Abstract
Dengue is a viral disease of expanding global incidence without cures. Here we present a drug repositioning system (DenguePredict) leveraging upon a unique drug treatment database and vast amounts of disease- and drug-related data. We first constructed a large-scale genetic disease network with enriched dengue genetics data curated from biomedical literature. We applied a network-based ranking algorithm to find dengue-related diseases from the disease network. We then developed a novel algorithm to prioritize FDA-approved drugs from dengue-related diseases to treat dengue. When tested in a de-novo validation setting, DenguePredict found the only two drugs tested in clinical trials for treating dengue and ranked them highly: chloroquine ranked at top 0.96% and ivermectin at top 22.75%. We showed that drugs targeting immune systems and arachidonic acid metabolism-related apoptotic pathways might represent innovative drugs to treat dengue. In summary, DenguePredict, by combining comprehensive disease- and drug-related data and novel algorithms, may greatly facilitate drug discovery for dengue.
Introduction
Dengue is the most common vector-born viral infection in humans and the most rapidly spreading viral disease globally. Over 40% of the world’s population live in dengue-endemic areas, and about 50 to 100 million people are infected with the dengue virus every year. Currently, there are no curative drugs for dengue [1–3]. Therefore, cost-effective approaches are needed to rapidly discover innovative drug treatments for it. Drug repositioning is a drug discovery strategy that seeks to renew failed drugs or expand indications for approved drugs [4]. Currently, computational drug repositioning has not yet been applied to the search for drug treatments for dengue [5].
Disease genetics provide strong evidence to connect genes to human diseases. Variations in several genes have been shown to influence susceptibility and resistance to the dengue virus, as well as disease progression and severity [6–9]. These genes are involved in multiple genetic pathways associated with dengue as well as many other diseases. We hypothesize that diseases that share high genetic relevance with dengue may offer insights into disease biological basis and provide unique opportunities in developing effective drug treatments for dengue. Here we present a drug repositioning system (DenguePredict) that first finds diseases that are genetically related to dengue and then use dengue-related diseases as a window into understanding the biology of dengue and discovering drug candidates to treat it. Our study is different from current disease genetics-based drug discovery studies, which often directly infer drug targets from disease-associated genes [10–11]. To directly translate disease genetics into therapeutics, we need to know that disease-associated genes are involved in disease pathogenesis. However, the genetic basis of many diseases, including dengue, still remains unknown and the effect size of many disease-associated genes, for instance disease-associated genes discovered through genome-wide association studies (GWAS), is generally modest. Here we present an alternative strategy to circumvent these obstacles. We use disease genetics data as merely a starting point to infer interconnections among thousands of diseases and then develop a novel drug repositioning strategy to infer drug treatments based on these genetically related diseases and their associated drug treatments. Our intuition is that if two diseases share high genetic relevance, it is likely that these two diseases are related in pathophysiology even though the exact biology may remain unknown, therefore drugs that are effective in treating one disease may treat the other.
DenguePredict is a computation-based drug repositioning system. Computational drug repositioning approaches can be classified as drug-based, disease-based, and both [12–14]. Drug-based approaches leverage upon known drug molecular structures or functions such as chemical structure and properties, molecular docking, gene expression and drug side effects [15–21]. It was recognized that drug screens based on existing drugs might fail to identify new therapeutic mechanisms [22]. On the other hand, disease-based approaches put less emphasis on existing drugs and focus more on disease mechanisms and interrelationships, therefore have potential in discovering truly innovative drugs. Disease-based approaches used disease-related data ranging from genome [10–11, 19–20] to phenome [23–27]. Many drug repositioning systems used well-established computational and statistical algorithms, including regression/classification, machine learning, network analysis, and text mining [14]. The keys to the success of a computational drug repositioning system include both the unique datasets included in the system as well as innovative ways in integrating various disease- and drug-related data towards specific problems (i.e. specific diseases or drugs).
There are three key components in DenguePredict. First DenguePredict contains a comprehensive drug-disease treatment relationship knowledge base (TreatKB) that we recently constructed from multiple heterogeneous and complementary data resources using advanced computational techniques including natural language processing, text mining and data mining [28–30]. TreatKB includes 9,216 drug-disease treatment pairs extracted from FDA drug labels, 111,862 pairs extracted from the FDA Adverse Event Reporting System (FAERS), a database supporting the FDA’s post-marketing drug safety surveillance, 34,306 pairs extracted from 22 million published biomedical literature abstracts, and 69,724 pairs extracted from 171,805 clinical trials. All together, TreatKB contains 208,330 drug-disease treatment pairs for 2484 drugs and 24,511 diseases. Second, we used disease genetics data from both the Online Mendelian Inheritance in Man (OMIM), a comprehensive database of human genes and genetic phenotypes [31], and the Catalog of Published Genome-Wide Association Studies from the US National Human Genome Research Institute (NHGRI), an exhaustive source containing the description of disease-/trait-associated single nucleotide polymorphisms (SNPs) from published GWAS data [32]. We then enriched these disease genetics data by manually curating dengue-associated genes from published biomedical literature. Third, we used a novel signal prioritization algorithm that we recently developed [25] to find candidate drugs from dengue-related diseases.
