Abstract
Background and Aims Carnivorous plants have developed strategies to enable growth in nutrient-poor soils. For the genus Nepenthes, this strategy represents producing pitcher-modified leaves that can trap and digest various prey. These pitchers produce a digestive fluid composed of proteins, including hydrolytic enzymes. The focus of this study was on the identification of these proteins.
Methods In order to better characterize and have an overview of these proteins, digestive fluid was sampled from pitchers at different stages of maturity from five species of Nepenthes (N. mirabilis, N. alata, N. sanguinea, N. bicalcarata and N. albomarginata) that vary in their ecological niches and grew under different conditions. Three complementary approaches based on transcriptomic resources, mass spectrometry and in silico analysis were used.
Key Results This study permitted the identification of 29 proteins excreted in the pitchers. Twenty of these proteins were never reported in Nepenthes previously and included serine carboxypeptidases, α- and β-galactosidases, lipid transfer proteins and esterases/lipases. These 20 proteins display sequence signals allowing their secretion into the pitcher fluid.
Conclusions Nepenthes pitcher plants have evolved an arsenal of enzymes to digest prey caught in their traps. The panel of new proteins identified in this study provides new insights into the digestive process of these carnivorous plants.
Keywords: Carnivorous plants, digestive fluid, enzymes, mass spectrometry, Nepenthes mirabilis, Nepenthes alata, Nepenthes sanguinea, Nepenthes bicalcarata, Nepenthes albomarginata, pitcher plants, proteome, transcriptomic analysis
INTRODUCTION
In nutrient-poor habitats, carnivorous plants have developed original feeding strategies based on the capture and digestion of arthropods and the assimilation of prey-derived nutrients by leaves highly specialized for trapping. Among the dozen genera of carnivorous plants recorded to date, Nepenthes is one of the richest, comprising nearly 160 species that are widespread in South-western Asia. The greatest diversity occurs in Borneo (Clarke, 1997), Sumatra (Clarke, 2001) and the Philippine islands (McPherson, 2009). From lowlands to mountain forests, these plants have colonized a large number of habitats comprising different substrates (e.g. sandy, peat, swampy, cliff or epiphytic substrates) (Clarke, 1997; McPherson, 2009). On these plants, leaves are extended by a tendril that bears a pitfall trap, also called a pitcher. Glandular structures have been described in the inner and basal part of the pitchers. These glands produce secretions that contain a cocktail of hydrolytic enzymes for the degradation of insects (An et al., 2002).
Proteins present in the secretions of different Nepenthes species have been identified. Most of the proteins are enzymes related to the digestion of various prey items. In addition to the aspartic acid proteases Nepenthesin 1 and 2 isolated from N. alata, N. distillatoria and N. gracilis (Athauda et al., 2004; Takahashi et al., 2005; Hatano and Hamada, 2008; Kadek et al., 2014), various hydrolytic enzymes have been identified, including S-like RNases (N. ventricosa) (Matthews, 1960; Stephenson and Hogan, 2006; Nishimura et al., 2014), esterases (N. hybrida) (Higashi et al., 1993), lipases (N. macfarlanei) (Tökés et al., 1974), acid or alkaline phosphatases (N. hybrida and N. macfarlanei) (Higashi et al., 1993; Płachno et al., 2006), phosphoaminases (N. hybrida) (Higashi et al., 1993), peroxidases (N. alata) (Hatano and Hamada, 2012) and chitinases (N. khasiana, N. alata, N. singalana, N. ventricosa, N. gracilis, N. thorelii, N. mirabilis, N. ampullaria, N. rafflesiana and the hybrid ‘Mizuho’) (Eilenberg, 2006; Hatano and Hamada, 2008; Rottloff et al., 2009, 2011; Ishisaki et al., 2012a, b).
Proteins that are not directly involved in prey degradation have also been reported. For example, a β-d-xylosidase in N. alata secretions could be involved in trap maturation according to Hatano and Hamada (2008). Other identified proteins are dedicated to plant defence against pathogens, and include a thaumatin-like protein (N. alata, N. thorelii, N. superba, N. gracilis, N. fusca, N. ampullaria, N. mirabilis, N. singalana and N. ventricosa) (Hatano and Hamada, 2008; Rottloff et al., 2009), a PR-1 protein (N. mirabilis) (Buch et al., 2014) and β-1,3-glucanases (N. alata) (Hatano and Hamada, 2008, 2012). These proteins may contribute to the inhibition of microbial growth in the pitcher fluid to prevent competition between the plant and microorganisms for the nutrients derived from the digested prey (Eilenberg, 2006; Hatano and Hamada, 2008, 2012; Mithofer; 2010; Buch et al., 2013).
Given their adaptation to different habitats and spectra of prey (Kato et al., 1993), the Nepenthes species have evolved different trapping strategies (Bonhomme et al., 2011b; Bazile et al., 2015). It is thus likely that each species has developed the ability to synthesize an arsenal of different and specific enzymes. Using SDS–PAGE approaches to analyse the pitcher fluids of 17 Nepenthes species, we previously suggested that the protein composition of the secretion might be specific to each species (Biteau et al., 2013). In order to complete the study carried out by Biteau and colleagues, we further investigated the protein compositions of the digestive fluids from five Nepenthes species, N. mirabilis, N. alata, N. sanguinea, N. bicalcarata and N. albomarginata, which were grown in different conditions and collected from greenhouse cultures (N. mirabilis, N. sanguinea and N. alata), in vitro cultures (N. alata and N mirabilis) and from their natural habitats (N. bicalcarata and N. albomaginata). Also N. bicalcarata and N. albomaginata differ in their diet from the other species. The first one lives with C. schmitzi and the second one uses food hairs on the peristome to capture hundreds to thousands of termites (literature: N. bicalcarata versus C. schmitzi: Clarke and Kitching, 1995; N. albomarginata: Merbach et al., 2002). We used a complementary three-step approach involving (1) deep sequencing of the transcriptome of tissues located in the digestive zone of an N. mirabilis pitcher; (2) mass spectrometry (MS)-based analyses of proteins present in the digestive fluid; and (3) an in silico analysis of the putative proteins.
MATERIALS AND METHODS
Plants
To construct the cDNA library, total RNA was extracted from Nepenthes mirabilis pitchers (1988.3.265) provided by the Montet Botanical Garden (Villers-Lès-Nancy, France).
For proteomic analyses, secretions from different Nepenthes species, states and cultivations were used. Ex vitro N. alata and N. sanguinea were purchased from Araflora (http://www.araflora.com) and cultivated in heated greenhouses with natural light at a temperature of 23 °C and a relative humidity of 75–85 %. Nepenthes bicalcarata and N. albomarginata were directly sampled in their natural sites in Brunei (Borneo). Nepenthes bicalcarata was sampled in lowlands in the peat swamp forest (4 °59'N, 114 °49'E), and N. albomarginata was sampled in heath forest (4 °56'N, 114 °49'E).
In vitro N. alata plants were supplied by Deroose Plants (www.derooseplants.com) and micropropagated in half-strength MS medium (Murashige and Skoog, 1962) containing 2-fold MS vitamin mixture, 2 % (w/v) sucrose, 0.05 % (w/v) casein hydrolysate, 0.07 % (w/v) MES, 0.2 % (w/v) active charcoal and 0.7 % (w/v) HP696 agar (Kalys, Bernin, France). The pH of the medium was adjusted to 5.8 before autoclaving.
In vitro stocks of N. mirabilis plants were established from fresh seeds supplied by Districarnivores (www.districarnivores.com). Seeds were sterilized by total immersion in a dilute commercial bleach solution of 0.25 % sodium hypochlorite for 5 min and washed three times in sterile water. After a drying step, they were sown on quarter-strength MS medium (Murashige and Skoog, 1962) containing 2-fold MS vitamin mixture, 2 % (w/v) sucrose, 0.05 % (w/v) casein hydrolysate, 0.07 % (w/v) MES, 6 mg L–1 ProClin® 200 (Sigma-Aldrich, St Louis, MO, USA) and 0.7 % (w/v) HP696 agar (Kalys). All cultures were cultivated under a 16 h/8 h day/night photoperiod provided by natural white fluorescent lamps at a temperature of 23 °C. Germinated plantlets were then transplanted in the same medium as N. alata described above, without active charcoal.
Construction of a Nepenthes RNA-seq library
An RNA sequencing (RNA-seq) library was constructed based on an RNA extraction performed on secreting tissues (from the bottom of pitchers) from an N. mirabilis specimen (N. mirabilis 1988.3.265) provided by the Montet Botanical Garden.
RNA preparation.
Total RNA (40 μg) was extracted from the glandular part of pitchers 8 d after opening (wrapped in perforated bags to prevent insect contamination) using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich) according to the manufacturer’s instructions. To remove genomic DNA efficiently, RNA was treated with the Turbo™ DNA Free Kit (AM1907M, Ambion, Thermo Fisher Scientific, Waltham, MA, USA).
Construction of a random primed cDNA library and high-throughput sequencing.
Construction of a de novo normalized 454-based RNA-seq library and deep sequencing were performed by Eurofins MWG Operon (Ebersberg, Germany) using GS FLX+ chemistry. Clustering and assembly of the reads were performed by the supplier.
Protein identification
Sample preparation.
The digestive fluid was collected from closed pitchers using a sterile syringe. When this was not possible, pitchers were wrapped in bags before they opened in order to avoid any insect contamination. For secretions from Nepenthes growing in their natural habitats (N. albomarginata and N. bicalcarata), samples were collected from closed and recently opened pitchers for which we verified visually the absence of prey. Proteins present in the digestive fluid were precipitated overnight at –20 °C with 4 vols of acetone. The proteins were collected by centrifugation at 12 000 g for 30 min at 4 °C and used directly for MS analyses or separated by 1-D or 2-D PAGE.
1-D SDS–PAGE: samples were resuspended in 40 μL of electrophoresis buffer [25 mm Tris–HCl, 192 mm glycine, 0.1 % (w/v) SDS, pH 8.3 at 25 °C] and supplemented with 10 μL of denaturation buffer [0.313 m Tris–HCl pH 6.8 at 25 °C, 10 % (w/v) SDS, 0.5 % (w/v) bromophenol blue, 50 % (v/v) glycerol, 2 M DTT (dithiothreitol)]. Proteins were heat denatured at 95 °C for 10 min and separated by 1-D SDS–PAGE on Mini-PROTEAN® TGX pre-cast gels, AnykD™ (10-well, 50 μL) (Bio-Rad, Hercules, CA, USA) for 1 h at 100 V. Protein bands were visualized by silver staining (Blum et al., 1987).
