xie G.Q., Zhou H.D., Xie H., Wu X.P., Luo X.H. miR-93/Sp7 function loop mediates osteoblast mineralization. J. Bone Miner. Res. 2012;27(7):1598–1606. doi: 10.1002/jbmr.1621. [DOI] [PubMed] [Google Scholar]
- 55.Liu H., Lin H., Zhang L., Sun Q., Yuan G., Zhang L., Chen S., Chen Z. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J. Biol. Chem. 2013;288(13):9261–9271. doi: 10.1074/jbc.M112.433730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leboy P.S. Regulating bone growth and development with bone morphogenetic proteins. Ann. N. Y. Acad. Sci. 2006;1068:14–18. doi: 10.1196/annals.1346.003. [DOI] [PubMed] [Google Scholar]
- 57.Feng X.H., Derynck R. Specificity and versatility in tgf-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 58.Li Z., Hassan M.Q., Volinia S., van Wijnen A.J., Stein J.L., Croce C.M., Lian J.B., Stein G.S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. USA. 2008;105(37):13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inose H., Ochi H., Kimura A., Fujita K., Xu R., Sato S., Iwasaki M., Sunamura S., Takeuchi Y., Fukumoto S., Saito K., Nakamura T., Siomi H., Ito H., Arai Y., Shinomiya K., Takeda S. A microRNA regulatory mechanism of osteoblast differentiation. Proc. Natl. Acad. Sci. USA. 2009;106(49):20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh T., Takeda S., Akao Y. MicroRNA-208 modulates BMP-2-stimulated mouse preosteoblast differentiation by directly targeting V-ets erythroblastosis virus E26 oncogene homolog 1. J. Biol. Chem. 2010;285(36):27745–27752. doi: 10.1074/jbc.M110.105080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raouf A., Seth A. Ets transcription factors and targets in osteogenesis. Oncogene. 2000;19(55):6455–6463. doi: 10.1038/sj.onc.1204037. [DOI] [PubMed] [Google Scholar]
- 62.Luzi E., Marini F., Sala S.C., Tognarini I., Galli G., Brandi M.L. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J. Bone Miner. Res. 2008;23(2):287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 63.Duan Z., Choy E., Harmon D., Liu X., Susa M., Mankin H., Hornicek F. MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 2011;10(8):1337–1345. doi: 10.1158/1535-7163.MCT-11-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu R., Liu W., Li H., Yang L., Chen C., Xia Z.Y., Guo L.J., Xie H., Zhou H.D., Wu X.P., Luo X.H. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J. Biol. Chem. 2011;286(14):12328–12339. doi: 10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bakhshandeh B., Soleimani M., Hafizi M., Paylakhi S.H., Ghaemi N. MicroRNA signature associated with osteogenic lineage commitment. Mol. Biol. Rep. 2012;39(7):7569–7581. doi: 10.1007/s11033-012-1591-2. [DOI] [PubMed] [Google Scholar]
- 66.Veeman M.T., Axelrod J.D., Moon R.T. A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell. 2003;5(3):367–377. doi: 10.1016/S1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 67.Arvidson K., Abdallah B.M., Applegate L.A., Baldini N., Cenni E., Gomez-Barrena E., Granchi D., Kassem M., Konttinen Y.T., Mustafa K., Pioletti D.P., Sillat T., Finne-Wistrand A. Bone regeneration and stem cells. J. Cell. Mol. Med. 2011;15(4):718–746. doi: 10.1111/j.1582-4934.2010.01224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kapinas K., Kessler C., Ricks T., Gronowicz G., Delany A.M. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 2010;285(33):25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kapinas K., Kessler C.B., Delany A.M. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 2009;108(1):216–224. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hassan M.Q., Maeda Y., Taipaleenmaki H., Zhang W., Jafferji M., Gordon J.A., Li Z., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J. Biol. Chem. 2012;287(50):42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Egea V., Zahler S., Rieth N., Neth P., Popp T., Kehe K., Jochum M., Ries C. Tissue inhibitor of metalloproteinase-1 (TIMP-1) regulates mesenchymal stem cells through let-7f microRNA and Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA. 2012;109(6):E309–E316. doi: 10.1073/pnas.1115083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang T., Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem. Biophys. Res. Commun. 