Materials and methods
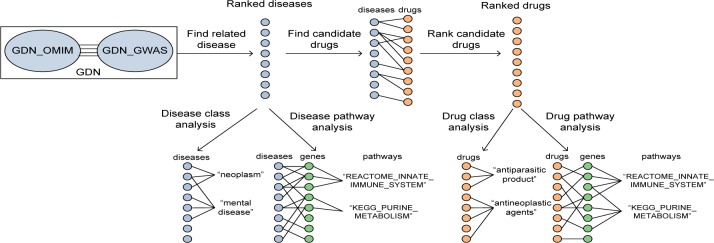
DenguePredict is depicted in Fig. 1 and consists of the following steps: (1) we constructed an integrated genetic disease network (GDN) using disease-gene associations and protein-protein interaction data from multiple large data resources. We applied a network-based ranking algorithm to find dengue-related diseases from GDN; (2) we examined what kinds of diseases and genetic pathways were enriched among top-ranked diseases; (3) we developed a drug repositioning approach to systematically transfer drugs from dengue-related diseases to treat dengue itself. We evaluated DenguePredict using the only two drugs that have been tested in clinical trials for the treatment of dengue; and (4) in order to better understand the top-ranked repositioned drug candidates, we determined which classes of drugs were enriched and which common genetic pathways these drug candidates target.
Fig. 1.
The drug repositioning and analysis pipeline of DenguePredict.
1. Construct an integrated genetic disease network (GDN) and find dengue-related diseases from GDN
1.1 Construct GDNs
We used disease-gene association data from three data resources to construct GDN. The first resource is the OMIM database [31]. We downloaded the OMIM database and mapped gene names to their corresponding approved human gene symbols as defined by the HUGO Gene Nomenclature Committee (HGNC) [33]. We extracted a total of 15,462 disease-gene pairs, representing 5,983 diseases and 8,831 genes.
The second source is the GWAS Catalog [32]. We mapped SNPs to their associated strongest genes, which were subsequently mapped to their corresponding approved human gene symbols as defined by the HGNC. In total, we obtained 22,470 disease/trait-gene pairs, representing 881 diseases/traits and 8,689 genes.
The third source is the published biomedical literature. We manually curated dengue-related biomedical literature and enriched dengue-associated genes included in OMIM and the GWAS Catalog. We classified curated dengue-gene pairs into genetics-based (to enrich data in OMIM) and genomics-based (to enrich data in the GWAS Catalog). Due to the intensive manual effort, we only curated dengue-associated genes from literature.
We first built two sub-networks separately using disease-gene associations from OMIM (GDN_OMIM) and the GWAS catalog (GDN_GWAS). We then integrated them into one network. On both sub-networks, two diseases were connected if their associated genes (proteins) interacted. The edge weights were determined by the numbers of protein-protein interaction (PPI) pairs between two diseases. The PPI data was obtained from the STRING database [34]. From the STRING database, we obtained a total of 4,137,054 human PPI pairs representing 17,756 human proteins. In building the integrated GDN, we mapped nodes in GDN_OMIM to the nodes in GDN_GWAS if the nodes represented the same diseases. The mapping was done based through the unified medical language system (UMLS) Concept Unique IDs (CUIs) [35]. The fact that only 29 diseases mapped between OMIM and GWAS Catalog demonstrate that the diseases in these two databases are largely complementary and that our mapping algorithm needs further improvements. For comparison, we also generated ten random networks by randomly shuffling the edges of the real GDN while maintaining the proportion of edges between diseases. The summary statistics of GDN and the two sub-networks are shown in Table 1.
Table 1.
Numbers of nodes and edges for two sub-networks and the integrated GDN.
| Network | Nodes (diseases) (n) | Edges (n) |
|---|---|---|
| GDN_OMIM | 4,848 | 882,751 |
| GDN_GWAS | 856 | 200,758 |
| GDN | 5,675 | 1,083,538, including 29 inter-network edges |
1.2 Find diseases that share high genetics with dengue from GDN
Recently we develop network-based ranking algorithms to prioritize genes for a given disease [27, 36], to prioritize diseases for a given disease [25], and to prioritize diseases for a given microbial metabolite [37]. In this study, we applied these network-based ranking algorithms to find diseases that share high genetics with dengue. The iterative ranking algorithm is defined as: pt+1 = (1 − r)Wpt + rp0, wherein W is the column-normalized adjacency matrix of the integrated GDN (the equal transition probability between GDN_OMIM and GDN_GWAS) and p is a vector in which the i-th element held the normalized ranking score of disease i at t-th iteration. The initial probability vector p0 contains dengue with a probability of 1.0. Other diseases are then ranked according to the steady-state probability vector, which is obtained by iterating the algorithm until the change between pt+1 and pt is less than 10−6.