2-D SDS–PAGE: to perform 2-D separation, we used the ReadyPrep™ 2D Starter Kit (Bio-Rad). To prepare samples, protein pellets were dissolved in 125 μL of Rehydration/Sample Buffer and loaded on 7 cm pH 3–6 IPG strips (ReadyStrip™ IPG Strips; Bio-Rad). The PROTEAN® i12™ IEF System (Bio-Rad) was used for isoelectric focusing with the following program: step 1 (250 V, 20 min, linear), step 2 (4000 V, 2 h, linear) and step 3 (4000 V, 5 h, rapid). After the IPG equilibration for SDS–PAGE described in the ReadyPrep™ 2D Starter Kit protocol, the strips were placed on Mini-PROTEAN® TGX™ pre-cast gels for 2-D electrophoresis (Bio-Rad). Separation was then performed for 45 min at 200 V. Spots were visualized by silver staining (Blum et al., 1987).
Spot sampling: proteins were excised from 1-D or 2-D gels with a scalpel and chopped into 1 mm3 pieces. Silver-stained spots were destained with a solution containing a 1:1 mixture of 30 mm potassium ferricyanide/100 mm sodium thiosulphate until the brown colour vanished. The samples were then washed three times with distilled water (Gharahdaghi et al., 1999). Next, the samples were incubated by shaking for 15 min in 200 μL of distilled water/acetonitrile (ACN) (1:1, v/v), 5 min in 200 μL of ACN, 5 min in 100 μL of 100 mm ammonium bicarbonate (BiCa) and 15 min in 100 μL of ACN. After removing the supernatant, the gel pieces were dried with a Speed Vac Concentrator Plus (Eppendorf, Hamburg, Germany). Peptides were reduced by adding 10 mm DTT in 100 mm BiCa to the gels for 45 min at 56 °C and then cooling on ice for 5 min. Peptides were alkylated by incubation with 55 mm iodoacetamide in 100 mm BiCa in the dark for 30 min. Iodoacetamide was replaced with 1 vol. of 100 mm BiCa and incubated for 15 min with shaking prior to the addition of 1 vol. of ACN. The samples were then incubated for an additional 15 min with shaking. The supernatant was removed, and the gel pieces were dried using an Eppendorf Speed Vac Concentrator Plus.
In-gel digestion with trypsin was performed as described previously (Jensen et al., 1999). Proteins were treated with 10 μL of trypsin (10 ng μL–1 in 50 mm BiCa) and incubated for 10 min on ice. This step was repeated until the gel pieces did not expand further. The trypsin solution was removed, 25 mm BiCa was added until the gel pieces were completely covered and the mixtures were incubated overnight at 37 °C. The samples were subjected to an Elmasonic S70 ultrasonic bath (Elma Schmidbauer GmbH, Singen, Germany) for 2 min, and the supernatant was transferred to a new tube. The remaining acrylamide was incubated in 1 vol. of 25 mm BiCa for 15 min with shaking. After the addition of 1 vol. of ACN, the sample was incubated for 15 min with shaking and then transferred to an ultrasonic bath for 2 min. The supernatant was removed and added to the previously collected supernatant. Finally, the sample was incubated for 15 min with shaking in 1 vol. of 5 % (v/v) formic acid/ACN (1:1, v/v), and the supernatant was removed and added to the previously pooled supernatant. The sample was concentrated using an Eppendorf Concentrator Plus at 60 °C, and the peptides were stored at –20 °C.
NanoLC-MS/MS analyses.
The peptide mixtures obtained from SDS–PAGE spots or crude protein extracts were resuspended in 15 μL of water containing 0.1 % formic acid (solvent A) and further analysed using a NanoLC-2DPlus system (with the nanoFlex ChiP module; Eksigent, ABSciex, Concord, Ontario, Canada) coupled to a TripleTOF 5600 mass spectrometer (ABSciex) operating in positive mode. A total of 5 μL of each sample (up to 1 μg for the crude extracts) was loaded on a ChIP C-18 pre-column (300 μm ID × 5 mm ChromXP; Eksigent) at 2 μL min–1 in solvent A. After 10 min of desalting and concentration in the trap, the pre-column was switched online with the analytical ChIP C-18 analytical column (75 μm ID × 15 cm ChromXP; Eksigent) equilibrated in 95 % solvent A and 5 % solvent B (0.1 % formic acid in ACN). Peptides were eluted using a 5–40 % gradient of solvent B at a flow rate of 300 nL min–1. The length of the LC gradient was adjusted depending on the sample complexity, from 45 min for the SDS–PAGE bands to 90 min for the crude protein extracts. The TripleTOF 5600 was operated in data-dependent acquisition mode (DDA) with Analyst software (v1.6, ABSciex). Survey MS scans were acquired during 250 ms in the 350–1250 m/z range. Up to 20 of the most intense multiply charged ions (2+ to 4+) with a threshold intensity exceeding 150 counts per second were selected for collision-induced dissociation (CID) fragmentation. Ions were fragmented using a rolling collision energy script within a 60 ms accumulation time and an exclusion time of 15 s. This so-called ‘Top20’ method, which had a constant cycle time of 1.5 s, was set in high-sensitivity mode.
Bioinformatics analyses
Protein data
The raw data files obtained for each nanoLC-MS/MS injection were first converted to .mgf files using the MS-Data converter script (version 1.3, AB Sciex) and subsequently used by the Mascot algorithm (version 2.2, Matrix Science, London, UK) in the ProteinScape package (version 3.1, Bruker). The Nepenthes sub-taxonomy from the UniProt database (release from 2012-07-12 containing 235 entries) was first searched using the following parameters: (1) allowed variable peptide modifications of N-acetyl (protein), carbamidomethylation (C) and oxidation (M); (2) mass tolerances in MS and MS/MS of 20 ppm and 0.5 Da, respectively; and (3) trypsin as enzyme specificity with a maximum of two missed cleavages sites. Peptides were validated using the Mascot individual ions score threshold to give a P-value <0.05. The protein identifications obtained from Mascot were validated using a false discovery rate (FDR) of 1 % by employing a decoy database strategy and the Protein Assessment Configuration from Proteinscape 3.1 package (Thiele et al., 2010). To create the decoy database, the makeDecoyDB.pl Pearl script from Bruker was used: a .fasta database was generated with the forward and the reversed sequences, with ‘rnd’ as the accession prefix associated with each decoy entry. Moreover, protein inference is taken into account by the Proteinscape package during the Protein Assessment and Protein compiling steps: for each accession number, a set of so-called Altenative Proteins is given, with the list of their shared peptides.
PEAKS Studio 5.3 (Bioinformatics Solutions Inc., Waterloo, Canada) was used in a second analysis to examine in greater depth the assignment of a peptide sequence to the maximum number of experimental MS/MS fragmentation spectra. A PEAKS identification workflow was established that consisted of an initial auto de novo step with the generation of ten tags per spectrum, parent and fragment mass tolerances set at 20 ppm and 0.5 Da, trypsin as the enzyme, and carbamidomethylation (C) and oxidation (M) as variable modifications. A PEAKS search was then launched with the same mass tolerances and variable modifications against three different databases: a whole SwissProt database (release from 2012-01-11 containing 533 657 entries) and the two sub-databases described above (Viridiplantae and Nepenthes taxonomic sub-divisions). Spectra that remained unassigned after the PEAKS DB searches were further submitted to PTM Finder and SPIDER searches. PTM Finder was configured to search 18 additional variable modifications (including acetylation, methylation, deamidation and phosphorylation), whereas SPIDER search allowed homology matches with L = I and Q = K. In these steps, the de novo score (ALC%) threshold was set at 90, and the peptide hit threshold (–10logP) was set at 30. Finally, peptides belonging to proteins validated at 1 % FDR were manually inspected to validate only high-quality MS/MS fragmentation spectra with a minimum of five consecutive amino acids, major peaks assigned to fragments and specific proline fragmentation patterns.
cDNA library analysis.
The FASTA file provided by MWG Eurofins Operon (Ebersberg, Germany) was imported as a library into the software BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The peptide sequences obtained by MS analysis were compared with the RNA data available in the Nepenthes cDNA library using the Basic Local Alignment Search Tool to identify full-length or partial coding sequences. The presence of an N-terminal signal peptide (SP), which is necessary for the secretion of proteins out of the cell in eukaryotic organisms and described before for other pitcher fluid proteins (Rottloff et al., 2011; Buch et al., 2014), was confirmed for all identified proteins. For these analyses, we used the SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) and the Signal-3 L (http://www.csbio.sjtu.edu.cn/bioinf/Signal-3 L/) servers. The latter contains the option to adjust the origin of the protein, e.g. plants. For proteins with a predicted SP, the molecular weight and isoelectric point (pI) of the mature protein (without SP) and the number of amino acids were calculated using the ExPASy compute pI/MW tool (http://web.expasy.org/compute_pi/). To predict potential amino acid sites for glycosylation, we used the NetNGlyc 1.0 and the NetOGlyc 4.0 servers (http://www.cbs.dtu.dk/services/NetNGlyc/ and http://www.cbs.dtu.dk/services/NetOGlyc/) at the Center for Biological Sequence Analysis website (http://www.cbs.dtu.dk/services/).
RESULTS
cDNA library description
To characterize proteins present in the digestive fluid of Nepenthes pitchers, we first identified peptides corresponding to trypsin-digested proteins using MS and then fished the complete corresponding coding sequence using a transcriptomic cDNA library. Because only a small number of coding sequences from Nepenthes species are available in public genomic libraries, we generated a 454-based de novo RNA-seq library of N. mirabilis pitchers cultivated in a greenhouse. This library was normalized to ensure representation of most of the genes expressed in these tissues.
After trap opening, the hydrolytic activity of N. mirabilis pitcher fluid requires time to become fully active. This delay is in contrast to other Nepenthes species and is related to the concentration of proteins detected in the pitcher fluid (Biteau et al., 2013). SDS–PAGE analysis of the secreted proteins revealed a strong increase in protein amounts 5 d after trap opening compared with the first day, with a peak 8 d after opening (Biteau et al., 2013). Based on these results, we used pitchers opened for 8 d as our experimental plant material because these pitchers should exhibit the greatest transcriptional activity. Total RNA was extracted from the lower part of pitchers (from a unique N. mirabilis specimen) covered by multicellular secretory glands that reportedly secrete the digestive enzymes (Rottloff et al., 2011). Nearly 248 ×106 bp were sequenced from the RNA sample distributed across 631 074 reads with an average length of 432 bp. Clustering and assembly of these singletons led to the identification of 43 026 contigs or 10 218 large contigs (>1000 bp). These contigs were annotated and functionally assigned using Blast2GO (https://www.blast2go.com/).
Protein analyses of in vitro N. mirabilis secretions
To establish an experimental procedure for identifying proteins present in pitcher fluid, we began our investigations with fluid sampled from 3-year-old in vitro cultivated N. mirabilis plants. Working with in vitro plants presents two main advantages. First, the precise culture conditions are known. Secondly, the sterile growth conditions ensured that no insect or pathogen was responsible for inducing proteins or enzymes involved in a defence reaction and that no foreign protein was present in the pitcher fluid. Whole proteins (600 μg of total proteins) were precipitated in the presence of acetone and separated by 2-D gel electrophoresis. Twenty-five different silver-stained protein spots were excised from the gel and analysed by nanoLC-MS/MS (Supplementary Data Table S1; Fig. S1). Unfortunately, this technique only allowed the identification of two different proteins, Nepenthesin 1 and a glucanase (NgNep1 and NkGluc1) detected in different spots (Table 1; Table S2). Conclusive results could not be obtained for other spots, possibly due to (1) a loss of material during the precipitation and purification steps prior to or after migration on the acrylamide gel; and/or (2) the low concentration of most proteins present in the pitcher fluid of in vitro cultivated N. mirabilis.