2010;402(2):186–189. doi: 10.1016/j.bbrc.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J., Tu Q., Bonewald L.F., He X., Stein G., Lian J., Chen J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J. Bone Miner. Res. 2011;26(8):1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu W., Ye Y., Zhang W., Wang J., Chen A., Guo F. miR‑142‑3p promotes osteoblast differentiation by modulating Wnt signaling. Mol. Med. Rep. 2013;7(2):689–693. doi: 10.3892/mmr.2012.1207. [DOI] [PubMed] [Google Scholar]
- 75.Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H.M., Teitelbaum S.L., Ross F.P., Kopan R., Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14(3):306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canalis E., Parker K., Feng J.Q., Zanotti S. Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology. 2013;154(2):623–634. doi: 10.1210/en.2012-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bae Y., Yang T., Zeng H.C., Campeau P.M., Chen Y., Bertin T., Dawson B.C., Munivez E., Tao J., Lee B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012;21(13):2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H., Xie H., Liu W., Hu R., Huang B., Tan Y.F., Xu K., Sheng Z.F., Zhou H.D., Wu X.P., Luo X.H. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Invest. 2009;119(12):3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y., Li L., Moore B.T., Peng X.H., Fang X., Lappe J.M., Recker R.R., Xiao P. MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One. 2012;7(4):e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G., Cao H., Wu H., Song J., Pan X., Sun Q., Ling S., Li Y., Zhu M., Zhang P., Peng S., Xie X., Tang T., Hong A., Bian Z., Bai Y., Lu A., Li Y., He F., Zhang G., Li Y. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19(1):93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 81.Yang N., Wang G., Hu C., Shi Y., Liao L., Shi S., Cai Y., Cheng S., Wang X., Liu Y., Tang L., Ding Y., Jin Y. TNF-alpha suppresses the mesenchymal stem cell osteogenesis promotor miR-21 in estrogen deficiency-induced osteoporosis. J. Bone Miner. Res. 2013;28(3):559–573. doi: 10.1002/jbmr.1798. [DOI] [PubMed] [Google Scholar]
- 82.You L., Gu W., Chen L., Pan L., Chen J., Peng Y. MiR-378 overexpression attenuates high glucose-suppressed osteogenic differentiation through targeting CASP3 and activating PI3K/Akt signaling pathway. Int. J. Clin. Exp. Pathol. 2014;7(10):7249–7261. [PMC free article] [PubMed] [Google Scholar]
- 83.Seeliger C., Karpinski K., Haug A.T., Vester H., Schmitt A., Bauer J.S., van Griensven M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014;29(8):1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 84.Zuo B., Zhu J., Li J., Wang C., Zhao X., Cai G., Li Z., Peng J., Wang P., Shen C., Huang Y., Xu J., Zhang X., Chen X. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone Miner. Res. 2015;30(2):330–345. doi: 10.1002/jbmr.2352. [DOI] [PubMed] [Google Scholar]
- 85.Shi C., Qi J., Huang P., Jiang M., Zhou Q., Zhou H., Kang H., Qian N., Yang Q., Guo L., Deng L. MicroRNA-17/20a inhibits glucocorticoid-induced osteoclast differentiation and function through targeting RANKL expression in osteoblast cells. Bone. 2014;68:67–75. doi: 10.1016/j.bone.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 86.Krzeszinski J.Y., Wei W., Huynh H., Jin Z., Wang X., Chang T.C., Xie X.J., He L., Mangala L.S., Lopez-Berestein G., Sood A.K., Mendell J.T., Wan Y. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512(7515):431–435. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Cao Z., Moore B.T., Wang Y., Peng X.H., Lappe J.M., Recker R.R., Xiao P. MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One. 2014;9(5):e97098. doi: 10.1371/journal.pone.0097098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murata K., Ito H., Yoshitomi H., Yamamoto K., Fukuda A., Yoshikawa J., Furu M., Ishikawa M., Shibuya H., Matsuda S. Inhibition of miR-92a enhances fracture healing via promoting angiogenesis in a model of stabilized fracture in young mice. J. Bone Miner. Res. 2014;29(2):316–326. doi: 10.1002/jbmr.2040. [DOI] [PubMed] [Google Scholar]