2 Analyze top-ranked diseases that share a high degree of genetic similarity with dengue
2.1 Analyze disease classes among ranked dengue-related diseases
To systematically understand dengue-related diseases, we examined disease classes enriched among top-ranked diseases retrieved from GDN with dengue as the input. We classified these diseases into sixteen categories using the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD10) [38]. The ICD10 includes 22 highest-level disease classes. We used 16 of the 22 chapters and excluded six non-specific disease classes such as “Codes for special purposes,” and “Injury, poisoning and certain other consequences of external causes.” Since the terms used in ICD10 are often different from those in GDN, we mapped disease terms in ICD10 to their synonyms through UMLS CUIs [35]. We retrieved a list of ranked dengue-related diseases from GDN. For diseases at 10 different ranking cutoffs (top 10%, 20%, … 100%), we calculated percentages of the sixteen disease classes among them.
2.2 Analyze genetic pathways shared among ranked dengue-related diseases
To gain insights into common mechanistic relationships shared among dengue-related diseases, we analyzed genetic pathways associated with them. Functions of highly enriched pathways might provide insights into common molecular mechanisms shared among dengue-related diseases. First, we retrieved disease-associated genes from OMIM and from the GWAS catalog. We ranked each gene based on how many of the dengue-related diseases it was associated with as well as the ranking scores of those diseases: , where n is the number of dengue-related diseases that the gene is associated with and Rdisease_i is the disease ranking score (as determined by the network-based disease ranking algorithm described above). We then analyzed gene-associated pathways using pathway data from the Molecular Signatures Database (MSigDB), a collection of 10,295 annotated genetic pathways or gene sets from multiple sources [39]. We ranked these pathways based on the number of genes associated with dengue-related diseases as well as the rank scores of those genes: , where n is the number of genes that the pathway contains and Rgene_i is the gene ranking score as determined above. We compared top-ranked pathways for GDN to those for random GDNs in order to identify pathways enriched for dengue-related diseases.
3 Reposition drugs associated with dengue-related diseases to treat dengue
3.1 Drug repositioning algorithm
We ranked FDA-approved drugs by the number of dengue-related diseases that they could treat as well as the ranking scores of those diseases. The drug prioritization algorithm is defined as: , wherein n is the number of dengue-related diseases that can be treated by a drug and Rdisease_i is the disease ranking score (as determined by the network-based disease ranking algorithm described above).
3.2 De-novo validation using dengue drugs tested in clinical trials
The inputs to the drug prioritization algorithm are a ranked list of dengue-related diseases (output from the disease ranking algorithm) and their associated drug treatments as determined by the drug-disease treatment pairs from four TreatKBs. Since the inputs contain neither dengue nor dengue-related treatment information, thereby our evaluation is in fact a de novo validation approach. Since no FDA-approved drugs are currently available for the treatment of dengue, we used drugs that have been tested in clinical trials for evaluation. We retrieved a total of 101 dengue-related clinical trials from ClinicalTrials.gov (www.clinicaltrials.gov). While most of these trials test vaccines, five trials tested five different drugs: chloroquine (NCT00849602), ivermectin (NCT020445069), balapiravir (NCT01096576), celgosivir (NCT01619969), and uv-4B (NCT020661358). Among these five drugs, only chloroquine and ivermectin are FDA-approved drugs: chloroquine approved for treating malaria and amebiasis, and ivermectin approved for treating onchocerciasis and strongyloidiasis. For our drug repositioning purpose, ue used the two FDA-approved drugs (chloroquine and ivermectin) as the gold standard. We calculated the rankings of these two drugs among all FDA-approved drugs. The higher these two gold standard drugs were ranked, the better the ranking algorithm was. We compared the rankings of these two drugs derived from GDN to those from ten randomly generated GDNs. In addition, we also compared the performances across the four different TreatKBs.
4 Analyze top-ranked drug candidates
4.1 Analyze drug classes among ranked drug candidates
We examined which classes of drugs were enriched among top-ranked repositioned drug candidates. We classified drugs using drug classes defined by the Anatomical Therapeutic Chemical (ATC) classification system [40]. The ATC system consists of 13 first-level codes, which were further classified into 94 second-level codes, 267 third-level codes, 882 fourth-level codes, and 4,580 fifth-level codes, which are individual drugs. We experimented classifying drugs using different level ATC codes and found that the third level ATC codes provided sufficient but not too fine-grained granularity.