Table 1.
Summary of the proteins identified from digestive fluids of five Nepenthes species and their putative activities
| Nomenclature | Accession number | Protein description | No. of peptides |
|---|---|---|---|
| Nepenthesins | |||
| NgNep1* | BAD07474 (N. gracilis) | Nepenthesin 1 (Athauda et al., 2004) | 3 |
| NgNep2* | Q766C (N. gracilis) | Nepenthesin 2 (Athauda et al., 2004) | 3 |
| NmNep2LP | Q766C2 (N. gracilis) | Nepenthesin 2-like protein | 3 |
| Carboxypeptidases | |||
| NmSCP3 | NP_197689 (A. thaliana) | Serine carboxypeptidase III | 8 |
| NmSCP20 | XP_004290128 (F. vesca) | Serine carboxypeptidase-like 20 | 3 |
| NmSCP47 | NP_197689 (A. thaliana) | Serine carboxypeptidase-like 47 | 3 |
| NmSCP51 | XP_002273519 (V. vinifera) | Serine carboxypeptidase-like 51 | 2 |
| Phosphatases | |||
| NmNPP1 | XP_003605731 (M. truncatula) | Nucleotide pyrophosphatase/phosphodiesterase | 1 |
| NmPP1 | XP_003603267 (M. truncatula) | Protein phosphatase 2C | 1 |
| Peroxidases | |||
| NmPRX27 | XP_002280216 (V. vinifera) | Peroxidase 27 | 3 |
| NmPRX-N | XP_002285642 (V. vinifera) | Peroxidase N | 3 |
| Lipid transfer proteins (LTPs) | |||
| NmLTP1 | XP_003531043 (G. max) | Lipid transfer protein 1 | 2 |
| NmLTP2 | ABK9681 (O. sativa) | Lipid transfer protein 2 | 2 |
| Galactosidases | |||
| NmAG1 | XP_004297698 (F. vesca) | α-Galactosidase 1 | 4 |
| NmAG2 | XP_002279730 (V. vinifera) | α-Galactosidase 2 | 2 |
| NmBG1 | ACC7825 (C. papaya) | β-Galactosidase 1 | 2 |
| NmBG2 | CAC44500 (F. ananassa) | β-Galactosidase 2 | 1 |
| NmBG3 | EOY3046 (T. cacao) | β-Galactosidase 3 | 2 |
| Other functions | |||
| NaChit1* | BAF98919 (N. alata) | Chitinase (class IV) (Hatano and Hamada, 2008; Ishisaki et al., 2012b) | 5 |
| NrChit1* | ABF7462 (N. rafflesiana) | Acidic chitinase (class III) (Rottloff et al., 2011; Hatano and Hamada, 2012; Ishisaki et al., 2012a) | 3 |
| NaGluc2* | BAM28606 (N. alata) | β-1,3-Glucanase (Hatano and Hamada, 2008) | 2 |
| NkGluc1* | ABB8952 (N. khasinana) | Glucanase (Eilenberg and Zilberstein, 2008) | 12 |
| NmAGluc1 | AGA82514 (C. sinensis) | α-Glucosidase | 1 |
| NmBGluc7 | NP_195174 (A. thaliana) | β-d-1,3-Glucosidase 7-like protein | 3 |
| NaPRX1* | BAM2860 (N. alata) | Cationic peroxidase 1 (Hatano and Hamada, 2012) | 3 |
| NgTLP* | ABC73397 (N. gracilis) | Thaumatin-like protein (Hatano and Hamada, 2008; Rottloff et al., 2009) | 13 |
| NmPR-1* | ACT9972 (N. mirabilis) | PR-1 protein (Buch et al., 2014) | 7 |
| NmXyl1* | AAX9296 (O. sativa) | β-Xylosidase (Hatano and Hamada, 2008) | 1 |
| NmLip1 | XP_004232991 (S. lycopersicum) | Esterase/lipase | 1 |
The protein names that are notated in bold are newly identified proteins.
Sequences identified in the NCBI database and used for protein identification were obtained from different species summarized in the ‘accession number’ column. The identity of peptides detected is given in Supplementary Data Table S2.
*Proteins, which were previously described in the pitcher fluid of Nepenthes.
We therefore modified our experimental procedure. Instead of analysing a single spot extracted from a polyacrylamide gel, we analysed a crude protein mixture followed by precipitation of all the proteins present in the pitcher fluid. Ten different proteins in N. mirabilis fluid were identified using this technique (Supplementary Data Table S2). The amino acid sequences of the peptides obtained were used to perform a tblastn search in public databases (GenBank) and/or the N. mirabilis RNA-seq database, which led to the identification of their corresponding full-length coding sequences. When a hit identified in a public library corresponded to a protein identified in other plants, we performed a second tblastn search on the N. mirabilis RNA-seq database. Four proteins had already been described in the literature: Nepenthesin 1 (Athauda et al., 2004), a thaumatin-like protein (Hatano and Hamada, 2008; Rottloff et al., 2009), a glucanase (Eilenberg and Zilberstein, 2008) and a class IV chitinase (Hatano and Hamada, 2008; Ishisaki et al., 2012a). The six remaining proteins were newly identified and were putatively related to a serine carboxypeptidase III (NmSCP3), a serine carboxypeptidase-like 47 (NmSCP47), α-galactosidases 1 and 2 (NmAG1 and NmAG2), a protein phosphatase 2 C (NmPP1) and a peroxidase N (NmPRX-N) (Table 1; Supplementary Data Table S2). This first set of experiments demonstrated that a global analysis of the proteins present in the pitcher is sufficient to obtain information on several new proteins. This approach allowed us to detect proteins present in small amounts in the pitcher fluid that might otherwise be below the detection threshold of an acrylamide gel. In addition, the direct digestion leads to the identification of a more exhaustive protein list since (1) it is not biased by the cutting of visible bands and (2) because proteins are directly exposed to the desired proteolytic digestion without the need to extract the peptides further from a gel.
In subsequent experiments, we performed both types of analyses: identification of proteins present in single protein bands sampled from polyacrylamide gels and global analyses of precipitated proteins (Supplementary Data Table S2). However, we restricted our analyses to 1-D separation, a technique that has already been efficiently used to identify new proteins from Nepenthes pitcher fluids (Hatano and Hamada, 2008, 2012). Although this method is not the best-adapted method for separating single proteins, it reduces the loss of protein during purification steps, a significant advantage over 2-D separation strategies.
Protein analyses of in vitro and ex vitro N. alata digestive fluids
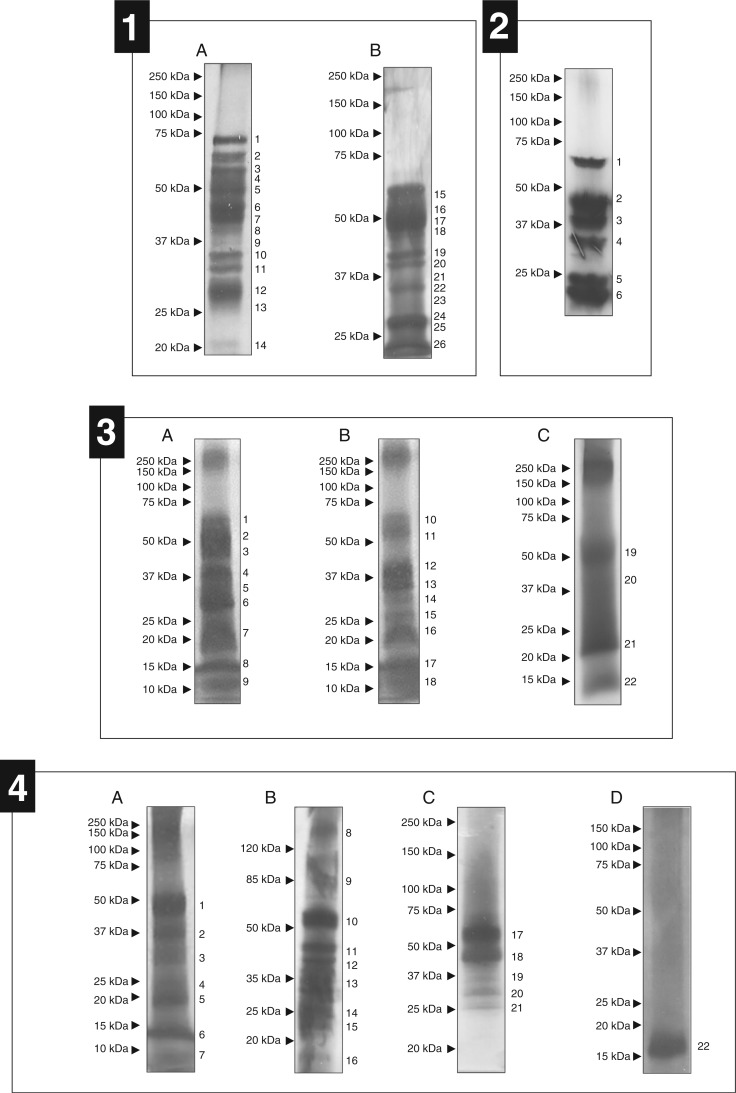
According to Biteau et al. (2013), the digestive fluid protein content in pitchers varies among Nepenthes species. After studying the proteins produced by N. mirabilis, we investigated N. alata. We separated the proteins present in four opened pitchers and one closed pitcher from N. alata grown on soil in a greenhouse (Supplementary Data Table S1). Because the electrophoresis patterns were rather similar from one gel to another, we performed the analyses on 26 bands collected from two independent gels (Fig. 1, panel 1). This investigation was completed by an analysis of a crude protein preparation collected from 4-year-old plants grown in vitro (one repetition) and from one opened and one closed pitcher of N. alata grown on soil (Table S1).
Fig. 1.