- 89.Deng Y., Bi X., Zhou H., You Z., Wang Y., Gu P., Fan X. Repair of critical-sized bone defects with anti-miR-31-expressing bone marrow stromal stem cells and poly(glycerol sebacate) scaffolds. Eur. Cell. Mater. 2014;27:13–24. doi: 10.22203/ecm.v027a02. [DOI] [PubMed] [Google Scholar]
- 90.Deng Y., Zhou H., Zou D., Xie Q., Bi X., Gu P., Fan X. The role of miR-31-modified adipose tissue-derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials. 2013;34(28):6717–6728. doi: 10.1016/j.biomaterials.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 91.Li Y., Fan L., Liu S., Liu W., Zhang H., Zhou T., Wu D., Yang P., Shen L., Chen J., Jin Y. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34(21):5048–5058. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 92.James E.N., Delany A.M., Nair L.S. Post-transcriptional regulation in osteoblasts using localized delivery of miR-29a inhibitor from nanofibers to enhance extracellular matrix deposition. Acta Biomater. 2014;10(8):3571–3580. doi: 10.1016/j.actbio.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yan J., Zhang C., Zhao Y., Cao C., Wu K., Zhao L., Zhang Y. Non-viral oligonucleotide antimiR-138 delivery to mesenchymal stem cell sheets and the effect on osteogenesis. Biomaterials. 2014;35(27):7734–7749. doi: 10.1016/j.biomaterials.2014.05.089. [DOI] [PubMed] [Google Scholar]
- 94.Deng Y., Zhou H., Gu P., Fan X. Repair of canine medial orbital bone defects with miR-31-modified bone marrow mesenchymal stem cells. Invest. Ophthalmol. Vis. Sci. 2014;55(9):6016–6023. doi: 10.1167/iovs.14-14977. [DOI] [PubMed] [Google Scholar]
- 95.Qureshi A.T., Doyle A., Chen C., Coulon D., Dasa V., Del Piero F., Levi B., Monroe W.T., Gimble J.M., Hayes D.J. Photoactivated miR-148b-nanoparticle conjugates improve closure of critical size mouse calvarial defects. Acta Biomater. 2015;12:166–173. doi: 10.1016/j.actbio.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 97.Aguda B.D., Kim Y., Piper-Hunter M.G., Friedman A., Marsh C.B. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc. Natl. Acad. Sci. USA. 2008;105(50):19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gordeladze J.O., Djouad F., Brondello J.M., Noël D., Duroux-Richard I., Apparailly F., Jorgensen C. Concerted stimuli regulating osteo-chondral differentiation from stem cells: phenotype acquisition regulated by microRNAs. Acta Pharmacol. Sin. 2009;30(10):1369–1384. doi: 10.1038/aps.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gordeladze J.O., Reseland J.E., Duroux-Richard I., Apparailly F., Jorgensen C. From stem cells to bone: phenotype acquisition, stabilization, and tissue engineering in animal models. ILAR J. 2009;51(1):42–61. doi: 10.1093/ilar.51.1.42. [DOI] [PubMed] [Google Scholar]
- 100.Tortelli F., Pujic N., Liu Y., Laroche N., Vico L., Cancedda R. Osteoblast and osteoclast differentiation in an in vitro three-dimensional model of bone. Tissue Eng. Part A. 2009;15(9):2373–2383. doi: 10.1089/ten.tea.2008.0501. [DOI] [PubMed] [Google Scholar]
- 101.Shimbo K., Miyaki S., Ishitobi H., Kato Y., Kubo T., Shimose S., Ochi M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem. Biophys. Res. Commun. 2014;445(2):381–387. doi: 10.1016/j.bbrc.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 102.Kooijmans S.A., Vader P., van Dommelen S.M., van Solinge W.W., Schiffelers R.M. Exosome mimetics: a novel class of drug delivery systems. Int. J. Nanomedicine. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]