4.2 Analyze genetic pathways targeted by repositioned drug candidates
To understand the common mechanisms of action underlying top-ranked repositioned drug candidates, we analyzed genetic pathways targeted by these drug candidates. The method for drug pathway analysis was similar to that used for disease pathway analysis as described above, except that the drug-target gene association data was from DrugBank [41]. We compared top-ranked pathways based on GDN to those based on random GDNs in order to identify pathways enriched for repositioned dengue drug candidates. Functions of these enriched pathways might provide insights into common molecular mechanisms targeted by drug candidates. We performed literature search for supporting evidence that these enriched pathways might be targeted for dengue treatments.
Results
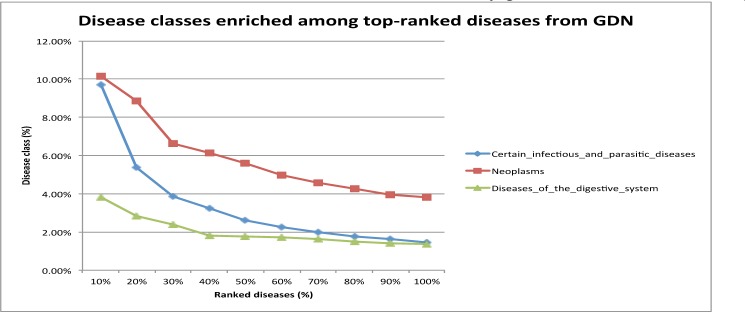
1. Infectious and parasitic diseases, neoplasms, and diseases of the digestive system are enriched among top-ranked dengue-related diseases
Using dengue as the input, we retrieved a ranked list of 4729 diseases from GDN. The disease class “Certain infectious and parasitic diseases” was highly enriched among top-ranked diseases: 9.73% among top 10% ranked diseases as compare to 1.46% among all diseases. This is expected, since dengue is known to share genetics with other infectious diseases such as malaria, mycobacterium tuberculosis, and HIV [42–44]. Therefore, the enrichment of this disease class among diseases retrieved from GDN roughly served as a positive control in validating both network construction and disease ranking algorithms. Interestingly, two other disease classes “Neoplasms” and “Diseases of the digestive system” were also significantly enriched among top-ranked diseases. For comparison, none of the sixteen disease classes were enriched when the randomly generated GDNs were used (data not shown).
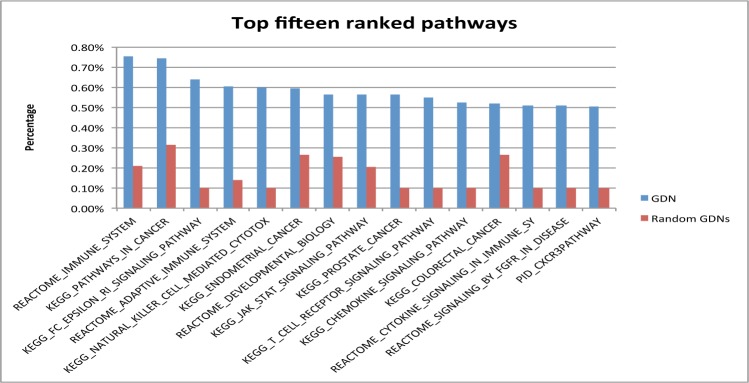
2. Immune-related pathways may be the common mechanisms underlying dengue and its related diseases
Fig. 3 shows the top fifteen enriched pathways and their percentages among all retrieved pathways. Among these enriched pathways, at least nine pathways are involved in human immune system, including “REACTOME IMMUNE SYSTEM,” “KEGG FC EPSILON RI SIGNALING PATHWAY,” and “REACTOME ADAPTIVE IMMUNE SYSTEM.” Experimental and observational findings in past years suggest that immune system-related mechanisms are involved in dengue pathophysiogenesis. Dengue fever is characterized by thrombocytopenia and vascular leak with altered plasma cytokine profiles. Suggested immune mechanisms include platelet activation and apoptosis modulating monocyte inflammatory responses, increased levels of mediators like tumor necrosis factor-α and interleukin-1β [45], interplay between plasmablasts, platelets, and complements [45], disruption of the interaction of Daxx and NF-kB to induce CD137-mediated apoptosis [47], immunodominance changes [48], the involvement of Notch signaling pathways to modulate host adaptive immune response and altered profiles of cytokines produced by cross-reactive T cells [49].