SDS–PAGE analyses of N. alata, N. sanguinea, N. albomarginata and N. bicalcarata secretions. (1) Nepenthes alata secretions from closed (A) and opened pitchers (B). 1, 3–8, 10, 12, 15–25: NgNep1; 2: NgNep1, NmSCP3 and NmXyl; 11: not detectable (ND); 13: NrChit1 and NgTLP; 14: NgTLP; 26: NgNep1 and NgTLP. (2) Nepenthes sanguinea secretions from closed pitchers. 1: NgNep1 and NaGluc2; 2: NgNep1, NgNep2, NmAG1 and/or NmAG2, and NkGluc1; 3: NgNep1, NgNep2 and NkGluc1; 4: NgNep2 and NkGluc1; 5: NgNep2, NgTLP and NkGluc1; 6: NgTLP, NkGluc1 and NaChit1. (3) Nepenthes albomarginata secretions from closed pitchers (A), just-opened pitchers (B) and pitchers open for an extended period (C). 1, 2, 4, 6, 8, 12, 14, 15, 18–22: ND; 3: NmAG2; 5, 13: NkGluc1; 7, 16: NgTLP; 9, 11, 17: no putative enzyme detected; 10: NmAG1 and/or NmAG2. (4) Nepenthes bicalcarata secretions from closed pitchers (A–C) and pitchers opened for an extended period and inhabited by C. schmitzi (D). 1–8, 11, 13–17, 20–22: ND; 9, 10, 12: NmPRX-N; 18: NgNep1; 19: NgNep1, NkGluc1 and NmLip1.
A total of 21 putative digestive enzymes were identified in N. alata secretions. If seven of these proteins have already been reported, others were never described in this botanical family. These proteins, identified as a lipid transfer protein 1 (NmLTP1), a serine carboxypeptidase-like 20 (NmSCP20), a serine carboxypeptidase-like 51 (NmSCP51), a PR-1 protein (NmPR-1) (Buch et al., 2014), a β-1,3-glucanase (NaGluc2) (Hatano and Hamada, 2008), a β-xylosidase (NmXyl1), α-galactosidases 1 and 2, β-galactosidases 1, 2 and 3 (NmBG1, 2 and 3), a β-d-1,3-glucosidase 7-like protein (NmBGluc7), a peroxidase 27 (NmPRX27) and a lipid transfer protein 2 (NmLTP2), were identified in ex vitro secretions (Table 1; Supplementary Data Table S2). NmXyl1 was previously described in N. alata by Hatano and Hamada (2008) using a proteomic approach, but complete coding cDNA sequence information was not available at the time of our study. The results of three replicates of digestive fluid collected from closed and opened pitchers highlighted a large number of different MS/MS spectra corresponding to peptides derived from Nepenthesin 1 and the thaumatin-like protein. This abundance might reflect a very high concentration of these proteins compared with others in the pitcher fluid.
Protein analyses of N. sanguinea digestive fluids
The whole protein mixes collected from five different pitchers were precipitated individually and separated by SDS–PAGE (Fig. 1, panel 2; Supplementary Data Table S1). The electrophoresis pattern was similar for each analysis, and six major bands were observed. Each band was excised from the gel, and the spots corresponding to proteins with the same apparent molecular weight were pooled. As observed previously, a single 1-D SDS–PAGE separation did not sufficiently resolve the proteins, and a mix of different proteins was detected in each spot. As described above, we also performed a de novo sequencing analysis of the fluid from opened and closed pitchers (Table S1). Using these two approaches, we detected 19 putative digestive enzymes (seven from SDS–PAGE spots and 12 from the crude extract), including a never previously described Nepenthesin 2-like protein (NmNep2LP), an α-glucosidase (NmAGluc1) and a nucleotide pyrophosphatase (NmNPP1). In addition to NgNep1, NkGluc1 and NrChit1 which were already available in databases, we could relate nine other N. sanguinea proteins to the N. mirabilis cDNA library proteins (NmSCP3, NmSCP47, NmPR-1, NgTLP, NmAG1, NmAG2, NmPRX27, NmPRX-N and NmLTP1) (Table 1; Supplementary Data Table S2).
Protein analyses of N. albomarginata digestive fluids
Pitcher fluid from 1–4 plants was collected from closed (three pitchers) and just-opened pitchers (two pitchers) (Supplementary Data Table S1). These samples were pooled and protein precipitated with acetone. The resulting mixes were separated using an SDS–PAGE approach. Because large smears encompassing more precise bands could be observed on the gel, we divided the gel into 22 parts that were analysed by nanoLC-MS/MS approaches. Nine gel pieces were collected from SDS–PAGE profiles generated with closed pitchers, and 13 spots were collected from two SDS–PAGE patterns corresponding to opened pitchers (Fig. 1, pnel 3). These experiments allowed us to identify four different proteins including three corresponding to proteins whose orthologues have already been reported in libraries (NgTLP and NkGluc1). The two remaining proteins could be identified as NmAG1 and NmAG2 in the N. mirabilis cDNA library. A complementary analysis was carried out on crude extracts of a single just-opened pitcher which allowed NmAG2, NkGluc1, NgTLP and an additional chitinase named NrChit1 to be discriminated (Table 1; Table S2).
Protein analyses of N. bicalcarata digestive fluids
We collected and pooled the contents of four pitchers. NanoLC-MS/MS analyses were performed on 22 bands collected from N. bicalcarata SDS–PAGE patterns obtained for relatively mature closed pitchers and one pitcher that had been open for an extended period and inhabited by C. schmitzi (Fig. 1, panel 4). In addition to these analyses, we performed an investigation of crude digestive fluid from just-opened pitchers (Supplementary Data Table S1). We identified peptides belonging to four different proteins in the analyses of the bands, and five additional proteins were identified using the global approach. Among those nine putative enzymes, seven had already been described for the other Nepenthes species (NgNep1, NkGkuc1, NmAG1, NMAG2, NmBGluc7, NmPRX-N and NmLTP2). The two remaining proteins were identified as an esterase/lipase (NmLip1) and a cationic peroxidase 1 (NaPRX1), and were observed only in this species so far (Table 1; Table S2). Interestingly, the secretions collected from opened pitchers inhabited by C. schmitzi exhibit a low amount of protein in comparison with closed pitchers without these ants.
Detailed in silico analysis of the protein sequences deduced from the N. mirabilis RNA-seq library
For detailed information on the identified proteins, we used various bioinformatical tools. Starting with identifying the complete coding sequence in the RNA-seq library, we were able to determine the predicted SP, the theoretical molecular weight ranging from 9.6 to 66.6 kDa, the pI, as well as putative post-translational modifications such as N- or O-glycosylation.
Nepenthesin.
We could identify three aspartic proteinases. NmNep1 and NmNep2 are similar to the two Nepenthesins that were already described in N. alata (97 % identity with NmNep1) and N. gracilis (97 % identity with NmNep2). They both harbour 12 cysteine residues, which could lead to the formation of six disulphide bonds contributing to the stability of this enzyme family. Their putative active sites are typical for aspartic proteases and contain Asp–Thr–Gly (at position 113–115 for NmNep1 and position 114–116 for NmNep1), Asp–Ser–Gly (at position 315–317) and the flap tyrosine (Tyr174 for NmNep1 and Tyr175 for NmNep2) (Table 2) (Athauda et al., 2004). NmNep2 contains three predicted sites for N-glycosylation and five for O-glycosylation.
Table 2.
Bioinformatic analyses of 20 putative new pitcher fluid proteins
| Nomenclature | Full-length protein | Signal peptide length (no. of amino acids) | No. of N-glycosylation sites | No. of O-glycosylation sites | No. of amino acids* | Mol. wt (kDa) | pI | Accession number |
|---|---|---|---|---|---|---|---|---|
| NmNep2LP | x | 25 | 3 | 5 | 417 | 45.7 | 4.9 | KT209998 |
| NmSCP3 | x | 21 | 7 | 4 | 474 | 52.5 | 4.3 | KT209999 |
| NmSCP20 | x | 22 | 3 | 10 | 473 | 52.4 | 5.6 | KT210000 |
| NmSCP47 | x | 36 | 1 | 8 | 496 | 55.3 | 5.2 | KT210001 |
| NmSCP51 | 26 | KT210002 | ||||||
| NmXyl1 | 26 | KT210017 | ||||||
| NmAGluc1 | KT210003 | |||||||
| NmAG1 | x | 28 | 2 | 1 | 377 | 42.2 | 6.6 | KT210004 |
| NmAG2 | x | 26 | 2 | 0 | 405 | 44.7 | 5.0 | KT210005 |
| NmBG1 | x | 27 | 0 | 0 | 196 | 22.5 | 6.8 | KT210006 |
| NmBG2 | 24 | KT210007 | ||||||
| NmBG3 | 2 | 6 | 577 | 64 | 6.2 | KT210008 | ||
| NmBGluc7 | 25 | KT210009 | ||||||
| NmPP1 | x | 16 | 2 | 1 | 293 | 32.0 | 5.0 | KT210010 |
| NmNPP1 | x | 36 | 5 | 3 | 604 | 66.6 | 5.1 | KT210011 |
| NmPRX27 | x | 24 | 5 | 8 | 302 | 33.3 | 4.6 | KT210012 |
| NmPRX-N | x | 31 | 5 | 5 | 303 | 32.5 | 4.9 | KT210013 |
| NmLTP1 | x | 20 | 1 | 20 | 165 | 16.4 | 6.1 | KT210014 |
| NmLTP2 | x | 26 | 0 | 3 | 91 | 9.6 | 9.6 | KT210015 |
| NmLip1 | KT210016 |
Sequence information was obtained from the cDNA library of N. mirabilis, and the corresponding peptides were identified in various Nepenthes species.
We determined the molecular weight and isoelectric point (pI) of the mature form of each protein, and predicted the size of the signal peptide and number of N- and O-glycosylation sites
The third N. mirabilis Nepenthesin identified was named Nepenthesin 2-like protein (NmNep2LP). Interestingly, this protein shares only 52 % identity with NmNep1 and 51 % with NmNep2, as detected in our cDNA library. NmNep2LP is composed of 442 amino acids, including a 417 amino acid mature enzyme and a 25 residue putative signal sequence (Table 2; Fig. 2A). The predicted cleavage site for the SP is located between Thr25 and Ser26. In contrast to NmNep1 and NmNep2, no putative propeptide was predicted for NmNep2LP. The calculated molecular weight was 45.7 kDa and the pI value 4.9. Like typical nepenthesin-aspartic proteases, NmNep2LP does not present a plant-specific insertion properly to plant vacuolar aspartic proteases, but contains ‘the nepenthesine-type AP (NAP)-specific insertion’ (Athauda et al., 2004) upstream of the flap tyrosine residue (Tyr173). It is made up by 20 residues (at position 149–168) and contains four cysteine residues (Cys151, Cys156, Cys163 and Cys167). Asp–Thr–Gly and Asp–Ser–Gly corresponding to putative active sites are located at positions 114–116 and 317–319 respectively.
Fig. 2.
Amino acid sequences of new putative digestive enzymes of N. mirabilis. Nepenthesin 2-like protein (NmN2LP) (A), serine carboxypeptidase III (NmSCP3) (B), serine carboxypeptidase-like 20 (NmSCP20) (C), serine carboxypeptidase-like 47 (NmSCP47) (D), protein phosphatase 2C (NmPP1) I, nucleotide pyrophosphatase (NmNPP1) (F), peroxidase 27 (NmPRX-27) (G), peroxidase N (NmPRX-N) (H), lipid transfer protein 2 (NmLTP2) (I), α-galactosidase 1 (NmAG1) (J) and α-galactosidase 2 (NmAG2) (K), β-galactosidase 1 (NmBG1) (L) and β-galactosidase 13 (NmBG3) (M). Matched peptides are shown in bold. Putative transit peptides are highlighted in black. Putative N-glycosylation sites are underlined, and putative O-glycosylation sites are double underlined. Cysteine residues are indicated by an asterisk. (A) The active site sequence motifs and flap tyrosine residue are boxed. (B) Residues indicated in italics correspond to putative propeptides, and conserved residues involved in the catalytic activity are boxed. (C and D) Conserved residues involved in the activity are boxed. (E) The Mn2+/Mg2+ aspartate-binding site is highlighted in grey, and conserved residues of the catalytic core related to metal co-ordination and phosphate binding are boxed. (F) Blocks of conserved amino acid residues required for metal co-ordination are boxed. (G and H) The peroxidase active site and peroxidase haem ligand site are highlighted in grey, and residues forming calcium-binding sites are boxed. (I) Conserved residues are boxed, and two consensus pentapeptides are highlighted in grey. (J and K) α-Galactosidase patterns are highlighted in grey, and catalytic aspartate residues specific to glycosyl hydrolase family 27 are boxed. (L and M) The putative active site-containing consensus sequences of the glycosyl hydrolase family 35 are highlighted in grey, with glutamate as a proton donor residue identified by a box.
Carboxypeptidase.
Four putative serine carboxypeptidases (NmSCP3, NmSCP20, NmSCP47 and NmSCP51) were identified in our study. The last, NmSCP51, is incomplete in our RNA-seq library. The molecular masses of NmSCP3, NmSCP20 and NmSCP47 range from 52.4 to 55.3 kDa, with a pI between 4.3 and 5.6 (Table 2). Each protein contains a 22–36 residue SP at the N-terminus (Table 2). Of the NmSCPs, only NmSCP3 has a 171 amino acid long sequence, which corresponds to a potential propeptide (Fig. 2.B). Each identified putative serine carboxypeptidase belongs to the S10 family of serine proteases, which are only active at an acidic pH. The catalytic activity of SCPs is induced through a charge relay system involving a catalytic triad composed of an aspartic acid residue linked to a histidine (hydrogen bond) and a serine (Liao and Remington, 1990). We identified these residues in each of the NmSCPs, with the exception of the incomplete Nm SCP51, using the peptidase database MEROPS, (http://merops.sanger.ac.uk): Ser227, Asp441 and His498 for NmSCP3 (Fig. 2B), Ser182, Asp419 and His472 for NmSCP20 (Fig.2C) and Ser253, Asp441 and His498 for NmSCP47 (Fig/ 2D). Various putative glycosylation sites could be predicted: nine for NmSCP47, 11 for NmSCP3 and 13 for NmSCP20 (Table 2).
Phosphatases.
NmPP1, which has a molecular mass of 32 kDa and a pI of 5 (Table 2), belongs to the 2 C family of serine/threonine phosphatases and may be an Mn2+- or Mg2+-dependent phosphatase. This protein has a 16 residue SP in the N-terminal position and three putative glycosylation sites (Table 2). An Mn2+/Mg2+ aspartate-binding site was detected at amino acids 64–72 (Fig. 2E). The predicted catalytic core harbours several amino acids that contribute to metal coordination (Asp48, Asp69–Gly70, Asp231 and Asp270) and phosphate binding (Arg43) (Table 2 Supplementary Data Fig. S1) (Shi, 2009) (Fig. 2E).
NmNPP1 is a putative digestive enzyme of 66.5 kDa with a pI of 5.1, a 36 residue SP and seven putative glycosylation sites (Table 2). This protein is an apparent acidic phosphatase of the metallophosphatase superfamily, which is known to hydrolyse phosphate esters and anhydrides under acidic conditions. NmNPP1 contains a putative active site consisting of five blocks of conserved residues required for metal coordination: Gly337–Asp–Met–Gly340, Gly379–Asp–Ile–Ser–Tyr383, Gly412–Asn–His–Glu415, His502 and Gly540–His–Val–His543 (bold residues are conserved metal-binding sites) (Fig. 2F) (Schenk et al., 2000).
Peroxidases.
Peroxidase 27 (33.4 kDa and pI 4.6) and peroxidase N-like protein (32.5 kDa and pI 4.9) belong to class III of plant secretory peroxidases and have an SP of 24 and 31 amino acids, respectively (Table 2). Class III plant peroxidases are glycoproteins. NmPrx-27 and NmPRX-N harbour 13 and 10 putative glycosylation sites, respectively (Table 2). NmPRX-27 contains a putative peroxidase active site between position 58 and 69 and a peroxidase haem ligand site between position 186 and 196 (Fig. 2G). NmPRX-N contains a putative peroxidase active site between position 64 and 75 and a peroxidase haem ligand site between position 189 and 199 (Fig. 2H). The general structure of class III peroxidases contains four conserved disulphide bonds (Hiraga et al., 2001), which are present in the peroxidases isolated from N. mirabilis: Cys36–Cys116, Cys69–Cys74, Cys122–Cys322, Cys201–Cys233 for NmPRX-27 (Fig. 2G); and Cys42–Cys119, Cys75–Cys80, Cys125–Cys329 and Cys125–Cys204 for NmPRX-N (Fig. 2H). The structure also includes two conserved calcium-binding sites in distal and proximal positions: the calcium-binding sites of NmPRX27 are formed by the distal cluster Asp68–Val71–Gly73–Asp75–Ser77 and the proximal cluster Thr195–Asp245–Thr248–Lys251–Asp253 (Fig. 2G). The binding sites of NmPRX-N are composed of the groups Asp74–Val77–Gly79–Asp81–Ser83 and Thr198–Asp249–Ser252–Leu255–Asp257 (Fig. 2H).
Lipid transfer protein (LTP)
According to Carvalho and Gomes (2007), the LTPs belonging to family 1 have molecular masses close to 10 kDa and are basic with pIs of 9–10. These 90–95 amino acid long LTPs feature cysteines conserved in similar positions along the primary structure. NmLTP2 seems to belong to this family of LTPs. Its molecular mass is 9.6 kDa, its pI is 9.6 (Table 2), and the amino acid sequence contains eight cysteines, which could form four disulphide bonds (Cys30–Cys77, Cys40–Cys54, Cys55–Cys99 and Cys75–Cys113) (Fig. 2I). Other residues are highly conserved, including Val33, Tyr43, Gly57, Leu78, Lys79, Ala93, Val101 and Ile103, as well as the two consensus pentapeptides Thr67–Thr68–Pro69–Asp70–Arg71 and Gly104–Phe105–Lys106–Ile107–Ala108 (Fig. 2I) (Carvalho and Gomes, 2007). As a member of the LTP family 1, NmLTP2 contains an internal hydrophobic cavity that may be the binding site for lipid-like molecules. This cavity is composed of 18 residues, including Leu37, Leu41, Leu44, Val58 or Ala61. Finally, this protein harbours a 26 residue SP.
Galactosidases.
NmAG1 and NmAG2 are putative α-galactosidases of 42.2 and 44.7 kDa with pI values of 6.6 and 5.0, respectively. They contain 28 or 26 amino acid long SPs, as well as three and two putative glycosylation sites, respectively (Table 2). NmAG1 features the α-galactosidase pattern from position 84 to 100 (α-galactosidase signature: G-[LIVMFY]-x-x-[LIVMFY]-x-[LIVM]-D-[DF]-x(1,2)-W-x(3,7)-[RV]-[DNSF]; x = any amino acid). NmAG1 belongs to the glycosyl hydrolase family 27 and contains two catalytic aspartate residues, which function as a nucleophile (Asp170) and an acid/base catalyst (Asp226) (Fig. 2J). The α-galactosidase consensus pattern of NmAG2 is located between positions 101 and 117, and two aspartate residues acting as a nucleophile and acid/base catalyst are located at positions 186 and 241, respectively (Fig. 2K) (http://prosite.expasy.org/PDOC00443).
β-Galactosidases, a widespread family of glycosyl hydrolases, can hydrolyse terminal, non-reducing β-d-galactosyl residues from β-d-galactosides. NmBG1 is a putative β-galactosidase of 22.5 kDa with a pI of 6.8 and a 22 residue SP (Table 2). It contains a putative active site containing the consensus sequence pattern G-G-P-[LIVM]-x-Q-x-E-N-E-[FY], which is found in glycosyl hydrolases. The active site is localized between positions 174 and 186, and Glu185 serves as a proton donor residue (Fig. 2L) (Smith and Gross, 2000). NmBG3 is also a putative β-galactosidase with a mol. wt of 64 kDa and a pI of 6.2 (Table 2). The sequence of its SP is incomplete in our RNA-seq database. However, it has the putative active site-containing consensus sequence pattern of the glycosyl hydrolase family 35 between positions 47 and 59, with Glu58 serving as the proton donor residue (Fig. 2M) (Smith and Gross, 2000).
DISCUSSION
To digest prey, Nepenthes species secrete an arsenal of enzymes which are mostly proteolytic enzymes. Some of these proteins have already been identified through many specific studies and their sequences are available in public libraries. In this report we extended the list of proteins through a study performed on five different Nepenthes species displaying different ecological behaviour. Since only a few accession are available in public databases (GenBank), we compared the peptide sequences obtained by a nano-LC/MS approach with a cDNA library based on RNA extracted from N. mirabilis. This study, however, is not exhaustive. It is likely that some proteins have not been identified by classical proteomics approaches because they are secreted in too low concentrations. The SDS–PAGE profile differences might also be linked to post-translational modifications. For example, a proteolytic digestion or maturation of proteins excreted in the pitcher fluid can lead to the finding of peptides of one protein in smaller bands than its actual size. This has been observed for Nepenthesin by Buch et al. (2015). Other post-translational modifications such as glycosylation also impact the SDS–PAGE pattern. Buch showed in 2014 that the highly glycosylated PR-1 protein migrates at double the size of its original molecular weight.