Fig. 3.
Fifteen top-ranked pathways and their percentages among all retrieved pathways from GDN or random GDNs.
Four cancer-related pathways were also enriched, including “KEGG PATHWAYS IN CANCER,” “KEGG ENDOMETRIAL CANCER,” “KEGG PROSTATE CANCER,” and “KEGG COLORECTAL CANCER.” The same enrichment for cancers was observed in above disease class enrichment analysis. However, we could not find literature evidence supporting the direct relationship between dengue and cancers. Since immunopathogeneis are known to be involved in many cancers and dengue, common immune mechanisms may underlie both dengue and cancers. Studies have shown that carica papaya leaves exhibits both anti-tumor activity and immunomodulatory effects in dengue [50–51]. In summary, by systematically analyzing genetic pathways involved with dengue-related diseases, we can gain deeper insights into molecular mechanisms underlying dengue and its related diseases.
3. DenguePredict found the two clinical trial dengue drugs and ranked them highly
As shown in Table 2, DenguePredict consistently ranked chloroquine highly (ranging from top 0.96% to 5.98%). Ivermectin was ranked lower than chloroquine, but still significantly higher than those derived with random GDNs. Comparing across four TreatKBs, DenguePredict using the MEDLINE-based TreatKB performed the best, with chloroquine ranked at 0.96% and ivermectine at 22.75%. Combining drug-disease treatment pairs from all four databases did not improve the performance (data not shown). The fact that we ranked both chloroquine and ivermectin highly demonstrated the validity of our repositioning strategy. The output of DenguePredict is a list of FDA-approved drugs ranked based on their likelihood for treating dengue.
Table 2.
Drug reposition evaluation across four different TreatKBs.
| TreatKB | GDN | GDN_Random | Improvment (%) | |||
|---|---|---|---|---|---|---|
| Chl | Iver | Chl | Iver | Chl | Iver | |
| FDA-approved | 4.65% | 54.85% | 20.85% | 77.32% | 348.38% | 40.97% |
| Post-market | 5.98% | 43.04% | 24.69% | 61.75% | 312.87% | 43.47% |
| ClinicalTrials | 5.83% | 56.76% | 18.56% | 72.91% | 218.35% | 28.45% |
| MEDLINE | 0.96% | 22.75% | 8.37% | 37.00% | 771.88% | 66.64% |
4. Immune system-related drugs are highly enriched among top-ranked repositioned drug candidate
The top 10 ranked repositioned drug candidates using the four TreatKBs are shown in Table 3. Drugs that were consistently ranked highly across four TreatKBs include many corticosteroids (i.e. methylprednisolone, dexamethasone, prednisone, and prednisolone).
Table 3.
Top 10 drug candidates repositioned from four TreatKBs.
| Rank | FDA-approved | Post-market | ClinicalTrials | MEDLINE |
|---|---|---|---|---|
| 1 | Methylprednisolone | Dexamethasone | Chlordiazepoxide | Nitric oxide |
| 2 | Betamethasone | Prednisolone | Bevacizumab | Heparin |
| 3 | Triamcinolone | Nitric oxide | Cisplatin | Methotrexate |
| 4 | Dexamethasone | Prednisone | Paclitaxel | Celecoxib |
| 5 | Prednisolone | Fentanyl | Carboplatin | Prednisolone |
| 6 | Prednisone | Methylprednisolone | Gemcitabine | Iron |
| 7 | Cortisone | Aspirin | Doxorubicin | Adenosine |
| 8 | Hydrocortisone | Methotrexate | Dexamethasone | Vitamin c |
| 9 | Fluorouracil, 5-fu | Acetaminophen | Cyclophosphamide | Indomethacin |
| 10 | Mometasone | Celecoxib | Prednisone | Resveratrol |
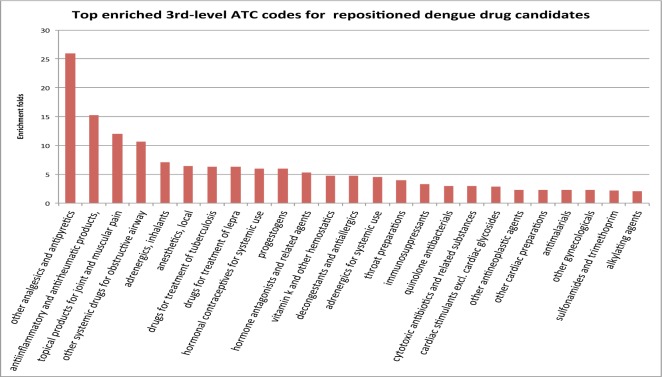
We examined which classes of drugs were enriched and analyzed genetic pathways associated with highly-ranked drug candidates. As shown in Fig. 4, the most enriched (more than 25-fold enrichment as compared to random GDNs) drug class was “Other analgesics and antipyretics.” This finding is consistent with the current symptomatic and supportive treatment for dengue using analgesic-antipyretic therapy for the relief of lethargy, malaise, and fever associated with the disease [2]. The second most enriched drug class was “anti-inflammatory and anti-rheumatic products” (15-fold enrichment), which includes corticosteroids. Corticosteroids are potent anti-inflammatory agents with a wide range of effects on the immune system. Observational studies have suggested that corticosteroids may benefit people with dengue-related shock and may prevent disease progression [52]. Our study provides independent mechanistic evidence supporting treatment benefits of corticosteroids in the treatment of dengue.