A total of 29 proteins were identified in the secretome of closed and opened pitchers harvested from five different Nepenthes species (N. mirabilis, N. alata, N. sanguinea, N. bicalcarata and N. albomarginata). Of these, nine hydrolytic enzymes had been previously reported: the aspartic proteases Nepenthesin 1 and 2 from N. gracilis and N. distillatoria (NgNep1 and NgNep2; Athauda et al., 2004), two glucanases isolated from N. khasiana (NkGluc1; Eilenberg and Zilberstein, 2008) and N. alata (NaGluc2; Hatano and Hamada, 2012), a cationic peroxidase (NaPRX1; Hatano and Hamada, 2012), and class I (NkChit1; Eilenberg, 2006), class III (NrChit1 or NaChit3; Rottloff et al., 2011) and class IV chitinases (NaChit1; Hatano and Hamada, 2008; Ishisaki et al., 2012b). Two additional previously identified putative antimicrobial proteins were also detected in our study: a PR-1 protein (NmPR-1; Buch et al., 2014) and a thaumatin-like protein (NgTLP; Hatano and Hamada, 2008; Rottloff et al., 2009). Beside these proteins, we could identify peptides which, when compared with the Nepenthes nucleotide database, correspond to 13 new putative digestive enzymes: a third aspartic protease (NmNep2LP), three serine carboxypeptidases of types 3, 20 and 47 (NmSCP3, NmSCP20 and NmSCP47), two α- (NmAG1 and NmAG2) and one β-galactosidase (NmBG1), a protein phosphatase (NmPP1), a nucleotide pyrophosphatase/phosphodiesterase (NmNPP1), two new peroxidases of type 27 (NmPRX27) and type N (NmPRX-N) and two lipid transfer proteins (NmLTP1 and NmLTP2). This work also led to the identification of partial sequences of seven additional putative enzymes: a serine carboxypeptidase (NmSCP51), a β-xylosidase (NmXyl1), an α-glucosidase (NmAGluc1), two β-galactosidases (NmBG2 and NmBG3), a β-d-1,3-glucosidase 7-like protein (NmBGluc7) and a lipase (NmLip1).
The carnivorous system, including trap formation, prey digestion and nutrient assimilation, is an important energetic investment for plants (Givnish et al., 1984; Ellison, 2006; Pavlovič and Saganová, 2015). The pool of digestive enzymes has to be efficient so that the benefits obtained from digested prey outweigh the cost of carnivory. A complex association of enzymes acting at different levels and favouring the gain of the greatest amount of nutrients from prey degradation is therefore expected. We demonstrated that the protease content of several of the studied species is composed of two classes of protein-degrading enzymes: aspartic proteases [Nepenthesin 1 and 2 and Nepenthesin 2-like protein (NmN2LP)] and serine carboxypeptidases (NmSCP3, NmSCP20, NmSCP47 and NmSCP51). The serine carboxypeptidases identified in this work putatively belong to the S10 family of serine proteases. These enzymes are only active at an acidic pH (Schulze et al., 2012), as is the case for the two Nepenthesin enzymes (with optimal activity at pH 2.6) (Athauda et al., 2004; Takahashi et al., 2005). A similar diversity of protease activities was found in the digestive fluid of the Venus flytrap, with cysteine proteases (Dionain), aspartic proteases and serine carboxypeptidases (Schulze et al., 2012; Libiakova et al., 2014). The frequency of proteases in carnivorous plants, such as Nepenthes and Dionaea, suggests a strong dependence on nutrients gained from digested prey.
The assimilation of products derived from protein degradation is an important step in the carnivory process. Our results support that N. mirabilis, N. alata and N. sanguinea absorb protein-derived nutrients by two different strategies. First, Nepenthesin 1 (in cooperation with Nepenthesin 2) degrades prey-derived proteins into peptides, which can then be directly absorbed by the digestive glands (Schulze et al., 1999). Alternatively, proteins may be degraded by serine carboxypeptidases starting at the C-terminus, and the resulting amino acids could be delivered into plant tissues by transporters expressed in the digestive glands (Schulze et al., 1999). Free radicals released during the generation of reactive oxygen species (ROS) by peroxidases increase the degradation rate of proteases (Chia et al., 2004; Hatano and Hamada, 2012). In addition to the cationic peroxidase 1 described by Hatano and Hamada, we identified two new sub-classes of putative peroxidases: peroxidase 27-like protein (NmPRX27) in N. alata and N. sanguinea fluids, and peroxidase N-like protein (NmPRX-N) in N. mirabilis, N. alata, N. sanguinea and N. bicalcarata fluids. These enzymes belong to the class III peroxidases.
Chitinases found in the fluids of Drosera sp., Dionaea muscipula and Triphyophyllum peltatum contribute to the degradation of prey exoskeletons (Matušíková et al., 2005; Schulze et al., 2012; Paszota et al., 2014). A two-step mechanism involving different types of chitinases may be implemented to degrade chitin present in the insect cuticle. First, chitin polymers derived from prey might be fragmented into shorter polymers by a class III endochitinase (NrChit1 or NaChit3). Secondly, the resulting shorter chitin chains might be hydrolysed to N-acetylglucosamine residues by the class I endochitinases (NkChit1; Eilenberg, 2006) and the class IV endochitinases (NaChit1; Hatano and Hamada, 2008). Chitinase activity has been detected in Nepenthes digestive fluid. Eilenberg (2006) reported chitinase activity in the digestive fluid of N. khasiana on both a long chitin polymer and a tetrameric substrate, 4MU-(GlcNAc)3. The latter was described to be hydrolysed by the chitinases isolated from N. khasiana. Another class III endochitinase (NrChit1) hydrolyses a long-chain chitin polymer (colloidal CM-chitin-RBV) (Rottloff et al., 2011), and ethylene glycol chitin or shorter chains of N-acetylglucosamine residues [(GlcNAc)n; n = 5, 6) (Ishisaki et al., 2012a)]. Hatano and Hamada (2012) identified a class IV chitinase (NaChit1) in N. alata secretions that also hydrolyses (GlcNAc)n and generates a chain length of 3–6 units. In this work, we reported the presence of class III and IV chitinases in the digestive fluid of N. sanguinea collected from closed and opened pitchers similar to those identified by Hatano and Hamada in N. alata secretions after chitin addition.
Phosphatases represent a broad group of enzymes that catalyse the hydrolysis of phosphate esters. They may play a role in phosphorus mobilization from prey and its uptake by the plant tissue (Płachno et al., 2006). Together with nitrogen, phosphorus limits the growth of carnivorous plants (Ellison, 2006). Recently, Pavlovič et al. (2014) showed that phosphatase activity was induced 24 h after prey capture in Drosera capensis. Therefore, phosphatases are crucial enzymes that enable prey-derived phosphorus acquisition in carnivorous plants (Plachno et al., 2009). We identified two putative phosphatases: protein phosphatase 2C (NmPP1) detected in N. mirabilis and a nucleotide pyrophosphatase/phosphodiesterase (NmNPP1) in N. sanguinea. These two enzymes represent the first putative phosphatases identified in Nepenthes species.
Given the high energy cost of the carnivorous strategy, it is likely that additional nutritional sources such as glucan-rich particles from pollen grains, fungal spores, seeds or plant detritus are utilized by members of the Nepenthes genus. This was previously observed in N. ampullaria (Moran et al., 2003; Adlassnig et al., 2011; Pavlovič et al., 2011) and other carnivorous plants such as Drosera (Michalko et al., 2013). A study of Drosera demonstrated that a β-1,3-glucanase is involved in the degradation of laminarin, a β-(1–3)-glucan polymer with β-(1–6)-branches, the products of which are taken up by the trap tissue. We observed putative glucanase activity in each Nepenthes species studied (NkGluc1 and NaGluc2). Other enzymes identified in Nepenthes secretions may also be involved in carbohydrate metabolism. For example, the thaumatin-like protein identified in N. mirabilis, N. alata and N. sanguinea (Hatano and Hamada, 2008; Rottloff et al., 2009, 2011) displays glucanase activity in other plants (Liu et al., 2010), although these proteins were also described to have osmotic function leading to cell wall degradation (Yun et al., 1997).
In addition to their role in food acquisition through degrading macromolecules, such enzymes might also be involved in the formation of a polysaccharidic network responsible for the viscoelasticity of the secretion. Although the exact composition of the Nepenthes polysaccharide remains to be elucidated, Gaume and Forterre (2007) speculated that the Nepenthes fluid could exhibit an acidic polysaccharide mucilage similar to that of the secretions produced by sundew. The polysaccharide of D. capensis is composed of l-arabinose, d-xylose, d-galactose, d-mannose and d-glucuronic acid in a molar ratio of 3.6:1.0:4.9:8.4:8.2 (Gowda et al., 1983) and constitutes 4 % of D. capensis adhesive (Rost and Schauer, 1977). Drosera binata polysaccharide has a different ratio, 8:1:10:18:17 (Gowda et al., 1982). Recently, Bauer and Bazile demonstrated that the viscoelastic properties of N. rafflesiana fluid decrease significantly after pitcher opening (Bauer and Federle, 2009; Bazile et al., 2015). These properties contribute significantly to the capture and retention of prey (Gaume and Forterre, 2007; Bonhomme et al., 2011b). Enzymes that may be involved in this process were highlighted in the present analyses. Two types of glucosidases were identified: an α-glucosidase (NmAGluc1 in N. sanguinea) and a β-d-1,3-glucosidase 7-like protein (NmBGluc7 in N. alata, N. sanguinea and bicalcarata). In addition to glucosidases, we also detected galactosidases such as α-galactosidases 1 and 2 (NmAG1 and NmAG2) in N. mirabilis, N. alata, N. sanguinea, N. bicalcarata and N. albomarginata secretions, and β-galactosidase 1, 2 and 3 (NmBG1, NmBG2 and NmBG3) in N. alata. At the end of pitcher life, the polysaccharide might be removed by saccharide-cleaving enzymes to facilitate the digestion and absorption of nutrients.
To recover most nutrients coming from prey digestion it is necessary to limit the proliferation of micro-organisms that could metabolize part of them. Many studies have highlighted the antimicrobial properties of the digestive fluid of Nepenthes after opening of the pitcher (Adlassnig et al., 2011; Chou et al., 2014). The study carried out by Buch and co-workers (2013) showed that the microbial proliferation is limited because of the naturally poor mineral content of the secretions, the presence of some secondary metabolites, the acidic pH and the presence of pathogenesis-related (PR) proteins. Six different PR protein families (PR-1, 3, 5, 7, 8 and 9) were identified in Nepenthes fluids by Mithöfer (2010). In our study we extended this list in N. alata, N. sanguinea and N. bicalcarata to other PR proteins belonging to the PR-14 family: NmLTP1 and NmLTP2. Although never described in Nepenthes, these proteins related to LTPs were previously detected in the secretions of another carnivorous plant, Dionaea muscipula (Schulze et al., 2012). Some LTPs were also described to exhibit antifungal and antibacterial activities (Van Loon et al., 2006) and might therefore be part of the antimicrobial strategy of Nepenthes. They might also be involved in the formation of cutin barriers in the digestive zone of the pitchers to prevent the entry of pathogens into the tissue (Heredia, 2003; Carvalho and Gomes, 2007).
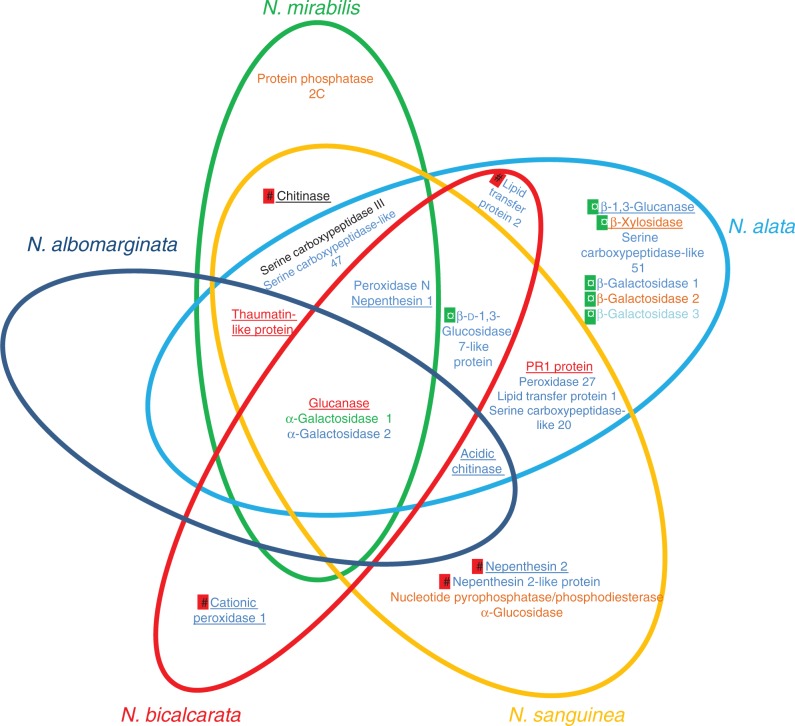
At the species level, there is an increasing body of evidence showing that species differ in their trapping strategies (Gaume and Di Giusto, 2009; Benz et al., 2011; Bonhomme et al., 2011a; Bauer et al., 2012) as well as in their nutrient sequestration strategies, with nitrogen derived from prey in truly carnivorous species (the majority of species), but also from vegetal detritus in more detritivorous species (Moran et al., 2003; Pavlovič et al., 2011), from faeces of mammals (Clarke et al., 2009; Chin et al., 2010) or from symbiotically associated ants (Bonhomme et al., 2011a; Bazile et al., 2012) in more coprophagous species. Our work demonstrated that although several enzymes were detected in all considered species (NkGluc1, NmAG1 and NmAG2), the protein composition in digestive fluid could differ according to the considered species and development stage (Fig. 3). These differences might reflect trapping strategies established by the plant. For example, N. bicalcarata develops an outstanding strategy of nutrient sequestration thanks to a mutualistic relationship with the symbiotic ant species, Camponotus schmitzi. These ants assist the plant by the catching and the digestion of the prey (Bonhomme et al., 2011a; Bazile et al., 2012), prevent nutrient export by fluid inhabitants (Scharmann et al., 2013), clean pitcher trapping surfaces (Thornham et al., 2011) or protect the host plant against damage caused by Alcidodes sp. (Merbach et al., 2007). This relationship considerably increases the foliar nitrogen in adult plants (Bazile et al., 2012). Surprisingly, ants can live and swim in the digestive fluid (Clarke and Kitching, 1995; Bohn et al., 2012) without being damaged by the hydrolytic enzymes. The analyses of N. bicalcarata pitcher fluids enabled us to visualize 22 SDS–PAGE bands which were characterized as only nine different putative proteins. Among all proteases, we detected only the presence of Nepenthesin 1 active only at an acidic pH (Athauda et al., 2004) and the pH of the N. bicalcarata fluid has been described to be at around pH 5 (Bazile et al., 2012). Therefore, Nepenthesin 1 might be less active, which could protect the ants against the proteolytic activity. These observations are correlated with a low amount of protein in secretions of opened pitchers inhabited by C. schmitzi in comparison with opened pitchers without ants. Another example concerns N. sanguinea, N. alata and N. mirabilis which display a conventional nitrogen acquisition strategy of carnivorous plants by trapping and digesting arthropods. According to our results, they produce a blend of digestive enzymes which, in conjunction with acidic pH conditions, ensures essential functions of prey degradation (protein degradation through abundant aspartic proteases, carboxypeptidases and peroxidases, and exoskeleton degradation through chitinases) and potential use of sugar-rich particles as a supplementary nutritional source (through sugar-cleaving enzymes) (Fig. 3).
Fig. 3.
Summary of the proteins identified from the digestive fluid of five Nepenthes species. Already identified proteins are underlined. Proteins found only in closed pitchers are marked by red symbols and proteins found only in open pitchers are marked by green symbols. Proteins identified thanks to one peptide are coloured in orange, two or three peptides in blue, four or five peptides in green, 5–10 peptides in black and >10 peptides in red.
The present characterization of the fluid protein content of Nepenthes based on deep sequencing of the transcriptome in combination with proteomic analyses provides new insights into the composition of Nepenthes digestive fluids. The Nepenthes secretome reveals an important diversity of hydrolytic enzymes that differ among species, hypothetically in response to particular ecological pressures. In addition to nine digestive enzymes described previously in the literature, 20 new putative proteins were identified as hydrolytic enzymes. The plant origin of enzymes identified in secretions coming from plants cultivated in greenhouses or living in natural sites remains to be confirmed because the experimental conditions did not exclude microbial contamination in most of the fluids. This study is, however, not exhaustive and needs to be extended to other species that vary in their nitrogen source and diet. A recent report published by Buch et al. (2015) describes the first evidence of direct interaction between Nepenthesin 2 and prey. Such an analysis might be extended to many other proteins and therefore provide valuable information on the evolution of digestive processes in these fascinating plant species.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: 2-D analysis of proteins present in the pitcher fluid of two in vitro N. mirabilis plants. Table S1: summary of growth conditions, origin of Nepenthes plant, studied pitcher development stage and the proteomic approach used for each condition. Table S2: summary of the proteins identified from digestive fluids of five Nepenthes species.
ACKNOWLEDGEMENTS
This project has been funded by the BIOPROLOR project (Région Lorraine, France). The authors wish to acknowledge Ms Cindy Michel for technical assistance. The authors would also like to thank the reviewers for the valuable comments during the evaluation process of the manuscript and would also like to acknowledge the Botanical Garden du Montet (Nancy, France) for providing Nepenthes plants.
LITERATURE CITED
- Adlassnig W, Peroutka M, Lendl T. 2011. Traps of carnivorous pitcher plants as a habitat: composition of the fluid, biodiversity and mutualistic activities. Annals of Botany 107: 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C-I, Takekawa S, Okazawa A, Fukusaki E-i, Kobayashi A. 2002. Degradation of a peptide in pitcher fluid of the carnivorous plant Nepenthes alata Blanco. Planta 215: 472–477. [DOI] [PubMed] [Google Scholar]
- Athauda SBP, Matsumoto K, Rajapakshe S, et al. 2004. Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochemical Journal 381: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer U, Clemente CJ, Renner T, Federle W. 2012. Form follows function: morphological diversification and alternative trapping strategies in carnivorous Nepenthes pitcher plants. Journal of Evolutionary Biology 25: 90–102. [DOI] [PubMed] [Google Scholar]
- Bauer U, Federle W. 2009. The insect-trapping rim of Nepenthes pitchers: surface structure and function. Plant Signaling and Behavior 4: 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazile V, Moran JA, Le Moguédec G, Marshall DJ, Gaume L. 2012. A carnivorous plant fed by its ant symbiont: a unique multi-faceted nutritional mutualism. PLoS One 7: e36179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazile V, Le Moguédec G, Marshall DJ, Gaume L. 2015. Fluid physico-chemical properties influence capture and diet in Nepenthes pitcher plants. Annals of Botany 115: 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz MJ, Gorb EV, Gorb SN. 2011. Diversity of the slippery zone microstructure in pitchers of nine carnivorous Nepenthes taxa. Arthropod-Plant Interactions 6: 147–158. [Google Scholar]
- Biteau F, Nisse E, Miguel S, et al. 2013. A simple SDS–PAGE protein pattern from pitcher secretions as a new tool to distinguish Nepenthes species (Nepenthaceae). American Journal of Botany 100: 2478–2484. [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99. [Google Scholar]
- Bohn H, Thornham D, Federle W. 2012. Ants swimming in pitcher plants: kinematics of aquatic and terrestrial locomotion in Camponotus schmitzi. Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology 198: 465–476. [DOI] [PubMed] [Google Scholar]
- Bonhomme V, Gounand I, Alaux C, Jousselin E, Barthelemy D, Gaume L. 2011a. The plant-ant Camponotus schmitzi helps its carnivorous host-plant Nepenthes bicalcarata to catch its prey. Journal of Tropical Ecology 27: 15–24. [Google Scholar]
- Bonhomme V, Pelloux-Prayer H, Jousselin E, Forterre Y, Labat JJ, Gaume L. 2011b. Slippery or sticky? Functional diversity in the trapping strategy of Nepenthes carnivorous plants. New Phytologist 191: 545–554. [DOI] [PubMed] [Google Scholar]
- Buch F, Rott M, Rottloff S, Paetz C, Hilke I, Raessler M, Mithöfer A. 2013. Secreted pitfall-trap fluid of carnivorous Nepenthes plants is unsuitable for microbial growth. Annals of Botany 111: 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch F, Pauchet Y, Rott M, Mithöfer A. 2014. Characterization and heterologous expression of a PR-1 protein from traps of the carnivorous plant Nepenthes mirabilis. Phytochemistry 100: 43–50. [DOI] [PubMed] [Google Scholar]
- Buch F, Kaman WE, Bikker FJ, Yilamujiang A, Mithofer A. 2015. Nepenthesin protease activity indicates digestive fluid dynamics in carnivorous Nepenthes plants. PLoS One 10: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AdO, Gomes VM. 2007. Role of plant lipid transfer proteins in plant cell physiology – A concise review. Peptides 28: 1144–1153. [DOI] [PubMed] [Google Scholar]
- Chia T, Aung H, Osipov A, Goh N, Chia L. 2004. Carnivorous pitcher plant uses free radicals in the digestion of prey. Redox Report 9: 255–261. [DOI] [PubMed] [Google Scholar]
- Chin L, Moran JA, Clarke C. 2010. Trap geometry in three giant montane pitcher plant species from Borneo is a function of tree shrew body size. New Phytologist 186: 461–470. [DOI] [PubMed] [Google Scholar]
- Chou LY, Clarke CM, Dykes GA. 2014. Bacterial communities associated with the pitcher fluids of three Nepenthes (Nepenthaceae) pitcher plant species growing in the wild. Archives of Microbiology 196: 709–717. [DOI] [PubMed] [Google Scholar]
- Clarke C. 1997. Nepenthes of Borneo. Kota Kinabalu, Sabah, Malaysia: Natural History Publications (Borneo) Sdn. Bhd. [Google Scholar]
- Clarke C. 2001. Nepenthes of Sumatra and Peninsular Malaysia. Kota Kinabalu, Sabah, Malaysia: Natural History Publications (Borneo) Sdn. Bhd. [Google Scholar]
- Clarke CM, Kitching RL. 1995. Swimming ants and pitcher plants: a unique ant–plant interaction from Borneo. Journal of Tropical Ecology 11: 589–602. [Google Scholar]
- Clarke CM, Bauer U, Lee CiC, Tuen AA, Rembold K, Moran JA. 2009. Tree shrew lavatories: a novel nitrogen sequestration strategy in a tropical pitcher plant. Biology Letters 5: 632–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilenberg H. 2006. Isolation and characterization of chitinase genes from pitchers of the carnivorous plant Nepenthes khasiana. Journal of Experimental Botany 57: 2775–2784. [DOI] [PubMed] [Google Scholar]