Fig. 4.
The 25 most enriched (>=2 fold enrichment) drug classes for repositioned drug candidates.
5. Repositioned drug candidates mainly target apoptosis- and immune-related pathways
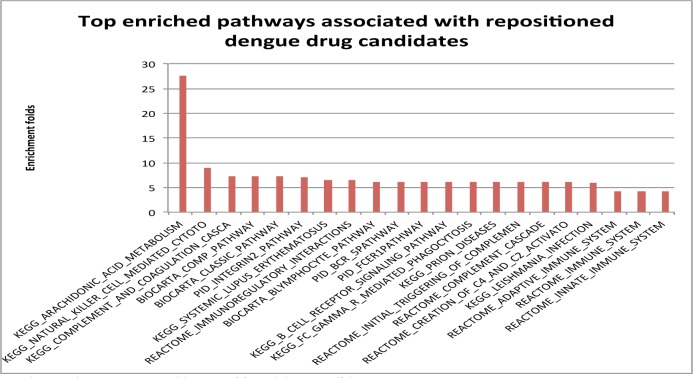
Fig. 5 shows top 21 enriched pathways for repositioned drug candidates. The pathway involved in arachidonic acid metabolism showed 28-fold enrichment as compared to random networks. Arachidonic acid (AA) is a lipid second messenger generated by hydrolysis of membrane phospholipids via phospholipase A2 (PLA2). Malewicz et al. reported that dengue virus was able to activate PLA2 and generate AA [53]. Jan et al. showed that AA, superoxide anion, and NF-kappa B are sequentially involved in dengue virus-triggered apoptotic pathways in human neuroblastoma cells and that inhibition of PLA(2) activity by the PLA(2) inhibitors diminished DEN-2 virus-induced apoptosis [54]. Many of these top enriched pathways are related to immune systems, including natural killer cell mediated cytotoxicity, complement pathway, immunoregulatory interactions, and B lymphocyte pathways. This is consistent with the pathway analysis for dengue-related diseases. In summary, our study indicates that repositioned drug candidates that target arachidonic acid metabolism and/or immune systems might benefit people with dengue. While we did not perform literature search for all other enriched pathways, further investigating these pathways may generate novel hypotheses for dengue drug discovery.
Fig. 5.
Top pathways that were targeted by repositioned drug candidates.
Discussion
By leveraging upon vast amount of knowledge of disease genetics, drug targets, protein interactions, and drug treatments, DenguePredict effectively ranked the only two drugs currently under clinical trial for the treatment of dengue highly. Our approach also suggested potential genetic pathways involved in disease mechanisms and mechanisms of actions of the drug repositioning candidates, which warrants further investigation. DenguePredict is highly generalizable and can easily be retargeted to find drug candidates for other genetic diseases. We expect that its performance will vary among specific diseases and critically depends on the data (both available disease genetics and drug treatments) included for each disease.
We were unable to compare DenguePredict to existing computational drug repositioning systems since these systems did not include dengue in their study. For example, in a recent study, Gottlieb et al used disease-disease similarities and drug-drug similarities from multiple databases to construct a classifier (PREDICT) to determine treatment associations between 593 drugs and 313 diseases [24]. While PREDICT is among currently most comprehensive drug repositioning systems, dengue was not included. On the other hand, DenguePredict included significantly more drugs and diseases: 5,675 diseases on GDN; 2,484 drugs and 24,511 diseases in TreatKBs.
Our study can be further improved in several aspects. First, we can further reprioritize the generated ranked list of repositioned drug candidates by their costs. Dengue is most prevalent in developing countries; therefore costs of drugs needs to be considered. Second, our study generated lists of putative genetic pathways that might be involved with dengue-related diseases and drug candidates, however these candidates will evolve as new disease-associated genes are discovered. For example, as new genes are discovered for dengue or other diseases, more diseases will be linked to dengue from the updated genetic network. Third, we validated our repositioning algorithm using the two clinical trial dengue drugs. Due to the small size of our testing data, we are still uncertain about the precision of top-ranked drug candidates. Experimental or clinical studies are needed to test those candidates.
Fig. 2.
Three ICD10AM disease classes were enriched among top-ranked diseases retrieved from GDN.
Acknowledgments
RX was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under the NIH Director’s New Innovator Award number DP2HD084068, the Training grant in Computational Genomic Epidemiology of Cancer (CoGEC) (R25 CA094186-06), and Case Western Reserve University/Cleveland Clinic CTSA Grant (UL1TR000439). The work done by QW was supported by above funding resources.
Footnotes
Competing interests
None.
Author’s contributions
RX and QW have jointly conceived the idea, designed and implemented the algorithms, and prepared the manuscript. All authors read and approved the final manuscript.
References
- 1.World Health Organization . Dengue and severe dengue. Fact sheet N 117. Geneva: WHO; 2012. [Google Scholar]
- 2.World Health Organization, Special Programme for Research, Training in Tropical Diseases, World Health Organization. Department of Control of Neglected Tropical Diseases, World Health Organization. Epidemic, & Pandemic Alert . Dengue: guidelines for diagnosis, treatment, prevention and control. World Health Organization; 2009. [Google Scholar]
- 3.Rajapakse S, Chaturaka R, Rajapakse A. Treatment of dengue fever. Infection and Drug Resistance. 2012;5:103–12. doi: 10.2147/IDR.S22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews Drug discovery. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 5.Selisko B, Guillemot JC, Alvarez K, Canard B. Opportunities in the development of ANTI-dengue drugs; Working paper for the Scientific Working Group on Dengue Research, convened by the Special Programme for Research and Training in Tropical Diseases; 1–5 October 2006.Geneva: 2006. [Google Scholar]
- 6.Sakuntabhai A, Turbpaiboon C, Casadémont I, Chuansumrit A, Lowhnoo T, et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nature genetics. 2005;37(5):507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, et al. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453(7195):672–676. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- 8.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458(7241):1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khor CC, Chau TNB, Pang J, Davila S, Long HT, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nature genetics. 2011;43(11):1139–1141. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nature Reviews Genetics. 2012;13(8):576–588. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 11.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Toes RE. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: from data to therapeutics. Clinical Pharmacology & Therapeutics. 2013;93(4):335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 13.Shameer K, Readhead B, T Dudley J. Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Current topics in medicinal chemistry. 2015;15(1):5–20. doi: 10.2174/1568026615666150112103510. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Zheng S, Chen B, Butte AJ, Swamidass SJ, Lu Z. A survey of current trends in computational drug repositioning. Briefings in bioinformatics. 2015;1:11. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 17.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 18.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science translational medicine. 2011;3(96):96ra77–96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Science translational medicine. 2011;3(96):96ra76–96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang AP, Butte AJ. Systematic evaluation of drug–disease relationships to identify leads for novel drug uses. Clinical Pharmacology & Therapeutics. 2009;86(5):507–510. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nature neuroscience. 2010;13(10):1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoehndorf R, Oellrich A, Rebholz-Schuhmann D, Schofield PN, Gkoutos GV. Linking PharmGKB to phenotype studies and animal models of disease for drug repurposing. Pac Symp Biocomput. 2012;17:388–399. [PubMed] [Google Scholar]
- 24.Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Molecular systems biology. 2011;7(1) doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R, Wang Q. PhenoPredict: a disease phenome-wide drug repositioning approach towards schizophrenia drug discovery. Journal of Biomedical Informatics. 2015 doi: 10.1016/j.jbi.2015.06.027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Zhang Xiang, Zhang GQ, Xu R. Comparative Analysis of A Novel Disease Phenotype Network Based on Clinical Manifestations. Journal of Biomedical Informatics. 2015 Feb;53:113–20. doi: 10.1016/j.jbi.201409.007. Epub 2014 Sep 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Xu R. Phenome-driven Disease Genetics Prediction Towards Drug Discovery. Bioinformatics. 2015;2015;31(12):i276–i283. doi: 10.1093/bioinformatics/btv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu R, Wang Q. Large-scale extraction of drug-disease treatment pairs from biomedical literature for drug repurposing. BMC Bioinformatics. 2013;14(1):181. doi: 10.1186/1471-2105-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu R, Li L, Wang Q. Towards building a disease-phenotype relationship knowledge base: large-scale extraction of disease-manifestation relationship from literature. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R, Wang Q. Automatic signal prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS) Journal of Biomedical Informatics. 2014:171–177. doi: 10.1016/j.jbi.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids research. 2005;33(suppl 1):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welter D, MacArthur J, Morales J, Burdett T, Hall P, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42(Database issue):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Povey S, Lovering R, Bruford E, Wright M, Lush M, et al. The HUGO gene nomenclature committee (HGNC) Human genetics. 2001;109(6):678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 34.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41(D1):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Xu R. Network-based Gene Prediction for Plasmodium falciparum Malaria Towards Genetics-based Drug Discovery. BMC Genomics. 2015 2015 Jun 11;16(Suppl 7):S9. doi: 10.1186/1471-2164-16-S7-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu R, Wang Q, Li L. sgenome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2015;16(Suppl 5):S6. doi: 10.1186/1471-2105-16-S5-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization . International statistical classification of diseases and related health problems. Vol. 1. World Health Organization; 2004. [Google Scholar]
- 39.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization . The anatomical therapeutic chemical classification system with defined daily doses (ATC/DDD) Norway: WHO; 2006. [Google Scholar]
- 41.Knox C, Law V, Jewison T, Liu P, Ly S, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic acids research. 2011;39(suppl 1):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carme B, Matheus S, Donutil G, Raulin O, Nacher M, et al. Concurrent dengue and malaria in cayenne hospital, French Guiana. Emerging infectious diseases. 2009;15(4):668. doi: 10.3201/eid1504.080891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geijtenbeek TB, van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. The Journal of experimental medicine. 2003;197(1):7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bashirova AA, Geijtenbeek TB, Van Duijnhoven GC, van Vliet SJ, Eilering JB, et al. dendritic cell– specific intercellular adhesion molecule 3–grabbing nonintegrin (DC-SIGN)–related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. The Journal of experimental medicine. 2001;193(6):671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hottz ED, Medeiros-de-Moraes IM, Vieira-de-Abreu A, de Assis EF, Vals-de-Souza R, et al. Platelet Activation and Apoptosis Modulate Monocyte Inflammatory Responses in Dengue. The Journal of Immunology. 2014:1400091. doi: 10.4049/jimmunol.1400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nascimento EJ, Hottz ED, Garcia-Bates TM, Bozza F, Marques ET, Jr, et al. Emerging Concepts in Dengue Pathogenesis: Interplay between Plasmablasts, Platelets, and Complement in Triggering Vasculopathy. Critical Reviews™ in Immunology. 2014;34(3) doi: 10.1615/critrevimmunol.2014010212. [DOI] [PubMed] [Google Scholar]
- 47.Netsawang J, Panaampon J, Khunchai S, Kooptiwut S, Nagila A, et al. Dengue virus disrupts Daxx and NF-κB interaction to induce CD137-mediated apoptosis. Biochemical and biophysical research communications. 2014 2014 Aug 8;450(4):1485–1491. doi: 10.1016/j.bbrc.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, et al. Immunodominance Changes as a Function of the Infecting Dengue Virus Serotype and Primary versus Secondary Infection. Journal of virology. 2014:JVI-01108. doi: 10.1128/JVI.01108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Wu S, Pu J, Huang X, Zhang P. Dengue Virus Upregulates Expression of Notch Ligands Dll1 and Dll4 Through IFN-β Signaling Pathway. Immunology. 2014 doi: 10.1111/imm.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otsuki N, Dang NH, Kumagai E, Kondo A, Iwata S, et al. Aqueous extract of Carica papaya leaves exhibits anti-tumor activity and immunomodulatory effects. Journal of Ethnopharmacology. 2010;127(3):760–767. doi: 10.1016/j.jep.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 51.Siddique O, Sundus A, Ibrahim MF. Effects of papaya leaves on thrombocyte counts in dengue-a case report. JPMA The Journal of the Pakistan Medical Association. 2014;64(3):364–366. [PubMed] [Google Scholar]
- 52.Sprung CL, Annane D, Keh D, Moreno R, Singer M. Hydrocortisone therapy for patients with septic shock. New England Journal of Medicine. 2008;358(2):111. doi: 10.1056/NEJMoa071366. [DOI] [PubMed] [Google Scholar]
- 53.Malewicz B, Parthasarathy S, Jenkin HM, Baumann WJ. Rapid phospholipase A 2 stimulation and diacylglycerol cholinephosphotransferase inhibition in baby hamster kidney cells during initiation of dengue virus infection. Biochemical and biophysical research communications. 1981;101(2):404–410. doi: 10.1016/0006-291x(81)91274-2. [DOI] [PubMed] [Google Scholar]
- 54.Jan JT, Chen BH, Ma SH, Liu CI, Tsai HP. Potential dengue virus-triggered apoptotic pathway in human neuroblastoma cells: arachidonic acid, superoxide anion, and NF-κB are sequentially involved. Journal of virology. 2000;74(18):8680–8691. doi: 10.1128/jvi.74.18.8680-8691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]