- Eilenberg H, Zilberstein A. 2008. Carnivorous pitcher plants – towards understanding the molecular basis of prey digestion. In: Teixeira Da Silva JA, ed. Floriculture, ornamental and plant biotechnology. Advances and topical issues, Vol. V. London, Global Science Books, 287–294. [Google Scholar]
- Ellison AM. 2006. Nutrient limitation and stoichiometry of carnivorous plants. Plant Biology 8: 740–747. [DOI] [PubMed] [Google Scholar]
- Gaume L, Di Giusto B. 2009. Adaptive significance and ontogenetic variability of the waxy zone in Nepenthes rafflesiana. Annals of Botany 104: 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume L, Forterre Y. 2007. A viscoelastic deadly fluid in carnivorous pitcher plants. PLoS One 2: e1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20: 601–605. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Burkhardt EL, Happel RE, Weintraub JD. 1984. Carnivory in the Bromeliad Brocchinia reducta, with a cost/benefit model for the general restriction of carnivorous plants to sunny, moist, nutrient-poor habitats. American Naturalist 124: 479–497. [Google Scholar]
- Gowda DC, Reuter G, Schauer R. 1982. Structural features of an acidic polysaccharide from the mucin of Drosera binata. Phytochemistry 21: 2297–2300. [Google Scholar]
- Gowda DC, Reuter G, Schauer R. 1983. Structural studies of an acidic polysaccharide from the mucin secreted by Drosera capensis. Carbohydrate Research 113: 113–124. [Google Scholar]
- Hatano N, Hamada T. 2008. Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. Journal of Proteome Research 7: 809–816. [DOI] [PubMed] [Google Scholar]
- Hatano N, Hamada T. 2012. Proteomic analysis of secreted protein induced by a component of prey in pitcher fluid of the carnivorous plant Nepenthes alata. Journal of Proteomics 75: 4844–4852. [DOI] [PubMed] [Google Scholar]
- Heredia A. 2003. Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochimica et Biophysica Acta 1620: 1–7. [DOI] [PubMed] [Google Scholar]
- Higashi S, Nakashima A, Ozaki H, Abe M, Uchiumi T. 1993. Analysis of feeding mechanism in a pitcher of Nepenthes hybrida. Journal of Plant Research 106: 47–54. [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. 2001. A large family of class III plant peroxidases. Plant and Cell Physiology 42: 462–468. [DOI] [PubMed] [Google Scholar]
- Ishisaki K, Arai S, Hamada T, Honda Y. 2012a. Biochemical characterization of a recombinant plant class III chitinase from the pitcher of the carnivorous plant Nepenthes alata. Carbohydrate Research 361: 170–174. [DOI] [PubMed] [Google Scholar]
- Ishisaki K, Honda Y, Taniguchi H, Hatano N, Hamada T. 2012b. Heterogonous expression and characterization of a plant class IV chitinase from the pitcher of the carnivorous plant Nepenthes alata. Glycobiology 22: 345–351. [DOI] [PubMed] [Google Scholar]
- Jensen ON, Wilm M, Shevchenko A, Mann M. 1999. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods in Molcular Biology 112: 513–530. [DOI] [PubMed] [Google Scholar]
- Kadek A, Tretyachenko V, Mrazek H, et al. 2014. Expression and characterization of plant aspartic protease nepenthesin-1 from Nepenthes gracilis. Protein Expression and Purification 95: 121–128. [DOI] [PubMed] [Google Scholar]
- Kato M, Hotta M, Tamin R, Itino T. 1993. Inter- and intra-specific variation in prey assemblages and inhabitant communities in Nepenthes pitchers in Sumatra. Tropical Zoology 6: 11–25. [Google Scholar]
- Liao DI, Remington SJ. 1990. Structure of wheat serine carboxypeptidase II at 3.5-Å resolution. A new class of serine proteinase. Journal of Biological Chemistry 265: 6528–6531. [DOI] [PubMed] [Google Scholar]
- Libiakova M, Flokova K, Novak O, Slovakova L, Pavlovic A. 2014. Abundance of cysteine endopeptidase dionain in digestive fluid of Venus flytrap (Dionaea muscipula Ellis) is regulated by different stimuli from prey through jasmonates. PLoS One 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Sturrock R, Ekramoddoullah AKM. 2010. The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Reports 29: 419–436. [DOI] [PubMed] [Google Scholar]
- Matthews REF. 1960. A ribonuclease from Nepenthes spp. Biochimica et Biophysica Acta 38: 552–553. [Google Scholar]
- Matušíková I, Salaj J, Moravčíková J, Mlynárová L, Nap J-P, Libantová J. 2005. Tentacles of in vitro-grown round-leaf sundew (Drosera rotundifoliaL.) show induction of chitinase activity upon mimicking the presence of prey. Planta 222: 1020–1027. [DOI] [PubMed] [Google Scholar]
- McPherson S. 2009. Pitcher plants of the Old Wold. Poole, UK: Redfern Natural History. [Google Scholar]
- Merbach MA, Merbach DJ, Maschwitz U, Booth WE, Fiala B, Zizka G. 2002. Mass march of termites into the deadly trap. Nature 415: 37. [DOI] [PubMed] [Google Scholar]
- Merbach MA, Zizka G, Fiala B, Merbach D, Booth WE, Maschwitz U. 2007. Why a carnivorous plant cooperates with an ant-selective defense against pitcher-destroying weevils in the myrmecophytic pitcher plant Nepenthes bicalcarata Hook. f. Ecotropica 13: 45–56. [Google Scholar]
- Michalko J, Socha P, Mészáros P, et al. 2013. Glucan-rich diet is digested and taken up by the carnivorous sundew (Drosera rotundifolia L.): implication for a novel role of plant β-1,3-glucanases. Planta 238: 715–725. [DOI] [PubMed] [Google Scholar]
- Mithofer A. 2010. Carnivorous pitcher plants: insights in an old topic. Phytochemistry 72: 1678–1682. [DOI] [PubMed] [Google Scholar]
- Moran JA, Clarke CM, Hawkins BJ. 2003. From carnivore to detritivore? Isotopic evidence for leaf litter utilization by the tropical pitcher plant Nepenthes ampullaria. International Journal of Plant Sciences 164: 635–639. [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15: 473–497. [Google Scholar]
- Nishimura E, Jumyo S, Arai N, et al. 2014. Structural and functional characteristics of S-like ribonucleases from carnivorous plants. Planta 240: 147–159. [DOI] [PubMed] [Google Scholar]
- Paszota P, Escalante-Perez M, Thomsen LR, et al. 2014. Secreted major Venus flytrap chitinase enables digestion of arthropod prey. Biochimica et Biophysica Acta 1844: 374–383. [DOI] [PubMed] [Google Scholar]
- Pavlovič A, Saganová M. 2015. A novel insight into the cost–benefit model for the evolution of botanical carnivory. Annals of Botany 115: 1075–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovič A, Slováková L, Šantrŭček J. 2011. Nutritional benefit from leaf litter utilization in the pitcher plant Nepenthes ampullaria. Plant, Cell and Environment 34: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Plachno BJ, Adamec L, Huet H. 2009. Mineral nutrient uptake from prey and glandular phosphatase activity as a dual test of carnivory in semi-desert plants with glandular leaves suspected of carnivory. Annals of Botany 104: 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płachno BJ, Adamec L, Lichtscheidl IK, Peroutka M, Adlassnig W, Vrba J. 2006. Fluorescence labelling of phosphatase activity in digestive glands of carnivorous plants. Plant Biology 8: 813–820. [DOI] [PubMed] [Google Scholar]
- Rost K, Schauer R. 1977. Physical and chemical properties of the mucin secreted by Drosera capensis. Phytochemistry 16: 1365–1368. [Google Scholar]
- Rottloff S, Müller U, Kilper R, Mithöfer A. 2009. Micropreparation of single secretory glands from the carnivorous plant Nepenthes. Analytical Biochemistry 394: 135–137. [DOI] [PubMed] [Google Scholar]
- Rottloff S, Stieber R, Maischak H, Turini FG, Heubl G, Mithofer A. 2011. Functional characterization of a class III acid endochitinase from the traps of the carnivorous pitcher plant genus, Nepenthes. Journal of Experimental Botany 62: 4639–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharmann M, Thornham DG, Grafe TU, Federle W. 2013. A novel type of nutritional ant–plant interaction: ant partners of carnivorous pitcher plants prevent nutrient export by dipteran pitcher infauna. PLoS One 8: e63556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Korsinczky MLJ, Hume DA, Hamilton S, DeJersey J. 2000. Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes. Gene 255: 419–424. [DOI] [PubMed] [Google Scholar]
- Schulze W, Frommer WB, Ward JM. 1999. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. The Plant Journal 17: 637–646. [DOI] [PubMed] [Google Scholar]
- Schulze WX, Sanggaard KW, Kreuzer I, et al. 2012. The protein composition of the digestive fluid from the Venus flytrap sheds light on prey digestion mechanisms. Molecular and Cellular Proteomics 11: 1306–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484. [DOI] [PubMed] [Google Scholar]
- Smith DL, Gross KC. 2000. A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiology, 123: 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P, Hogan J. 2006. Cloning and characterization of a ribonuclease, a cysteine proteinase, and an aspartic proteinase from pitchers of the carnivorous plant Nepenthes ventricosa Blanco. International Journal of Plant Sciences 167: 239–248. [Google Scholar]
- Takahashi K, Athauda SBP, Matsumoto K, et al. 2005. Nepenthesin, a unique member of a novel subfamily of aspartic proteinases: enzymatic and structural characteristics. Current Protein and Peptide Science 6: 513–525. [DOI] [PubMed] [Google Scholar]
- Thiele H, Glandorf J, Hufnagel P. 2010. Bioinformatics strategies in life sciences: from data processing and data warehousing to biological knowledge extraction. Journal of Integrative Bioinformatics 7: 141. [DOI] [PubMed] [Google Scholar]
- Thornham DG, Smith JM, Ulmar Grafe T, Federle W. 2011. Setting the trap: cleaning behaviour of Camponotus schmitzi ants increases long-term capture efficiency of their pitcher plant host, Nepenthes bicalcarata. Functional Ecology 26: 11–19. [Google Scholar]
- Tökés ZA, Woon WC, Chambers SM. 1974. Digestive enzymes secreted by the carnivorous plant Nepenthes macferlanei L. Planta 119: 39–46. [DOI] [PubMed] [Google Scholar]
- Van Loon L, Rep M, Pieterse C. 2006. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology 44: 135–162. [DOI] [PubMed] [Google Scholar]
- Yun DJ, Zhao Y, Pardo JM, et al. 1997. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proceedings of the National Academy of Sciences, USA 94: 7082–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.